Abstract
Crucial aspects of the foamy virus (FV) replication strategy have so far only been investigated for the prototypic FV (PFV) isolate, which is supposed to be derived from nonhuman primates. To study whether the unusual features of this replication pathway also apply to more-distantly related FVs, we constructed feline FV (FFV) infectious molecular clones and vectors. It is shown by quantitative RNA and DNA PCR analysis that FFV virions contain more RNA than DNA. Full-length linear DNA was found in extracellular FFV by Southern blot analysis. Similar to PFV, azidothymidine inhibition experiments and the transfection of nucleic acids extracted from extracellular FFV indicated that DNA is the functional relevant FFV genome. Unlike PFV, no evidence was found indicating that FFV recycles its DNA into the nucleus.
The family Retroviridae is taxonomically divided into the subfamilies Spumaretrovirinae, which is made up of only one genus, the foamy viruses (FVs), and Orthoretrovirinae, which is made up of the remaining six genera (International Committee on Taxonomy of Viruses, http://www.danforthcenter.org/iltab/ICTVnet). Both subfamilies are reverse transcribing, have the same principal genome order of long terminal repeat (LTR), gag, pol, env (accessory genes), LTR, and require integration into the host cell genome (15, 16, 24). However, FVs display some very unusual features, such as the presence of an internal promoter, which directs the expression of accessory genes, Gag-independent translation of Pol from a spliced transcript, or a highly unusual way of particle assembly and egress, in which the leader peptide of Env plays a crucial role (3, 4, 14, 17, 33, 34). The most distinctive feature between orthoretroviruses and FVs resides in the nature of the virion nucleic acid. Only very small fractions of orthoretroviruses contain partially reverse-transcribed DNA genomes, which appear to be irrelevant for the maintenance of the infectious process (18, 31). In contrast, full-length linear DNA has been shown to be a major component of the FV virion (34). Furthermore, studies on the time point of reverse transcription with the inhibitor of reverse transcription, zidovudine (AZT), the ability to recycle a cDNA genome into the host cell genome, and the rescue of infectious virus after transfection of cells with DNA extracted from virions strongly suggested that the functionally relevant FV nucleic acid is DNA (11, 16, 21, 24, 35). Most of these findings were made by analyzing a single FV isolate derived from primates, which is now called prototypic FV (PFV) (16, 24). Based on nucleic acid and protein homology, it is likely that these PFV-specific features do apply to other primate FVs as well. However, the group of FVs also comprises members from feline (FFV), bovine, and equine species, which are more distantly related to the primate viruses (26). We therefore wished to analyze crucial aspects of the FFV replication pathway to elucidate the distinctive features common to all FVs more precisely.
MATERIALS AND METHODS
Cells and viruses.
Mus dunni cells, BHK/LTR(PFV)lacZ cells (29), BHK/LTR(SFV-1)lacZ cells, Crandell feline kidney cells (CrFK), CrFK/LTR(FFV)lacZ cells (2), HeLa, HT1080, 293, and 293T cells (7) were cultivated in Dulbecco's modified Eagle's minimal essential medium supplemented with 10% fetal calf serum, antibiotics, and 1 mg of G418 (Invitrogen)/ml or 0.4 mg of zeocin (Cayla)/ml when appropriate. FVs were usually derived by calcium phosphate coprecipitation-mediated transfection of 293T cells, and the cell-free supernatant was passaged to the appropriate indicator cell line. The determination of FV titers was done as described previously by using 1 × 104 to 4 × 104 indicator cells seeded in 12-well plates (29). Friend murine leukemia virus (FrMuLV) was amplified on M. dunni cells and titrated as reported previously (6).
Recombinant DNA.
Established techniques in molecular biology were used to generate recombinant DNAs (1, 27). The subgenomic FFV plasmids 94, Env1, and F3 (12), the full-length simian FV type 1 (SFV-1) clone (pSFV-1) (20), the infectious PFV clone pcHSRV2 (21), the murine leukemia virus (MuLV) vectors pSFG-NLSlacZ (9) and pcAMS/U3EN (11), the MuLV packaging plasmid pHIT60 (30), and the vesicular stomatitis virus G protein expression plasmid pcVG-wt (23) have been described previously.
To generate the infectious FFV molecular clone pChatul-1, we assembled step by step the subgenomic FFV clones 94, Env1, and F3 (12), starting with the human cytomegalovirus (CMV) immediate early gene promoter to drive the initial transcription as described previously for PFV (21). The insertion of the simian virus 40 (SV40) origin of replication as a 0.2-kb PCR fragment into the vector backbone of pChatul-1 led to pChatul-2. Cell-free titers that were obtained following transfection of cells with either pChatul-1 or pChatul-2 were initially low (102 to 103).
