Abstract
DNA sequencing analyses have demonstrated relatively limited polymorphisms within the fungal internal transcribed spacer (ITS) regions among Trichophyton spp. We sequenced the ITS region (ITS1, 5.8S, and ITS2) for 42 dermatophytes belonging to seven species (Trichophyton rubrum, T. mentagrophytes, T. soudanense, T. tonsurans, Epidermophyton floccosum, Microsporum canis, and M. gypseum) and developed a novel padlock probe and rolling-circle amplification (RCA)-based method for identification of single nucleotide polymorphisms (SNPs) that could be exploited to differentiate between Trichophyton spp. Sequencing results demonstrated intraspecies genetic variation for T. tonsurans, T. mentagrophytes, and T. soudanense but not T. rubrum. Signature sets of SNPs between T. rubrum and T. soudanense (4-bp difference) and T. violaceum and T. soudanense (3-bp difference) were identified. The RCA assay correctly identified five Trichophyton species. Although the use of two “group-specific” probes targeting both the ITS1 and the ITS2 regions were required to identify T. soudanense, the other species were identified by single ITS1- or ITS2-targeted species-specific probes. There was good agreement between ITS sequencing and the RCA assay. Despite limited genetic variation between Trichophyton spp., the sensitive, specific RCA-based SNP detection assay showed potential as a simple, reproducible method for the rapid (2-h) identification of Trichophyton spp.
Members of the genus Trichophyton are the commonest agents of dermatophytoses. They are especially significant in onychomycosis but also invade skin and hair, causing infection associated with substantial morbidity (2, 32, 39). Although Trichophyton rubrum and Trichophyton mentagrophytes are the most frequent pathogens (27 to 76% and 4 to 41%, respectively) (2, 20), Trichophyton tonsurans, Trichophyton violaceum, and Trichophyton soudanense are also important. Furthermore, there are differences in clinical associations and in geographic distribution between species (20, 29). Because Trichophyton comprise anthropophilic and zoophilic species, accurate identification is important, not only to guide clinical management but for epidemiological purposes, case clusters have occurred in long-term care facilities and in response to increased public participation in fitness-related activities (19, 25). Standard culture-based identification methods, however, are slow (2 to 4 weeks) and imprecise due to fungal phenotypic pleomorphism (11, 24). Species identification within the genus Trichophyton is further confused by the use of synonyms for the same species (8, 29).
To address these deficiencies, PCR-based tools have been developed to provide for rapid, accurate species determination. In particular, the internal transcribed spacer (ITS) regions, ITS1 and ITS2, of the fungal ribosomal DNA (rDNA) gene complex, have shown promise as targets for species identification (8, 15, 24, 28, 35). Among the many techniques, including PCR-restriction enzyme analysis (13, 16-18), multiplex PCR (1), PCR fingerprinting (4, 9), and arbitrarily primed PCR (22), successfully applied in this context, DNA sequencing of the ITS region has emerged as one of the more discriminatory tools for species delineation (8, 9, 24).
Despite these efforts however, species assignment remains difficult due to the close phylogenetic relationship between anthropophilic Trichophyton spp. Furthermore, the main species, T. mentagrophytes, T. rubrum, and T. tonsurans, are actually species complexes containing many members (8, 9, 24). ITS-based analyses have found sequence variation to be limited to only a few, or single nucleotide polymorphisms (SNPs) between certain species, such as between T. tonsurans and T. equinum and between T. rubrum and T. soudanense (35). This limited genetic variation suggests the development of alternative rapid, sequencing-independent methods to detect signature SNPs between species. Approaches with good specificity include probe-based reverse line blot (21) or microarray methods (14). The efficiency of these platforms is best suited to “batch-testing” of large numbers of isolates.
Recently, the utility of circularizable oligonucleotide or padlock probes has been demonstrated for the detection of target nucleic acid sequences, including SNPs, with high specificity (5, 26, 27). Such probes comprise two sequences complementary to the 5′ and 3′ termini of the target sequence joined by a genetic linker region. Upon hybridization to the target, the two probe ends become juxtaposed and are joined by DNA ligase to form a closed molecule. The intensity of the signal generated by the probe is then increased exponentially by hyperbranching or rolling-circle amplification (RCA), capable of a 109-fold signal amplification of each circle within 90 min (5, 38). The application of padlock probes in the detection of polymorphisms in fungi has not yet been examined.
In the present study, we developed a sensitive, RCA-based SNP detection method using real-time PCR for species determination of clinically important Trichophyton spp. We firstly analyzed Trichophyton isolates by standard ITS sequencing to identify signature polymorphisms between species. We then tested the ability of the RCA assay to provide species identification. Probes targeting either ITS1, ITS2, or both ITS1 and ITS2 polymorphism(s) between species were designed.
MATERIALS AND METHODS
Dermatophyte isolates.
Thirty-six Trichophyton isolates were studied (Table 1). Three Epidermophyton floccosum, two Microsporum canis, and one Microsporum gypseum, strains were included to aid in determining the specificity of the RCA assay. The Trichophyton isolates comprised three American Type Culture Collection (ATCC; Rockville, MD) strains (T. tonsurans ATCC 28942, T. mentagrophytes ATCC 28185, and T. rubrum ATCC 28188) and 33 clinical isolates, including T. mentagrophytes (n = 16), T. rubrum (n = 10), T. tonsurans (n = 5), and T. soudanense (n = 2). All clinical isolates (strains 1 to 39; Table 1) were recovered from different patients, were collected on separate occasions, and were obtained from the Clinical Mycology laboratory at Westmead Hospital, Sydney, Australia, and the Women's and Children's Hospital, Adelaide, Australia. Isolates were identified by using standard colonial and microscopic characteristics (3).
TABLE 1.
