Abstract
In anticipation of rotavirus vaccine introduction in Saudi Arabia, this study was undertaken to determine the distribution of the G and P genotypes of rotaviruses in order to examine whether there was any emerging serotype or unusual strain circulating in children in Saudi Arabia. Of 984 stool specimens collected between 17 April 2004 and 16 April 2005, rotavirus was detected by an enzyme-linked immunosorbent assay in 187 (19%) diarrheal children less than 5 years of age. Of these, 160 (86%) were classified into G and P genotypes as follows: G1P[8] (44%), G2P[4] (20%), G9P[8] (11%), G12P[8] (4%), and G3P[8] (4%). RNA polyacrylamide gel electrophoresis identified 94 (50%) specimens as long RNA patterns, 30 (16%) specimens as short RNA patterns, and 1 mixed infection. Only a single long RNA electropherotype was identified for seven specimens containing G12P[8] rotavirus. RNA-RNA hybridization demonstrated that the G12P[8] strains were similar in their genomic constellation to locally cocirculating strains and to a Nepalese G12P[8] strain. The Saudi Arabian G12 VP7 gene had a 99% nucleotide sequence identity with Nepalese and Indian G12 VP7 genes and belonged to the third lineage. This study is the first to describe the distribution of rotavirus G and P types and also the first to identify G9P[8] and G12P[8] strains in the country.
Rotavirus is the single most important etiological agent causing severe diarrhea in infants and young children in both developing and developed countries (6). While virtually all children experience rotavirus infection by the age of 3 to 5 years wherever they live (12), the majority of an estimated 527,000 rotavirus-associated deaths occur in developing countries (41). Two, live, orally administrable rotavirus vaccines are currently licensed in many countries after they had gone through large-scale safety and efficacy trials, each involving more than 60,000 children in both developing and developed countries (32, 40).
The genome of rotavirus comprises 11 segments of double-stranded RNA, which are encased within a triple-layered capsid. The outermost capsid of the virion is composed of two independent neutralization antigens, VP7 and VP4, which define the G serotype and the P serotype, respectively (6). Since molecular assays are more routinely used than serological assays to determine these serotypes, the term genotype is more often used to define the VP7 and the VP4 specificity of a given strain. Since G and P type-specific immunity is believed to play a role in protection against disease, the epidemiology of G and P serotypes (and genotypes) of circulating strains forms a critical knowledge base for the development and implementation of rotavirus vaccines (19).
Rotavirus nonstructural protein NSP4, encoded by genome segment 10, has been shown to function as a viral enterotoxin (28). Extensive sequence analysis has shown that human and animal NSP4 genes fall into five genetic groups (A to E), and strains tend to cluster within genotypes according to their species of origin (4, 7, 15-17). Thus, examination of the NSP4 genes provides useful information in analyzing unusual isolates.
In addition, identification of electropherotypes provides complementary information on the genomic diversity of rotavirus strains circulating in the region because the migration pattern of 11 segments of double-stranded RNA on a polyacrylamide gel helps define an individual strain of rotavirus (21, 25, 26).
Saudi Arabia is a country characterized by extreme heat and aridity, and it occupies ca. 80% of the Arabian Peninsula. The World Health Organization estimates that there were 462 rotavirus-associated deaths in 2004 (41), and the Ministry of Health reported that there were 267,000 to 439,000 health center visits and approximately 8,000 hospitalizations due to diarrhea of any cause in 2001 (20). However, few studies have addressed the epidemiology of rotavirus in Saudi Arabia. Particularly, information about the distribution of G and P types is completely lacking (20).
The aim of the present study was therefore to determine the distribution of G and P genotypes and electropherotypes of rotaviruses circulating in children less than 5 years of age in Saudi Arabia, in order to examine whether there was any emerging serotype or unusual strain in the region that might challenge the effectiveness of rotavirus vaccines that would be considered for use in Saudi Arabia.
MATERIALS AND METHODS
Detection of rotavirus positive stool specimens by ELISA.
Stool specimens were collected from children with acute diarrhea who were referred to the oral rehydration unit (outpatients, n = 423) and who were admitted to the hospital (inpatients, n = 561) in Maternity and Children's Hospital and Ohod Hospital during a 1-year period between 17 April 2004 and 16 April 2005. The two hospitals are the only large general pediatric hospitals providing services for the community of Maddina. The city is one of the largest cities in Saudi Arabia and is situated in the northwest region of the kingdom. A commercially available enzyme-linked immunosorbent assay (ELISA) kit (Rotaclone; Meridian Bioscience. Inc., Cincinnati, OH) was used to detect group A rotavirus antigen.
Extraction of genomic RNA.