FFV derived from transfection of cells with pChatul-2 was serially passaged cell-free on CrFK cells until cell-free titers were in the range of 106 to 107. Two separate long-range PCRs were performed on DNA extracted from cells infected with fast-replicating FFV with the oligonucleotide primer pairs 788 (5′ TGT ACG GGA GCT CTT CTC ACA GAC TTG GC)-791 (5′ TGC AAG TTC TTC TTC ACG CGT CCA GTC CAA GG) and 790 (5′ TCC TTG GAC TGG ACG CGT GAA GAA GAA CTT GC)-789 (5′ TCC CCA CGT GTA GAG AAA CAC CAC TC) as previously described for the chimpanzee FV SFVcpz (13). These primers incorporate a 5′ SacI site (underlined in primer 788), a 3′ PmlI site (underlined in primer 789), and a pol gene-located MluI site (underlined in primers 790 and 791). The amplimers were assembled downstream of a CMV promoter by using the unique SacI, MluI, and PmlI restriction sites to give rise to pChatul-3. The FFV vectors pWR6 and pWR7 were made by the insertion of a 2.06-kb HindIII fragment comprising the spleen focus-forming virus U3 promoter-directed expression cassette for an enhanced green fluorescent protein (EGFP)-neo fusion protein and a 3.69-kb Eco47III/Bsp68I fragment containing an SSFV U3 nuclear localization signal lacZ expression cassette, respectively, into KpnI-digested pChatul-2 by blunt-end ligation.
The competitor plasmid pJR40 (ΔLTR) was made by separately amplifying two regions from the FFV LTR of pChatul-3 with primer pairs 1158 (5′ TCT TCT CAC AGA CTT GGC TG)-1160 (5′ TCC AAT TTA TTT GAT CTG CTA GCA AGG) and 1161 (5′ TGA GTG GTG TTT CGC TAG CCC TGG GG)-1159 (5′ TAC CTG GGA TAG GTT AGT CCT TC). The amplimers leave an unamplified region of 172 bp after assembly in the pCRII-TOPO vector (Invitrogen) downstream of the SP6 promoter. Likewise, pJR41 (ΔGAG) was generated with primer pairs 991 (5′ TAA TAG GGC CAG TAA TAC CG)-993 (5′ TGT ATA TTA AGGGCT AGC ATA GTG) and 994 (5′ TCG CTA GCA GCC TCA GCG CTA CGG)-1236 (5′ TTC CTC CTC CAT TTC TTG GG). The genomic region missing in pJR41 is 232 bp in length. The LightCycler (LC) plasmid standards pJR36 and pJR38 were derived by amplifying pChatul-3 plasmid DNA with primer pair 1164 (5′ TGA ATA GCC CTG GGT TGT TTA CTG)-1202 (5′ TAT CCT GTT GTA TAA CTA GC) and total DNA from FrMuLV-infected M. dunni cells with primer pair 1367 (5′ TAT CGC CAG TTG CTC CTA GCG GGT CTC)-1368 (5′ TTC TTT GCA GTA GGC ACA CTG GTC GTG). The amplimers (544 bp from the FFV pol gene and 533 bp from the FrMuLV gag gene) were inserted into the pCRII-TOPO vector downstream of the SP6 promoter.
To establish BHK/LTR(SFV-1)lacZ cells, the LTR from pSFV-1LTR/CAT (19) was amplified from position −1295 to +30 relative to the start of transcription with primers 532 (5′ GCG CGA GAT CTT GTG GCA GAC AGC CAC TA) and 497 (5′ GCG CTG CAG ACT CTC GGC GCA GCG AGT), which incorporate BglII and PstI restriction sites (underlined), respectively. The amplimer was ligated with the pClacZ/neo vector (A. Rethwilm, unpublished data) after digestion of both DNAs with BglII and PstI. BHK-21 cells were transfected with BglII-linearized plasmid DNA and selected in 1 mg of G418/ml. Single-cell clones were obtained by limiting dilution, and one clone displaying no background and high inducible β-galactosidase activity upon SFV-1 infection was used in titration experiments.
Sequence analysis.
The viral sequences of pChatul-2 and pChatul-3 were determined on both strands with the help of automated DNA analysis devices (ABI3100 and ABI377) and the Big Dye sequencing kit (Applied Biosystems). Importantly, an additional C nucleotide was found after position 2874 of the published FFV sequence (12). This insertion results in an altered gag reading frame, which is 25 triplets longer than the published sequence and overlaps with the pol frame.
AZT inhibition experiments.
293T cells were transfected with plasmid DNA as described above. The cell-free supernatant (0.45-μm-pore-size filtrate) was transferred to the indicator cells at 48 h posttransfection. HT1080 cells (2.5 × 104 cells per well of a 12-well plate) received supernatant from cells transfected with the MuLV vector or with pWR7. BHK/LTR(PFV)lacZ and BHK/LTR(SFV-1)lacZ cells were seeded at a density of 1 × 104 cells per well and CrFK/LTR(FFV)lacZ cells were seeded at a density of 4 × 104 cells per well and received supernatants from cells transfected with PFV, SFV-1, and FFV infectious molecular clones, respectively. The indicator cells were stained at 72 h after transduction. When indicated, AZT was used at a concentration of 5 μM, except for cells incubated with Chatul-3 virus, for which AZT was used at a 50 μM concentration.
Competitive PCR and RT-PCR.
