Abstract
The prospect that rotavirus diarrhea in children may soon be prevented by vaccines has placed a new priority on understanding the diversity of rotavirus strains and the mechanism by which these strains evolve over time. We have characterized a total of 465 rotavirus strains collected in North India from 2000 to 2007 for G and P types by reverse transcription-PCR and sequencing. The novel G12 rotavirus strains recently detected in other countries were first detected in India in 2001 and have emerged as the predominant strains in Delhi, India, during 2005 to 2007. While the VP7 sequence was highly homologous among G12 strains isolated in Delhi, suggesting recent emergence from a common ancestor, the strains had a diverse constellation of other gene segments, demonstrating substantial reassortment. For the entire period, the common rotavirus G types G1 (26%), G2 (25%), and G9 (14%) comprised 65% of the strains, and common P types, P[4] (19%), P[6] (22%), and P[8] (35%), comprised 76% of the total P types. Of note, we detected a high percentage of unusual (17%) strains and fecal specimens with mixed (12% G and 15% P) rotavirus infections having a variety of genomic constellations. For the first time, we identified two novel rotavirus strains with unusual G/P combinations, G2P[11] and G3P[11], in patients with diarrhea. The study highlights the great diversity among rotaviruses isolated from Indian children, the opportunity for genetic reassortment between strains, and the emergence of a novel G12 strain in our country. Due to the demonstrated effect of antigenic diversity on rotavirus vaccines, it will be important to continue careful monitoring of these strains as rotavirus vaccine programs are implemented in India.
Rotavirus is the single most important etiological agent of severe diarrheal illness of infants and young children worldwide, claiming between 454,000 and 705,000 lives each year (37). Approximately 17% (100,000) of the world's estimated rotavirus-associated deaths, together with about 400,000 hospitalizations, occur each year in India (25). This large disease burden and the high mortality associated with rotavirus gastroenteritis in children emphasize the urgent need for rotavirus vaccines (8, 19). Two viral surface proteins, VP4, a protease-cleaved, or P, protein, and VP7, a glycoprotein, or G protein, are the targets of neutralizing antibodies. These proteins may mediate protection induced by a rotavirus vaccine and form the basis for the dual molecular classification scheme that indicates the G and P genotypes of the viruses. To date, 15 G and 27 P genotypes have been described (28, 30, 42). Worldwide surveillance of rotavirus strains has demonstrated G1 to G4 to be the most common circulating G types (1, 9, 48). In the 1990s, a new genotype, G9, emerged in India and is now quite common worldwide (22, 27, 29, 31, 36). Several recent international studies have demonstrated a novel G12 rotavirus to be emerging as well (2, 4, 9, 38, 43, 49, 56). Studies from India, Bangladesh, and Nepal have recently shown a high prevalence of G12 and its association with multiple VP4 genotypes (40, 41, 47, 53). In addition, several other rotavirus strains have demonstrated regional predominance. G5 rotaviruses, previously found only in pigs and horses, were detected in Brazilian children in 1983 and have since been regularly reported from Brazil at a high frequency (20). Similarly, G8 rotaviruses have been detected all over the African continent at a high frequency, especially in Malawi (10, 33).
Two live oral vaccines have recently been licensed to protect children against rotavirus diarrhea: Rotateq, a human-bovine pentavalent vaccine, and Rotarix, a G1P[8] monovalent human rotavirus vaccine (6, 46, 55). Clinical trials of these vaccines among children in rich and middle-income countries have demonstrated high efficacies against the most common rotavirus strains, although Rotarix was less effective against G2P[4] strains (35). This raised the question of whether strains that share neither serotype antigen with the vaccine may evade immunity. Rotavirus vaccines are currently in development in India, and the candidate 116E (G9P[11]) vaccine is analogous to Rotarix in having a monovalent formulation (7). Therefore, it will be important to continuously monitor strains before, during, and after vaccine implementation in India to gauge the impact of vaccines on strains and help understand the effect of strain variation on the efficacies of these vaccines.
MATERIALS AND METHODS
Stool specimens.
In the present study, we characterized rotaviruses identified in 1,524 fecal specimens obtained from children hospitalized with diarrhea. Of these specimens, 1,051 were collected between 2000 and 2002 from surveillance at six major hospitals in South Delhi (1), and 473 were collected between 2002 and 2003 and 2004 to 2007 from another study at the All India Institute of Medical Sciences (AIIMS), New Delhi, India (J. Malik et al., unpublished data). For both studies, monitoring of rotavirus strains was performed from August to July. Rotavirus G and P typing data for 2000 to 2001 were described previously (1), but the nontypeable strains for which samples were still available (53/66) were further characterized by improved reverse transcription-PCR (RT-PCR) and sequencing, and the revised data have been used in the present analysis.