Rotavirus genomic RNAs were extracted from an ca. 10% suspension of rotavirus-positive specimens in phosphate-buffered saline according to the guanidine isothiocyanate-silica method as previously described (11).
Determination of G and P genotypes.
The G types and P types were determined by the method described by Das et al. (8) and Gentsch et al. (11), respectively. Samples that were found G nontypeable were repeated with additional primers, including an alternative G1 typing primer NAC 9 (5), a G5 primer (13) and Beg9 degenerate and End9-degenerate primers to obtain full-length VP7 gene product (14). Samples that were found P nontypeable were repeated with additional primers, including an alternative P[8] typing primer NAC 10 (5) and a degenerate P[8] primer (18). Finally, if the remaining G or P nontypeable samples produced a visible band of an expected size after 30 cycles of reverse transcription-PCR, the product was sent for sequencing.
Culture adaptation.
Samples representing different combinations of rotavirus G and P genotypes, as well as major electropherotypes, were chosen for culture adaptation in MA104 cells by using a previously described method (22).
Polyacrylamide gel electrophoresis of rotavirus genomic RNA.
Rotavirus genomic RNAs were separated on a 10% polyacrylamide gel by electrophoresis for 18 h at a constant current of 8 mA per gel in a Laemmli buffer system using an SE600 Ruby gel apparatus (GE Health Care Bioscience, Piscataway, NJ) as described previously (21). Electropherotypes were determined by comparison of the individual RNA migration patterns of genome segments on the gel. Short and long RNA patterns were defined by the relative migration of genome segments 10 and 11, with slower-moving genome segments 10 and 11, indicating a short RNA pattern.
Nucleotide sequencing.
The double-stranded RNA gene segments 9, 4, and 10 encoding proteins VP7, VP4, and NSP4, respectively, were denatured, reverse transcribed, and then amplified by PCR. For the gene encoding VP7, primers Beg9 and End 9 (14) were used for amplification to obtain a full-length genome segment 9 (Table 1). For the gene encoding VP4, primers con2 and con3 were used to amplify an 877-bp fragment covering the VP8* fragment of genome segment 4 (11). A 739-bp fragment of the NSP4 gene (genome segment 10) was amplified by using primers described previously (7).
TABLE 1.
Oligonucleotide primers used for sequencing of VP7, VP4, and NSP4 genes of MD844
| Primer | Gene | Sense | Sequence (5′-3′) |
|---|---|---|---|
| End9G12R | VP7 | - | TAACGCTAATGAATTTTGGTACTG |
| AKG12F-01 | VP7 | + | TTTTCCGCGTTTGCTACC |
| AKG12F-02 | VP7 | + | CGCTTCATCTGTTTGTTG |
| JRG30 | NSP4 | + | GGCTTTTAAAAGTTCTGTT |
| JRG31 | NSP4 | - | ACCATTCCTTCCATTAAC |
| AKNSF | NSP4 | + | GACGTCAGCTAGGATATGA |
| AKNSR | NSP4 | - | CTAGCTGACGTCTCATCTC |
| AKP8F-01 | VP4 | + | GCAGTGTTTGAGCTATCG |
| AKP8F-02 | VP4 | + | GTAGCCCTCGGTGTTTCA |
| AKP8R-01 | VP4 | - | GTAGCCCTCGGTGTTTCA |
| AKP8R-02 | VP4 | - | CTATCTACTGGATCGACG |
Amplicons obtained for each genome segment were purified by using a QIAquick Spin kit (Qiagen, Inc., Valencia, CA). In order to obtain the full sequence in both directions for each genome segment, a series of internal primers was designed (Table 1). Amplicons and primers were sent for sequencing to Lark Technology, Inc., Takeley, Essex, United Kingdom.
Phylogenetic analysis.
The nucleotide sequences thus obtained were compared to sequences deposited in the DNA databases by using the BLAST program and the genotypes were determined based on the deduced amino acid sequences that showed the highest identity. Phylogenetic trees were constructed based on the neighbor-joining method (33) in the CLUSTAL W software package (38).
Genogroup analysis.
RNA-RNA hybridization was performed as previously described (27). Briefly the 32P-labeled single-stranded RNA probes from reference strains were hybridized to the denatured genomic RNAs from a panel of human rotavirus strains. The human rotavirus strains used in these experiments were Wa (G1P1A[8]), AU-l (G3 P3[9]), Se585 G12(P2A[6]), L26 (G12P[4]G12), 04S027 (G12P[8]G12), MD1166 (G2P[6]), MD 713 (G3P[8]), MD409 (G2P[4]), and MD28 (G9P[8]).
