Abstract
The new Abbott RealTime hepatitis B virus (HBV) assay was compared to the Cobas AmpliPrep/Cobas TaqMan assay with 128 serum samples from patients with chronic hepatitis B. There was an excellent correlation (r = 0.961) between the two assays, with the Abbott RealTime test showing at least equivalent sensitivity and a slightly wider dynamic range than the Cobas TaqMan assay. By coupling high sensitivity with a large dynamic range, the Abbott RealTime HBV assay is useful in monitoring the response to antiviral therapy.
Hepatitis B virus (HBV) is a small partially double-stranded virus belonging to the family of the Hepadnaviridae that can cause acute or chronic hepatitis (2). Millions of people around the world are infected by HBV, and it is estimated that about 350 million people worldwide have developed a persistent infection (6). Measurement of HBV levels in serum is a reliable marker for prognosis of acute and chronic infection and plays an important role in the management of patients receiving antiviral drugs (7, 8). Therefore, diagnostic assays for the accurate quantitation of HBV DNA are essential for optimal patient management. Monitoring of HBV DNA allows to predict the success of antiviral therapy and to identify the development of drug resistance. Real-time PCR is a new molecular tool that is progressively replacing endpoint PCR systems for monitoring patients with chronic hepatitis B since it allows for high sensitivity, a broad dynamic range, accuracy, and rapid results (1, 9).
Quantification by real-time PCR is based on the determination of the threshold cycle (CT) when the amplified product is detected for the first time (4) and the PCR is still in the exponential phase. In this case the quantification of the viral load is much more accurate and reliable than that measured with endpoint PCR systems (3).
The aim of our study was to evaluate the Abbott RealTime HBV assay for the detection and quantification of HBV-DNA in serum samples and to compare these results with those obtained using the Cobas AmpliPrep/Cobas TaqMan HBV test (Roche Diagnostics). The study was carried out on 128 serum samples collected from patients with chronic HBV infections attending our hepatology clinic at the University Hospital Tor Vergata, Rome, Italy.
Blood samples were collected in a BD Vacutainer tube containing a separation gel. After centrifugation, the serum was divided into two aliquots of 1 ml each and tested by the Abbott and Cobas TaqMan Real-time PCR assays, respectively. Both systems use a hybridization probe with a fluorescent moiety covalently linked to the 5′ end of the probe (reporter) and a quenching moiety bound to the 3′ end of the probe (quencher). In the presence of target, probe hybridization and primer extension occur simultaneously. The fluorescent signal is generated by removing the reporter through the 5′→3′ exonuclease activity of a thermostable Taq DNA polymerase (4, 5). In the TaqMan and Abbott RealTime PCR assays an additional probe is used to detect genotype G or C, respectively. While the Cobas TaqMan system targets the pre-core region, the Abbott RealTime assay amplifies a highly conserved region of the HBV genome which is represented by the N-terminal portion of the S gene.
With the Abbott assay, HBV DNA was extracted from 500 μl of serum by using the m2000sp, an automated sample preparation system designed to use magnetic microparticle-based reagents for the purification of nucleic acids from samples. An internal control DNA is introduced into the sample preparation procedure and is processed along with the calibrators, controls, and specimens. The presence of the amplified HBV-DNA and internal control is detected during the extension and annealing step. The amplification cycle at which a normalized fluorescent signal is detected by the Abbott m2000rt is proportional to the log of the HBV DNA concentration present in the original sample. Each sample is then quantitated by an external calibration curve.
The Cobas AmpliPrep extracts HBV-DNA from 850 μl of serum, and during the extraction a known number of HBV quantitation standard DNA molecules are introduced in each specimen (5). The quantitation of HBV-DNA is performed using the HBV quantitation standard. The system quantifies the amplicons during the exponential phase of amplification. The PCR cycle at which the increase in normalized fluorescence of a sample exceeds the background noise is considered the critical CT and marks the beginning of the exponential growth phase of this signal.
