Abstract
Although Streptococcus mutans has been implicated as a major etiological agent of dental caries, our cross-sectional preliminary study indicated that 10% of subjects with rampant caries in permanent teeth do not have detectable levels of S. mutans. Our aims were to use molecular methods to detect all bacterial species associated with caries in primary and permanent teeth and to determine the bacterial profiles associated with different disease states. Plaque was collected from 39 healthy controls and from intact enamel and white-spot lesions, dentin lesions, and deep-dentin lesions in each of 51 subjects with severe caries. 16S rRNA genes were PCR amplified, cloned, and sequenced to determine species identities. In a reverse-capture checkerboard assay, 243 samples were analyzed for 110 prevalent bacterial species. A sequencing analysis of 1,285 16S rRNA clones detected 197 bacterial species/phylotypes, of which 50% were not cultivable. Twenty-two new phylotypes were identified. PROC MIXED tests revealed health- and disease-associated species. In subjects with S. mutans, additional species, e.g., species of the genera Atopobium, Propionibacterium, and Lactobacillus, were present at significantly higher levels than those of S. mutans. Lactobacillus spp., Bifidobacterium dentium, and low-pH non-S. mutans streptococci were predominant in subjects with no detectable S. mutans. Actinomyces spp. and non-S. mutans streptococci were predominant in white-spot lesions, while known acid producers were found at their highest levels later in disease. Bacterial profiles change with disease states and differ between primary and secondary dentitions. Bacterial species other than S. mutans, e.g., species of the genera Veillonella, Lactobacillus, Bifidobacterium, and Propionibacterium, low-pH non-S. mutans streptococci, Actinomyces spp., and Atopobium spp., likely play important roles in caries progression.
Dental caries is one of the most common chronic infectious diseases in the world (2, 39). There are three major hypotheses for the etiology of dental caries: the specific plaque hypothesis, the nonspecific plaque hypothesis, and the ecological plaque hypothesis (24, 26, 37). The specific plaque hypothesis has proposed that only a few specific species, such as Streptococcus mutans and Streptococcus sobrinus, are actively involved in the disease. On the other hand, the nonspecific plaque hypothesis maintains that caries is the outcome of the overall activity of the total plaque microflora, which is comprised of many bacterial species (37). The ecological plaque hypothesis suggests that caries is a result of a shift in the balance of the resident microflora driven by changes in local environmental conditions (26).
Caries-associated bacteria traditionally have been identified by using culture-based methods, which exclude not-yet-cultivated species. Molecular methods for bacterial identification and enumeration now are performed routinely to more precisely study bacterial species that are associated with dental caries, including those that are not presently cultivable (32, 33). In a previous study, Becker et al. (3) compared the bacterial species found in early childhood caries to those found in caries-free children. Some species, such as Streptococcus sanguinis, were associated with health, while others, such as S. mutans, other Streptococcus spp., Veillonella spp., Actinomyces spp., Bifidobacterium spp., and Lactobacillus fermentum, were associated with caries (3). These data also suggested that Actinomyces gerencseriae and other Actinomyces spp. play an important role in caries initiation. Munson et al. (29) used cultural and molecular techniques similar to ours to determine those species associated with the middle and advancing front of dental caries in adults. The authors demonstrated a diverse bacterial community, including S. mutans, Lactobacillus spp., Rothia dentocariosa, and Propionibacterium spp. (29). They also found that numerous novel taxa were present in carious lesions. Chhour et al. (8) used similar molecular techniques to determine the microbial diversity in advanced caries in adults. They demonstrated an abundance of species of the genera Lactobacillus, Prevotella, Selenomonas, Dialister, Fusobacterium, Eubacterium, Olsenella, Bifidobacterium, Propionibacterium, and Pseudoramibacter. S. mutans was not commonly detected. Corby et al. (10) examined the bacteria associated with dental caries and health in a subset of 204 twins aged 1.5 to 7 years old. A strain of an Actinomyces species, S. mutans, and Lactobacillus spp. were associated with disease. In contrast, bacterial species, including Streptococcus parasanguinis, Abiotrophia defectiva, Streptococcus mitis, Streptococcus oralis, and S. sanguinis, predominated in the indigenous bacterial flora of caries-free subjects (10). These findings concurred with those of our earlier study (1), demonstrating that there is a distinctive microbiota of the healthy oral cavity that is different from that associated with oral disease.
The purposes of this study were twofold: (i) to determine all bacterial species, cultivable as well as not yet cultivable, that are associated with health and dental caries of permanent teeth in children and young adults; and (ii) to describe the changes in bacterial profiles associated with the different states of this disease in primary and permanent teeth. Our major goal is to identify all of the species associated with health and disease, especially early on in the infection, that would provide alternative targets for biological intervention.
MATERIALS AND METHODS
Subject population.
Subjects with severe dental caries and age-matched, caries-free controls were recruited from two different groups: (i) 15 subjects with caries in primary teeth and 14 controls and (ii) 36 subjects with caries in secondary teeth and 25 controls. Subjects were recruited from The Columbus Children's Hospital Dental Clinic, Columbus, OH, and The Ohio State University College of Dentistry Dental Clinic, Columbus. The subjects ranged in age from 2 to 21 years. Matching control subjects had no caries or existing restorations. Samples from primary teeth were previously analyzed (3) for 23 bacterial species or species groups. The actual samples were included in this study for comparisons and for analysis in an expanded panel of 110 bacterial species.
Sampling.
Plaque samples from the 39 healthy subjects were pooled from a minimum of four sites, including anterior and posterior teeth. From the 51 caries subjects, plaque was collected separately from each of the following four different types of sites: (i) surfaces of intact enamel, (ii) surfaces of white-spot lesions, (iii) cavitated dentin lesions, and (iv) deep-dentin lesions. From the enamel, white-spot lesions, and cavitated lesions, the plaque was obtained by swiping the tooth surface with a dental explorer and transferring it onto a coarse sterile endodontic paper point. The carious dentin was excavated with either a spoon excavator or a round burr at slow speed using a dental drill. Each sample was obtained by pooling material collected from at least three different teeth. Samples were placed in a sterile 1.5-ml microcentrifuge tube and transported to the laboratory, where they were frozen until further analysis. Samples were collected in a cross-sectional fashion, representing all of the defined disease states, to describe the changes in the bacterial profiles of predominant bacteria.