Results of species identification by culture, ITS sequencing, and RCA analysis for 42 dermatophyte isolates
| Strain no. | GenBank accession no. | Species identification:
|
Base pair (%) identity with GenBank sequence | Polymorphisms identified (other than 100% identity to GenBank sequence) | Species identification by RCA | |
|---|---|---|---|---|---|---|
| Upon receipta | Obtained by ITS sequencing (GenBank accession no.) | |||||
| Reference strains | ||||||
| ATCC 28942 | EU200367 | T. tonsurans | T. tonsurans (AF170479) | 660/661 (99.9) | 1-bp difference at position 565 (A to G) | T. tonsurans |
| ATCC 28185 | EU200369 | T. mentagrophytes | A. vanbreuseghemii (AF170466) | 656/657 (99.9) | 1-bp difference at position 10 (G to A) | T. mentagrophytes |
| ATCC 28188 | EU200370 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| Clinical isolates | ||||||
| 1 | EU200408 | M. gypseum | A. incurvatum (AB193687) | 613/613 (100) | M. gypseum | |
| 2 | EU200371 | M. canis | M. canis (AB193649) | 711/711 (100) | M. canis | |
| 3 | EU200368 | M. canis | M. canis (AB193649) | 711/711 (100) | M. canis | |
| 4 | EU200372 | E. floccosum | E. floccosum (AY213646) | 754/754 (100) | E. floccosum | |
| 5 | EU200373 | E. floccosum | E. floccosum (AY213646) | 754/754 (100) | E. floccosum | |
| 6 | EU200374 | E. floccosum | E. floccosum (AY213646) | 754/754 (100) | E. floccosum | |
| 7 | EU200375 | T. mentagrophytesb | A. vanbreuseghemii (AF170466) | 652/657 (99.2) | Position 165 (G to A), position 260 (A to C), position 291 (C to T), position 628 (C to T), position 780 (G to A) | T. mentagrophytes |
| 8 | EU200376 | T. mentagrophytes | A. vanbreuseghemii (AF170466) | 652/657 (99.2) | Position 165 (G to A), position 260 (A to C), position 291 (C to T), position 628 (C to T), position 780 (G to A) | T. mentagrophytes |
| 9 | EU200377 | T. mentagrophytes var. interdigitale | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 10 | EU200378 | T. mentagrophytes var. interdigitale | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 11 | EU200379 | T. mentagrophytes var. interdigitale | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 12 | EU200380 | T. mentagrophytes var. interdigitale | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 13 | EU200381 | T. mentagrophytes var. mentagrophytes | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 14 | EU200382 | T. mentagrophytes var. mentagrophytes | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 15 | EU200383 | T. mentagrophytes var. interdigitale | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 16 | EU200384 | T. mentagrophytes var. mentagrophytes | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 17 | EU200385 | T. mentagrophytes var. interdigitale | A. vanbreuseghemii (AF170466) | 657/658 (99.9) | 1-bp difference (insertion of G at position 440) | T. mentagrophytes |
| 18 | EU200386 | T. mentagrophytes var. mentagrophytes | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 19 | EU200387 | T. mentagrophytes var. mentagrophytes | A. vanbreuseghemii (AF170466) | 656/657 (99.9) | 1-bp difference at position 10 (G to A) | T. mentagrophytes |
| 20 | EU200388 | T. mentagrophytes var. mentagrophytes | A. vanbreuseghemii (AF170466) | 656/657 (99.9) | 1-bp difference at position 10 (G to A) | T. mentagrophytes |
| 21 | EU200389 | T. mentagrophytes var. interdigitale | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 22 | EU200390 | T. mentagrophytes var. interdigitale | A. vanbreuseghemii (AF170466) | 657/657 (100) | T. mentagrophytes | |
| 23 | EU200391 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 24 | EU200392 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 25 | EU200393 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 26 | EU200394 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 27 | EU200395 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 28 | EU200396 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 29 | EU200397 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 30 | EU200398 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 31 | EU200399 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 32 | EU200400 | T. rubrum | T. rubrum (AF170472) | 666/666 (100) | T. rubrum | |
| 33 | EU200401 | T. soudanense | T. soudanense (AM049997) | 563/571 (98.6)c | 8-bp difference of a (TA)4 deletion at positions 546 to 553 | T. soudanense |
| 34 | EU200402 | T. soudanense | T. soudanense (AM049997) | 571/575 (99.3)d | 4-bp difference of a (TA)2 insertion at position 546 | T. soudanense |
| 35 | EU200403 | T. tonsurans | T. tonsurans (AF170479) | 661/661 (100) | T. tonsurans | |
| 36 | EU200404 | T. tonsurans | T. tonsurans (AF170479) | 661/661 (100) | T. tonsurans | |
| 37 | EU200405 | T. tonsurans | T. tonsurans (AF170479) | 661/661 (100) | T. tonsurans | |
| 38 | EU200406 | T. tonsurans | T. tonsurans (AF170479) | 661/661 (100) | T. tonsurans | |
| 39 | EU200407 | T. tonsurans | T. tonsurans (AF170479) | 661/661 (100) | T. tonsurans | |
That is, the original identification of the species.
Unless otherwise stated, the identification “T. mentagrophytes” is used to represent “T. mentagrophytes complex.”
The ITS of this strain was shorter than the sequence of T. soudanense (GenBank accession no. AM049997) by a (TA)4 deletion but otherwise 100% identical.
The ITS of this strain was longer than the sequence of T. soudanense (GenBank accession no. AM049997) by a (TA)2 insertion but otherwise 100% identical.
DNA extraction.
For all dermatophyte isolates, DNA extraction was performed as described previously (30).
Artificial DNA template for T. violaceum.
Since we were unable to obtain T. violaceum isolates, we synthesized an artificial T. violaceum DNA template to test the ability of a T. violaceum-specific padlock probe (Tv-ITS1; see below) to identify this species. This single-stranded oligonucleotide template (5′-261-CGCGCCCGCCGGAGGACAGACACCAAGGAAAATTCTCTGAAGGGCTGTCAGTCT-314-3′) was derived after examination of the ITS sequences of T. violaceum contained in the GenBank database (accession no. AJ270811) and synthesized commercially (Sigma-Aldrich, Castle Hill, Australia). Single-stranded artificial templates have previously been shown used to establish molecular models of probe-based RCA assays with good results (38).