Based on a positive Cobas TaqMan result, 128 serum samples were selected from patients with chronic hepatitis B, including samples with values below the limit of detection (LOD), but with viral target detected (<12 IU/ml). All but one sample (<12 IU/ml as determined by TaqMan) were detected by the Abbott RealTime assay, with a relative sensitivity of 99.2% (127 of 128; confidence interval = 97.69 to 100.00%).
As shown in Table 1, both assays provided a result within their dynamic range for 100 samples, while 14 samples were below the LOD for both assays. Of the remaining eight samples with a TaqMan result below LOD, seven were quantifiable by the Abbott RealTime assay (range, 12 to 35 IU/ml), and one produced a negative result. Of the two samples quantified by Roche TaqMan, but below its linearity range (54 IU/ml), one was quantified (84 IU/ml) and one was <10 IU/ml with the target detected by Abbott. Finally, four samples with a value exceeding the upper limit of linearity with Roche (>8.04 log IU/ml) had reportable results with the Abbott assay, whose upper limit of linearity reaches 9.0 log IU/ml. For three of the samples, the Abbott assay result was consistent with the TaqMan assay off-scale limit.
TABLE 1.
Comparison between Abbott RealTime HBV and Cobas TaqMan 48 HBV real-time PCR assays
| Abbott RealTime assay result | No. of results obtained by COBAS AmpliPrep/ Cobas TaqMan
|
||||
|---|---|---|---|---|---|
| <12 IU/ml, target detected | 12-54 IU/ml | Within dynamic range | >8.04 log IU/ml | Total | |
| Negative | 1 | 1 | |||
| <10 IU/ml, target detected | 14 | 1b | 15 | ||
| Within dynamic range | 7a | 1c | 100 | 4d | 112 |
| >9.0 log IU/ml | 0 | ||||
| Total no. | 22 | 2 | 100 | 4 | 128 |
Abbott RealTime results ranged from 12 to 34 IU/ml.
Roche TaqMan, 34 IU/ml.
Roche TaqMan, 12 IU/ml; Abbott RealTime, 84 IU/ml.
Abbott RealTime results: 7.91, 8.15, 8.16, and 8.90 log IU/ml.
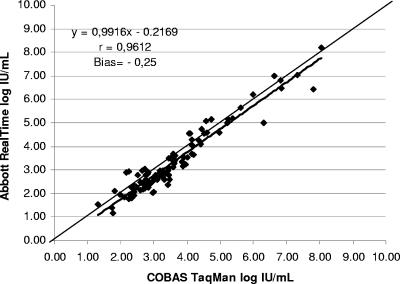
Linear regression analysis was carried out for the 100 paired quantitative results available for both tests (Fig. 1). An excellent correlation was found (r = 0.9612), with the regression line showing the following equation: Abbott RealTime (log IU/ml) = 0.9916 Roche TaqMan (log IU/ml) − 0.2169. The mean assay result difference (Abbott − Roche) was −0.246 log IU/ml (range, −1.364 to +0.712; standard deviation = 0.381 log IU/ml) with 81 and 97% of the samples differing by less 0.5 and 1 log, respectively.
FIG. 1.
Linear regression analysis performed on 100 paired quantitative results obtained with the two assays.
Interestingly, if expressed in log copies/ml, the bias would be −0.479, implying that the adoption of international units is helpful in reducing the differences and making the assays results interchangeable, although in this case the standardization seems to have worked partially. Worthy of note, the observed bias doesn't impact the Abbott assay sensitivity when the low-level viremia results are taken into account. Indeed, the Abbott assay was able to quantify one-third of the values below LOD with the Cobas TaqMan assay, and this was probably due to a better precision at the low-end range.
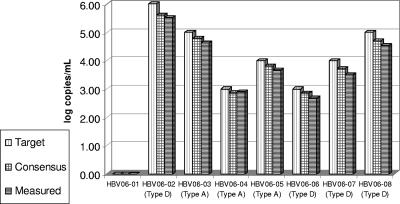
In order to further verify the assay accuracy, the QCMD HBV06-01 panel (a past program proficiency panel, organized by the Quality Control for Molecular Diagnostics) was tested with the Abbott RealTime HBV assay. The panel consisted of eight members targeted at 3, 4, 5, and 6 log copies/ml (type D) and 3, 4, and 5 log copies/ml (type A) plus a negative sample (Fig. 2).