Isolation of bacterial DNA.
The DNA was isolated (3) and purified (22) as previously described and frozen at −20°C for later analysis.
Clonal analysis.
The samples from permanent teeth of the following subjects were selected for bacterial clonal analysis: three caries subjects with detectable levels of S. mutans, two caries subjects without detectable levels of S. mutans, and two subjects from the healthy control group. Each of the disease states (as described above) was represented among the five selected caries subjects and one pooled sample from the healthy subjects, for a total of 22 samples. The selection of subjects for clonal analysis was based on results from preliminary checkerboard analyses (see below).
Amplification of 16S rRNA genes by PCR and purification of PCR products.
The 16S rRNA gene was amplified using a universal primer set (1) under standardized conditions, and the purification of PCR products was performed as previously described (1).
Cloning procedures.
The cloning of PCR-amplified DNA was performed with the TOPO TA cloning kit (Invitrogen, San Diego, CA) according to the instructions of the manufacturer. Correct sizes of the inserts were determined in a PCR with an M13 (−20) forward primer and an M13 reverse primer (Invitrogen). Prior to the sequencing of the fragments, the PCR-amplified 16S rRNA gene fragments were purified and concentrated according to the methods of Paster et al. (33).
16S rRNA gene sequencing.
Purified PCR-amplified 16S rRNA gene inserts were sequenced using an ABI Prism cycle sequencing kit (BigDye Terminator cycle sequencing kit with AmpliTaq DNA polymerase FS; ABI, Foster City, CA) and a GeneAmp PCR system 9700 (ABI). The primers and sequencing reactions used for sequencing have been described previously (33).
16S rRNA gene sequencing and data analysis of unrecognized inserts.
Inserts of the correct size of approximately 1,500 bases were analyzed from 46 to 70 clones per sample (an average of 58.4 clones with a standard deviation of 6.3) for a total of 1,285 clones. Healthy controls and each of the four disease states (described above) were represented in the 22 samples analyzed. A sequence of approximately 500 bases was obtained first to determine the identity or approximate phylogenetic position. Full sequences of about 1,500 bases were achieved by using five to six additional sequencing primers (19) for those species deemed novel. Sequences of inserts were analyzed according to Aas et al. (1). The similarity matrices were corrected for multiple-base changes at single positions (18) and were constructed from the aligned sequences by using only those sequence positions for which data were available for 90% of the strains tested. The construction (34) and drawing (38) of phylogenetic trees were done according to Paster et al. (33). We are aware of the potential creation of 16S rRNA gene chimera molecules assembled during the PCR (23). The percentage of chimeras in 16S rRNA gene libraries ranged from 1 to 15%. Chimeric sequences were identified by using the Chimera Check program in The Ribosomal Database Project (RDP-II; release 9.55; Center for Microbial Ecology, Michigan State University; http://rdp.cme.msu.edu/), by treeing analysis, or by base signature analysis. The species identification of the chimeras was obtained, but the sequences were not examined for the phylogenetic analysis.
Reverse-capture checkerboard assay.
Based on the sequencing data from clonal analyses and previous results (3), 16S rRNA gene-based probes were designed for the most prevalent bacterial species associated with caries. Sequences selected for use as probes were identified by the alignment of 16S rRNA gene sequences to detect regions where mismatches occurred (32). 16S rRNA gene probes were designed for a hybridization temperature of 55°C, and the standard probe length was 18 to 20 nucleotides. New probes were quality tested against an extended clone library for cross-reactivity. From the 90 subjects, 1,000 clinical samples were analyzed for 110 bacterial supragingival species in a DNA-DNA hybridization assay (Table 1). The reverse-capture checkerboard assay was carried out as previously described (3, 32). DNA probes were synthesized with multiple thymidines (T) at the 5′ end of the oligonucleotide. The poly(T) tails were cross-linked to a nylon membrane support via UV irradiation, leaving the specific probe available for hybridization. The 16S rRNA genes from clinical samples were amplified by PCR with universal primers; the forward primer was labeled at the 5′ end with digoxigenin. The digoxigenin-labeled amplicon was hybridized to the capture probes bound to the membrane, and the digoxigenin residues were complexed to anti-digoxigenin antibody covalently bound to alkaline phosphatase. The levels of chemifluorescence were measured using a Storm system (Storm Technology Inc., Mountain View, CA).
TABLE 1.