PCR amplification of ITS region.
The entire ITS (ITS1, 5.8S rDNA, and ITS2) region of all 42 isolates was amplified (i) for ITS sequence analysis and (ii) in preparation for hybridization with padlock probes and subsequent RCA (see below). The primers SR6RL and LR1L (40) (Sigma-Aldrich) were used to amplify the ITS1 and ITS2 regions.
ITS sequence analysis.
The primers ITS1 and ITS4 (6) were used as the inner sequencing primers (Sigma-Aldrich). PCRs were performed as described previously (30). Each reaction contained 1.0 μl of template DNA, 0.25 μl (50 pmol/μl) each of forward primer and reverse primer, 1.25 μl of deoxynucleoside triphosphates (2.5 mM concentrations of each deoxynucleoside triphosphate; Roche Diagnostics, Mannheim, Germany), 0.1 μl of HotStar Taq polymerase (5 U/μl), 2.5 μl of 10× PCR buffer (catalogue no. 203203; Qiagen, Doncaster, Victoria, Australia), and water to a total volume of 25 μl. Amplification was performed on a MasterCycler gradient thermocycler (Eppendorf AG, North Ryde, Australia). The thermal cycling conditions were 95°C for 15 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s, with a final extension step at 72°C for 10 min. PCR product (8 μl) loaded on a 1.5% agarose gel was visualized under UV illumination to verify the amplicon quantity prior to sequence analysis.
PCR products were purified by using a PCR product pre-sequencing kit (USB Corp., Cleveland, OH) and sequenced by using the ITS1 and ITS4 primer and the BigDye Terminator (version 3.1) cycle sequencing kit in an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Sequences were entered into a BLASTn sequence analysis search (33) and analyzed by using editing and analyses programs in the BioManager (ANGIS) facility (accessed via http://biomanager.angis.org.au/).
Primer and padlock probe design for RCA.
Relevant DNA sequences spanning the fungal rDNA gene complex (18S, 5.8S, and 28S) and the intervening ITS1 and ITS2 regions of major dermatophyte species accessed from GenBank were compared by using the CLUSTAL W program in BioManager (ANGIS). These data were further compared to ITS results of the isolates to identify informative nucleotide polymorphisms between species.
Nine padlock probes (six specific for Trichophyton spp., one for E. floccosum, and one each for M. gypseum and M. canis) were designed targeting the ITS1 region, the ITS2 region, or both ITS regions after multiple sequence alignment of the above results (Table 2). Briefly, the probes were 96 to 102 nucleotides in length, consisting of two adjacent target complementary sequences with a linker) region (48 nucleotides) (38). To optimize binding to target DNA, the probes were designed with a minimum secondary structure and with a Tm of the 5′-end probe binding arm greater than the temperature used for probe ligation (62°C in the present study, see below). To increase the specificity, the 3′-end binding arm was designed to have a Tm (51 to 56°C) below the ligation temperature (see below). In addition, the linker region for each Trichophyton species-specific probe was carefully designed to (i) minimize similarity to other closely related Trichophyton spp. and (ii) allow primer binding during RCA and amplification of the probe-specific signal. The sequences of the two primers (RCA primer 1 and RCA primer 2, Tm = 55°C) used for RCA are shown in Table 2. These two primers were designed to specifically bind the linker region of the probes (26, 27).
TABLE 2.
Oligonucleotide padlock probes and primers used for RCA
| Probe or primer | Target species | GenBank accession no. | Sequence and locations of the two binding arms on comparison with relevant reference GenBank sequences (5′-3′)a |
|---|---|---|---|
| Probes | |||
| Tr-ITS2 | T. rubrum | Z97993 | p-593TTGGCTGCCCATTCGCCTAG574 |
| GATCATGCTTCTTCGGTGCCCATTACGAGGTGCGGATAGCTACCGCGCAGACACGATAGTCTA | |||
| 606TGAGGGCGCTGAA594 | |||
| Tt-ITS2 | T. tonsurans | AY213690 | p-552AAGCCGGAATCGCGGCCTGG533 |
| GATCATGCTTCTTCGGTGCCCATCCTAGATCAGACGTTCCTGTCCGCGCAGACACGATAGTCTA | |||
| 576CCCATTCGCCTAGA553 | |||
| Ts/v-ITS2 | T. soudanense/T. violaceum | AM049997 | p-502CTGCCCATTCGCCCAGGAAGC482 |
| GATCATGCTTCTTCGGTGCCCATTCGTGGCTAGTCGAATCTTAGCGCGCAGACACGATAGTCTA | |||
| 514GCGCTGGTTTGG503 | |||
| Ts/r-ITS1 | T. soudanense/T. rubrum | AM049997 | p-172CTTGGTGTCTGTCCTCCGGC153 |
| GATCATGCTTCTTCGGTGCCCATGCTAACCTGGTACCGTCATTCGCGCAGACACGATAGTCTA | |||
| 191GCTCTTCAGAGAATTTTTT173 | |||
| Tv-ITS1 | T. violaceum | AJ270811 | p-287CTTGGTGTCTGTCCTCCGGCG267 |
| GATCATGCTTCTTCGGTGCCCATTGACCGTGCTATGAATGCATCGCGCAGACACGATAGTCTA | |||
| 304CCCTTCAGAGAATTTTC288 | |||
| Tm-ITS1 | T. mentagrophytes | Z98001 | p-215AGCCACTAAAGAGAGGCTCGC195 |
| GATCATGCTTCTTCGGTGCCCATGCTTAGCTTGGCATGTCACTCGCGCAGACACGATAGTCTA | |||
| 228CGGTCCAGCGTTT216 | |||
| Mg-ITS2 | M. gypseum | AB193687 | p-482CCATTCGCCCAGGAGCCGGAATC460 |
| GATCATGCTTCTTCGGTGCCCATCCTACTAGTTGCACGCTGTTCCGCGCAGACACGATAGTCTA | |||
| 506CGTTGGTTTGTTGC483 | |||
| Mc-ITS1 | M. canis | AY213657 | p-218GAGTCCCCCTCAGGCGTCCC198 |
| GATCATGCTTCTTCGGTGCCCATGACTCTCGCTCGACACAGTAGCGCGCAGACACGATAGTCTA | |||
| 232TGGCCTAGGAAACAA219 | |||
| Ef-ITS2 | E. floccosum | AY213646 | p-562CTTTCTCCTCTCCCGGTGGAACG539 |
| GATCATGCTTCTTCGGTGCCCATAATGCCACGTTAACAGTCAGCGCGCAGACACGATAGTCTA | |||
| 574GCGTCCCCTCCA563 | |||
| Primersb | |||
| RCA primer 1 | ATGGGCACCGAAGAAGCA | ||
| RCA primer 2 | CGCGCAGACACGATA |
At the 5′ end of the probe, “p-”, indicates 5′ phosphorylation. The underlined sequences are the binding arms of the padlock probes (derived from reference GenBank sequences), which are joined by the nonspecific linker region.