Rotavirus detection.
Fecal specimens were individually screened for rotavirus antigen using a monoclonal antibody-based enzyme immunoassay kit (Premier Rotaclone; Meridian Bioscience Inc., Cincinnati, OH) according to the manufacturer's guidelines.
Polyacrylamide gel electrophoresis (PAGE).
Rotavirus double-stranded RNA was extracted from 10% fecal suspensions or rotavirus-infected culture lysates by the Trizol method (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's instructions and was electrophoresed on a 10% polyacrylamide gel for 16 h at 8 mA, as described previously (34).
G and P typing by multiplex RT-PCR.
G and P types of the strains were determined by RT-PCR with the Qiagen OneStep RT-PCR kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol (44). For G typing, we used two consensus primer pairs (9con1/9con2 and beg9/end9) and G-type-specific primers such as G1 to G4, G9, and G12 (11, 47). For P typing, consensus primer pair con2/con3 was used along with P[4]-, P[6]-, P[8]-, and P[11]-type-specific primers (16). Specimens that did not react with either G- or P-type-specific primers were recorded as nontypeable.
Sequence determination.
Sequences were determined from amplicons with primer pair Beg9/End9 following PCR. The amplification product was purified by GeneClean purification method (Q-biogene, Cambridge, United Kingdom) and sequenced on an automated sequencer (45). Phylogenetic analysis was performed with MEGA software, version 3.1, based on the neighbor-joining method. Phylogenetic distances were measured by the Kimura two-parameter model. Phylogenetic trees were statistically supported by bootstrapping with 1,000 replicates (51).
Nucleotide sequence accession numbers.
The Delhi G12 VP7 gene sequences described in this study were submitted to the GenBank database, and sequence accession numbers are as follows: EU179533 for strain DB029, EU179534 for strain DS061, EU179535 for strain DS142, EU179536 for strain DA227, EU179537 for strain DA428, EU179538 for strain Dan15, EU179539 for strain Dan412, EU179540 for strain Dan516, and EU179541 for strain Dan297.
RESULTS
G and P genotypes of circulating rotavirus strains.
Of the 500 rotavirus-positive specimens (259 from the multicenter study and 241 from the AIIMS hospital), G/P genotyping could be performed for 465 (93%) strains for which an adequate quantity of sample was available (Table 1). Rotavirus genotypes G1, G2, and G9 were observed to be widely circulating in Delhi, with overall incidences of 25.8%, 24.9%, and 14.4%, respectively. Additionally, for the first time, we detected the emergence of G12 strains in Northern India, with a high overall prevalence (14.4%), identical to that of G9 strains, while G3 (2.4%) and G4 (0.4%) strains were rarely detected. Among the P types, P[4], P[6], and P[8] accounted for 19.3%, 22.4%, and 34.8% of the total rotavirus-positive samples, respectively. The most common rotavirus strains were G1P[8], G2P[4], G12P[6], and G9P[8], with prevalences of 19.3%, 17.2%, 9.7%, and 6.7%, respectively. Interestingly, the present study demonstrated a high incidence (17.2%) of unusual G/P combinations. Of note, two novel and unusual rotavirus strains with G2P[11] and G3P[11] specificity that have not previously been observed were detected in this study. Furthermore, a high percentage of mixed infections was also present, including 11.6% among G types and 15.5% for P types. However, throughout the study, nontypeable rotavirus strains (G and P) were detected at low frequencies. Comparison of G and P type data between AIIMS and other hospitals in South Delhi did not show any significant difference.
TABLE 1.
G/P distribution of rotavirus strains circulating in Delhi during 2000 to 2007
| Human rotavirus type | No. (%) of strainse
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G9 | G12 | Gmixa | Gntb | Total | |
| P[4] | 4 (0.9) | 80 (17.2) | 3 (0.6) | 2 (0.4) | 1 (0.2) | 90 (19.3) | |||
| P[6] | 11 (2.4) | 6 (1.3) | 9 (1.9) | 1 (0.2) | 12 (2.6) | 45 (9.7) | 15 (3.2) | 5 (1.1) | 104 (22.4) |
| P[8] | 90 (19.3) | 6 (1.3) | 31 (6.7) | 14 (3.0) | 14 (3.0) | 7 (1.5) | 162 (34.8) | ||
| P[11] | 1 (0.2) | 1 (0.2) | 4 (0.9) | 2 (0.4) | 8 (1.7) | ||||
| P[mix]c | 7 (1.5) | 20 (4.3) | 1 (0.2) | 14 (3.0) | 5 (1.1) | 24 (5.2) | 1 (0.2) | 72 (15.5) | |
| P[nt]d | 8 (1.7) | 3 (0.6) | 1 (0.2) | 3 (0.6) | 1 (0.2) | 13 (2.8) | 29 (6.2) | ||
| Total | 120 (25.8) | 116 (24.9) | 11 (2.4) | 2 (0.4) | 67 (14.4) | 67 (14.4) | 54 (11.6) | 28 (6.0) | 465 |
Gmix, G1/G2/G3/G4/G9/G12.