Cell-free supernatant from CrFK cells infected with Chatul-3 virus was concentrated by centrifugation in an SW28 rotor (Beckman) at 28,000 rpm and 4°C for 2 h. The sediment was resolved in 1 ml of phosphate-buffered saline (PBS) and incubated with 20 U of RNase-free DNase (RQ1-DNase from Promega) at 37°C overnight. After centrifugation through a 20% sucrose cushion (SW40 rotor, 30,000 rpm, 4°C, 2 h), the virion DNA was extracted with the QIAamp DNA blood mini kit (Qiagen) from the remaining sediment. RNA was extracted from separately prepared virions with the QIAamp viral RNA mini kit (Qiagen). After treatment of DNA with RNase (0.2 mg/ml) and treatment of RNA with RQ1-DNase (1 U/50 μl) at 37°C for 1 h, nucleic acids were resolved in 1/1,000 of the original volume. The RNA standards were generated by in vitro transcription with the MAXIscript in vitro transcription kit (Ambion), SP6 polymerase, and XbaI-linearized pJR40 or pJR41 competitor plasmid. Following transcription, the template DNAs were digested with DNase I (2 U/20 μl) at 37°C for 15 min. For reverse transcriptase PCR (RT-PCR), diluted competitor RNA, 1 μl of viral RNA probe, and the Qiagen OneStep RT-PCR kit with 20 pmol of each oligonucleotide primer (1158-1159 for the LTR and 991-1263 for gag) were used. The incubation conditions were 42°C for 50 min and 95°C for 15 min followed by 35 cycles of 95°C for 15 s, 57°C for 45 s, and 72°C for 60 s. A final elongation step was carried out at 72°C for 5 min. For PCR, we used 20 pmol of the primers, 4 mM MgCl2, 2.5 U of Taq polymerase (Promega), diluted competitor plasmid, 1 μl of viral DNA probe, and identical cycle conditions as described above.
Real-time PCR and RT-PCR.
Cell-free FFV or FrMuLV supernatant (10.5 ml) from CrFK or M. dunni cells, respectively, was concentrated by centrifugation through a 20% sucrose cushion. The sedimented material was resolved in 200 μl of PBS and used for the isolation of DNA and RNA with the Qiagen kits. This was followed by treatment with nucleases and resolution in 1/1,000 of the original volume as described above. For standardization, SP6 transcripts were generated from pJR36 and pJR38 plasmids. The reverse transcription step was carried out in a 20-μl reaction mixture containing 200 U of SuperscriptII RNaseH− RT (Invitrogen), 20 pmol of primer 1202 (for FFV RNA) or 1368 (for FrMuLV RNA), 1 μl of RNA probe, and buffer at 42°C for 50 min. After heat inactivation of the enzyme (70°C for 15 min) 2 μl was loaded into the LC reaction mixture, which also contained 3 mM MgCl2, 10 pmol of each primer (1164-1202 for the amplification of FFV and 1367-1368 for the detection of FrMuLV), and 2 μl of FastStart DNA Master SYBR Green I (Roche). For the amplification of viral DNA, 5 μl of probe and 4 mM MgCl2 were used. After an initial denaturation step at 95°C for 10 min, 40 cycles consisting of 95°C for 15 s, 57°C for 5 s, and 72°C for 60 s were run. The results were evaluated with the LC software, version 3.5, package from Roche.
Southern blotting.
The supernatant from FFV-infected CrFK cells was cleared by low-speed centrifugation and passed through a 0.45-μm-pore-size filter (Schleicher & Schuell). After ultracentrifugation as described above and overnight treatment with RNase-free DNase, the virions were fractionated on a 20 to 66% sucrose gradient prepared in TNE buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA). Following centrifugation in an SW40 rotor at 30,000 rpm at 4°C for 2 h, 18 fractions of 400 μl were drawn from the bottom of the gradient. Twenty microliters of each fraction was used for protein analysis (as described below), and 350 μl was further concentrated by centrifugation. The sediments were resolved in PBS, and DNA was extracted from 175 μl with a commercial kit (Qiagen). After separation in agarose gel, nucleic acids were blotted onto a nylon membrane. The blot was hybridized to a 1.5-kb Eco47III/NsiI FFV pol gene fragment that was labeled randomly with the Megaprime DNA labeling system (Amersham Biosciences) and [α-32P]dATP. Washing at stringent conditions and exposure to X-ray films were by standard procedures (1, 27).
Protein detection.
Twenty microliters of the gradient fractions was separated on sodium dodecyl sulfate-containing 8% polyacrylamide gels. FFV antigen was analyzed by reaction with a polyclonal anti-Gag serum as detailed elsewhere (J. Roy, D. Westphal, W. Rudolph, O. Herchenröder, A. Berg, M. Rammling, M. Heinkelein, D. Lindemann, and A. Rethwilm, unpublished data).
Transfection of virion DNA.
DNA was extracted from 37 ml of cell-free virus, which was passed through a 0.45-μm-pore-size filter and concentrated by centrifugation (Beckman SW28 rotor, 28,000 rpm, 4°C, 3 h). The sediment was resolved in 200 μl of PBS and treated with 4 U of DNase I (Promega) at 37°C overnight. The virion DNA was extracted by using the QIAamp blood mini kit (Qiagen) and resolved in 24 μl of H2O. CrFK/LTR(FFV)lacZ cells seeded in 12-well plates were transfected with 20 μl of eluate by using Polyfect (Qiagen). Two days after transfection, the cells were split and the cell-free supernatant was transferred to fresh indicator cells after two additional days. These cells were then fixed and stained. For a control, we spiked the resolved FFV sediment with 5 μg of the infectious FV plasmid pcHSRV2 or pSFV-1 before DNase digestion.
Assay for intracellular retrotransposition (IRT).
