Abstract
Infection load and the integration of human papillomaviruses (HPV) have been implicated as determinants for oncogenesis, but whether variation among different HPV types exists remains unclear. We investigated 91 women infected with HPV type 52 (HPV-52), a type that is rare worldwide but common in East Asia. The median viral load increased with the severity of the lesion (248 copies/cell equivalent for normal/cervical intraepithelial neoplasia [CIN] grade 1, 402 copies/cell equivalent for CIN 2, 523 copies/cell equivalent for CIN 3, and 1,435 copies/cell equivalent for invasive cancer). The proportion of specimens with integration increased significantly with the severity of the lesion (P < 0.001). The viral load was associated with the physical status of the viral genome, with higher levels for the pure episomal form (P = 0.001). Infection status should be considered when interpreting viral load data for HPV-52, as single infections with this HPV type were found to have marginally higher viral loads than coinfections (P = 0.051). All except one sample had E2 disruption restricted to only a part of the gene. Integration is a critical step in HPV-52-induced carcinogenesis. The profile of E2 disruption was different from that described for HPV-16, with the amino-terminal region being most frequently involved. Selecting the appropriate E2 region for amplification is critical in studying the integration of HPV-52. In summary, the HPV-52 viral load and the integrated proportion increased with the severity of the cervical lesions but had a different pattern than that of HPV-16.
Infection with human papillomavirus (HPV) is known to play a pivotal role in the development of cervical cancer (4, 37, 41). Some previous studies have suggested that not the presence of viral DNA per se but the viral load of HPV is an important determinant for the development of high-grade lesions (18, 25, 27, 34, 35, 36, 40). However, a substantial number of studies did not observe a correlation between viral load and the severity of cervical disease (5, 8, 26, 29, 38). The techniques that have been used to measure HPV infection load vary across studies. Some studies measured the overall viral load across a panel of high-risk HPV types without considering the specific type of HPV actually harbored in the specimen. Although coinfections with multiple types are commonly found throughout different grades of cervical lesion, previous studies often did not consider single infection or coinfection status. It is now recognized that the measurement and interpretation of the viral load in HPV infection are far more complex than previously perceived (39).
The HPV genome can exist in two physical states: it can either be a circular episomal form or integrated into the human genome (10). The integration of viral DNA is considered to result in transformation of the infected cells, as integration usually disrupts the E2 gene whose normal function is to down-regulate the expression of the E6 and E7 viral oncogenes (11, 16, 17, 33). It has therefore been proposed that viral integration could be an adjunctive diagnostic or prognostic marker (19, 21). Studies based on HPV type 18 (HPV-18)-positive clinical specimens showed that integration is virtually completed in high-grade lesions and invasive cancers (3, 30). However, the results obtained from HPV-16-positive specimens are more diverse (5, 8, 20). Furthermore, it has recently been demonstrated that the integration of the HPV-16 genome does not invariably result in high levels of oncogene E6/E7 expression (12).
Infection load and viral integration are interrelated, as the replication of HPV can only occur in the presence of the episomal form of the genome. At present, only a few studies have examined these two events in the same cohort of subjects (1, 13, 20, 31). Furthermore, most studies have targeted the type that is the most common worldwide, HPV-16, whereas data on other HPV types are very limited (6, 13, 32). Whether the association of infection load and viral integration with cervical cancer development varies with the type of HPV infection remains unknown. HPV-52, though generally regarded as an uncommon type, has been found to exist with an unusually high prevalence in a few Chinese populations, including the population in Hong Kong (7, 9, 15, 22, 23, 24). In some of these areas, HPV-52 was the second- or third-most-common HPV type found in cervical cancer (7, 13, 15, 24). We took advantage of this special epidemiological feature in our local population to examine the two key viral parameters, infection load and viral genome integration, of HPV-52 infection in a cohort of southern Chinese women with various degrees of cervical lesions.
MATERIALS AND METHODS
Study subjects.
This study was conducted in Hong Kong, where cervical cancer ranks ninth for new cancers in women, with an age-standardized incidence of 7.8 per 100,000 (14). The cervical samples for this study were obtained from women attending the colposcopy clinic at the Prince of Wales Hospital. Women with normal colposcopic findings were classified into the “normal” group provided that their subsequent 6-month follow-up cervical cytology results did not reveal abnormalities. The cervical disease status of the specimens was defined histologically and classified as cervical intraepithelial neoplasia (CIN) grade 1, 2, or 3 or invasive cervical cancer (ICC). Pregnant or immunocompromised women or those with a previous history of cervical disease or surgery were excluded. Consent was obtained from all study subjects, and the study was approved by the institutional ethics committee.