Gnt, nontypeable for the G genotype.
Pmix, P[4]/P[6]/P[8]/P[11].
P[nt], nontypeable for the P genotype.
Data in boldface type represent the globally common rotavirus strains. Data in both boldface and italic type represent unusual G/P combinations, while data in italic type represent regionally common P[6] strains.
Temporal distribution of rotavirus strains.
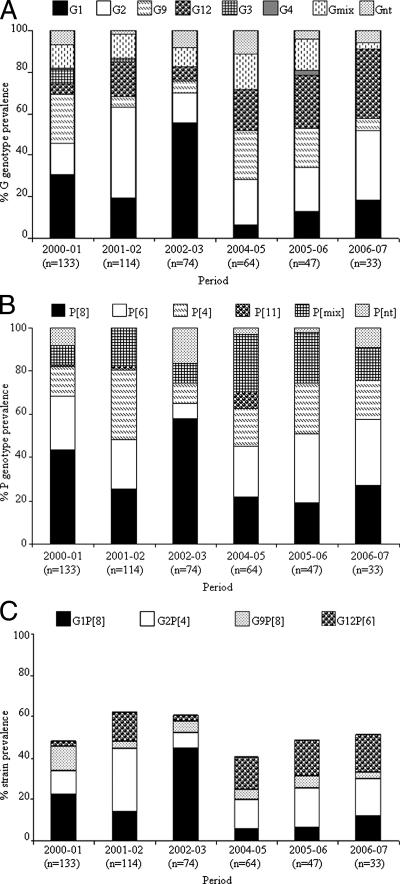
We examined the yearly distribution of rotavirus genotypes throughout the period (Fig. 1A to C). From 2004 to 2007, G12 strains were detected more frequently, with a concomitant decrease in G1 strains (Fig. 1A). Interestingly, other common strains, such as G3 and G4 strains, were rarely detected, while G9 strains were detected consistently throughout the study period. The novel G12 rotaviruses were first observed with low prevalence (5.3%) in 2000 to 2001, and their prevalence increased to 33.3% by 2006 to 2007. However, no such seasonal distribution was seen in the case of P genotypes (Fig. 1B). The common rotavirus strains (G1P[8], G2P[4], and G9P[8]) and the novel G12P[6] strain constituted 40 to 60% of the total rotavirus strains annually (Fig. 1C).
FIG. 1.
(A) Yearly percentage of prevalence of G genotypes (G1 to G4, G9, G12, mixed [Gmix], and nontypeable [Gnt]) among children hospitalized with acute rotavirus diarrhea in Delhi. (B) Yearly percentage of prevalence of P genotypes (P[4], P[6], P[8], P[11], mixed, and nontypeable [nt]) among children hospitalized with acute rotavirus diarrhea in Delhi. (C) Temporal shift and percentage of incidence of predominant rotavirus strains circulating in Delhi during 2000 to 2007.
Phylogenetic analysis of G12.
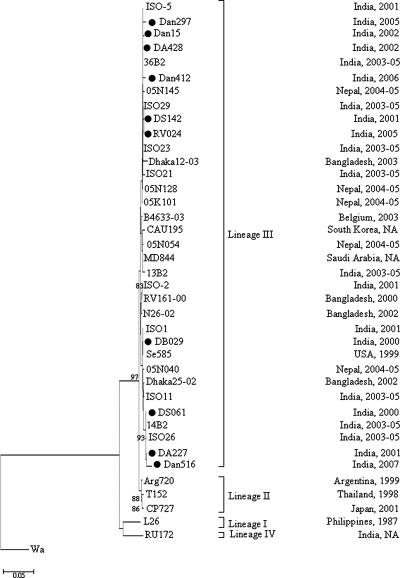
We sequenced the VP7 gene (nucleotides 54 to 484) of G12 strains from Delhi collected during 2000 to 2007 to determine their relationship to previously reported G12 strains (Fig. 2). The sequence homology between the Delhi strains and other G12 strains was high (∼95.2 to 99.5%), and all of them clustered in one lineage (lineage III) (Fig. 2), showing no significant temporal or geographical variation.
FIG. 2.
Phylogenetic analysis of the VP7 gene (nucleotides 54 to 484) of G12 rotavirus strains from Delhi (shown in closed circles). The tree was constructed using the neighbor-joining method with 1,000 bootstrap resamplings and rooted with rotavirus strain Wa (G1 genotype). Percent bootstrap support is indicated by the value at each node, and values of <80 are omitted. Reference sequences were obtained from the GenBank/EMBL database. Origin and year of isolation for each strain are shown. The bar indicates the variation scale. NA, data not available.