Hybridization was allowed to occur at 65°C for 16 h in a buffer containing 25 mM Tris-HCl, 100 mM NaCl, 1 mM EDTA, and 0.1% sodium dodecyl sulfate (pH 8.0). The resulting hybrids were then precipitated with ethanol and separated on a 10% polyacrylamide gel. Re-annealed genomic RNAs were visualized by staining with ethidium bromide under UV illumination. Autoradiographs were prepared by exposing dried gels to BioMax MS films (Eastman Kodak Co., Rochester, NY).
Nucleotide accession numbers.
The nucleotide sequences for strains MD844 and MD28 have been submitted to the DNA databases, DDBJ, GenBank, and EMBL. The accession numbers of the genes encoding NSP4, VP7, and VP4 of MD844 are AB269688, AB269689, and AB269690, respectively. The accession numbers of the genes encoding VP7 and VP4 of MD28 are AB297791 and AB297792, respectively.
RESULTS
Epidemiologic features of rotavirus diarrhea in Saudi Arabia.
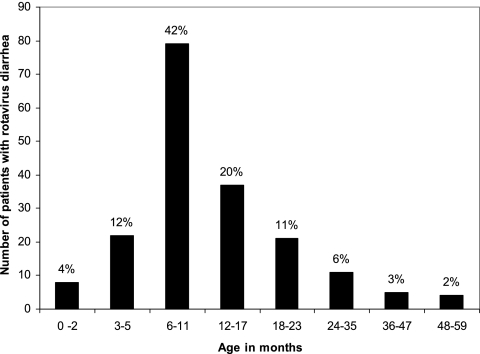
Of 984 stool specimens collected from children less than 5 years of age with acute diarrhea between 17 April 2004 and 16 April 2005, rotavirus was detected in 187 (19%) specimens by ELISA. Rotavirus was detected in 158 (28%) of 561 specimens collected from hospitalized children, whereas it was detected in only 29 (7%) of 423 children treated in the oral rehydration unit (outpatients). When the distribution of cases was examined among children less than age 5 years, 159 (83%) occurred in those aged between 4 months and 23 months, whereas only 8 cases (4.3%) occurred in the first 3 months of life (Fig. 1).
FIG. 1.
Distribution of rotavirus diarrhea cases among children less than 5 years of age.
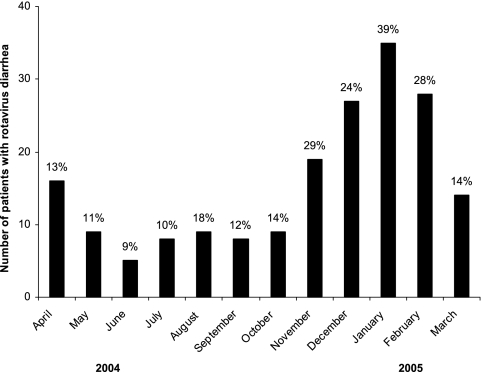
Although rotavirus diarrhea occurred throughout the year, there was a seasonal variation in the occurrence of rotavirus diarrhea, with peaks between November and February, where the detection rate of rotavirus-positive cases was 29% on average (Fig. 2). Thus, rotavirus diarrhea was most prevalent in the cooler months of the year in Saudi Arabia.
FIG. 2.
Temporal distribution of rotavirus diarrheal cases in children less than 5 years of age in Maddina, Saudi Arabia. The percentage shown above each bar indicates the detection rate of rotavirus among all diarrheal cases in the indicated month.
Molecular characterization of rotavirus genotypes circulating in Saudi Arabia.
Of 187 rotavirus-positive specimens, 160 (86%) were successfully classified into G and P genotypes (Table 2): G1P[8] (44%), G2P[4] (20%), G9P[8] (11%), G12P[8] (4%), and G3P[8] (4%). In five specimens (3%) neither the G type nor the P type was detected. In 16 specimens (8%) more than one G or P type was detected, suggesting that they represented a mixed infection of two or more rotavirus strains in a single child. In six specimens (3%) neither G nor P type was determined despite repeated reverse transcription-PCR experiments with different sets of primers, including a G5-specific primer and degenerated primers for genes encoding VP7 or VP4. However, repeated ELISA examination of these specimens confirmed the presence of rotavirus antigen, suggesting the degradation of rotavirus particles in the specimens rather than errors in the initial ELISA testing.
TABLE 2.