HPV-52 detection and genotyping.
Total DNA was extracted from cervical scrape samples by using a QIAamp DNA mini kit (Qiagen). HPV was genotyped by using a linear array HPV genotyping test (Roche Molecular Systems, Inc., CA) which can detect 37 HPV types. Samples found to contain HPV-52 either as a single infection or as a coinfection with other HPV types were subjected to further examination for viral load and E2 gene disruption status.
Viral load, host gene quantitation, and integration.
The total (episomal plus integrated) crude viral load was determined by real-time PCR targeting the E7 gene of HPV-52. The E7 gene was used as it is retained both in the episomal and integrated forms. To account for variations in the amount of cells collected in each sample, the housekeeping gene beta-actin was also quantified. The normalized viral load was obtained with the equation (E7copy/beta-actincopy) × 2 and expressed as viral copies/cell equivalent.
The disruption in the E2 gene was inferred by comparing the levels of the E2 and E7 gene copies in the viral load. E2 gene disruption was taken as a marker of HPV integration. To account for the possibility that disruption might involve only part of the E2 gene, four sets of nonoverlapping real-time PCR primers were designed to encompass the whole E2 gene, with the first two sets covering the amino (N)-terminal region and the other two sets targeting, respectively, the hinge (H) region and the carboxy (C)-terminal region. When any of the E2 gene real-time PCR assays revealed an undetectable gene copy number, the specimen was regarded as harboring the “pure integrated form” of the HPV genome. An E2/E7 gene copy number ratio of 1 indicated “pure episomal form”, whereas ratios of less than 1 were taken as having “concomitant forms.”
The six real-time PCRs targeting HPV-52 E7; the E2 C-terminal, E2 H, E2 N-terminal 1, and E2 N-terminal 2 regions; and beta-actin were performed by using power Sybr green PCR master mix (Applied Biosystems, Foster City, CA) with a 7900HT sequence detection system (Applied Biosystems, Foster City, CA). The standard curve for each viral target was generated by plotting the threshold cycle values against 10-fold serial dilutions of plasmids containing the whole genome of HPV-52 (American Type Culture Collection, Rockville, MD). Quantified preparations from a commercial kit (TaqMan DNA template reagent; Applied Biosystems, Foster City, CA) were used to generate a standard curve for beta-actin gene quantification. During the process of DNA extraction, a 200-μl aliquot of each cervical sample was eluted to a final volume of 100 μl. This concentration factor was taken into account when calculating the gene copy numbers. Five microliters of the extracted preparation was amplified in a 25-μl reaction mix containing 0.125 to 1.0 μmol of primers (Table 1). The cycling conditions were 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, 58°C for 15 s (except that 56°C for 15 s was used for beta-actin), and 72°C for 30 s. All real-time PCR assays showed a wide linear range that covered at least 10 to 10,000,000 copies/μl, and all showed high amplification efficiencies. The specificity of each amplification was confirmed by checking the dissociation curve against the expected melting temperature of the amplification product.
TABLE 1.
Primer sets for real-time PCRs
| Amplification target | Primer sequences (nta) | Annealing temp (°C) | Concn (μM) | Product size (bp) |
|---|---|---|---|---|
| E2 N-terminal region 1 | 5′-GATACCGGCACGTTTAAATG-3′ (2751-2772) and 5′-CTTTGCCTTAGACACTGCCA-3′ (2937-2946) | 58 | 0.75 | 196 |
| E2 N-terminal region 2 | 5′-AGTCTAGAAATGTGGCGTGC-3′ (3034-3053) and 5′-CAATTGTACATTCACACTCACC-3′ (3163-3184) | 58 | 1.0 | 151 |
| E2 H region | 5′-GTCCACCTATGCACCGAAAC-3′ (3373-3392) and 5′-TCAGTTGCAGTGACGAGTCC-3′ (3544-3563) | 58 | 0.75 | 191 |
| E2 C-terminal region | 5′-GTTCAAATTTCATCTACCTGGC-3′ (3685-3706) and 5′-GACATGACACCTTGTATAACTTGC-3′ (3819-3843) | 58 | 1.0 | 158 |
| E7 | 5′-GCAACCTGAAACAACTGACCT-3′ (597-617) and 5′-TCCGTCGCAGTGCTATGAAT-3′ (763-782) | 58 | 0.75 | 186 |
| Beta-actin | 5′-GCACGGCATCGTCACCAACT-3′ (1283-1303) and 5′-CATCTTCTCGCGGTTGGCCT-3′ (1404-1424) | 56 | 0.125 | 142 |
nt, nucleotide position numbering according to the HPV52 reference strain (GenBank accession no. X74481).