16S rRNA gene probes used in a reverse-capture checkerboard assay to detect predominant species and groups of species of bacteriaa
| Probe designation |
|---|
| Abiotrophia defectiva |
| Actinomyces georgiae |
| Actinomyces gerencseriae |
| Actinomyces israelii |
| Actinomyces naeslundii serotype II |
| Actinomyces odontolyticus sp. cl. DR002 |
| Actinomyces sp. GU067 |
| Actinomyces sp. oral cl. AG004 |
| Actinomyces sp. oral cl. AP064 |
| Actinomyces sp. oral cl. EP005 |
| Actinomyces sp. strain ATCC 49338 |
| Actinomyces sp. strain B19SC |
| Actinomyces sp. strain B27SC |
| Atopobium genomospecies C1 |
| Bacteroides sp. cl. AU126 |
| Bifidobacterium allb |
| Bifidobacterium sp. oral cl. CX010 |
| Campylobacter concisus |
| Campylobacter gracilis |
| Campylobacter showae |
| Capnocytophaga gingivalis |
| Capnocytophaga granulosum |
| Capnocytophaga ochracea ρ 089 |
| Capnocytophaga sp. cl. DS022 |
| Capnocytophaga sputigena |
| Cardiobacterium sp. strain A/Cardiobacterium hominis |
| Cardiobacterium sp. cl. BU084/BM058 |
| Catonella morbi |
| Corynebacterium matruchotii |
| Corynebacterium sp. cl. AK153 |
| Eubacterium saburreum sp. cl. GT038 |
| Eubacterium sp. cl. BB124 |
| Eubacterium sp. cl. BE088 |
| Eubacterium sp. cl. C27KA |
| Eubacterium sp. cl. DO008 |
| Eubacterium sp. cl. DO016 |
| Eubacterium sp. cl. EI074 |
| Eubacterium yurii |
| Fusobacterium all |
| Fusobacterium animalis |
| Fusobacterium nucleatum subsp. vincentii |
| Fusobacterium periodonticum |
| Fusobacterium nucleatum subsp. polymorphum |
| Gemella haemolysans |
| Gemella morbillorum |
| Granulicatella adiacens |
| Haemophilus parainfluenzae |
| Kingella sp. cl. DE012/Neisseria polysaccharea |
| Kingella denitrificans |
| Kingella oralis |
| Lactobacillus all |
| Lactobacillus fermentum |
| Lactobacillus gasseri |
| Lactobacillus sp. cl. CX036 |
| Lautropia mirabilis |
| Leptotrichia all |
| Leptotrichia sp. cl. BU064 |
| Leptotrichia sp. cl. DE081 |
| Leptotrichia sp. cl. DR011 |
| Leptotrichia sp. cl. DS049 |
| Leptotrichia sp. cl. DT031 |
| Leptotrichia sp. cl. GT020 |
| Leptotrichia sp. cl. GT018 |
| Leptotrichia sp. cl. A39FD |
| Neisseria mucosa/Neisseria flavescens |
| Peptostreptococcus sp. cl. CK035 |
| Porphyromonas sp. cl. DS033 |
| Prevotella sp. cl. GU069/Prevotella sp. cl. GU027 |
| Prevotella loescheii |
| Prevotella melaninogenica |
| Prevotella oris |
| Prevotella sp. cl. AO036 |
| Propionibacterium sp. strain FMA5 |
| Rothia dentocariosa |
| Selenomonas infelix |
| Selenomonas noxia |
| Selenomonas sp. cl. AA024 |
| Selenomonas sp. cl. GT010 |
| Selenomonas sp. cl. GT052 |
| Selenomonas sp. cl. EY047 |
| Selenomonas sp. oral cl. Cs024 |
| Selenomonas sp. caries cl. DS051 |
| Selenomonas sp. caries cl. DS071 |
| Selenomonas sputigena |
| Streptococcus anginosus |
| Streptococcus australis T1-E5 |
| Streptococcus cristatus |
| Streptococcus gordonii |
| Streptococcus intermedius |
| Streptococcus mitis |
| Streptococcus mitis biovar 2 |
| Streptococcus mitis/Streptococcus oralis |
| Streptococcus mutans |
| Streptococcus parasanguinis |
| Streptococcus salivarius |
| Streptococcus sanguinis |
| Streptococcus sp. cl. CH016 |
| Streptococcus sp. group H6 |
| Streptococcus sp. oral cl. AA007 |
| Streptococcus sp. strain 7A/Streptococcus sp. strain H6 |
| Streptococcus sp. strain T4-E3 |
| Tannerella forsythia-like cl. BU063 |
| Treponema socranskii subsp. paredis |
| Veillonella atypica |
| Veillonella dispar/parvula |
| Veillonella sp. oral cl. BU083 |
Combination probes are named after the pairs of species (e.g., “Cardiobacterium sp. strain A/Cardiobacterium hominis”) or groups of species (e.g., “Bifidobacterium all”) of bacteria detected.
all, all species of the genus.
Statistical analysis.
Bacterial signals were measured by the intensity of spots on checkerboard membranes with ImageQuant TL (Amersham Biosciences, Piscataway, NJ) for array analysis as previously described (10). Statistical comparisons were made by pairwise comparisons of the healthy controls to each of the sample groups (intact enamel, white-spot lesions, cavitated dentin lesions, and deep-dentin lesions) in subjects with caries by the PROC MIXED procedure (SAS, version 8; SAS Institute Inc., Cary, NC). The variance between the disease states is defined with the random statement of disease and dentition association for each of the bacteria tested (data not shown).
Nucleotide sequence accession numbers.
The complete 16S rRNA gene sequences of clones representing novel phylotypes defined in this study, sequences of known species not previously reported, and published sequences are available for electronic retrieval from the EMBL (http://www.ebi.ac.uk/embl/), GenBank (http://www.ncbi.nlm.nih.gov/GenBank/), and DDBJ (http://www.ddbj.nig.ac.jp/) nucleotide sequence databases under the accession numbers shown in Fig. 1, 2, and 3.
FIG. 1.
Phylogenetic relationships of species detected from dental caries in permanent teeth and from healthy controls. Subjects with (w Sm) and without (w/o Sm) detectable levels of S. mutans (Sm) were selected for clonal analysis. The distribution and levels of bacterial species/phylotypes among the five subjects and two healthy controls are shown by the columns of boxes to the right of the tree as either not detected (clear box), <15% of the total number of clones assayed (shaded box), or ≥15% of the total number of clones assayed (darkened box). Fifteen percent was chosen as the arbitrary level. GenBank accession numbers are provided. The marker bar represents a 5% difference in nucleotide sequences.
FIG. 2.
Mean levels of health-associated species or species groups in primary teeth determined by a reverse-capture checkerboard assay for 15 subjects with caries (green bars) and 14 healthy controls (white bars). *, P ≤ 0.05 for pairwise comparisons of the healthy group to other groups by PROC MIXED. P indicates the overall P value comparing all groups and sites by PROC MIXED. Strep., Streptococcus; Porphyrom., Porphyromonas; Eubact., Eubacterium; Capnocyto., Capnocytophaga; Fusobact., Fusobacterium; cl., clone.
FIG. 3.