293, HeLa, and CrFK cells were transfected with plasmid DNAs expressing the EGFP marker protein by CaPO4 coprecipitation. The percentage of EGFP-positive cells was followed over time by flow cytometry on a FACSCalibur (Becton Dickinson) as described previously (11, 11a).
Nucleotide sequence accession number.
The pChatul-2 and pChatul-3 sequences were submitted to EMBL under accession numbers AJ564745 and AJ564746, respectively.
RESULTS AND DISCUSSION
Characterization of FFV infectious molecular clones.
The subgenomic FFV plasmids described by Helps and Harbour (12) were assembled downstream of a CMV promoter to generate the full-length pChatul-1 proviral clone. To enhance transient virus production after transfection of SV40 large-T-antigen-expressing cells, we constructed pChatul-2 by insertion of an SV40 origin of replication into the vector backbone of pChatul-1. However, after transfection of 293T cells and transfer of the cell-free supernatant to CrFK/LTR(FFV)lacZ cells, both plasmids gave rise to only relatively small amounts of infectious FFV (data not shown). The serial passage of supernatant from pChatul-2-infected CrFK cells gave rise to an efficiently replicating virus. DNA from such cells was the source for the establishment of pChatul-3 by long-range PCR. Transient transfection of 293T cells with pChatul-3 resulted in cell-free virus titers of 105 to 106. pChatul-2 and pChatul-3 were completely sequenced on both strands. Comparison of the sequences revealed that pChatul-3 had acquired a number of nonsilent mutations predominantly located in the env gene and in the accessory open reading frame 2 (data not shown). The significance of individual mutations in changing the phenotype of pChatul-2 was not analyzed.
Characterization of virion nucleic acids.
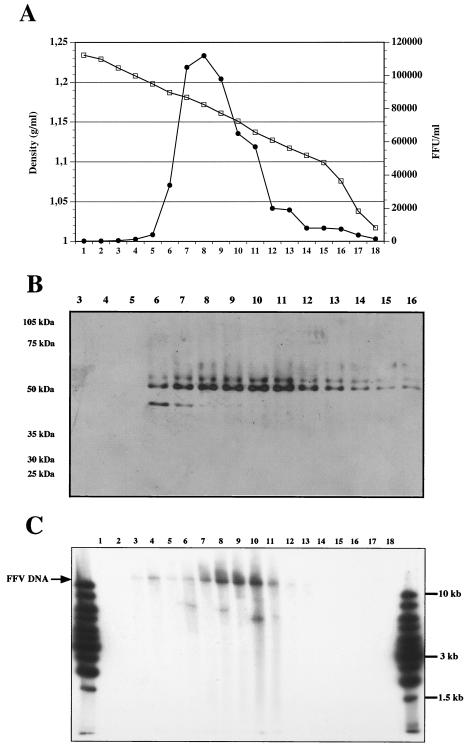
Concentrated FFV was separated by sucrose gradient centrifugation. Fractions from the gradient were collected and analyzed for infectivity on CrFK/LTR(FFV)lacZ indicator cells. As shown in Fig. 1A, the peak infectivity was found in fractions 7 to 11. Immunoblots revealed that most of the Gag protein was enriched in the same fractions (Fig. 1B). By Southern blotting we detected genome-length (approximately 11.7 kb) virion DNA, the majority of which was found in fractions 7 to 11, thus, in the same fractions with peak infectivity and viral protein (Fig. 1C).
FIG. 1.
Characterization of gradient-purified FFV. FFV was first concentrated by ultracentrifugation and then loaded onto a 20 to 66% sucrose gradient. Four hundred-microliter fractions were drawn from the bottom. (A) Ten microliters of each fraction was diluted with 990 μl of Dulbecco's modified Eagle's minimal essential medium and used for the determination of viral infectivity on CrFK/LTR(FFV)lacZ cells. (B) Twenty microliters of each fraction was loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, which was blotted, and stained with a rabbit anti-FFV Gag serum. The migration of molecular mass markers is indicated. (C) DNA was extracted from each fraction and analyzed by Southern blotting with a labeled FFV pol gene probe.
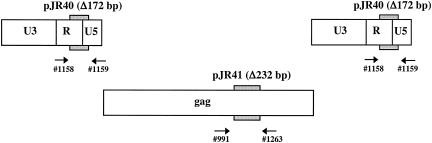
To more exactly determine the relative amounts of nucleic acids in FFV virions we used competitive DNA PCR, RNA RT-PCR, and real-time PCR and RT-PCR. The methods used for competitive PCR and RT-PCR were analogous to those described by Yu et al. for respective analyses on PFV (35). Nucleic acids were extracted from virions sedimented through a sucrose cushion and incubated with DNase to avoid contamination with traces of DNA not incorporated into the virus particles. Competitor DNA and RNA were generated from subgenomic FFV plasmids, which contained small deletions as shown in Fig. 2. The LTR primers (1158-1159) and pJR40-derived competitor were used to detect early steps of reverse transcription while the gag probes (primer pair 991-1263 and competitor plasmid pJR41) were expected to detect the later steps. The results are summarized in Table 1. Since the LTR is detected twice by the R-U5-located primers by PCR but only once by RT-PCR, the DNA PCR values were divided by 2. The calculation of the results from 8 experiments revealed a mean relative ratio of virion LTR DNA to RNA of 1:1.1, with a range from 5:1 to 1:4. For the gag region, the mean DNA/RNA ratio was determined to be 1:2.3 (range, 1:1 to 1:5) for 4 experiments. Thus, these results obtained for FFV are very similar to those reported previously for PFV (35).