Characterization of rotavirus-positive specimens detected in Maddina, Saudi Arabia, into G and P types and RNA patterns
| Parameter | P[8]
|
P[4]
|
P[6]
|
PNT
|
P[MIX]
|
Total | % | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G3 | G4 | G9 | G12 | GMIX | GNT | G2 | GNT | G1 | G2 | G4 | G4 | GMIX | GNT | G1 | G2 | G4 | GMIX | |||
| No. of specimens | |||||||||||||||||||||
| Long | 50 | 5 | 2 | 16 | 7 | 8 | 3 | 1 | 1 | 1 | 94 | 50 | |||||||||
| Short | 1 | 27 | 1 | 1 | 30 | 16 | |||||||||||||||
| UDa | 17 | 1 | 2 | 7 | 1 | 1 | 1 | 2 | 32 | 17 | |||||||||||
| Negative | 15 | 1 | 1 | 2 | 1 | 3 | 1 | 6 | 30 | 16 | |||||||||||
| Mix | 1 | 1 | 1 | ||||||||||||||||||
| Total | 82 | 7 | 3 | 20 | 7 | 10 | 3 | 38 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 1 | 2 | 1 | 1 | ||
| % | 44 | 4 | 2 | 11 | 4 | 5 | 2 | 20 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 1 | 187 | 100 |
UD, undetermined.
RNA polyacrylamide gel electrophoresis identified 94 (50%) specimens as long RNA patterns, 30 (16%) specimens as short RNA patterns, and one mixed infection, whereas RNA patterns were not assignable in 32 (17%) specimens or were totally undetectable in 30 (16%) specimens (Table 2). As to the relationships between RNA patterns and genotypes (Table 2), all specimens containing short RNA pattern were classified as either G2P[4] (four electropherotypes) or G2P[6] (one electropherotype), whereas all specimens containing long RNA pattern were classified as any one of G1P[8] (four electropherotypes), G3P[8] (two electropherotypes), G4P[8] (1 electropherotype), G9P[8] (two electropherotypes), and G12P[8] (one electropherotype) (data not shown for electropherotypes).
Molecular characterization of the G12 rotavirus detected in Saudi Arabia.
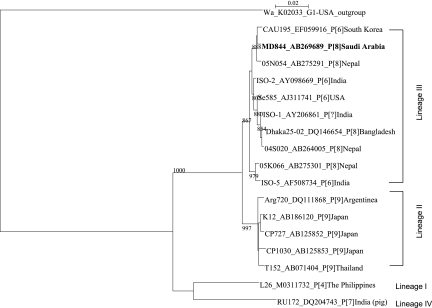
Among the specimens that were initially found G nontypeable and P[8], some were isolated in MA104 cells for further characterization. One of these cell-culture-adapted strains, designated MD844, was determined to possess a G12 VP7 gene after sequencing. This prompted us to examine the rest of nontypeable specimens with the G12-specific primer (29, 34), resulting in the identification of six additional G12P[8] specimens, all of which showed an identical long electropherotype (data not shown). The VP7 gene of one of such specimen was sequenced and found to be 100% identical with that of MD844, confirming its G12 specificity (data not shown). Based on these results, it was concluded that there were seven G12 rotavirus specimens of a single strain origin. When the gene encoding VP7 of strain MD844 was analyzed phylogenetically, it was found to belong to the third lineage of G12 VP7 gene, according to Das et al.'s classification (9), which comprise the VP7 genes of G12P[8] and G12P[6] human rotaviruses (Fig. 3).
FIG. 3.
Phylogenetic tree for the G12 VP7 genes constructed by the neighbor-joining method. The horizontal distance connecting two VP7 sequences is proportional to the genetic distance between these two VP7 sequences. The distance is expressed as the number of the nucleotide substitutions per site. Strain Wa (G1P[8]) was used as an outgroup. The bootstrap probabilities equal or greater than 800 after 1,000 replications are provided at each branching point.
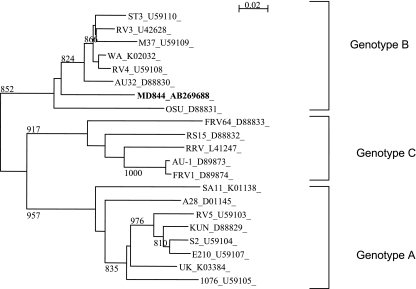
To further characterize the Saudi Arabian G12 rotavirus, the nucleotide sequences for the genes encoding VP4 and NSP4 of strain MD844 were also determined. The gene encoding VP4 of MD844 was 99% identical to that of a G9P[8] strain, designated MD28, detected and also culture-adapted in the present study, as well as those of many G12P[8] strains reported elsewhere (Table 3). The NSP4 gene of MD844 was determined to belong to genotype B or the Wa-like NSP4 (Fig. 4).
TABLE 3.