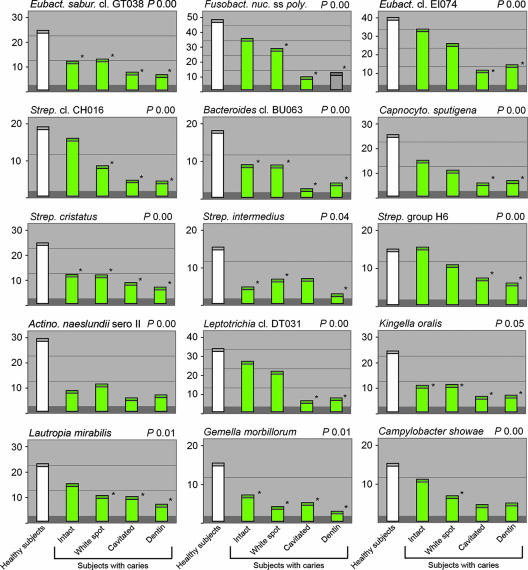
Mean levels of health-associated species or species groups in permanent teeth determined by a reverse-capture checkerboard assay for 36 subjects with caries (green bars) and 25 healthy controls (white bars). *, P ≤ 0.05 for pairwise comparisons of the healthy group to other groups by PROC MIXED. P indicates the overall P value comparing all groups and sites by PROC MIXED. Strep., Streptococcus; Eubact., Eubacterium; Eubact. sabur., Eubacterium saburreum; Capnocyto., Capnocytophaga; Fusobact. nuc. ss poly., Fusobacterium nucleatum subsp. polymorphum; Actino, Actinomyces; cl., clone.
RESULTS AND DISCUSSION
In our preliminary study, supragingival plaque from each of the four defined disease states in 42 subjects with severe dental caries in the secondary dentition was analyzed, screening for a selection of 28 caries-associated bacterial species using a reverse-capture DNA-DNA hybridization assay. The results of the study demonstrated that 10% of subjects with rampant caries in the secondary dentition do not have detectable levels of S. mutans. In that study, S. mutans was not detected in intact enamel and white-spot lesions of diseased subjects, while most dentin lesions or deep-dentin lesions harbored S. mutans.
The second step of the study was to assess those bacterial species associated with the progression of dental caries in permanent teeth using pooled samples representing the disease states; intact enamel, white-spot lesions, dentin lesions, and deep-dentin lesions in each of the five subjects with severe caries and pooled plaque from two healthy controls were selected for cloning and sequencing. Samples from subjects with and without detectable levels of S. mutans were included for comparison, and the selection of subjects for the clonal analysis was based on results from preliminary checkerboard analyses. Sequences from a total of 1,285 16S rRNA gene clones were used to identify species and the closest relatives in 22 samples. Sequence analysis demonstrated a striking bacterial diversity of 197 species or phylotypes, representing eight bacterial phyla. Of this total, 99 (50%) strains have not yet been cultivated, 12 were previously uncharacterized strains, and 86 were known species or phylotypes. Twenty-two novel phylotypes were identified in this study. From each lesion, the number of taxa ranged from 8 to 37 (Fig. 1). The phylogenetic relationships of species detected in health and disease are shown in Fig. 1, and the differences in bacterial profiles between healthy samples and those from each of the four stages of the disease are summarized in Table 2.
TABLE 2.
Progression of dental caries
| Species | Dental caries progression fora:
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy subject no.
|
Diseased subject no.
|
|||||||||||||||||||||
| 1 | 2 | 1 (with S. mutans)
|
2 (with S. mutans)
|
3 (with S. mutans)
|
4 (without S. mutans)
|
5 (without S. mutans)
|
||||||||||||||||
| a | b | c | d | a | b | c | d | a | b | c | d | a | b | c | d | a | b | c | d | |||
| Streptococcus mutans | − | − | + | + | − | + | − | − | + | + | − | − | + | + | − | − | − | − | − | − | − | − |
| Abiotrophia and Granulicatella spp. | − | + | − | − | − | − | + | + | + | − | − | + | − | − | − | − | − | − | + | + | + | + |
| Actinomyces spp. | + | + | + | + | − | + | − | − | − | − | + | + | − | − | + | + | − | − | + | + | + | − |
| Actinobaculum spp. | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Atopobium spp. | − | − | − | − | − | − | − | − | + | − | − | − | − | + | − | − | − | − | − | − | − | − |
| Bifidobacterium dentium | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + |
| Capnocytophaga spp. | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| Corynebacterium spp. | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Eubacterium spp. | − | + | − | − | + | + | − | − | − | + | + | − | + | + | − | − | − | − | − | − | − | − |
| Fusobacterium spp. | + | − | − | − | − | − | + | + | − | − | + | − | − | − | − | − | − | − | − | + | − | − |
| Lactobacillus spp. | − | − | − | − | − | − | − | − | + | + | − | − | − | + | − | − | + | + | − | − | + | − |
| Leptotrichia spp. | + | + | + | − | − | − | + | + | − | − | + | − | − | − | − | − | − | − | − | − | − | − |
| Prevotella spp. | − | + | − | − | − | + | + | + | − | − | + | + | − | − | − | − | − | − | − | − | − | − |
| Selenomonas spp. | + | + | − | − | + | − | − | − | + | − | + | − | + | − | − | − | − | − | + | − | + | − |
| Streptococcus anginosus | − | − | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Streptococcus gordonii | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − |
| Streptococcus intermedius | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − |
| Streptococcus parasanguinis | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | + | + | − | − | − | − |
| Streptococcus salivarius | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | + | + |
| Streptococcus sanguinis | − | + | − | − | − | − | − | − | − | − | + | + | + | + | − | − | − | − | + | + | + | + |
| Veillonella spp. | + | + | + | + | + | + | − | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + |
The presence (+) or absence (−) of predominant species at specific sites is indicated. Boldface indicates species detected in >15% of the total number of clones. a, intact enamel; b, white spots; c, dentin cavities; d, deep-dentin cavities.