FIG. 2.
QCMD HBV06-01 panel tested with the Abbott m2000 HBV assay. The Abbott RealTime HBV results (measured) are compared to expected sample concentrations (target) and the mean results obtained by all 109 participants (consensus).
The difference between the Abbott RealTime and the overall consensus results (the mean of the reported results from all 109 program participants) ranged from −0.22 to +0.04 (mean = −0.13 log copies/ml) when the results were expressed in log copies as requested by the quality control scheme. It is conceivable that the international unit result expression would reduce this difference by normalizing the assay-specific copies to the commonly adopted international units.
The assay reproducibility was assessed by testing in five separate different runs two four-member panels created by diluting two high-titer patient samples.
The coefficient of variation ranged from 0.77 to 2.93% (log copies/ml) for concentrations between 4.27 and 8.12 log copies/ml. The corresponding standard deviation (range, 0.06 to 0.19) was always below 0.25 log copies, a value that is recognized as the maximum allowed for analytical variability.
In conclusion, this evaluation demonstrated that in comparison to the Cobas AmpliPrep/Cobas TaqMan, the newly developed Abbott RealTime HBV-DNA assay has at least equivalent sensitivity and has a slightly wider dynamic range, and its results correlate extremely well. A small bias between the two assays was observed, most probably due to a different standardization process. Both assays, by coupling an exquisite sensitivity to a large linear range, are useful in monitoring the therapy of patients with chronic hepatitis B.
Acknowledgments
We thank Carolyn Mullen and James Rhoads (Abbott Molecular, Des Plaines, IL) for manuscript revision and F. Renato Pulvirenti (Abbott Molecular Italy) for providing free kits for our study.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Chen, R. W., H. Piiparinen, M. Seppanen, P. Koskela, S. Sarna, and M. Lappalainen. 2001. Real-time PCR for detection and quantitation of hepatitis B virus DNA. J. Med. Virol. 65250-256. [DOI] [PubMed] [Google Scholar]
- 2.Ganem, D., and A. M. Prince. 2004. Hepatitis B virus infection: natural history and clinical consequences. N. Engl. J. Med. 3501118-1129. [DOI] [PubMed] [Google Scholar]
- 3.Gibosn, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative real-time PCR. Genomes Res. 6995-1001. [DOI] [PubMed] [Google Scholar]
- 4.Gordillo, R. F., J. Gutierrez, and M. Casal. 2005. Evaluation of the COBAS TaqMan 48 real-time PCR system for quantitation of hepatitis B virus DNA. J. Clin. Microbiol. 4335043507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hochberger, S., D. Althof, R. Gallegos de Schrott, N. Nachbaur, H. Rock, and H. Leying. 2006. Fully automated quantitation of hepatitis B virus (HBV) DNA in human plasma by the COBAS TaqMan system. J. Clin. Virol. 35373-380. [DOI] [PubMed] [Google Scholar]
- 6.Lee, W. 1997. Hepatitis B infection. N. Engl. J. Med. 3371733-1745. [DOI] [PubMed] [Google Scholar]
- 7.Locarnini, S., A. Hatzakis, J. Heathcote, E. B. Keefe, J. J. Liang, D. Mutimer, J. M. Pawlotsky, and F. Zoulim. 2004. Management of antiviral resistance in patients with chronic hepatitis B. Antiviral Ther. 9679-693. [PubMed] [Google Scholar]
- 8.Ranki, M., H. M. Schatzl, and R. Zachoval. 1995. Quantification of hepatitis B virus DNA over a wide range from serum for studying viral replicative activity in response to treatment and in recurrent infection. Hepatology 211492-1499. [PubMed] [Google Scholar]
- 9.Zanella, I., A. Rossini, D. Domeneghini, A. Alberini, and E. Cariani. 2002. Quantitative analysis of hepatitis B virus DNA by real-time amplification. Eur. J. Clin. Microbiol. Infect. Dis. 2122-26. [DOI] [PubMed] [Google Scholar]