Comparison of the gene encoding VP4 of Saudi Arabian rotavirus strain MD844 (accession no. AB269690) with other human VP4 genes
| Strain | Genotype | Source | Country of origin | % Identitya
|
Accession no. | |
|---|---|---|---|---|---|---|
| nt | aa | |||||
| MD28 | G9P[8] | Human | Saudi Arabia | 99 | 98 | AB297792 |
| B4633-03 | G12P[8] | Human | Belgium | 98 | 99 | DQ146641 |
| Dahka25-02 | G12P[8] | Human | Bangladesh | 98 | 98 | DQ146652 |
| Hun9 | G9P[8] | Human | Hungary | 99 | 98 | AJ605320 |
| DRC88 | G8P[8] | Human | Congo | 98 | 99 | DQ005111 |
| OP351 | G1P[8] | Human | Malawi | 99 | 98 | AJ302147 |
| OP601 | G1P[8] | Human | Malawi | 98 | 98 | AJ302153 |
| CAU202 | G9P[8] | Human | South Korea | 98 | 98 | EF059923 |
| TF101 | G1P[8] | Human | Taiwan | 98 | 97 | AF183870 |
| RV161 | G12P[6] | Human | Belgium | 69 | 78 | DQ490548 |
| Dhaka4-03 | G2P[4] | Human | Bangladesh | 85 | 88 | DQ482714 |
Abbreviations: nt, nucleotide; aa, amino acid.
FIG. 4.
Phylogenetic tree for the NSP4 genes constructed by the neighbor-joining method.
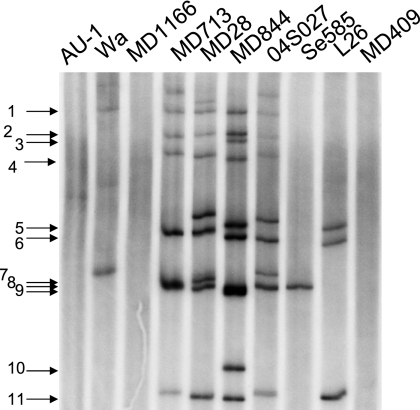
To determine to what extent the G12P[8] isolates were similar in their overall genomic RNA constellation to G12 strains isolated previously elsewhere and to other concurrently circulating genotypes in Saudi Arabia (denoted as strains with the prefix MD), RNA-RNA hybridization was performed with probes made from MD844 (Fig. 5). When the MD844 probe was allowed to hybridize with the genomic RNAs from Se585 (G12P2A[6]), L26 (G12P[4]), newly emerging Nepalese strain 04S027 (G12P[8]) (39), Wa (G1P1A[8]), AU-1 (G1P3[9]), MD1166 (G2P[6]), MD713 (G3P[8]), MD409 (G2P[4]), and MD28 (G9P[8]), the MD844 probe formed seven and nine hybrid bands with the genomic RNAs from MD713 and MD28, respectively. The probe also formed seven hybrid bands with the genomic RNAs from strain 04S027, a Nepalese G12P[8] strain. However, the probe formed only one and three hybrid bands with Se585 and L26, respectively. Unexpectedly, the MD844 probe formed only one hybrid band with the genomic RNA of strain Wa, the prototype of the Wa genogroup. The probe formed no hybrid band with AU-1 (the prototype of the AU-1 genogroup), MD1166, or MD409 (both strain most likely to belong to the DS-1 genogroup since they possess a short RNA pattern).
FIG. 5.
Result of genogrouping by RNA-RNA hybridization assays. The genomic RNAs from the indicated virus strains were hybridized with the 32P-labeled probe made from MD844. The positions of the RNA segments of the MD844 strain are indicated to the left.
DISCUSSION
The characteristics of rotavirus diarrhea among children less than 5 years of age in Maddina, Saudi Arabia, included a relatively low detection rate (19%) and marked seasonal variation with peaks of infection in the cooler months of the year. However, the rotavirus detection rate obtained in the present study fell well within the expected range since a review of 22 studies reported in this country showed that the detection rate ranged between 10 and 46% (20). Approximately 60% of rotavirus infections were identified in the first year of life, and 89% were identified by the end of the second year of life. When rotavirus vaccine is introduced into the routine childhood immunization schedule, the first vaccine dose will be given to infants at 2 months of age, meaning that infants younger than this age will not be protected by the vaccine. The observation that only 4% rotavirus infections occurred in the first 3 months of age (Fig. 2) will assure a higher effectiveness of the vaccine in this country according to the proposed immunization schedule.
While this is the first study to describe the distribution of G and P types of rotavirus circulating in Saudi Arabia, there are at least two important observations that merit particular attention. First, we identified here the circulation of G9 strains for the first time in Saudi Arabia at a relative frequency of 11%, confirming and extending the previous observation made in Iran, Iraq, and Kuwait that globally spreading G9 strains have entered the Middle East (1, 10, 24). Although the G9 strains detected in Iran, Iraq, and Kuwait were not sequenced, the VP7 gene that was carried by the G9 strain isolated in Saudi Arabia (MD28) was shown to belong to lineage III (data not shown), a finding consistent with the globally emerging G9 strains (23, 37).