In subjects with S. mutans (subjects 1, 2, and 3) (Table 2), additional species, such as Atopobium genomospecies C1 or Lactobacillus spp., were present at significantly higher levels. In subject 1, Atopobium genomospecies C1 predominated among all species, constituting 40% of the clones, while S. mutans was detected only at a low level (Fig. 1). In subjects without S. mutans, subject 4 had a predominance of Lactobacillus spp., whereas subject 5 had a predominance of Bifidobacterium dentium and low-pH non-S. mutans streptococci, including S. mitis, S. sanguinis, Streptococcus gordonii, Streptococcus sp. clone BW009, Streptococcus sp. clone EK048, and Streptococcus salivarius (Fig. 1). The bacterial profiles during the initiation of caries (i.e., on intact enamel and white-spot lesions) (Fig. 1, sites a and b) were more distinct and complex than those in established caries (i.e., dentin and deep-dentin lesions) (Fig. 1, sites c and d, and Table 2). Actinomyces spp. and non-S. mutans streptococci were detected at high levels in caries initiation (Fig. 1, sites a and b, and Table 2) compared to those of other disease states. Bacterial profiles of caries in the secondary dentition varied from subject to subject (Fig. 1, Table 2).
The goals of the clonal analyses were to first determine the breadth of bacterial diversity in dental plaque samples and then assess specific bacterial associations with health and disease. However, to better assess bacterial associations with health status, the final step of our study was to use a reverse-capture DNA-DNA hybridization assay using oligonucleotide probes targeting 110 prevalent oral bacterial taxa. A total of 243 clinical samples from primary and secondary teeth in a total of 90 selected subjects were analyzed. Bacteria in pooled plaque samples from the enamel of healthy subjects (n = 39) were compared to those in samples from intact enamel, white-spot lesions, and dentin and deep-dentin lesions from each of the subjects with severe decay (n = 51). Pooled plaque samples from a minimum of four sites in healthy individuals and pooled material collected from at least three different teeth representing four states of caries may dilute the number of bacteria that dominate and alter the proportion of species present at a single site. Nevertheless, the predominant species present still will be detected.
Based on these checkerboard analyses, differences in bacterial profiles between primary and secondary teeth, as well as those of a number of health-associated and disease-associated species, were revealed (Fig. 2 to 5, Table 3). Statistical analyses also showed that bacterial profiles of the intact enamel of healthy subjects differed significantly from the bacterial profiles of intact enamel from diseased subjects in both age groups (data not shown). In primary teeth, health-associated species such as Capnocytophaga granulosa, Eubacterium sp. clone DO016, and Streptococcus cristatus were detected at the same levels in plaque from intact enamel of diseased subjects and plaque from healthy subjects (Fig. 2). Even though S. mutans was barely present in the permanent teeth of healthy subjects, it was well represented in the bacterial flora of the healthy enamel of diseased subjects (Fig. 5). The bacterial flora of healthy sites in primary teeth was characterized by 15 significant health-associated species, which was in contrast to the 30 health-associated species that we found at healthy sites in permanent teeth. As many as 13 of the 15 significant health-associated species in primary teeth also were significant in permanent teeth (data not shown). Species such as Kingella oralis, Eubacterium saburreum clone GT038, Gemella morbillorum, S. cristatus, and Streptococcus intermedius were found at a high level in healthy subjects and were at significantly reduced levels in the healthy enamel of diseased subjects in permanent teeth (Fig. 3). Other health-associated species in permanent teeth, such as Streptococcus sp. clone CH016, Streptococcus sp. group H6, Leptotrichia sp. clone DT031, Eubacterium sp. clone EI074, Campylobacter showae, Fusobacterium nucleatum subsp. polymorphum, and Capnocytophaga sputigena, were detected at reduced levels in plaque from white-spot or dentin lesions of diseased subjects compared to the levels detected in healthy subjects (Fig. 3). Previous studies support the presence of these species in the healthy oral cavity, which is different from those of oral disease (1, 10). Among the health-associated species, we also noted that F. nucleatum subsp. polymorphum, E. saburreum clone GT038, Eubacterium sp. clone EI074, Eubacterium sp. clone DO016, Bacteroides sp. clone BU063, Leptotrichia sp. clone DE084, Leptotrichia sp. clone DT031, G. morbillorum, Streptococcus spp., and Kingella spp. were more pronounced in the secondary dentition than in the primary dentition (Fig. 2 and 3 and data not shown). Furthermore, we observed an overall greater diversity associated with caries in permanent teeth based on bacteria present at significant levels.
FIG. 5.
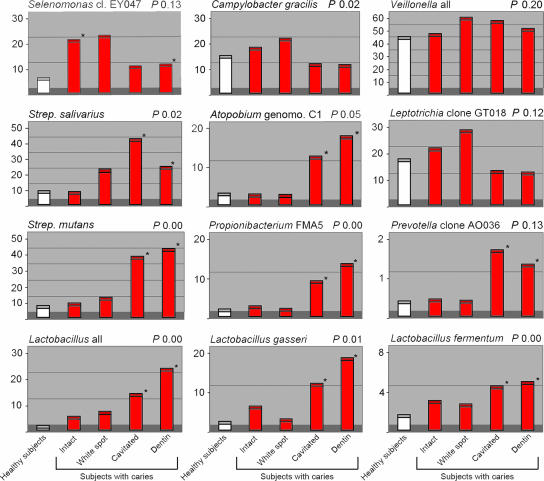
Mean levels of caries-associated species or species groups in permanent teeth determined by a reverse-capture checkerboard assay for 36 subjects with caries (red bars) and 25 healthy controls (white bars). *, P ≤ 0.05 for pairwise comparisons of the healthy group to other groups by PROC MIXED. P indicates the overall P value comparing all groups and sites by PROC MIXED. Strep., Streptococcus; genomo., genomospecies.
TABLE 3.