The RCA primer-1 binds to the padlock probe, generating a long ssDNA. Its sequence is the same as that of the underlined italicized segments, in reverse. The RCA primer-2 binds to nascent single-stranded DNA as their binding sites become available. Its sequence is he same as that of the segments shown in italics.
Ligation of padlock probe.
Approximately 1011 copy numbers of standard DNA template (estimated by DNA calculator accessed through http://www.uri.edu/research/gsc/resources/cndna.html) was mixed with 2 U of PFU DNA ligase (catalogue no. 600191; Stratagene) and 0.1 μM padlock probe in 20 mM Tris-HCl (pH 7.5), 20 mM KCl, 10 mM MgCl2, 0.1% Igepal CA-360 [tert-octylphenoxy poly(oxyethylene) ethanol; Sigma-Aldrich], 0.01 mM rATP, and 1 mM dithiothreitol in a 10-μl reaction volume. Multiple cycle ligation was conducted with one cycle of denaturation at 94°C of 5 min, followed by five cycles at 94°C for 30 s and 4 min of ligation at 62°C (37, 38).
Exonucleolysis.
Exonucleolysis was performed to remove unligated padlock probe and template PCR product and so reduce subsequent ligation-independent amplification events. It was performed in 20-μl volumes by adding 10 U each of exonuclease I and exonuclease III (New England Biolabs, United Kingdom) to the ligation mixture, followed by incubation at 37°C for 30 to 60 min and then 95°C for 3 min.
Hyperbranched or rolling circle amplification reaction.
After exonucleolysis of the now-circularized probes, RCA reactions was performed in a 50-μl volume containing 8 U of Bst DNA polymerase (New England Biolabs), a 400 μM deoxynucleoside triphosphate mix, 10 pmol of each RCA primer (Table 2), 5% of dimethyl sulfoxide (vol/vol), and 10× Sybr green I (Sigma-Aldrich). Probe signals were amplified by incubation at 65°C for 30 min, and the accumulation of double-stranded DNA products was monitored by using a Corbett RotorGene 3000 real-time PCR machine (Corbett Research, Mortlake, Australia).
Nucleotide sequence accession numbers.
New sequence data generated in the present study have been deposited in the GenBank database (accession numbers EU200367 to EU200408; see Table 1) after editing of the sequences using the edit and revseq programs in BioManager (ANGIS).
RESULTS
ITS sequence analysis and interspecies variation.
The universal fungal primers SR6RL and LR1L amplified the ITS1, 5.8S, and ITS2 regions for all 42 isolates and in each case, satisfactory sequencing results were obtained. Species identification, as determined by phenotype-based methods, ITS analysis, and RCA are shown in Table 1.
The major Trichophyton species complexes—T. rubrum, T. mentagrophytes, and T. tonsurans—were readily distinguished from each other by ITS sequence analysis and from the two other genera of dermatophytes (Table 1). Despite the high sequence identity, T. rubrum was distinguished from T. soudanense (4-bp difference), as well as from T. violaceum (compared to T. violaceum sequences in GenBank). There was a 3-bp ITS polymorphism that allowed the differentiation of T. soudanense from T. violaceum.
There was no genetic variation in either the ITS1 or the ITS2 regions among 11 T. rubrum strains. However, the sequences of two clinical T. soudanense isolates differed from each other and from reference ITS sequences of T. soudanense in GenBank (GenBank accession numbers AM049997, AF170474, AF170473, and AJ270809; Table 1). Sequencing was repeated to exclude sequence errors introduced during the initial PCR and sequencing, with the same results as before. Strain 33 yielded a sequence with 100% identity to the T. soudanense strain with the closest sequence match (GenBank accession no. AM049997) with the exception of a (TA)4 nucleotide deletion in the ITS2 region between positions 546 and 553. Strain 34 also showed otherwise 100% sequence identity with the same GenBank sequence but with a (TA)2 insertion polymorphism in the ITS2 region beginning at position 546 (Table 1).
There were four genetic types within T. mentagrophytes. Eleven of sixteen clinical isolates had identical ITS sequences which showed 100% identity to a reference Arthroderma vanbreuseghemii (anamorph Trichophyton interdigitale) sequence in GenBank (GenBank accession no. AF170466; Table 1); the sequences of four others (strains 17, 19, 20, and T. mentagrophytes ATCC 28185) demonstrated a single base pair difference (Table 1). The remaining two clinical isolates (strains 7 and 8) yielded a sequence with a 5-bp difference from that obtained for the majority (n = 11) of strains. Both had identical sequences with 99% identity to A. vanbreuseghemii (GenBank accession no. AF170466). Of note, seven isolates received as T. mentagrophytes var. interdigitale and four received as T. mentagrophytes var. mentagrophytes had identical ITS sequences (Table 1). The sequence of T. tonsurans ATCC 28942 showed a SNP with the sequence of the clinical strains (Table 1). Concordant culture- and sequence-based identification was observed for 100% of T. tonsurans, T. rubrum, and T. soudanense isolates.