Second, we also identified the circulation of G12 strains for the first time in Saudi Arabia. Although the first detection in the Middle East was in Iran in 2004 (10), there was only one specimen found to contain G12 virus. In this regard it is noteworthy that G12 genotype occurred on multiple occasions, accounting for 4% of rotavirus-positive specimens. Despite the fact the seven specimens containing G12P[8] rotavirus were detected in May 2004 (two specimens), January 2005 (two specimens), and February 2005 (three specimens), there was only a single long RNA electropherotype present in these specimens. This suggests that the introduction of G12 strain into Saudi Arabia was a rather recent event. This observation is in sharp contrast to the observation in Nepal made by Uchida et al. (39) that there were seven electropherotypes for G12P[8] and G12P[6] rotaviruses. Although it is difficult to speculate how such G12P[8] strains were introduced into the country, RNA-RNA hybridization assays provided further information to aid our understanding of the overall genomic RNA relatedness to strains concurrently circulating in the country, as well as to another G12P[8] strain isolated in Nepal. An interesting observation in this context was that strain MD844 was similar in overall genomic constellation to strain MD28, a G9P[8] strain circulating in the region, and, to a lesser degree, to strain 04S027, a G12P[8] strain isolated in Nepal, although no cocirculating G1P[8] strain was included in this comparison. Given that the G12 VP7 gene obtained in the present study showed a high degree of nucleotide sequence identity with those detected in Nepal and India, it may be that the G12 VP7 gene originating in Asia was introduced into locally cocirculating strains by the mechanism of genetic reassortment. Because G12P[8] strains in Nepal and probably in other South Asian countries have a range of genetic diversity (39), it is also likely that the G12P[8] strain detected in Saudi Arabia could result from the direct introduction of one of the G12P[8] strains circulating in South Asia. In this regard, it may merit mention that there is significant population movement from South Asian countries to Saudi Arabia, which may facilitate the transmission of G12P[8] strains.
Interestingly, G12 strains have now been detected with a variety of gene constellations, including long and short electropherotypes, and in association with three different P genotypes and two different genogroups (2, 3, 9, 30, 31, 35, 36). Such diversity observed in G12 strains strongly suggests the versatility of this strain to spread efficiently in the population through reassortment. Since the study period was limited to 1 year, consideration of the introduction of rotavirus vaccine into Saudi Arabia in the near future requires the need for continuing and broader surveillance of rotavirus strains in circulation.
Acknowledgments
We thank Mahr Al-Mokhalfi and Abdualrahman T. Saadi for their help in sample collection.
A.M.K. is supported by Ministry of Health, Saudi Arabia. O.N. and T.N. are honorary members of The University of Liverpool and participated in this study according to the Agreement on Academic Partnership between The University of Liverpool and Nagasaki University.
We regret the recent and untimely death of our coauthor, Tony Hart, who encouraged A.M.K. throughout this study.
Footnotes
Published ahead of print on 30 January 2008.
REFERENCES
- 1.Ahmed, H. M., J. B. Coulter, O. Nakagomi, C. A. Hart, J. M. Zaki, A. A. Al-Rabaty, W. Dove, and N. A. Cunliffe. 2006. Molecular characterization of rotavirus gastroenteritis strains, Iraqi Kurdistan. Emerg. Infect. Dis. 12824-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banyai, K., A. Bogdan, P. Kisfali, P. Molnar, I. Mihaly, B. Melegh, V. Martella, J. R. Gentsch, and G. Szucs. 2007. Emergence of serotype G12 rotaviruses, Hungary. Emerg. Infect. Dis. 13916-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castello, A. A., M. H. Arguelles, R. P. Rota, A. Olthoff, B. Jiang, R. I. Glass, J. R. Gentsch, and G. Glikmann. 2006. Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J. Clin. Microbiol. 442046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciarlet, M., F. Liprandi, M. E. Conner, and M. K. Estes. 2000. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch. Virol. 145371-383. [DOI] [PubMed] [Google Scholar]