Predominant species in bacterial profiles related to caries progression in primary and permanent dentitionsa
| Species detected in primary dentition
|
Species detected in permanent dentition
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | Intact enamel | White-spot lesion | Dentin lesion | Deep-dentin lesion | Control | Intact enamel | White-spot lesion | Dentin lesion | Deep-dentin lesion |
| Leptotrichia allb | Actinomyces sp. cl. GU067 | Veillonella all | Veillonella all* | Streptococcus mutans* | Actinomyces sp. cl. GU067 | Actinomyces sp. cl. GU067 | Veillonella all | Veillonella all | Veillonella all |
| Veillonella all | Veillonella all | Actinomyces sp. cl. GU067* | Streptococcus mutans* | Veillonella all | Fusobacterium nucleatum subsp. polymorphum | Veillonella all | Actinomyces sp. cl. GU067 | Actinomyces sp. cl. GU067* | Actinomyces sp. cl. GU067 |
| Streptococcus anginosus | Leptotrichia all | Streptococcus sanguinis* | Actinomyces sp. cl. GU067 | Actinomyces sp. cl. GU067 | Veillonella all | Streptococcus sanguinis | Streptococcus sanguinis | Streptococcus salivarius* | Streptococcus mutans* |
| Actinomyces sp. cl. GU067 | Streptococcus sanguinis* | Leptotrichia all | Streptococcus sanguinis | Lactobacillus all* | Streptococcus sanguinis | Fusobacterium nucleatum subsp. polymorphum | Leptotrichia sp. cl. GT018 | Streptococcus mutans* | Streptococcus salivarius* |
| Capnocytophaga granulosa | Streptococcus mitis* | Streptococcus salivarius | Streptococcus salivarius* | Streptococcus salivarius | Eubacterium sp. cl. EI074 | Eubacterium sp. cl. EI074 | Fusobacterium nucleatum subsp. polymorphum* | Streptococcus sanguinis | Lactobacillus all* |
| Fusobacterium all | Capnocytophaga sputigena | Capnocytophaga sputigena | Veillonella sp. cl. BU083 | Streptococcus sanguinis | Leptotrichia sp. cl. DT031 | Leptotrichia sp. cl. DT031 | Eubacterium sp. cl. EI074 | Streptococcus mitis | Streptococcus sanguinis |
| Streptococcus sanguinis | Capnocytophaga granulosa | Streptococcus mitis | Streptococcus mitis | Streptococcus mitis | Fusobacterium animalis | Streptococcus mitis | Fusobacterium animalis | Fusobacterium animalis | Streptococcus mitis |
| Campylobacter gracilis | Streptococcus cristatus | Streptococcus mutans | Selenomonas sp. cl. EY047 | Veillonella sp. cl. BU083 | Leptotrichia all | Leptotrichia sp. cl. GT018 | Leptotrichia all | Veillonella sp. cl. BU083 | Streptococcus anginosus |
| Streptococcus cristatus | Actinomyces sp. strain B19SC | Corynebacterium sp. cl. AK153 | Leptotrichia all* | Actinomyces sp. strain B19SC | Actinomyces naeslundii serotype II | Selenomonas sp. cl. EY047* | Selenomonas sp. cl. EY047 | Lactobacillus all* | Lactobacillus gasseri* |
| Capnocytophaga sputigena | Streptococcus sp. cl. CH016 | Actinomyces gerencseriae* | Actinomyces gerencseriae | Selenomonas sp. cl. EY047 | Capnocytophaga sputigena | Capnocytophaga granulosa | Streptococcus mitis | Atopobium genomospecies C1* | Atopobium genomospecies C1* |
%The results are based on the signal level in a reverse-capture checkerboard hybridization assay. *, P ≤ 0.05 for pairwise comparisons of the healthy group to other groups by PROC MIXED tests. cl., clone.
all, all species of the genus.
There were several disease-associated species with distinct bacterial profiles at each stage of caries progression (Fig. 4 and 5). In white-spot and dentin lesions, species like S. parasanguinis and S. salivarius were observed at high levels in both dentitions. In addition, Corynebacterium sp. clone AK153 and A. gerencseriae were detected at high levels in primary dentition, as were Leptotrichia sp. clone GT018 (data not shown), Campylobacter gracilis, and Selenomonas sp. clone EY04 in permanent dentition. The microflora of deep-dentin lesions was dominated by S. mutans, Lactobacillus spp., Propionibacterium sp. strain FMA5, and Atopobium genomospecies C1 (permanent) and Bifidobacterium spp. (primary) (Fig. 4 and 5). S. mutans seems to have a more dominating role in dentin and deep-dentin caries of primary teeth than in those of secondary teeth, in which additional species like Propionibacterium sp. strain FMA5 were commonly detected at higher levels (Fig. 4 and 5). All of these species are acid producers and may be involved in the etiology of caries. As illustrated in Fig. 4 and 5, these species were found at the highest levels only at the late stages of disease. Notably, 10% of the subjects with dental caries did not have detectable or low levels of S. mutans. Other studies also report about 10 to 15% of caries-active subjects that do not have detectable levels of S. mutans, and therefore the presence of S. mutans does not necessarily indicate caries activity (4). These data clearly demonstrated that the bacterial etiology of caries involves other potential acid-producing species, some of which have not yet been cultivated. Further longitudinal studies with animal models are necessary to assess other cariogenic species in addition to S. mutans (13, 14). As in our study, additional studies based on molecular techniques performed by Munson et al. (29), Chhour et al. (8), and Corby et al. (10) found S. mutans and Lactobacillus spp. to be dominant in advanced caries. The results from the previous study on primary teeth (3) were confirmed, and additional species were detected according to the expanded panel of species. Our results also confirmed those of previous studies (3, 5, 8, 10, 15, 17, 25, 27, 29), indicating that a diversity of bacterial species, primarily gram-positive species, is involved in caries.
FIG. 4.
Mean levels of caries-associated species or species groups in primary teeth determined by a reverse-capture checkerboard assay for 15 subjects with caries (red bars) and 14 healthy controls (white bars). *, P ≤ 0.05 for pairwise comparisons of the healthy group to other groups by PROC MIXED. P indicates the overall P value comparing all groups and sites by PROC MIXED. Strep., Streptococcus; Actino, Actinomyces; cl., clone.