Statistical analysis.
The differences in viral load levels between groups were compared with the Mann-Whitney U test or the Kruskal-Wallis test using SPSS software (version 14.0; SPSS Inc., Chicago, IL). The distributions of categorical variables, including integration status and location of disruption among groups, were assessed with the chi-squared test or Fisher's test as appropriate. The linear trend in proportions was examined with the chi-squared test. These statistical tests were conducted by using the Statcalc program (Epi Info; Centers for Disease Control and Prevention, Atlanta, GA). P values of less than 0.05 were regarded as significant.
RESULTS
Ninety-one women with HPV-52 infections were included in this study. Their ages ranged from 22 to 88 (mean, 50.0; standard deviation, 15.2) years. Twelve women had normal cervices, 12 had CIN 1, 20 had CIN 2, 27 had CIN 3, and 20 had ICC. All invasive cancers were of the squamous-cell type, except for one case of adenocarcinoma.
Viral load.
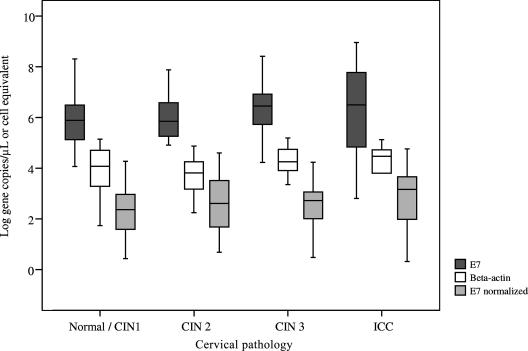
The crude viral load of the 91 specimens spread over a wide range of from 644 to 9 × 108 copies/μl (median, 1,669,812; interquartile range, 267,937 to 8,384,753) (Table 2). When normalized to the beta-actin DNA level, the viral load ranged from 0.03 to 89,344 copies/cell equivalent (median, 462; interquartile range, 41 to 1,564). The distribution of viral loads and beta-actin levels with respect to the degree of cervical lesion is shown in Fig. 1. The normalized median viral load appeared to increase with the severity of the cervical lesion (248 copies/cell equivalent for normal/CIN 1, 402 copies/cell equivalent for CIN 2, 523 copies/cell equivalent for CIN 3, and 1,435 copies/cell equivalent for ICC) (Table 2). However, the interquartile ranges overlapped, and the differences were not statistically significant (P = 0.308 by Kruskal-Wallis test).
TABLE 2.
Viral load and physical status of the viral genome in HPV-52 infection according to the degree of cervical lesion
| Viral characteristics | Cervical pathology
|
|||
|---|---|---|---|---|
| Normal/CIN 1 (n = 24) | CIN 2 (n = 20) | CIN 3 (n = 27) | ICC (n = 20) | |
| Median viral load (interquartile range) | ||||
| Crude total viral load (copies/μl) | 777,734 (129,962-3,744,271) | 712,453 (166,251-3,995,568) | 2,817,698 (525,172-9,348,353) | 3,325,361 (87,655-100,000,000) |
| Beta-actin level (copies/μl) | 11,909 (1,827-51,890) | 6,564 (1,461-18,151) | 18,008 (7,898-62,012) | 29,201 (6,341-53,428) |
| Normalized viral load (copies/cell equivalent) | 248 (38-1,051) | 402 (45-3,318) | 523 (91-1,410) | 1,435 (86-4,980) |
| Viral genome physical status (no. [%]) | ||||
| Pure episomal form | 15 (62.5) | 11 (55.0) | 12 (44.4) | 2 (10.0) |
| Pure integrated form | 5 (20.8) | 2 (10.0) | 2 (7.4) | 5 (25.0) |
| Concomitant forms | 4 (16.8) | 7 (35.0) | 13 (48.1) | 13 (65.0) |
| HPV-52 infection status (no. [%]) | ||||
| Single infection | 15 (62.5) | 13 (65.0) | 16 (59.3) | 9 (45.0) |
| Coinfection with other HPV type(s) | 9 (37.5) | 7 (35.0) | 11 (40.7) | 11 (55.0) |
| E2 gene region involved in disruption (no. [%a]) | ||||
| N-terminal region 1 | 7 (77.8) | 4 (44.4) | 10 (66.7) | 18 (100.0) |
| N-terminal region 2 | 9 (100.0) | 7 (77.8) | 13 (86.7) | 10 (55.6) |
| H region | 2 (22.2) | 0 (00.0) | 0 (00.0) | 1 (5.6) |
| C-terminal region | 4 (44.4) | 2 (22.2) | 2 (13.3) | 1 (5.6) |
| N-terminal region 1 or 2 | 9 (100.0) | 9 (100.0) | 15 (100.0) | 18 (100.0) |
| Whole E2 gene | 0 (00.0) | 0 (00.0) | 0 (00.0) | 1 (5.0) |
| Pattern of E2 disruption (no. [%]a) | ||||
| Single | 5 (55.6) | 7 (77.8) | 12 (80.0) | 15 (83.3) |
| Multiple | 4 (44.4) | 2 (22.2) | 3 (20.0) | 3 (16.7) |
The number of specimens with integration was the denominator. Twelve specimens showed mixed patterns of disruption.