- 5.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantyre, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunliffe, N. A., and O. Nakagomi. 2005. A critical time for rotavirus vaccines: a review. Expert. Rev. Vaccines 4521-532. [DOI] [PubMed] [Google Scholar]
- 7.Cunliffe, N. A., P. A. Woods, J. P. Leite, B. K. Das, M. Ramachandran, M. K. Bhan, C. A. Hart, R. I. Glass, and J. R. Gentsch. 1997. Sequence analysis of NSP4 gene of human rotavirus allows classification into two main genetic groups. J. Med. Virol. 5341-50. [PubMed] [Google Scholar]
- 8.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 321820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, S., V. Varghese, S. Chaudhury, P. Barman, S. Mahapatra, K. Kojima, S. K. Bhattacharya, T. Krishnan, R. K. Ratho, G. P. Chhotray, A. C. Phukan, N. Kobayashi, and T. N. Naik. 2003. Emergence of novel human group A rotavirus G12 strains in India. J. Clin. Microbiol. 412760-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farahtaj, F., C. I. Gallimore, M. Iturriza-Gomara, M. Taremi, M. R. Zali, H. Edalatkhah, A. Fayaz, and J. J. Gray. 2007. Rotavirus VP7, VP4 and VP6 genotypes co-circulating in Tehran, Iran, between 2003 and 2004. Epidemiol. Infect. 135834-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 301365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass, R. I., J. R. Gentsch, and B. Ivanoff. 1996. New lessons for rotavirus vaccines. Science 27246-48. [DOI] [PubMed] [Google Scholar]
- 13.Gouvea, V., L. de Castro, M. C. Timenetsky, H. Greenberg, and N. Santos. 1994. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J. Clin. Microbiol. 321408-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horie, Y., O. Masamune, and O. Nakagomi. 1997. Three major alleles of rotavirus NSP4 proteins identified by sequence analysis. J. Gen. Virol. 78(Pt. 9)2341-2346. [DOI] [PubMed] [Google Scholar]
- 16.Horie, Y., O. Nakagomi, Y. Koshimura, T. Nakagomi, Y. Suzuki, T. Oka, S. Sasaki, Y. Matsuda, and S. Watanabe. 1999. Diarrhea induction by rotavirus NSP4 in the homologous mouse model system. Virology 262398-407. [DOI] [PubMed] [Google Scholar]
- 17.Iturriza-Gomara, M., E. Anderton, G. Kang, C. Gallimore, W. Phillips, U. Desselberger, and J. Gray. 2003. Evidence for genetic linkage between the gene segments encoding NSP4 and VP6 proteins in common and reassortant human rotavirus strains. J. Clin. Microbiol. 413566-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iturriza-Gomara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J. Clin. Microbiol. 38898-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapikian, A. Z., and Y. Hoshino. 2007. To serotype or not to serotype: that is still the question. J. Infect. Dis. 195611-614. [DOI] [PubMed] [Google Scholar]
- 20.Kheyami, A. M., N. A. Cunliffe, and C. A. Hart. 2006. Rotavirus infection in Saudi Arabia. Ann. Saudi. Med. 26184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshimura, Y., T. Nakagomi, and O. Nakagomi. 2000. The relative frequencies of G serotypes of rotaviruses recovered from hospitalized children with diarrhea: a 10-year survey (1987-1996) in Japan with a review of globally collected data. Microbiol. Immunol. 44499-510. [DOI] [PubMed] [Google Scholar]
- 22.Kutsuzawa, T., T. Konno, H. Suzuki, A. Z. Kapikian, T. Ebina, and N. Ishida. 1982. Isolation of human rotavirus subgroups 1 and 2 in cell culture. J. Clin. Microbiol. 16727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laird, A. R., J. R. Gentsch, T. Nakagomi, O. Nakagomi, and R. I. Glass. 2003. Characterization of serotype G9 rotavirus strains isolated in the United States and India from 1993 to 2001. J. Clin. Microbiol. 413100-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmash, R. W., A. K. Dalwai, G. Szucs, A. M. Molla, A. S. Pacsa, W. Al-Nakib, and M. J. Albert. 2007. Genotypic characterization of rotaviruses and prevalence of serotype-specific serum antibodies in children in Kuwait. Epidemiol. Infect. 1351331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagomi, T., K. Akatani, N. Ikegami, N. Katsushima, and O. Nakagomi. 1988. Occurrence of changes in human rotavirus serotypes with concurrent changes in genomic RNA electropherotypes. J. Clin. Microbiol. 262586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagomi, T., J. R. Gentsch, B. K. Das, R. Kumar, M. K. Bhan, R. I. Glass, and O. Nakagomi. 2002. Molecular characterization of serotype G2 and G3 human rotavirus strains that have an apparently identical electropherotype of the short RNA pattern. Arch. Virol. 1472187-2195. [DOI] [PubMed] [Google Scholar]