Electropherotype of G12 and other rotavirus strains.
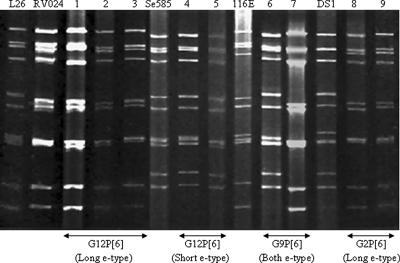
The overall genomic profile of the novel G12 strains and several other P[6] rotavirus strains from Delhi was analyzed by RNA PAGE (Fig. 3). Both long and short electropherotypes were observed among 25 of 67 G12 strains. All G12P[8] (n = 4) and a majority of the G12P[6] (n = 16) strains including RV024, a strain detected in Delhi, previously had a long electropherotype (Fig. 3, lanes 1 to 3), while a short electropherotype (n = 5) was found exclusively among G12[P6] strains (lanes 4 and 5). Genomic heterogeneity was observed among G9P[6] (Fig. 3, lanes 6 and 7) and G2P[6] (lanes 8 and 9) rotavirus strains. It is noteworthy that G12P[6], G9P[6], and G2P[6] strains with long electropherotypes had a much smaller segment 5, encoding NSP1, than other strains. Electropherotyping for many unusual rotavirus strains (G9P[4], G2P[11], G3P[11], and others) detected in the present study could not be performed because of the unavailability of sample with enough quantity.
FIG. 3.
Genomic profiles of G12 and other P[6] rotavirus strains having long (L) and short (S) electropherotypes. Lanes 1 to 3 (long) and 4 and 5 (short) represent G12P[6] strains; lanes 6 (short) and 7 (long) were G9P[6] strains; lanes 8 and 9 represent G2P[6] strains with a long RNA migration pattern. Reference strains, L26 (G12P[4]), RV024 (G12P[6]), Se585 (G12P[6]), 116E (G9P[11]), and DS1 (G2P[4]), were also included for PAGE.
DISCUSSION
This study documents the first detection of G12 in Delhi in 2000 and its emergence by 2005 to 2007 to become a predominant strain. We sequenced the VP7 gene to identify the degree of homology among G12 rotavirus strains, which suggested a common and recent ancestor (41, 47, 50). However, like recent G9 strains, these strains were reassortants, and we identified G12 strains with all common P types (i.e., P[4], P[6], and P[8]) and also with both short and long electropherotypes, confirming the potential of G12 strains to undergo rapid evolution (12, 40, 43). Despite this variety, the ages of children infected with G12 versus other strains were not significantly different. In addition, G1, G2, and G9 genotypes were still prevalent in Delhi, while G3 and G4 strains were very uncommon.
The prevalence of genotypes G1, G2, and G9 is in agreement with numerous reports published worldwide, including in India (1, 3, 17, 48). An earlier study conducted during 1996 to 1998 demonstrated the predominance of G2P[4], followed by G9P[8] and G1P[8] strains in Delhi (26). Our previous study with samples collected in 2000 to 2001 demonstrated that G1 rotaviruses were the predominant genotype, followed by G9 and G2 (1). The low detection rates of G/P-nontypeable strains compared to those reported by some other studies, particularly compared to those in our previous report, may be attributed to differences in the RT-PCR assay that we employed, such as the use of more sensitive enzymes and the addition of both G12-specific primers and an alternate set of consensus primers (1, 15). Our observation of a high percentage of unusual rotavirus strains was in agreement with data from several other studies from the Indian subcontinent (3, 13, 18, 26, 27). Additionally, genomic diversity among the regionally common rotavirus strains (G9P[6] and G2P[6]), particularly in regard to the migration pattern of various gene segments, could be attributed to a close association of humans with domesticated animals and/or a frequent occurrence of mixed infections (5, 14, 17, 21-24, 27, 32, 54). The introduction of G12 strains with a P[6] combination in large numbers may also be attributed to mixed infections. This is evident from our recent finding that both G12P[6] and G9P[6] neonatal strains were circulating simultaneously in the AIIMS nursery (45). Of note, the G3P[11] rotavirus that we recently detected in a child with diarrhea was first detected a decade ago among newborns in hospitals in New Delhi (11).