Species identification by RCA: sensitivity and specificity.
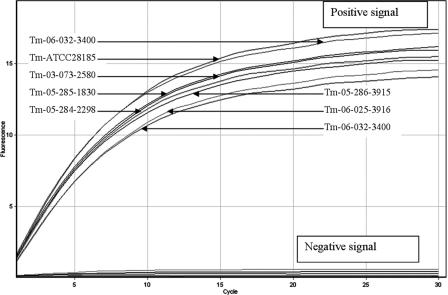
Individual species-specific or “group-specific” probes (for example, the T. soudanense/T. rubrum probe) were designed based on signature SNPs identified by alignment of the ITS sequence data (Table 2). The T. violaceum-specific probe (Tv-ITS1) was designed on the basis of GenBank sequence data alone. To assess the sensitivity of the assay, RCA was initially performed on serial dilutions of the target template (1011 copies) containing 100, 10, 1, 0.1, and 0.01% of the template. For all dermatophytes studied, a clearly measurable RCA signal was observed with a sensitivity of detection of 107 copies (0.01% template in reaction mixture); the signal was weak below this copy number (data not shown). Figure 1 illustrates a typical padlock probe-RCA reaction using the Tm-ITS1 probe to detect T. mentagrophytes DNA. For all T. mentagrophytes strains, exponential increases in fluorescence signals were readily interpretable. The duration of the RCA procedure was 2 h; however, a positive signal was usually evident 15 min after commencement of the RCA reaction.
FIG. 1.
RCA results monitored by the RotorGene 3000 real-time PCR machine (Corbett Research). The accumulation of double-stranded DNA was detected by staining with Sybr green I. Positive signals (labeled as “positive signal”) are shown as exponential increases in fluorescence signal. The experiment was conducted using the T. mentagrophytes-specific RCA probe (Tm-ITS1) and tested on eight T. mentagrophytes and other dermatophyte isolates. Ligation-mediated RCA with matched templates of T. mentagrophytes containing the targeted SNPs produced positive signals. In contrast, other species (e.g., T. tonsurans, T. rubrum, T. violaceum, E. floccosum, and M. canis) showed an absence of signal (labeled as “negative signal”).
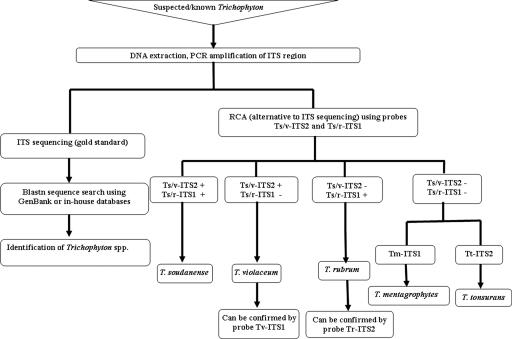
The RCA assay was also highly specific. All isolates were correctly identified (Table 1). Of note, amplification of probe signals were seen only with matched template-probe mixtures; DNA from species that contained SNPs not targeted by the padlock probe in use produced no signal (Fig. 1). Among Trichophyton spp., SNPs selectively targeted by the relevant probes correctly identified T. rubrum, T. mentagrophytes, T. tonsurans, and T. violaceum DNA (Table 1); these species were identified by single probes, targeted at either the ITS1 or the ITS2 region. However, the system was unable to identify T. soudanense in a single reaction since the design of a T. soudanense-specific probe was not possible. Although a signature SNP was identified in the ITS2 region (position 503) of T. soudanense (GenBank accession no. AM049997) to differentiate this species from T. rubrum, this same SNP was found in T. violaceum. Similarly, SNPs that allowed differentiation of T. soudanense from T. violaceum were found in T. rubrum (for example, position 173 in the ITS1 region of T. soudanense (GenBank accession no. AM049997). The combined use of two probes, an ITS1-based T. soudanense/T. rubrum probe and an ITS2-based T. soudanense/T. violaceum probe (Fig. 2) enabled the differentiation of T. soudanense from T. rubrum and from T. violaceum.
FIG. 2.
Algorithm for identification of Trichophyton spp. using a padlock probe RCA assay.
Hybridization of the Trichophyton probes used in the present study may occur with template DNA from species other than the target species (Table 3). However, these expected “cross-reactions” would be observed for species now reduced to synonymy with the target species (Table 3) (10) with one exception: cross-hybridization of the T. tonsurans probe, Tt-ITS2, with T. equinum DNA (a single ITS1 base polymorphism) (35). We were unable to identify strains of T. equinum among our culture collection to test this hypothesis.
TABLE 3.
Specificity of padlock probes for the identification of Trichophyton spp.
| Padlock probe | Trichophyton and Arthroderma species expected to hybridize with the probea |
|---|---|
| Tr-ITS2 | T. rubrum, T. raubitschekii, T. kanei, T. fischeri, T. pedis, T. rodhaini |
| Tt-ITS2 | T. equinumb |
| Ts/v-ITS2 | Target species T. soudanense: T. gourvilii*, T. megninii†, T. fluviomuniense†, T. circonvolutum†; target species T. violaceum: T. yaoundei*, T. kuryangei†, and T. glabrum* |
| Ts/r-ITS1 | Target species T. soudanense: T. gourvilii*, T. megninii†, T. fluviomuniense†, and T. circonvolutum†; target species T. rubrum: T. raubitscheki, T. kanei, T. fischeri, T. pedis, and T. rodhaini |
| Tv-ITS1 | T. yaounde*, T. glabrum* |
| Tm-ITS1 | A. vanbreuseghemiib, A. benhamiae, T. interdigitale, A. simii, T. krajdenii, T. abissinicum |
T. raubitschekii is considered to be synonymous with T. rubrum (11). For T. equinum there is a single base polymorphism at position 18 (ITS1 region) from C (T. tonsurans) to T (T. equinum). *, Conspecific with T. violaceum (11); †, doubtful identity but considered by some to be synonymous with T. rubrum (11).