Veillonella spp., identified by a probe specific for all Veillonella spp., dominated the bacterial population in all stages, from intact enamel to deep-dentin cavities, in both the primary and secondary dentitions (Fig. 4 and 5). Other members of the Acidaminococcaceae family, such as Selenomonas spp., also were well represented in healthy and diseased sites, as seen from both clonal and checkerboard analyses. These findings are consistent with the findings of Munson et al. (29) and Chhour et al. (8), who detected Veillonella, albeit at lower levels. These organisms use lactate as a metabolic carbon source (12) but have a limited ability to adhere to host tissue. Veillonella spp., however, participate in coaggregation with many oral bacteria, e.g., Streptococcus spp., Eubacterium saburreum, and Actinomyces viscosus (12). A number of observations have associated Veillonella spp. with caries. Bradshaw and Marsh (6) observed that Veillonella dispar was the most prevalent organism found after all glucose-pulsing regimens used, especially at low pH, and Noorda et al. (31) showed in mixed bacterial plaques with S. mutans and Veillonella alcalescens that the production of acid was higher than that in plaque containing only one of these species. The present study confirmed the coactive relationship between Veillonella spp. and acid-producing bacteria (28). Veillonella spp. also may be important for acid-producing bacteria in caries through nitrate reduction (11). Silva Mendez et al. (36) demonstrated that acidity levels below pH 7 and low concentrations of nitrite (0.2 mM) caused the complete killing of S. mutans, with similar effects on the other organisms tested.
Actinomyces spp. play a major role in coaggregation interactions (9). Burt et al. (7) reported that Actinomyces-like organisms usually predominate in noncariogenic plaques with high levels of S. mutans streptococci and lactobacilli. Actinomyces spp. are heterofermenters but tend to become homolactic producers under anaerobic conditions (16), thus contributing to enamel demineralization. Strong coaggregation between Actinomyces odontolyticus and Actinomyces israelii with strains of either Veillonella parvula or Prevotella prevotii has been reported (35). Studies on coaggregation interactions between Streptococcus spp. and Actinomyces spp. have revealed the complementary adhesion-receptor mechanisms among the latter organisms (9). A large proportion of positively coaggregating pairs between either Prevotella intermedia ND8-9A or Campylobacter gracilis ND9-8A and strains of Streptococcus, Gemella, Peptostreptococcus, Lactobacillus, and Actinomyces indicate that the gram-negative obligate anaerobic rods play an important role in the interactions leading to root caries (35). Nadkarni et al. (30) and Chhour et al. (8) found that novel and uncultured Prevotella and Prevotella-like bacteria dominate the diverse polymicrobial community in some cases of caries, suggesting an active role of Prevotella in caries progression. Furthermore, coaggregation between Fusobacterium nucleatum NT6-6A and six other bacterial species, i.e., Streptococcus bovis II/2 ND2-2, Streptococcus constellatus ND10-13A, Streptococcus sanguinis II ND7-3, Lactobacillus acidophilus ND7-2A, C. sputigena ND2-12A, and P. intermedia ND8-9A, shows that these bacteria have the ability to coaggregate with a large number of oral bacteria and possibly act as key organisms in dental plaque formation during the later stages of plaque maturation and modulation of the climax community (20, 21). With further evidence that a large number of bacterial species are involved in the progression of dental caries, the interactions within different bacterial communities representing different stages of the disease would be of considerable interest.
In light of recent efforts to develop vaccines specific for S. mutans, the study of the potential etiologic role of all species associated with caries and caries progression is essential. The significance of our work is that the identification of additional caries pathogens provides alternative targets for biological interventions, and the identification of beneficial health-associated species could provide the basis for therapeutic interventions to establish caries-resistant microbial communities.
Although we have shown that specific bacterial species are associated with health, caries initiation, and caries production, there is subject-to-subject variation in the bacterial composition. Munson et al. (29) demonstrated that many species of Lactobacillus were found overall in carious lesions, but only one or two Lactobacillus spp. were detected in each lesion in a subject. We and others have shown that 10 to 20% of subjects with severe caries may not have detectable levels of S. mutans but rather exhibit other acid-producing species. Furthermore, in some carious lesions S. mutans may be a minor bacterial component of the dental plaque. These results support the ecological plaque hypothesis (26), which suggests that caries is a result of a shift in the balance of the resident microflora driven by changes in local environmental conditions, e.g., acid production by any number of species that cause tooth decay.
In summary, we have shown that half of the bacteria associated with dental caries have not yet been cultivated. Species in addition to S. mutans, e.g., species of Veillonella, Lactobacillus, Bifidobacterium, Propionibacterium, low-pH non-S. mutans streptococci, Actinomyces, and Atopobium, also may play an important role in caries production. Actinomyces spp. and non-S. mutans streptococci may be involved in the initiation of the disease. Several specific species are associated with health, while others are associated with caries. Bacterial profiles change with the progression of disease and differ from the primary to the secondary dentition. The present findings support the ecological plaque hypothesis in caries disease, in that changes in ecologic factors require different bacterial qualities and stimulate alterations in the bacterial composition. Further studies of the potential etiologic roles of these diverse bacterial communities, including additional and novel species, are recommended. Identified cultivable and not-yet-cultivable organisms might provide additional targets for caries intervention.
Acknowledgments
We thank Melvin Moeschberger and Kevin Tordoff for their contributions to the statistical analyses.
The study was supported by NIH grants DE11443 and DE016125, Omni Products, and the Faculty of Dentistry, University of Oslo, Norway.