G12 rotaviruses were first detected in the Philippines in 1987 (52) and in many other places since then (12, 39, 43, 49, 56). Regular detection of G12 has been reported in Bangladesh, Nepal, Japan, South Korea, the United Kingdom, the United States, Belgium, Argentina, Brazil, Hungary, and Slovenia, similar to the emergence of G9 strains previously (4, 9, 23, 38, 40, 41, 50). The high level of VP7 sequence homology within the recently detected G12 strains indicates a common Se585-like ancestor (strains having a genetic makeup similar to that of Se585). However, great strain diversity among G12 rotavirus strains with different VP4 types, similar to G9 strains within the Indian subcontinent, has been due to reassortment with other gene segments (12, 43, 47, 53). Recently, Rahman et al. made a similar observation with very high diversity among G12 strains; additionally, they reported that the first G12 rotavirus prototype strain, L26, has porcine-like NSP2 and NSP5 genes, indicative of G12 having an animal origin (41).
One limitation of this study was our inclusion of only a single city (Delhi), so the prevalence of G12 in other parts of India, both north and south, could not be ascertained. In addition, since this study was restricted to hospitalized children, representing only severe cases, the overall incidence of G12 infections within the community needs to be determined. Future rotavirus surveillance studies should involve more sites from both hospital and community settings. The study emphasizes the inclusion of G12 primers for all future rotavirus strain surveillance. It is now important to evaluate the antigenic cross-reactivity of these novel G12 strains with other common global strains including vaccine strains.
In conclusion, the present study documents the first detection of G12 strains in Delhi and their high frequency in the latest year. Rich genomic diversity along with high prevalences of unusual and mixed strains in India further increase the chances of evolution of more novel strains. Hence, efforts to survey prevalent and newly evolving rotavirus strains in communities should continue. Additional study needs to be carried out to assess the cross-neutralization potential of G12 in the context to other rotavirus strains, which will facilitate further evaluation of vaccines.
Acknowledgments
This work was supported by the Department of Biotechnology, Government of India. S.S. has received a senior research fellowship from the Council of Scientific and Industrial Research, India.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Bahl, R., P. Ray, S. Subodh, P. Shambharkar, M. Saxena, U. Parashar, J. Gentsch, R. Glass, M. K. Bhan, and the Delhi Rotavirus Study Group. 2005. Incidence of severe rotavirus diarrhea in New Delhi, India, and G and P types of the infecting rotavirus strains. J. Infect. Dis. 192S114-S119. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, I., S. Ramani, B. Primrose, M. Iturriza-Gomara, J. J. Gray, D. W. Brown, and G. Kang. 2007. Modification of rotavirus multiplex RT-PCR for the detection of G12 strains based on characterization of emerging G12 rotavirus strains from South India. J. Med. Virol. 791413-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, I., S. Ramani, B. Primrose, P. Moses, M. Iturriza-Gomara, J. J. Gray, S. Jaffar, B. Monica, J. P. Muliyil, D. W. Brown, M. K. Estes, and G. Kang. 2006. Comparative study of the epidemiology of rotavirus in children from a community-based birth cohort and a hospital in South India. J. Clin. Microbiol. 442468-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banyai, K., A. Bogdan, P. Kisfali, P. Molnar, I. Mihaly, B. Melegh, V. Martella, J. R. Gentsch, and G. Szucs. 2007. Emergence of serotype G12 rotaviruses, Hungary. Emerg. Infect. Dis. 13916-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banyai, K., J. R. Gentsch, R. I. Glass, M. Uj, I. Mihaly, and G. Szucs. 2004. Eight-year survey of human rotavirus strains demonstrates circulation of unusual G and P types in Hungary. J. Clin. Microbiol. 42393-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, D. I., D. A. Sack, K. Reisinger, E. Rothstein, and R. L. Ward. 2002. Second year follow-up evaluation of live attenuated human rotavirus vaccine 89-12 in healthy infants. J. Infect. Dis. 1861487-1489. [DOI] [PubMed] [Google Scholar]