- 27.Nakagomi, T., and O. Nakagomi. 1989. RNA-RNA hybridization identifies a human rotavirus that is genetically related to feline rotavirus. J. Virol. 631431-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez, J. F., M. C. Ruiz, M. E. Chemello, and F. Michelangeli. 1999. Characterization of a membrane calcium pathway induced by rotavirus infection in cultured cells. J. Virol. 732481-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pun, S. B., T. Nakagomi, J. B. Sherchand, B. D. Pandey, L. E. Cuevas, N. A. Cunliffe, C. A. Hart, and O. Nakagomi. 2007. Detection of G12 human rotaviruses in Nepal. Emerg. Infect. Dis. 13482-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahman, M., J. Matthijnssens, X. Yang, T. Delbeke, I. Arijs, K. Taniguchi, M. Iturriza-Gomara, N. Iftekharuddin, T. Azim, and M. Van Ranst. 2007. Evolutionary history and global spread of the emerging g12 human rotaviruses. J. Virol. 812382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray, P., S. Sharma, R. K. Agarwal, K. Longmei, J. R. Gentsch, V. K. Paul, R. I. Glass, and M. K. Bhan. 2007. First detection of g12 rotaviruses in newborns with neonatal rotavirus infection at all India institute of medical sciences, New Delhi, India. J. Clin. Microbiol. 453824-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Palacios, G. M., I. Perez-Schael, F. R. Velazquez, H. Abate, T. Breuer, S. C. Clemens, B. Cheuvart, F. Espinoza, P. Gillard, B. L. Innis, Y. Cervantes, A. C. Linhares, P. Lopez, M. Macias-Parra, E. Ortega-Barria, V. Richardson, D. M. Rivera-Medina, L. Rivera, B. Salinas, N. Pavia-Ruz, J. Salmeron, R. Ruttimann, J. C. Tinoco, P. Rubio, E. Nunez, M. L. Guerrero, J. P. Yarzabal, S. Damaso, N. Tornieporth, X. Saez-Llorens, R. F. Vergara, T. Vesikari, A. Bouckenooghe, R. Clemens, B. De Vos, and M. O'Ryan. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 35411-22. [DOI] [PubMed] [Google Scholar]
- 33.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 34.Samajdar, S., V. Varghese, P. Barman, S. Ghosh, U. Mitra, P. Dutta, S. K. Bhattacharya, M. V. Narasimham, P. Panda, T. Krishnan, N. Kobayashi, and T. N. Naik. 2006. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. J. Clin. Virol. 36183-188. [DOI] [PubMed] [Google Scholar]
- 35.Shinozaki, K., M. Okada, S. Nagashima, I. Kaiho, and K. Taniguchi. 2004. Characterization of human rotavirus strains with G12 and P[9] detected in Japan. J. Med. Virol. 73612-616. [DOI] [PubMed] [Google Scholar]
- 36.Steyer, A., M. Poljsak-Prijatelj, T. L. Bufon, N. Marcun-Varda, and J. Marin. 2007. Rotavirus genotypes in Slovenia: unexpected detection of G8P[8] and G12P[8] genotypes. J. Med. Virol. 79626-632. [DOI] [PubMed] [Google Scholar]
- 37.Stupka, J. A., G. I. Parra, J. Gomez, and J. Arbiza. 2007. Detection of human rotavirus G9P[8] strains circulating in Argentina: phylogenetic analysis of VP7 and NSP4 genes. J. Med. Virol. 79838-842. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchida, R., B. D. Pandey, J. B. Sherchand, K. Ahmed, M. Yokoo, T. Nakagomi, L. E. Cuevas, N. A. Cunliffe, C. A. Hart, and O. Nakagomi. 2006. Molecular epidemiology of rotavirus diarrhea among children and adults in Nepal: detection of G12 strains with P[6] or P[8] and a G11P[25] strain. J. Clin. Microbiol. 443499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vesikari, T., D. O. Matson, P. Dennehy, P. Van Damme, M. Santosham, Z. Rodriguez, M. J. Dallas, J. F. Heyse, M. G. Goveia, S. B. Black, H. R. Shinefield, C. D. Christie, S. Ylitalo, R. F. Itzler, M. L. Coia, M. T. Onorato, B. A. Adeyi, G. S. Marshall, L. Gothefors, D. Campens, A. Karvonen, J. P. Watt, K. L. O'Brien, M. J. DiNubile, H. F. Clark, J. W. Boslego, P. A. Offit, and P. M. Heaton. 2006. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 35423-33. [DOI] [PubMed] [Google Scholar]
- 41.WHO. 2006. Global and national estimates of deaths under age five attributable to rotavirus infection: 2004. World Health Organization, Geneva, Switzerland. www.who.int/entity/immunization_monitoring/burden/Global_national_estimates_2004_deaths_under_age_five_attributable_to_rotavirus_infection_2004.pdf.