Footnotes
Published ahead of print on 23 January 2008.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 435721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anusavice, K. J. 2002. Dental caries: risk assessment and treatment solutions for an elderly population. Compend. Contin. Educ. Dent. 2312-20. [PubMed] [Google Scholar]
- 3.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 401001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beighton, D. 2005. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent. Oral Epidemiol. 33248-255. [DOI] [PubMed] [Google Scholar]
- 5.Bjørndal, L., and T. Larsen. 2000. Changes in the cultivable flora in deep carious lesions following a stepwise excavation procedure. Caries Res. 34502-508. [DOI] [PubMed] [Google Scholar]
- 6.Bradshaw, D. J., and P. D. Marsh. 1998. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res. 32456-462. [DOI] [PubMed] [Google Scholar]
- 7.Burt, B. A., W. J. Loesche, and S. A. Eklund. 1985. Stability of selected plaque species and their relationship to caries in a child population over 2 years. Caries Res. 19193-200. [DOI] [PubMed] [Google Scholar]
- 8.Chhour, K. L., M. A. Nadkarni, R. Byun, F. E. Martin, N. A. Jacques, and N. Hunter. 2005. Molecular analysis of microbial diversity in advanced caries. J. Clin. Microbiol. 43843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corby, P. M., J. Lyons-Weiler, W. A. Bretz, T. C. Hart, J. A. Aas, T. Boumenna, J. Goss, A. L. Corby, H. M. Junior, R. J. Weyant, and B. J. Paster. 2005. Microbial risk indicators in early childhood caries. J. Clin. Microbiol. 435753-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doel, J. J., N. Benjamin, M. P. Hector, M. Rogers, and R. P. Allaker. 2005. Evaluation of bacterial nitrate reduction in the human oral cavity. Eur. J. Oral Sci. 11314-19. [DOI] [PubMed] [Google Scholar]
- 12.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 10116917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald, R. J., and P. H. Keyes. 1960. Demonstration of the etiologic role of streptococci in experimental caries in the hamster. J. Am. Dent. Assoc. 619-19. [DOI] [PubMed] [Google Scholar]
- 14.Gibbons, R. J., and J. van Houte. 1975. Dental caries. Annu. Rev. Med. 25121-134. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, C.-L., W. A. J. Falkler, and G. E. Minah. 1991. Microbiological studies of carious dentine from human teeth with irreversible pulpitis. Arch. Oral Biol. 36147-153. [DOI] [PubMed] [Google Scholar]
- 16.Hogg, S. D. 1992. The lactic microflora of the oral cavity, p. 115-132. In B. J. B. Wood (ed.), The lactic acid bacteria. The lactic acid bacteria in health and disease, vol. 1. Elsevier Applied Science, London, United Kingdom. [Google Scholar]
- 17.Hoshino, E. 1985. Predominant obligate anaerobes in human carious dentin. J. Dent. 641195-1198. [DOI] [PubMed] [Google Scholar]
- 18.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York, NY. [Google Scholar]
- 19.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54413-437. [DOI] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E., Y. Inouye, and L. V. Holdeman. 1983. New Actinomyces and Streptococcus coaggregation groups among human oral isolates from the same site. Infect. Immun. 41501-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leys, E. J., A. L. Griffen, S. J. Strong, and P. A. Fuerst. 1994. Detection and strain identification of Actinobacillus actinomycetemcomitans by nested PCR. J. Clin. Microbiol. 321288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liesack, W., H. Weyland, and E. Stackebrandt. 1991. Potential risk of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb. Ecol. 21191-198. [DOI] [PubMed] [Google Scholar]
- 24.Loesche, W. J. 1992. The specific plaque hypothesis and the antimicrobial treatment of periodontal disease. Dent. Update 1968-74. [PubMed] [Google Scholar]
- 25.Loesche, W. J., and S. A. Syed. 1973. The predominant cultivable flora of carious plaque and carious dentine. Caries Res. 7201-216. [DOI] [PubMed] [Google Scholar]
- 26.Marsh, P. D. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 8263-271. [DOI] [PubMed] [Google Scholar]
- 27.Martin, F. E., M. A. Nadkarni, N. A. Jacques, and N. Hunter. 2002. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 401698-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikx, F. H., and J. S. Van der Hoeven. 1975. Symbiosis of Streptococcus mutans and Veillonella alcalescens in mixed continuous cultures. Arch. Oral Biol. 20407-410. [DOI] [PubMed] [Google Scholar]
- 29.Munson, M. A., A. Banerjee, T. F. Watson, and W. G. Wade. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 423023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadkarni, M. A., C. E. Caldon, K. L. Chhour, I. P. Fisher, F. E. Martin, N. A. Jacques, and N. Hunter. 2004. Carious dentine provides a habitat for a complex array of novel Prevotella-like bacteria. J. Clin. Microbiol. 425238-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noorda, W. D., D. J. Purdell-Lewis, A. M. van Montfort, and A. H. Weerkamp. 1988. Monobacterial and mixed bacterial plaques of Streptococcus mutans and Veillonella alcalescens in an artificial mouth: development, metabolism, and effect on human dental enamel. Caries Res. 22342-347. [DOI] [PubMed] [Google Scholar]
- 32.Paster, B. J., I. M. Bartoszyk, and F. E. Dewhirst. 1998. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 20223-231. [Google Scholar]
- 33.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 1833770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 35.Shen, S., L. P. Samaranayake, and H. K. Yip. 2005. Coaggregation profiles of the microflora from root surface caries lesions. Arch. Oral Biol. 5023-32. [DOI] [PubMed] [Google Scholar]
- 36.Silva Mendez, L. S., R. P. Allaker, J. M. Hardie, and N. Benjamin. 1999. Antimicrobial effect of acidified nitrite on cariogenic bacteria. Oral Microbiol. Immunol. 14391-392. [DOI] [PubMed] [Google Scholar]
- 37.Theilade, E. 1986. The non-specific theory in microbial etiology of inflammatory periodontal diseases. J. Clin. Periodontol. 13905-911. [DOI] [PubMed] [Google Scholar]
- 38.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10569-570. [DOI] [PubMed] [Google Scholar]
- 39.World Health Organization. 2002. The world health report. Reducing risks, promoting healthy life. http://www.who.int/whr/2002/en/.