- 7.Bhandari, N., P. Sharma, R. I. Glass, P. Ray, H. Greenberg, S. Taneja, M. Saksena, C. D. Rao, J. R. Gentsch, U. Parashar, Y. Maldonado, R. L. Ward, and M. K. Bhan. 2006. Safety and immunogenicity of two live attenuated human rotavirus vaccine candidates, 116E and I321, in infants: results of a randomised controlled trial. Vaccine 245817-5823. [DOI] [PubMed] [Google Scholar]
- 8.Bresee, J. S., R. I. Glass, B. Ivanoff, and J. R. Gentsch. 1999. Current status and future priorities for rotavirus vaccine development, evaluation and implementation in developing countries. Vaccine 172207-2222. [DOI] [PubMed] [Google Scholar]
- 9.Castello, A. A., M. H. Arguelles, R. P. Rota, A. Olthoff, B. Jiang, R. I. Glass, J. R. Gentsch, and G. Glikmann. 2006. Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J. Clin. Microbiol. 442046-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunliffe, N. A., J. S. Gondwe, S. M. Graham, B. D. Thindwa, W. Dove, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Rotavirus strain diversity in Blantrye, Malawi, from 1997 to 1999. J. Clin. Microbiol. 39836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 321820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das, S., V. Varghese, S. Chaudhury, P. Barman, S. Mahapatra, K. Kojima, S. K. Bhattacharya, T. Krishnan, R. K. Ratho, G. P. Chhotray, A. C. Phukan, N. Kobayashi, and T. N. Naik. 2003. Emergence of novel human group A rotavirus G12 strains in India. J. Clin. Microbiol. 412760-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, S., V. Varghese, S. Chaudhuri, P. Barman, K. Kojima, P. Dutta, S. K. Bhattacharya, T. Krishnan, N. Kobayashi, and T. N. Naik. 2004. Genetic variability of human rotavirus strains isolated from Eastern and Northern India. J. Med. Virol. 72156-161. [DOI] [PubMed] [Google Scholar]
- 14.Esona, M. D., G. E. Armah, A. Geyer, and A. D. Steele. 2004. Detection of an unusual rotavirus strain with G5P[8] specificity in a Cameroonian child with diarrhea. J. Clin. Microbiol. 42441-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer, T. K., and J. R. Gentsch. 2004. Rotavirus typing methods and algorithms. Rev. Med. Virol. 1471-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 301365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentsch, J. R., A. R. Laird, B. Bielfelt, D. D. Griffin, K. Banyai, M. Ramachandran, V. Jain, N. A. Cunliffe, O. Nakagomi, C. D. Kirkwood, T. K. Fischer, U. D. Parashar, J. S. Bresee, B. Jiang, and R. I. Glass. 2005. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J. Infect. Dis. 192S146-S159. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, S., V. Varghese, S. Samajdar, S. K. Bhattacharya, N. Kobayashi, and T. N. Naik. 2006. Molecular characterization of a porcine group A rotavirus strain with G12 genotype specificity. Arch. Virol. 1511329-1344. [DOI] [PubMed] [Google Scholar]
- 19.Glass, R. I., M. K. Bhan, P. Ray, R. Bahl, U. D. Parashar, H. Greenberg, C. D. Rao, N. Bhandari, Y. Maldonado, R. L. Ward, D. I. Bernstein, and J. R. Gentsch. 2005. Development of candidate rotavirus vaccines derived from neonatal strains in India. J. Infect. Dis. 192S30-S35. [DOI] [PubMed] [Google Scholar]
- 20.Gouvea, V., L. de Castro, M. C. Timenetsky, H. Greenberg, and N. Santos. 1994. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J. Clin. Microbiol. 321408-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gouvea, V., and M. Brantly. 1995. Is rotavirus a population of reassortants? Trends Microbiol. 3159-162. [DOI] [PubMed] [Google Scholar]
- 22.Grassi, T., A. De Donno, M. Guido, G. Gabutti, and the Collaborative Group for the Surveillance of Rotavirus Infection. 2006. G-genotyping of rotaviruses in stool samples in Salento, Italy. J. Prev. Med. Hyg. 47138-141. [PubMed] [Google Scholar]
- 23.Griffin, D. D., T. Nakagomi, Y. Hoshino, O. Nakagomi, C. D. Kirkwood, U. D. Parashar, R. I. Glass, J. R. Gentsch, and the National Rotavirus Surveillance System. 2002. Characterization of nontypeable rotavirus strains from the United States: identification of a new rotavirus reassortant (P2A[6], G12) and rare P3[9] strains related to bovine rotaviruses. Virology 294256-269. [DOI] [PubMed] [Google Scholar]
- 24.Iturriza-Gomara, M., B. Isherwood, U. Desselberger, and J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 753696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain, V., U. D. Parashar, R. I. Glass, and M. K. Bhan. 2001. Epidemiology of rotavirus in India. Indian J. Pediatr. 68855-862. [DOI] [PubMed] [Google Scholar]