Proposed algorithm for RCA-based identification of Trichophyton spp.
Each of the Trichophyton-specific probes designed in the present study may be used in parallel for species identification with good sensitivity and specificity. However, for practical purposes, a simple guide to species identification of suspected Trichophyton isolates is proposed as summarized in Fig. 2. Using this algorithm, the sequential use of two, or at most three padlock probes, enables identification and differentiation of the major human Trichophyton spp.
DISCUSSION
Given the increase in Trichophyton infections and their public health importance (2, 7), there is a need to develop reliable molecular tools for species delineation. To demonstrate its potential application, we developed a novel, simple, highly specific RCA-based SNP detection assay for identification of clinically important Trichophyton spp. The present study is the first to utilize this approach to identify fungal pathogens. Features of the RCA-based assay include high sensitivity, rapidity (2 h), and ability to detect signature SNPs between T. rubrum, T. soudanense, and T. violaceum. Both ITS1 and ITS2 polymorphisms were useful for species identification.
The validity of the RCA assay was established by determining inter- and intraspecies polymorphisms among the study isolates by ITS sequencing in order to design padlock probes, which would ensure the sensitivity and specificity of the assay. Since a significant proportion of sequences in public databases such as GenBank contain read errors (12, 24, 34, 35), we did not only rely on these as a resource for probe design. Furthermore, there is still debate over the correctness of taxonomic changes made in the classification of Trichophyton on the basis of molecular data (8, 11, 35). As noted previously, we observed minimal ITS sequence variation among T. rubrum and T. tonsurans isolates (8, 29, 35). However, small sets of SNPs were identified within T. soudanense and T. mentagrophytes, suggesting there is greater genetic variation for these species or species complexes (10, 23; the present study). Since there are few data describing genetic variation within T. soudanense, study of greater numbers of this species is indicated. The generation of new sequences and determination of sequence variation for all species is critical to improving sequence-based species identification processes, in particular, to any method choosing to exploit signature SNPs.
Species identification as determined by the RCA system was concordant with sequencing data for all study isolates and the T. violaceum template. The use of padlock probes was first of all able to distinguish Trichophyton spp. from other dermatophyte genera. This may guide medical decisions, since Trichophyton infections often persist and require longer periods of antifungal therapy (2, 32). Signature polymorphisms between the major Trichophyton spp., with the exception of T. soudanense, were also successfully targeted by individual RCA probes. To identity T. soudanense, however, the sequential use of two probes that were selective for T. rubrum-T. soudanense and T. soudanense-T. violaceum was required (Fig. 2). This was not unexpected since there was a high degree of genetic identity (a 4-bp difference between T. rubrum and T. soudanense and a 3-bp difference between T. violaceum and T. soudanense) in the ITS region between T. rubrum, T. soudanense, and T. violaceum (11, 24, 35; the present study). Although SNPs were identified that could be exploited to differentiate between T. rubrum and T. soudanense, one or more of these same SNPs were shared either between T. rubrum and T. violaceum or between T. soudanense and T. violaceum. Similarly, when we attempted to distinguish T. soudanense from T. violaceum, we found that the same informative SNPs were shared by T. rubrum.
The status of T. soudanense as a valid species appears controversial. Based on ITS sequence and clinical data, the species has been synonymized as T. violaceum (11). Other researchers, using microsatellite markers, have noted that T. soudanense is clearly distinguishable from T. violaceum and, instead, groups closely with T. rubrum (8, 29). Despite the close genetic relationship of these two organisms, T. soudanense is morphologically, physiologically, and ecologically distinct from T. rubrum. The former is endemic in sub-Saharan Africa and causes endothrix tinea capitis, whereas the cosmopolitan T. rubrum does not (2, 39). These differences meet the requirements for significance at the species level for T. soudanense since they satisfy both ecological and genetic criteria (35, 36). The same criteria validate T. violaceum (endemic in North Africa-Middle East) as a species distinct from T. soudanense. Given that these species are no longer restricted to their areas of endemicity due to increased migration and travel, ongoing study to optimize methods for species identification is warranted.
Sequence polymorphisms selectively targeted by the T. mentagrophytes probe (Tm-ITS1) allowed the identification of this species complex. No RCA signal was produced by the other Trichophyton spp. (Tables 1 and 3). Although not addressed in the present study, the probe cannot be expected to differentiate between individual members or genotypes (A. vanbreuseghemii, Arthroderma simii, Arthroderma benhamiae, and the anamorphic species) of the complex (10, 24) (see Table 3). Given the flexibility of the RCA assay, however, genotype-specific probes targeting signature SNPs that detect a particular member of interest, e.g., T. interdigitale, can be designed. T. interdigitale is one of the more well-recognized taxonomic entities (10, 23). In our study, ITS sequencing identified only four genetic types among 17 T. mentagrophytes isolates, one corresponding to A. vanbreuseghemii and the other two showing close genetic identity (Table 1). This suggests that Australian T. mentagrophytes strains may exhibit little genetic variation. Of note, the ITS sequences of several strains submitted as T. mentagrophytes var. mentagrophytes or T. mentagrophytes var. interdigitale (synonym T. interdigitale) were identical. It is likely the culture-based identification of the former was incorrect.
The T. tonsurans-specific (Tt-ITS2) probe correctly distinguished T. tonsurans from other human Trichophyton but may be expected to hybridize with the horse-associated T. equinum. ITS polymorphisms between these two species are limited to a single base pair at position 18 (35). However, since T. equinum is zoophilic and T. tonsurans is anthropophilic, any cross-reaction can be resolved except in the uncommon circumstance when human infection has resulted from contact with an infected horse or its fomites.