FIG. 2.
Competitive PCR and RT-PCR. Locations of primers in the LTR, gag gene, and deleted regions in the competitor plasmids pJR40 and pJR41 are shown.
TABLE 1.
Summary of results of competitive PCR and RT-PCR experiments
| Region | Expt no. | Amt of DNA (pg)a | Amt of RNA (pg) | DNA/RNA ratio |
|---|---|---|---|---|
| LTR | 1 | 10 (5) | 1 | 5:1 |
| 2 | 5 (2.5) | 1 | 2.5:1 | |
| 3 | 0.1 (0.05) | 0.1 | 1:2 | |
| 4 | 1,000 (500) | 500 | 1:1 | |
| 5 | 1,000 (500) | 1,000 | 1:2 | |
| 6 | 500 (250) | 1,000 | 1:4 | |
| 7 | 5,000 (2,500) | 5,000 | 1:2 | |
| 8 | 5,000 (2,500) | 5,000 | 1:2 | |
| gag | 6 | 100 | 500 | 1:5 |
| 7 | 500 | 500 | 1:1 | |
| 8 | 500 | 500 | 1:1 | |
| 9 | 500 | 1,000 | 1:2 |
The corrected DNA amounts are shown in parentheses (see text for details).
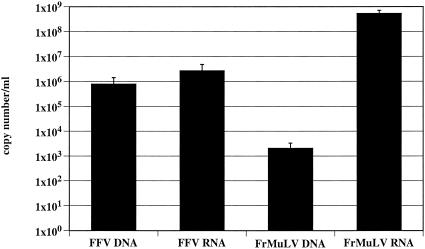
To analyze the nucleic acid composition of FFV by a different method, we applied real-time PCR and RT-PCR by using the LC technology with the orthoretrovirus FrMuLV as a control. The LC standard plasmids cover 0.5-kb regions from the gag and pol genes of FrMuLV and FFV, respectively. The results of the determination of the copy numbers are summarized in Fig. 3. For FFV, the mean ratio of DNA to RNA was determined from 5 independent experiments to be 1:5.6, with a range from 1:1 to 1:17. For FrMuLV, a DNA/RNA ratio of 1:4.1 × 105, with range from 1:2.2 × 105 to 1:1.8 × 106 was calculated from 6 independent experiments. The mean titer of centrifugation-concentrated FrMuLV, which was used for the extraction of nucleic acids, was determined from 3 experiments to be 1.3 × 104/ml (Table 2). The mean DNA copy number from the same probes was only 2.6 × 103/ml. The fact that more infectious units than DNA copies were found emphasizes the role of genomic RNA for FrMuLV replication. The respective values for FFV were 1.7 × 104 infectious units/ml compared to 6.1 × 105 DNA copies/ml. Thus, even if a great loss in infectivity by centrifugation is assumed, many more DNA copies per infectious units were found for FFV. However, the mean RNA copy number from these particular experiments was 1.2 × 106, which limits conclusions as to the direction for which FFV nucleic acid is required for replication.
FIG. 3.
Copy numbers of DNA and RNA in FFV and FrMuLV as determined by real time PCR on the LC. Mean copy numbers of all experiments are shown.
TABLE 2.
DNA and RNA copy numbers of individual experiments
| Expt no. | No. of copies of FFV:
|
FFV titer (IU/ml) | No. of copies of FrMuLV:
|
FrMuLV titer (IU/ml) | ||
|---|---|---|---|---|---|---|
| DNA | RNA | DNA | RNA | |||
| 1 | 124,000 | 750,000 | 14,000 | 1,660 | 709,000,000 | 5,750 |
| 2 | 640,000 | 600,000 | 15,000 | 2,800 | 616,000,000 | 7,000 |
| 3 | 1,070,000 | 2,335,000 | 23,250 | 3,400 | 737,000,000 | 27,000 |
The quantification of RNA and gag/pol reverse cDNA transcripts in partially purified FFV revealed approximately two to six times as much RNA, whereas in a typical orthoretrovirus we found several 100,000-fold-more RNA copies than DNA. Concerning the LTR reverse transcripts in FFV, we found a ratio that is close to 1:1. The analysis of the integrity of the nucleic acids, which were extracted from gradient-purified virions, showed genome-length DNA in the fractions of peak infectivity and viral capsid protein. Our results indicate that, in FFV, reverse transcription has taken place to a significantly different extent than in orthoretroviruses in virus-producing cells and corroborate previous reports on PFV (35). A physically detectable DNA genome appears to be a common feature of FV. However, the findings also indicate that FV reverse transcription is not necessarily complete. This view is supported by the observation that huge amounts of RNA are still present in FFV particles. Moreover, it is hard to imagine a virus that carries more genetic information stored in an irrelevant form (RNA) for further rounds of replication than in the relevant form (DNA).
Analysis of the time point of reverse transcription, IRT, and the infectivity of virion DNA.