- 26.Jain, V., B. K. Das, M. K. Bhan, R. I. Glass, J. R. Gentsch, and Indian Strains Surveillance Collaborating Laboratories. 2001. Great diversity of group A rotavirus strains and high prevalence of mixed rotavirus infections in India. J. Clin. Microbiol. 393524-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang, G., S. D. Kelkar, S. D. Chitambar, P. Ray, and T. Naik. 2005. Epidemiological profile of rotaviral infection in India: challenges for the 21st century. J. Infect. Dis. 192S120-S126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khamrin, P., N. Maneekarn, S. Peerakoma, W. Cha-it, F. Yagyu, S. Okitso, and H. Ushijima. 2007. Novel porcine rotavirus of genotype P[27] shares new phylogenetic lineage with G2 porcine rotavirus strain. Virology 361243-252. [DOI] [PubMed] [Google Scholar]
- 29.Kirkwood, C. D., D. Cannan, N. Bogdanovic-Sakran, R. F. Bishop, G. L. Barnes, and the National Rotavirus Surveillance Group. 2006. National Rotavirus Surveillance Program annual report, 2005-06. Commun. Dis. Intell. 30434-438. [DOI] [PubMed] [Google Scholar]
- 30.Martella, V., M. Ciarlet, K. Banyai, E. Lorusso, A. Cavalli, M. Corrente, G. Elia, S. Arista, M. Camero, C. Desario, N. Decaro, A. Lavazza, and C. Buonavoglia. 2006. Identification of a novel VP4 genotype carried by a serotype G5 porcine rotavirus strain. Virology 346301-311. [DOI] [PubMed] [Google Scholar]
- 31.Martella, V., V. Terio, G. Del Gaudio, M. Gentile, P. Fiorente, S. Barbuti, and C. Buonavoglia. 2003. Detection of the emerging rotavirus G9 serotype at high frequency in Italy. J. Clin. Microbiol. 413960-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthijnssens, J., M. Rahman, V. Martella, Y. Xuelei, S. De Vos, K. De Leener, M. Ciarlet, C. Buonavoglia, and M. Van Ranst. 2006. Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission. J. Virol. 803801-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matthijnssens, J., M. Rahman, X. Yang, T. Delbeke, J. Arijs, J. P. Kabue, J. J. Muyembe, and M. Van Ranst. 2006. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1 like genogroup. J. Clin. Microbiol. 441801-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakagomi, T., K. Akatani, N. Ikegami, N. Katsushima, and O. Nakagomi. 1988. Occurrence of changes in human rotavirus serotypes with concurrent changes in genomic RNA electropherotypes. J. Clin. Microbiol. 262586-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Ryan, M., and D. O. Matson. 2006. New rotavirus vaccines: renewed optimism. J. Pediatr. 149448-451. [DOI] [PubMed] [Google Scholar]
- 36.Palombo, E. A., P. J. Masendycz, H. C. Bugg, N. Bogdanovic-Sakran, G. L. Barnes, and R. F. Bishop. 2000. Emergence of serotype G9 human rotaviruses in Australia. J. Clin. Microbiol. 381305-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parashar, U. D., C. J. Gibson, J. S. Bresse, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietruchinski, E., F. Benati, F. Lauretti, J. Kisielius, M. Ueda, E. M. Volotao, C. C. Soares, Y. Hoshino, R. E. Linhares, C. Nozawa, and N. Santos. 2006. Rotavirus diarrhea in children and adults in a southern city of Brazil in 2003: distribution of G/P types and finding of a rare G12 strain. J. Med. Virol. 781241-1249. [DOI] [PubMed] [Google Scholar]
- 39.Pongsuwanna, Y., R. Guntapong, M. Chiwakul, R. Tacharoenmuang, N. Onvimala, M. Wakuda, N. Kobayashi, and K. Taniguchi. 2002. Detection of a human rotavirus with G12 and P[9] specificity in Thailand. J. Clin. Microbiol. 401390-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pun, S. B., T. Nakagomi, S. B. Sherchand, B. D. Pandey, L. E. Cuevas, N. A. Cunliffe, C. A. Hart, and O. Nakagomi. 2007. Detection of G12 human rotaviruses in Nepal. Emerg. Infect. Dis. 13482-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman, M., J. Matthijnssens, X. Yang, T. Delbeke, I. Arijs, K. Taniguchi, M. Iturriza-Gomara, N. Iftekharuddin, T. Azim, and M. Van Ranst. 2007. Evolutionary history and global spread of the emerging G12 human rotaviruses. J. Virol. 812382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahman, M., J. Matthijnssens, S. Nahar, S. Podder, D. A. Sack, T. Azim, and M. Van Ranst. 2005. Characterization of a novel P[25],G11 human group A rotavirus. J. Clin. Microbiol. 