FIG. 1.
Distribution of viral load according to degree of cervical lesion. The middle line indicates the median. The box represents the interquartile range. The lines extending from each box represent the upper and lower limit. The normalized value for E7 was determined by the equation E7 normalized = (E7copy/beta-actincopy) × 2.
Viral genome status.
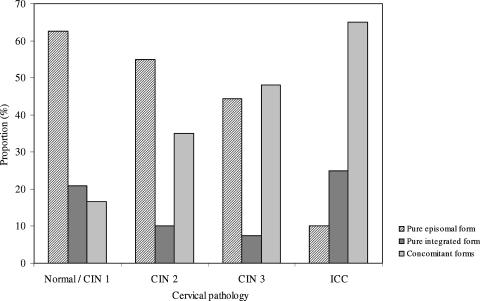
Of the 91 specimens examined, 40 (44.0%) did not show any disruption of the E2 gene. These specimens were regarded as harboring the pure episomal form of the viral genome. Fourteen specimens (15.4%) showed an undetectable gene copy number for one or more of the E2 regions and, hence, were regarded as harboring the pure integrated form of the viral genome. The remaining 37 specimens (40.7%) contained both the episomal and integrated form (i.e., concomitant forms). The proportion of specimens harboring integrated genomes in either the pure or concomitant form increased significantly with the increase in severity of the cervical lesion (chi-squared value for linear trend in proportions, 11.62; P < 0.001) (Fig. 2). The pure integrated form was found in 25.0% of ICC cases, as well as in a similar proportion (20.8%) of normal/CIN 1 cases. The proportion of the pure episomal form in ICC cases (10.0%) was significantly lower than its proportion in the other groups (44.4 to 62.5%) (P value of <0.05 by Fisher's exact test in comparison to each group) (Table 2).
FIG. 2.
Physical status of HPV-52 genome according to degree of cervical lesion.
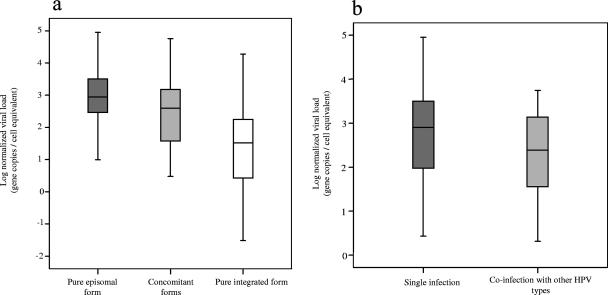
The levels of normalized viral load were significantly different among specimens harboring viral genomes with different physical statuses (P value of 0.001 by Kruskal-Wallis test) (Fig. 3a). Specimens harboring the pure episomal form had higher viral loads with a median (interquartile range) of 877 (281 to 3,208) copies/cell equivalent, whereas those harboring the pure integrated form had a lower viral load of 33 (3 to 260) copies/cell equivalent.
FIG. 3.
Distribution of viral load according to infection characteristics. (a) Physical status of viral genome. (b) Infection status. The middle line indicates the median. The box represents the interquartile range. The lines extending from each box represent the upper and lower limit. The normalized viral load was calculated by the equation (E7copy/beta-actincopy) × 2.
Single infection and coinfection.