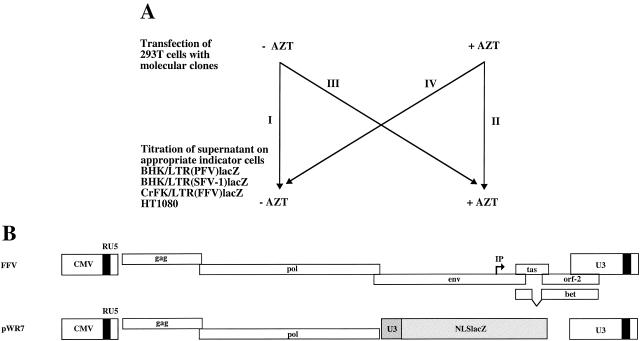
To address the question of the time point of FFV reverse transcription, i.e., early in the viral replication cycle directly after infection of a cell, as in orthoretroviruses (32), versus late in the replication cycle before new virus buds from an infected cell, as in hepadnaviruses (10, 22), we used a functional test. The AZT inhibition assay that is illustrated in Fig. 4A has already been instrumental in the analysis of PFV reverse transcription (21). AZT is the only RT inhibitor that is known to work reliably against FV RT (25). The advantage of the assay lies in the relative synchronicity of virus production and also infection, since transfection of cells with replication-deficient vectors can be used (Fig. 4A). Its disadvantage results from the use of a limited number of cell types used for virus production, which prevents a generalization of the results.
FIG. 4.
AZT inhibition assay. (A) Outline of the assay. 293T cells were transfected with viral vectors or infectious molecular clones. Production and analysis of virus were either in the absence (−) (I) or presence (+) (II) of AZT, or the drug was only present 24 h before and during virus titration (III) or production (IV). (B) Schematic presentation of the pChatul-3 infectious molecular clone and the pWR7 FFV vector. IP, internal promoter.
The transfection of 293T cells with an orthoretroviral vector system (pcSFG-NLSlacZ, which was packaged by pHIT60 and pseudotyped with the vesicular stomatitis virus glycoprotein G), the FFV vector (pWR7, pseudotyped with the envelope protein of PFV) (Fig. 4B), or FV infectious molecular clones (pcHSRV2, pSFV-1, or pChatul-3) (Fig. 4B) resulted in decent viral titers provided virus production and the analysis on the appropriate target cells was done in the absence of AZT (Table 3). Virus production and analysis in the presence of AZT reduced viral titers to almost zero (Table 3). We used AZT at 5 μM for all assays, except with pChatul-3, which required us to raise the AZT concentration to 50 μM to achieve a significant block of virus replication. The reason may be some intrinsic drug resistance of the FFV RT. This issue, however, was not further analyzed. The distinctive features of ortho- and spumaretroviruses appear in Table 3. The addition of AZT to the virus-producing cells had only a minor consequence for the assay of the MuLV titer (Table 3), whereas AZT in the target cells inhibited the establishment of infection (Table 3). The spumaretroviral plasmids behaved exactly the other way around (Table 3). This result indicates that, at least in 293T cells, FFV reverse transcription essentially occurs late in the viral replication cycle. Initiation of cDNA synthesis before viral budding appears to be a common feature of primate and nonprimate FV, at least in this cell line (21).
TABLE 3.
AZT inhibition assay results
| Construct | Target cell type | Results under conditiona:
|
|||
|---|---|---|---|---|---|
| I | II | III | IV | ||
| pcSFG-NLSlacZ/pHIT60/pcVG-wt | HT1080 | (1.32 ± 0.3) × 106 | 0 | 0 | (1.84 ± 1) × 105 |
| pcHSRV2 | BHK/LTR (PFV)lacZ | (9.25 ± 6) × 105 | 0 | (3.01 ± 3) × 105 | 0 |
| pSFV-1 | BHK/LTR(SFV-1)lacZ | (1.19 ± 0.2) × 106 | 0 | (4.11 ± 0.8) × 105 | 0.89 ± 1 |
| pChatul-3 | CrFK/LTR(FFV)lacZ | (1.68 ± 0.7) × 105 | 20 ± 20 | (9.17 ± 4) × 103 | 58.3 ± 30 |
| pWR7/pCenv-1 (PFV) | HT1080 | (4.2 ± 3) × 103 | 1 ± 1 | (2.31 ± 0.4) × 103 | 1 ± 1 |
| pWR7/pcDNA | HT1080 | 0 | 0 | 0 | 0 |
Results are means ± standard deviations from three independent experiments. Conditions: I, production and analysis of virus in the absence of AZT; II, production and analysis of virus in the presence of AZT; III, AZT was only present 24 h before and during virus titration; IV, AZT was only present 24 h before and during virus production.