At present, ITS sequence analysis is the gold standard for dermatophyte identification. We confirm its accuracy in this capacity. However, this approach is expensive, has a turnaround time of 2 to 3 days, and may be impractical for analyzing large numbers of isolates. Moreover, when applied to clinical specimens, sequencing is unable to simultaneously identify more than one pathogen. The rapid RCA-based SNP detection system described here is sensitive, reproducible, and flexible in that probes can be custom-made to meet specific requirements; this places it favorably as an option for high-throughput analysis. Although the expenses associated with the setup of the assay (AUD$300 per probe) are not insubstantial, the probes can be used numerous (up to 5000) times. Running costs are estimated at no more than AUD$2 per reaction compared to AUD$15 for DNA sequencing. All Trichophyton probes in the present study had good specificity and can be used in parallel to identify a presumptive “Trichophyton.” However, by following the recommended algorithm (Fig. 2) and using only three probes, expenses may be minimized.
In conclusion, species identification of Trichophyton is important for epidemiological and phylogenetic purposes and for genotype delineation. Despite the shortcomings of current molecular identification systems, there is strong impetus in continuing to use ITS polymorphisms as the common denominator to generate identification barcodes (35). This process makes use of species-specific SNPs or other “minor” sequence differences which are unlikely to yield resolution using traditional phylogenetic analyses (31). The role of techniques that selectively target SNPs in contributing to this goal warrants exploration. In its current format, the RCA-based assay showed promise as an alternative to DNA sequencing for species identification. Study of larger numbers of different Trichophyton spp. and other dermatophytes, as well as its utility when applied to clinical specimens, is required to validate the RCA assay for routine diagnosis.
Acknowledgments
We thank Ok Cha Lee for help with the culture-based identification of dermatophyte isolates, Maryann Pincevic for her assistance in sequencing, and Ping Zhu for help with the figures.
Footnotes
Published ahead of print on 30 January 2008.
REFERENCES
- 1.Brillowska-Dabrowska, A., D. M. Saunte, and M. C. Arendrup. 2007. Five-hour diagnosis of dermatophyte nail infections with specific detection of Trichophyton rubrum. J. Clin. Microbiol. 451200-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elewski, B. E., and M. A. Charif. 1997. Prevalence of onychomycosis in patients attending a dermatology clinic in northeastern Ohio for other conditions. Arch. Dermatol. 1331172-1173. [PubMed] [Google Scholar]
- 3.Ellis, D., S. Davis, H. Alexiou, R. Handke, and R. Bartley. 2007. Descriptions of medical fungi, 2nd ed. Nexus Print Solutions, Adelaide, South Australia, Australia.
- 4.Faggi, E., G. Pini, and E. Campisi. 2002. PCR fingerprinting for identification of common species of dermatophytes. J. Clin. Microbiol. 404804-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faruqi, A. F., S. Hosono, M. D. Driscoll, F. B. Dean, O. Alsmadi, R. Bandaru, G. Kumar, B. Grimwade, Q. Zong, Z. Sun, Y. Du, S. Kingsmore, T. Knott, and R. S. Lasken. 2001. High-throughput genotyping of single nucleotide polymorphisms with rolling circle amplification. BMC Genomics 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Mol. Ecol. 2113-118. [DOI] [PubMed] [Google Scholar]
- 7.Ghannoum, M., N. Isham, R. Hajjeh, M. Cano, F. Al Hasawi, D. Yearick, J. Warner, L. Long, C. Jessup, and B. Elewski. 2003. Tinea capitis in Cleveland: survey of elementary school students. J. Am. Acad. Dermatol. 48189-193. [DOI] [PubMed] [Google Scholar]
- 8.Graser, Y., S. De Hoog, and R. C. Summerbell. 2006. Dermatophytes: recognizing species of clonal fungi. Med. Mycol. 44199-209. [DOI] [PubMed] [Google Scholar]
- 9.Graser, Y., J. Frohlich, W. Presber, and S. de Hoog. 2007. Microsatellite markers reveal geographic population differentiation in Trichophyton rubrum. J. Med. Microbiol. 561058-1065. [DOI] [PubMed] [Google Scholar]
- 10.Graser, Y., A. F. Kuijpers, W. Presber, and G. S. De Hoog. 1999. Molecular taxonomy of Trichophyton mentagrophytes and T. tonsurans. Med. Mycol. 37315-330. [DOI] [PubMed] [Google Scholar]
- 11.Graser, Y., A. F. Kuijpers, W. Presber, and G. S. de Hoog. 2000. Molecular taxonomy of the Trichophyton rubrum complex. J. Clin. Microbiol. 383329-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkesworth, D. L. 2004. Fungal biodiversity and its implications for genetic resource collections. Stud. Mycol. 509-18. [Google Scholar]
- 13.He, G., J. Li, J. Ding, and Z. Tan. 2005. Identification of common species of dermatophytes by PCR-RFLP. J. Huazhong. Univ. Sci. Technolog. Med. Sci. 25458-460. [DOI] [PubMed] [Google Scholar]
- 14.Huang, A., J. W. Li, Z. Q. Shen, X. W. Wang, and M. Jin. 2006. High-throughput identification of clinical pathogenic fungi by hybridization to an oligonucleotide microarray. J. Clin. Microbiol. 443299-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, C. J., R. C. Barton, and E. G. Evans. 1999. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J. Clin. Microbiol. 37931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamiya, A., A. Kikuchi, Y. Tomita, and T. Kanbe. 2004. PCR and PCR-RFLP techniques targeting the DNA topoisomerase II gene for rapid clinical diagnosis of the etiologic agent of dermatophytosis. J. Dermatol. Sci. 3435-48. [DOI] [PubMed] [Google Scholar]
- 17.Kanbe, T., Y. Suzuki, A. Kamiya, T. Mochizuki, M. Fujihiro, and A. Kikuchi. 2003. PCR-based identification of common dermatophyte species using primer sets specific for the DNA topoisomerase II genes. J. Dermatol. Sci. 32151-161. [DOI] [PubMed] [Google Scholar]