Of the 91 subjects, 53 had single infections, with HPV-52 alone. The other 38 had coinfections with HPV-52 and other HPV type(s) (Table 2). The most common coinfection was with HPV-16 (12/38; 31.6%), followed by HPV-18 (18.4%) and HPV-58 and HPV-66 (5.3% each). Four cases had triple infections, three with HPV-16 and -18 and one with HPV-18 and -33. Other coinfecting types, with each found in one case, included HPV-33, -51, -53, -54, -55, -56, -61, -69, -81, -CP6108, and -MM8. The viral loads in specimens with single infections were marginally higher than in those with coinfections (median [interquartile range] of 798 [93 to 3,191] versus 255 [36 to 1,383] copies/cell equivalent; P value of 0.051 by Mann-Whitney U test) (Fig. 3B). When the physical status of the viral genome was correlated with the infection status of single infection or coinfection, no significant association was observed.
E2 disruption patterns.
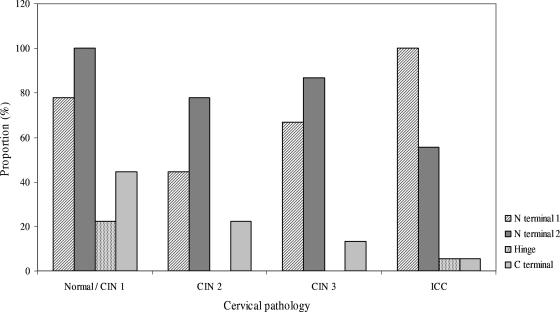
The E2 gene copy number profiles of 12 specimens suggested the presence of multiple E2 gene disruption patterns. These multiple disruption patterns were more commonly found in normal/CIN 1 cases than in the other groups (44.4% versus 16.7 to 22.2%) (Table 2). All 12 of these specimens contained a disruption that occurred across the N-terminal 1 and N-terminal 2 regions and which coexisted with a disruption that involved only the N-terminal 1, N-terminal 2, or C-terminal region. Overall, the N-terminal 1 and N-terminal 2 regions were the two most frequently disrupted regions, with each found to be disrupted in 76.5% (39/51) of specimens harboring integrated (pure or concomitant forms) viral genomes, whereas the less-involved C-terminal and H regions were found to be disrupted in 17.6% and 5.9% of specimens (Table 2).
The proportion of specimens with disruption involving the N-terminal 1 region increased significantly from CIN 2 to ICC (chi-squared value for linear trend of proportions, 4.2; P = 0.04). However, the proportion was paradoxically high for the normal/CIN 1 group (77.8%) (Fig. 4).
FIG. 4.
E2 gene disruption involvement according to the degree of cervical lesion.
The proportion of specimens with disruption involving the C-terminal region appeared to decrease with an increase in cervical disease severity. However, the trend was not significant when subjected to statistical analysis (chi-squared value for linear trend, 1.8; P = 0.18).
DISCUSSION
While at least 15 HPV types are known to cause cervical cancer (28), the model of oncogenesis for HPV is mainly established from studies of HPV-16 and HPV-18 (41). Whether these observations and hypotheses can be generalized to other high-risk HPV types remains to be verified. Studies of the viral characteristics of non-16/-18 HPV types and their correlation with the progression of infected cervical lesions are very limited. The current study provides data on an HPV type, HPV-52, that is associated with a substantial proportion of cervical cancers in East Asia.
Several studies have reported a positive association between the viral load of HPV-16 and current cervical disease status or prognosis (18, 25, 27, 34, 35, 36, 40). At present, there has been only one study reporting the viral load of HPV-52, which showed a higher viral load for invasive cancers than for CIN 2/3 (13). Results from the current study also suggested that, though not reaching statistical significance, the viral load of HPV-52 increased with the degree of cervical lesion. However, it should be noted that the wide range of variation and the overlaps in the viral load levels between normal and high-grade lesions do not favor a clinical role for viral load determination for HPV-52. We also observed that the interpretation of viral load can be complicated by its association with the viral genome status (episomal or integrated) and infection status (single or coinfection). Our finding that specimens harboring the episomal form of the HPV-52 genome were associated with a higher viral load can be explained, at least partly, by the fact that only the episomal form of the viral genome is capable of vegetative replication. On the other hand, the reason for the observed tendency for a higher viral load among cases of single infection than among cases of coinfection is not as obvious. One hypothesis could be that in the presence of other HPV types during a coinfection, HPV-52 becomes less successful in maintaining the infection and, thus, might be less transmissible. This raises a question about the possible type replacement effect following the widespread use of vaccines targeting HPV-16, but not HPV-52. Nonetheless, it should be noted that the viral load levels in cases of single infection with HPV-52 were only marginally higher than those in cases of coinfection. More studies are needed to confirm the current observation.