433208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramani, S., I. Banerjee, B. P. Gladstone, R. Sarkar, D. Selvapandian, A. M. Le Fevre, S. Jaffar, M. Iturriza-Gomara, J. J. Gray, M. K. Estes, D. W. Brown, and G. Kang. 2007. Geographic information systems and genotyping in identification of rotavirus G12 infections in residents of an urban slum with subsequent detection in hospitalized children: emergence of G12 genotype in South India. J. Clin. Microbiol. 45432-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray, P., M. Fenaux, S. Sharma, J. Malik, S. Subodh, S. Bhatnagar, H. Greenberg, R. I. Glass, J. Gentsch, and M. K. Bhan. 2006. Quantitative evaluation of rotaviral antigenemia in children with acute rotaviral diarrhea. J. Infect. Dis. 194588-593. [DOI] [PubMed] [Google Scholar]
- 45.Ray, P., S. Sharma, R. K. Agarwal, K. Longmei, J. R. Gentsch, V. K. Paul, R. I. Glass, and M. K. Bhan. 2007. First detection of G12 rotaviruses in newborns with neonatal rotavirus infection at All India Institute of Medical Sciences, New Delhi, India. J. Clin. Microbiol. 453824-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruiz-Palacios, G. M., I. Perez-Schael, F. R. Velazquez, H. Abate, T. Breuer, S. C. Clemens, B. Cheuvart, F. Espinoza, P. Gillard, B. L. Innis, Y. Cervantes, A. C. Linhares, P. Lopez, M. Macias-Parra, E. Ortega-Barria, V. Richardson, D. M. Rivera-Medina, L. Rivera, B. Salinas, N. Pavia-Ruz, J. Salmeron, R. Ruttimann, J. C. Tinoco, P. Rubio, E. Nunez, M. L. Guerrero, J. P. Yarzabal, S. Damaso, N. Tornieporth, X. Saez-Llorens, R. F. Vergara, T. Vesikari, A. Bouckenooghe, R. Clemens, B. De Vos, M. O'Ryan, and the Human Rotavirus Vaccine Study Group. 2006. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 35411-22. [DOI] [PubMed] [Google Scholar]
- 47.Samajdar, S., V. Varghese, P. Barman, S. Ghosh, U. Mitra, P. Dutta, S. K. Bhattacharya, M. V. Narasimham, P. Panda, T. Krishnan, N. Kobayashi, and T. N. Naik. 2006. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. J. Clin. Virol. 36183-188. [DOI] [PubMed] [Google Scholar]
- 48.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 1529-56. [DOI] [PubMed] [Google Scholar]
- 49.Shinozaki, K., M. Okada, S. Nagashima, I. Kaiho, and T. Taniguchi. 2004. Characterization of human rotavirus strains with G12 and P[9] detected in Japan. J. Med. Virol. 73612-616. [DOI] [PubMed] [Google Scholar]
- 50.Steyer, A., M. Poljsak-Prijatelj, T. L. Bufon, N. Marcun-Varda, and J. Marin. 2007. Rotavirus genotypes in Slovenia: unexpected detection of G8P[8] and G12P[8] genotypes. J. Med. Virol. 79626-632. [DOI] [PubMed] [Google Scholar]
- 51.Subodh, S., M. K. Bhan, and P. Ray. 2006. Genetic characterization of VP3 gene of group A rotaviruses. Virus Genes 33143-145. [DOI] [PubMed] [Google Scholar]
- 52.Taniguchi, K., T. Urasawa, N. Kobayashi, M. Gorziglia, and S. Urasawa. 1990. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication for new G serotype specificity. J. Virol. 645640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uchida, R., B. D. Pandey, S. B. Sherchand, K. Ahmed, M. Yokoo, T. Nakagomi, L. E. Cuevas, N. A. Cunliffe, C. A. Hart, and O. Nakagomi. 2006. Molecular epidemiology of rotavirus diarrhea among children and adults in Nepal: detection of G12 strains with P[6] or P[8] and a G11P[25] strain. J. Clin. Microbiol. 443499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Unicomb, L. E., G. Podder, J. R. Gentsch, P. A. Woods, K. Z. Hasan, A. S. Faruque, M. J. Albert, and R. I. Glass. 1999. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J. Clin. Microbiol. 371885-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vesikari, T., D. O. Matson, P. Dennehy, P. Van Damme, M. Santosham, Z. Rodriguez, M. J. Dallas, J. F. Heyse, M. G. Goveia, S. B. Black, H. R. Shinefield, C. D. Christie, S. Ylitalo, R. F. Itzler, M. L. Coia, M. T. Onorato, B. A. Adeyi, G. S. Marshall, L. Gothefors, D. Campens, A. Karvonen, J. P. Watt, K. L. O'Brien, M. J. DiNubile, H. F. Clark, J. W. Boslego, P. A. Offit, P. M. Heaton, and the Rotavirus Efficacy and Safety Trial (REST) Study Team. 2006. Safety and efficacy of a pentavalent human-bovine (WC3) reassortant rotavirus vaccine. N. Engl. J. Med. 35423-33. [DOI] [PubMed] [Google Scholar]
- 56.Wakuda, M., S. Nagashima, N. Kobayashi, Y. Pongsuwanna, and K. Taniguchi. 2003. Serologic and genomic characterization of a G12 human rotavirus in Thailand. J. Clin. Microbiol. 415764-5769. [DOI] [PMC free article] [PubMed] [Google Scholar]