- 18.Kanbe, T., Y. Suzuki, A. Kamiya, T. Mochizuki, M. Kawasaki, M. Fujihiro, and A. Kikuchi. 2003. Species-identification of dermatophytes Trichophyton, Microsporum, and Epidermophyton by PCR and PCR-RFLP targeting of the DNA topoisomerase II genes. J. Dermatol. Sci. 3341-54. [DOI] [PubMed] [Google Scholar]
- 19.Kane, J., E. Leavitt, R. C. Summerbell, S. Krajden, and S. S. Kasatiya. 1988. An outbreak of Trichophyton tonsurans dermatophytosis in a chronic care institution for the elderly. Eur. J. Epidemiol. 4144-149. [DOI] [PubMed] [Google Scholar]
- 20.Kardjeva, V., R. Summerbell, T. Kantardjiev, D. Devliotou-Panagiotidou, E. Sotiriou, and Y. Graser. 2006. Forty-eight-hour diagnosis of onychomycosis with subtyping of Trichophyton rubrum strains. J. Clin. Microbiol. 441419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong, F., and G. L. Gilbert. 2006. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB): a practical epidemiological and diagnostic tool. Nat. Protoc. 12668-2680. [DOI] [PubMed] [Google Scholar]
- 22.Liu, D., L. Pearce, G. Lilley, S. Coloe, R. Baird, and J. Pedersen. 2002. PCR identification of dermatophyte fungi Trichophyton rubrum, T. soudanense, and T. gourvilii. J. Med. Microbiol. 51117-122. [DOI] [PubMed] [Google Scholar]
- 23.Makimura, K., T. Mochizuki, A. Hasegawa, K. Uchida, H. Saito, and H. Yamaguchi. 1998. Phylogenetic classification of Trichophyton mentagrophytes complex strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J. Clin. Microbiol. 362629-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makimura, K., Y. Tamura, T. Mochizuki, A. Hasegawa, Y. Tajiri, R. Hanazawa, K. Uchida, H. Saito, and H. Yamaguchi. 1999. Phylogenetic classification and species identification of dermatophyte strains based on DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J. Clin. Microbiol. 37920-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mochizuki, T., H. Tanabe, M. Kawasaki, H. Ishizaki, and C. J. Jackson. 2003. Rapid identification of Trichophyton tonsurans by PCR-RFLP analysis of ribosomal DNA regions. J. Dermatol. Sci. 3225-32. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson, M. 2006. Lock and roll: single-molecule genotyping in situ using padlock probes and rolling-circle amplification. Histochem. Cell Biol. 126159-164. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson, M., F. Dahl, C. Larsson, M. Gullberg, and J. Stenberg. 2006. Analyzing genes using closing and replicating circles. Trends Biotechnol. 2483-88. [DOI] [PubMed] [Google Scholar]
- 28.Ninet, B., I. Jan, O. Bontems, B. Lechenne, O. Jousson, R. Panizzon, D. Lew, and M. Monod. 2003. Identification of dermatophyte species by 28S ribosomal DNA sequencing with a commercial kit. J. Clin. Microbiol. 41826-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohst, T., S. de Hoog, W. Presber, V. Stavrakieva, and Y. Graser. 2004. Origins of microsatellite diversity in the Trichophyton rubrum-T. violaceum clade (dermatophytes). J. Clin. Microbiol. 424444-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Playford, E. G., F. Kong, Y. Sun, H. Wang, C. Halliday, and T. C. Sorrell. 2006. Simultaneous detection and identification of Candida, Aspergillus, and Cryptococcus species by reverse line blot hybridization. J. Clin. Microbiol. 44876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savolainen, V., R. S. Cowan, A. P. Vogler, G. K. Roderick, and R. Lane. 2005. Towards writing the encyclopedia of life: an introduction to DNA barcoding. Philos. Trans. R. Soc. Lond. B Biol. Sci. 3601805-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scher, R. K., and R. Baran. 2003. Onychomycosis in clinical practice: factors contributing to recurrence. Br. J. Dermatol. 149(Suppl. 65)5-9. [DOI] [PubMed] [Google Scholar]
- 33.States, D. J., and P. Agarwal. 1996. Compact encoding strategies for DNA sequence similarity search. Proc. Int. Conf. Intell. Syst. Mol. Biol. 4211-217. [PubMed] [Google Scholar]
- 34.Summerbell, R. C., R. A. Haugland, A. Li, and A. K. Gupta. 1999. rRNA gene internal transcribed spacer 1 and 2 sequences of asexual, anthropophilic dermatophytes related to Trichophyton rubrum. J. Clin. Microbiol. 374005-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summerbell, R. C., M. K. Moore, M. Starink-Willemse, and A. Van Iperen. 2007. ITS barcodes for Trichophyton tonsurans and T. equinum. Med. Mycol. 45193-200. [DOI] [PubMed] [Google Scholar]
- 36.Templeton, A. R. 1989. The meaning of species and speciation: a genetic perspective, p. 3-27. In D. Otte and J. A. Endler (ed.), Speciation and its consequences. Sinauer Associates, Inc., Sunderland, MA.
- 37.Tong, Z., F. Kong, B. Wang, X. Zeng, and G. L. Gilbert. 2007. A practical method for subtyping of Streptococcus agalactiae serotype III, of human origin, using rolling circle amplification. J. Microbiol. Methods 7039-44. [DOI] [PubMed] [Google Scholar]
- 38.Wang, B., S. J. Potter, Y. Lin, A. L. Cunningham, D. E. Dwyer, Y. Su, X. Ma, Y. Hou, and N. K. Saksena. 2005. Rapid and sensitive detection of severe acute respiratory syndrome coronavirus by rolling circle amplification. J. Clin. Microbiol. 432339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weitzman, I., and R. C. Summerbell. 1995. The dermatophytes. Clin. Microbiol. Rev. 8240-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng, X., F. Kong, C. Halliday, S. Chen, A. Lau, G. Playford, and T. C. Sorrell. 2007. Reverse line blot (RLB) hybridization assay for the identification of medically important fungi from culture and clinical specimens. J. Clin. Microbiol. 452872-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]