Studies of HPV-16 have repeatedly shown that clinical samples, regardless of lesion grade, often contain a mixture of the episomal and integrated forms of the viral genome (1, 8, 20). Our study also revealed this phenomenon for HPV-52. Given this feature, techniques employed for determining integration status should be capable of detecting integration over the episomal genome background. Real-time PCR comparing the levels of the E2 and E6 or E7 genes as applied in this study is a commonly used method to achieve this purpose. However, real-time PCR has a technical limitation in that the target amplification product should be around 100 to 150 bp in length to provide an efficient amplification. Our results showed that only one specimen had disruption involving the whole E2 gene, implying that studies using amplification targets not covering the whole E2 gene could have underestimated the integration of HPV-52 (13). Our results showed that the N-terminal region of E2 was most frequently involved in disruption and, thus, the most sensitive target for identifying integration of HPV-52. In contrast to the results of a report on HPV-16 (2), the H region is rarely disrupted in HPV-52. These results suggest that the E2 gene disruption pattern of HPV could be type specific.
The concept that viral integration is a prerequisite for the oncogenic progression of an HPV-infected cell has been challenged by a few recent studies using HPV-16 as the study model (5, 8, 26, 29). It has been shown that the integration of HPV-16 does not invariably result in high levels of E6/E7 expression (12). The only such study of HPV-52, which was reported by Ho et al. (13), used the H region of E2 as the amplification target to detect integration in specimens from 13 invasive cancers and 18 CIN 2/3 cases. It was found that 92% of CIN 2/3 cases but only 25% of cancers harbored integrated genomes (either as the pure integrated or concomitant form). In contrast, our data show a significant correlation between integrated viral genomes and lesion severity. Although we observed two invasive cancers harboring the pure episomal form of the viral genome, the overall picture indicates that viral integration is still a key event leading to the oncogenic progression of an HPV-52-infected cervical lesion. Furthermore, our results show that the integration event of HPV-52 occurred early in the course of infection and might be one of the key initiators of oncogenic progression.
The two invasive cancers that harbored the pure episomal form of the viral genome were both instances of HPV-52 single infection, without the involvement of other HPV types. Although we cannot exclude the possibility that viral genome disruption had occurred outside the E2 region, it is worthwhile to investigate whether events such as methylation had bypassed viral integration in the development of invasive cancers in these patients.
The correlation of infection load and integration of HPV with oncogenic progression and its clinical usefulness may vary with the type of HPV in question. More studies on HPV types other than HPV-16 are warranted.
Acknowledgments
The work described in this paper was fully supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region, People's Republic of China (project no. CUHK4429/03 M).
The authors declare no conflict of interest.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Anderssen, S., H. Safari, M. Mints, I. Lewensohn-Fuchs, U. Gyllensten, and B. Johansson. 2005. Type distribution, viral load and integration status of high-risk human papillomaviruses in pre-stages of cervical cancer (CIN). Br. J. Cancer 922195-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias-Pulido, H., C. L. Peyton, N. E. Joste, H. Vargas, and C. M. Wheeler. 2006. Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J. Clin. Microbiol. 441755-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badaracco, G., A. Venuti, A. Sedati, and M. L. Marcante. 2002. HPV16 and HPV18 in genital tumors: significantly different levels of viral integration and correlation to tumor invasiveness. J. Med. Virol. 67574-582. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, F. X., A. Lorinzc, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castle, P. E., M. Schiffman, D. R. Scott, M. E. Sherman, A. G. Glass, B. B. Rush, J. E. Schussler, S. Wacholder, and A. T. Lorincz. 2005. Semiquantitative human papillomavirus type 16 viral load and the prospective risk of cervical precancer and cancer. Cancer Epidemiol. Biomarkers Prev. 141311-1314. [DOI] [PubMed] [Google Scholar]