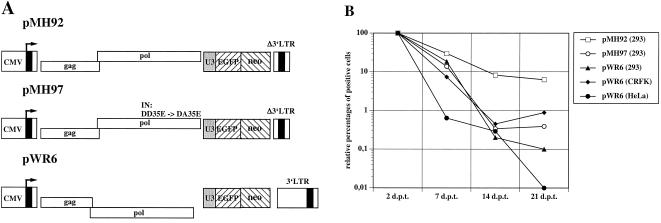
It has been demonstrated recently that PFV can recycle its cDNA genome into the nucleus, where it integrates into the host cell genome (11). The ability of FFV to reverse transcribe the RNA pregenome late in the replication cycle raises the question whether FFV can perform IRT. To analyze this, we transfected different cell lines with PFV and FFV vectors that express the gag and pol genes and a cassette with a marker gene, which can be easily monitored (Fig. 5A). The persistence of the EGFP marker encoded by the PFV vector pMH92 in transfected 293 cells is indicative for IRT (11). pMH97 is a negative control plasmid that contains a disabled PFV integrase (11). The gag- and pol-expressing FFV vector pWR6 (Fig. 5A) did not persist in transfected 293, HeLa, or CrFK cells (Fig. 5B). Thus, IRT could not be demonstrated to take place in FFV at a rate similar to that in PFV. Since the pChatul-2-derived pWR6 vector is functional in extracellular marker gene transfer (Roy et al., unpublished), it appears to be unlikely that some defect in the gag and pol genes is responsible for the lack of IRT.
FIG. 5.
Assay to determine FV IRT. (A) Schematic representation of pMH92, pMH97, and pWR6. In pMH97, the conserved DD35E motif of the integrase (IN) was converted to DA35E (8, 11). (B) Cells were transfected with the positive and negative control plasmids pMH92 and pMH97, respectively. The persistent expression of the EGFP marker by cells transfected with pMH92 is indicative for IRT (11). Values below 1% were regarded as negative (11a). The FFV plasmid pWR6, which is the homologue to the PFV plasmid pMH92, was found to be unable to lead to persistent EGFP expression after transfection of 293, HeLa, or CrFK cells. d.p.t., days posttransfection.
PFV Gag protein is transiently localized to the nucleus (28), whereas a Gag localization restricted to the cytoplasm has been reported for FFV (5). When we analyzed the intracellular FFV Gag distribution by indirect immunofluorescence, we also found the protein to be located outside the nucleus around the nuclear membrane (data not shown). These findings suggest that nuclear transport of the PFV capsid protein is required for IRT and that those FVs which do not encode a Gag nuclear localization signal are unable to perform IRT.
The infectious nature of virion DNA extracted from PFV virions after transfection of susceptible cells is probably the most distinctive features of FVs (35). To analyze this for FFV, we extracted DNA from concentrated virus that had been preincubated with DNase to destroy nucleic acids not protected by a capsid shell. Following transfection of virion DNA ranging from 1.4 to 6.8 μg, we recovered infectious cell-free virus in 6 of 6 attempts. Since control experiments revealed the complete destruction of infectious FV plasmids which were spiked into the virus solution (data not shown), we can exclude the possibility that traces of nonpackaged viral DNA sticking from outside the concentrated particles were responsible for the successful recovery of virus from transfected virion DNA.
Taken together, our results revealed the following similarities between FFV and PFV. (i) Genome-length DNA is present in viral particles. (ii) Transfection of cells with this DNA leads to infectious virus. (iii) AZT inhibition experiments point to the functional relevance of virion DNA. Despite these strong indications that all FVs are DNA viruses, we found more RNA than DNA in FFV particles. This has also been reported previously for PFV (35). These findings can be explained in several ways. (i) The virion RNA may be relevant for the completion of reverse transcription after infection of naive cells, i.e., early in the replication cycle. (ii) The virion RNA may be irrelevant for viral replication, because it was packaged supposedly in the absence of sufficient amounts of Pol protein (24). Therefore, the packed RNA could not be successfully reverse transcribed. It remains, however, physically detectable, and these virions are noninfectious. Both possibilities do not per se require genome-length RNA in the virion. (iii) It is possible that FVs can, dependent on the particular cell used to replicate the virus, switch their genome to be either RNA or DNA. In this case, the general definition of viruses would be wrong. However, the experimental evidence presented here and previously (35) argues that this possibility is unlikely, whereas possibilities i and ii require further investigation.
Acknowledgments
We thank Sylvia Kanzler for invaluable technical help throughout the study, C. R. Helps and D. A. Harbor (Bristol, United Kingdom) for the gift of FFV plasmids, A. Mergia (Gainesville, Fla.) for the SFV-1 plasmids, and U. Dittmer (Bochum, Germany) for FrMuLV and helpful discussion.
Grants from DFG (Re627/6-2 and -3), BMBF (BEO 0312191), Sächsisches Staatsministerium für Umwelt und Landwirtschaft (13-8811.61/142), Bayerische Forschungsstiftung (Forgen), and EU (BMH4-CT97-2010) supported this work.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley, New York, N.Y.