- 6.Chan, P. K., J. L. Cheung, T. H. Cheung, K. W. K. Lo, S. F. Yim, S. S. N. Siu, and J. W. Tang. 2007. Profile of viral load, integration and E2 disruption of HPV58 in normal cervix and cervical neoplasia. J. Infect. Dis. 196868-875. [DOI] [PubMed] [Google Scholar]
- 7.Chan, P. K., T. H. Cheung, A. O. Tam, K. W. Lo, S. F. Yim, M. M. Yu, K. F. To, Y. F. Wong, J. L. Cheung, D. P. Chan, M. Hui, and M. Ip. 2006. Biases in human papillomavirus genotype prevalence assessment associated with commonly used consensus primers. Int. J. Cancer 118243-245. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, J. L., K. W. Lo, T. H. Cheung, J. W. Tang, and P. K. Chan. 2006. Viral load, E2 gene disruption status and lineage of human papillomavirus type 16 infection in cervical neoplasia. J. Infect. Dis. 1941706-1712. [DOI] [PubMed] [Google Scholar]
- 9.Dai, M., Y. P. Bao, G. M. Clifford, S. Vaccarella, P. J. Snijders, R. D. Huang, L. X. Sun, C. J. Meijer, Y. L. Qiao, and S. Franceschi. 2006. Human papillomavirus infection in Shanxi Province, People's Republic of China: a population-based study. Br. J. Cancer 9596-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doorbar, J. 2006. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 110525-541. [DOI] [PubMed] [Google Scholar]
- 11.el Awady, M. K., J. B. Kaplan, S. J. O'Brien, and R. D. Burk. 1987. Molecular analysis of integrated human papillomavirus 16 sequences in the cervical cancer cell line SiHa. Virology 159389-398. [DOI] [PubMed] [Google Scholar]
- 12.Häfner, N., C. Driesch, M. Gajda, L. Jansen, R. Kirchmayr, I. B. Runnebaum, and M. Dürst. 10 September 2007. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene. doi: 10.1038/sj.onc.1210791. [DOI] [PubMed]
- 13.Ho, C. M., T. Y. Chien, S. H. Huang, B. H. Lee, and S. F. Chang. 2005. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol. Oncol. 10254-60. [DOI] [PubMed] [Google Scholar]
- 14.Hong Kong Cancer Registry, Hospital Authority. Fast stats on cervix cancer Stat 2004. Hong Kong Cancer Registry, Hospital Authority. http://www3.ha.org.hk/cancereg/eng/cx.pdf.
- 15.Huang, S., I. Afonina, B. A. Miller, and A. M. Beckmann. 1997. Human papillomavirus types 52 and 58 are prevalent in cervical cancers from Chinese women. Int. J. Cancer 70408-411. [DOI] [PubMed] [Google Scholar]
- 16.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 692989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeon, S., and P. F. Lambert. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 921654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Josefsson, A. M., P. K. Magnusson, N. Ylitalo, P. Sorensen, P. Qwarforth-Tubbin, P. K. Andersen, M. Melbye, H. Q. Adami, and U. B. Gyllensten. 2000. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet 3552189-2193. [DOI] [PubMed] [Google Scholar]
- 19.Kalantari, M., F. Karlsen, G. Kristensen, R. Holm, B. Hagmar, and B. Johansson. 1998. Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int. J. Gynecol. Pathol. 17146-153. [DOI] [PubMed] [Google Scholar]
- 20.Kumala, S. M., S. M. Syrjänen, U. B. Gyllensten, I. P. Shabalova, N. Petrovichev, P. Tosi, K. J. Syrjänen, and B. C. Johansson. 2006. Early integration of high copy HPV16 detectable in women with normal and low grade cervical cytology and histology. J. Clin. Pathol. 59513-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazo, P. A. 1997. Papillomavirus integration: prognostic marker in cervical cancer? Am. J. Obstet. Gynecol. 1761121-1122. [DOI] [PubMed] [Google Scholar]
- 22.Li, L. K., M. Dai, G. M. Clifford, W. Q. Yao, A. Arslan, N. Li, J. F. Shi, P. J. Snijders, C. J. Meijer, Y. L. Qiao, and S. Franceschi. 2006. Human papillomavirus infection in Shenyang City, People's Republic of China: a population-based study. Br. J. Cancer 951593-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liaw, K. L., A. W. Hsing, M. H. Schiffman, S. L. You, T. Zhang, R. Burk, and C. J. Chen. 1997. Human papillomavirus types 52 and 58 are prevalent in cervical cancer from Chinese women. Int. J. Cancer 73775-776. [DOI] [PubMed] [Google Scholar]
- 24.Lo, K. W., Y. F. Wong, M. K. Chan, J. C. Li, J. S. Poon, V. W. Wang, S. N. Zhu, T. M. Zhang, Z. G. He, O. L. Wu, G. D. Li, J. S. Tam, T. Kahn, P. Lam, T. H. Cheung, and T. K. Chung. 2002. Prevalence of human papillomavirus in cervical cancer: a multicenter study in China. Int. J. Cancer 100327-331. [DOI] [PubMed] [Google Scholar]
- 25.Lo, K. W., S. W. Yeung, T. H. Cheung, N. S. Siu, T. Kahn, and Y. F. Wong. 2005. Quantitative analysis of human papillomavirus type 16 in cervical neoplasm: a study in Chinese population. J. Clin. Virol. 3476-80. [DOI] [PubMed] [Google Scholar]
- 26.Lorincz, A. T., P. E. Castle, M. E. Sherman, D. R. Scott, A. G. Glass, S. Wacholder, B. B. Rush, P. E. Gravitt, J. E. Schussler, and M. Schiffman. 2002. Viral load of human papillomavirus and risk of CIN3 or cervical cancer. Lancet 360228-229. [DOI] [PubMed] [Google Scholar]
- 27.Moberg, M., I. Gustavsson, E. Wilander, and U. Gyllensten. 2005. High viral loads of human papillomavirus predict risk of invasive cervical carcinoma. Br. J. Cancer 92891-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, C. J. Meijer, et al. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348518-527. [DOI] [PubMed] [Google Scholar]
- 29.Onan, M. A., C. Taskiran, G. Bozdayi, A. Biri, O. Erdem, A. Acar, G. Gunaydin, S. Rota, O. Ataoglu, and H. Guner. 2005. Assessment of human papilloma viral load of archival cervical intraepithelial neoplasia by real-time polymerase chain reaction in a Turkish population. Eur. J. Gynaecol. Oncol. 26632-635. [PubMed] [Google Scholar]
- 30.Park, J. S., E. S. Hwang, S. N. Park, H. K. Ahn, S. J. Um, C. J. Kim, S. J. Kim, and S. E. Namkoong. 1997. Physical status and expression of HPV genes in cervical cancers. Gynecol. Oncol. 65121-129. [DOI] [PubMed] [Google Scholar]
- 31.Peitsaro, P., B. Johansson, and S. Syrjanen. 2002. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J. Clin. Microbiol. 40886-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirami, L., V. Giache, and A. Becciolini. 1997. Analysis of HPV16, 18, 31, and 35 DNA in pre-invasive lesions of the uterine cervix. J. Clin. Pathol. 50600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romanczuk, H., and P. W. Howley. 1992. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl. Acad. Sci. USA 893159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snijders, P. J., C. J. Hogewoning, A. T. Hesselink, J. Berkhof, F. J. Voorhorst, M. C. Bleeker, and C. J. Meijer. 2006. Determination of viral load thresholds in cervical scrapings to rule out CIN 3 in HPV 16, 18, 31, 33-positive women with normal cytology. Int. J. Cancer 1191102-1107. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, H. T., C. H. Wu, H. L. Lai, R. N. Li, Y. C. Tung, H. Y. Chuang, T. N. Wu, L. J. Lin, C. K. Ho, H. W. Liu, and M. T. Wu. 2005. Association between quantitative high-risk human papillomavirus DNA load and cervical intraepithelial neoplasm risk. Cancer Epidemiol. Biomarkers Prev. 142544-2549. [DOI] [PubMed] [Google Scholar]
- 36.van Duin, M., P. J. Snijders, H. F. Schrijnemakers, F. J. Voorhorst, L. Rozendaal, M. A. Nobbenhuis, A. J. van den Brule, R. H. Verheijen, T. J. Helmerhorst, and C. J. Meijer. 2002. Human papillomavirus 16 load in normal and abnormal cervical scrapes: an indicator of CIN II/III and viral clearance. Int. J. Cancer 98590-595. [DOI] [PubMed] [Google Scholar]
- 37.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189121-129. [DOI] [PubMed] [Google Scholar]
- 38.Wensveen, C. W., M. J. Kagie, N. J. Nagelkerke, R. W. Veldhuizen, and J. B. Trimbos. 2005. Can viral load, semi-quantitatively evaluated, of human papillomavirus predict cytological or histological outcome in women with atypical squamous or glandular cells of undetermined significance cytology? Eur. J. Gynaecol. Oncol. 26393-397. [PubMed] [Google Scholar]
- 39.Woodman, C. B., S. L. Collins, and L. S. Young. 2006. The natural history of cervical HPV infection: unresolved issues. Nat. Rev. Cancer 711-12. [DOI] [PubMed] [Google Scholar]
- 40.Ylitalo, N., P. Sorensen, A. M. Josefsson, P. K. Magnusson, P. K. Andersen, J. Ponten, H. O. Adami, U. B. Gyllensten, and M. Melbye. 2000. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case-control study. Lancet 3552194-2198. [DOI] [PubMed] [Google Scholar]
- 41.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2342-350. [DOI] [PubMed] [Google Scholar]