- 2.Bock, M., M. Heinkelein, D. Lindemann, and A. Rethwilm. 1998. Cells expressing the human foamy virus (HFV) accessory bet protein are resistant to productive HFV superinfection. Virology 250:194-204. [DOI] [PubMed] [Google Scholar]
- 3.Bodem, J., M. Löchelt, H. Delius, and R. M. Flügel. 1998. Detection of subgenomic cDNAs and mapping of feline foamy virus mRNAs reveals complex patterns of transcription. Virology 244:417-426. [DOI] [PubMed] [Google Scholar]
- 4.Bodem, J., M. Löchelt, I. Winkler, R. P. Flower, H. Delius, and R. M. Flügel. 1996. Characterization of the spliced pol transcript of feline foamy virus: the splice acceptor site of the pol transcript is located in gag of foamy viruses. J. Virol. 70:9024-9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodem, J., M. Zemba, and R. M. Flügel. 1998. Nuclear localization of the functional bel 1 transactivator but not of the gag proteins of the feline foamy virus. Virology 251:22-27. [DOI] [PubMed] [Google Scholar]
- 6.Dittmer, U., D. Brooks, and K. Hasenkrug. 1998. Characterization of a live-attenuated retroviral vaccine demonstrates protection via immune mechanisms. J. Virol. 72:6554-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DuBridge, R. B., P. Tang, H. C. Hsia, P.-M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enssle, J., A. Moebes, M. Heinkelein, M. Panhuysen, B. Mauer, M. Schweizer, D. Neumann-Haefelin, and A. Rethwilm. 1999. An active human foamy virus integrase is required for viral replication. J. Gen. Virol. 80:1445-1452. [DOI] [PubMed] [Google Scholar]
- 9.Ferry, N., O. Duplessis, D. Houssin, O. Danos, and J. M. Heard. 1991. Retroviral-mediated gene transfer into hepatocytes in vivo. Proc. Natl. Acad. Sci. USA 88:8377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 11.Heinkelein, M., T. Pietschmann, G. Jármy, M. Dressler, H. Imrich, J. Thurow, D. Lindemann, M. Bock, A. Moebes, J. Roy, O. Herchenröder, and A. Rethwilm. 2000. Efficient intra-cellular retrotransposition of an exogenous primate retrovirus genome. EMBO J. 19:3436-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Heinkelein, M., M. Rammling, T. Juretzek, D. Lindemann, and A. Rethwilm. 2003. Retrotransposition and cell-to-cell transfer of foamy viruses. J. Virol. 77:11855-11858 [DOI] [PMC free article] [PubMed]
- 12.Helps, C. R., and D. A. Harbour. 1997. Comparison of the complete sequence of feline spumavirus with those of the primate spumaviruses reveals a shorter gag gene. J. Gen. Virol. 78:2549-2564. [DOI] [PubMed] [Google Scholar]
- 13.Herchenröder, O., R. Turek, D. Neumann-Haefelin, A. Rethwilm, and J. Schneider. 1996. Infectious proviral molecular clones of foamy virus from chimpanzee (SFVcpz) generated by “long PCR” provide evidence for functional relationship with human foamy virus (HFV). Virology 214:685-689. [DOI] [PubMed] [Google Scholar]
- 14.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. Particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linial, M., and R. A. Weiss. 2001. Other human and primate retroviruses, p. 2123-2139. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 16.Linial, M. L. 1999. Foamy viruses are unconventional retroviruses. J. Virol. 73:1747-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löchelt, M., W. Muranyi, and R. M. Flügel. 1993. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc. Natl. Acad. Sci. USA 90:7317-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lori, F., F. di Marzo Veronese, A. L. de Vico, P. Lusso, M. S. Reitz, Jr., and R. C. Gallo. 1992. Viral DNA carried by human immunodeficiency virus type 1 virions. J. Virol. 66:5067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mergia, A., K. E. S. Shaw, E. Pratt-Lowe, P. A. Barry, and P. A. Luciw. 1990. Simian foamy virus type 1 is a retrovirus which encodes a transcriptional transactivator. J. Virol. 64:3598-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mergia, A., and M. Wu. 1998. Characterization of provirus clones of simian foamy virus type 1. J. Virol. 72:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moebes, A., J. Enssle, P. D. Bieniasz, M. Heinkelein, D. Lindemann, M. Bock, M. O. McClure, and A. Rethwilm. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nassal, M., and H. Schaller. 1993. Hepatitis B virus replication. Trends Microbiol. 1:221-228. [DOI] [PubMed] [Google Scholar]
- 23.Pietschmann, T., M. Heinkelein, M. A. Heldmann, H. Zentgraf, A. Rethwilm, and D. Lindemann. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rethwilm, A. 2003. Foamy virus replication strategy. Curr. Top. Microbiol. Immunol. 277:1-26. [DOI] [PubMed] [Google Scholar]
- 25.Rinke, C. S., P. L. Boyer, M. D. Sullivan, S. H. Hughes, and M. L. Linial. 2002. Mutation of the catalytic domain of the foamy virus reverse transcriptase leads to loss of processivity and infectivity. J. Virol. 76:7560-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saïb, A. 2003. Non-primate foamy viruses. Curr. Top. Microbiol. Immunol. 277:197-211. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schliephake, A. W., and A. Rethwilm. 1994. Nuclear localization of foamy virus gag precursor protein. J. Virol. 68:4946-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt, M., and A. Rethwilm. 1995. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology 210:167-178. [DOI] [PubMed] [Google Scholar]
- 30.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffith, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trono, D. 1992. Partial reverse transcripts in virions from human immunodeficiency and murine leukemia virus. J. Virol. 66:4893-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogt, P. K. 1997. Historical introduction to the general properties of retroviruses, p. 1-25. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 33.Wilk, T., V. Geiselhart, M. Frech, S. D. Fuller, R. M. Flügel, and M. Löchelt. 2001. Specific interaction of a novel foamy virus env leader protein with the N-terminal gag domain. J. Virol. 75:7995-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, S. F., D. N. Baldwin, S. R. Gwynn, S. Yendapalli, and M. L. Linial. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579-1582. [DOI] [PubMed] [Google Scholar]
- 35.Yu, S. F., M. D. Sullivan, and M. L. Linial. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]