Abstract
In this work we report on a high-throughput mass spectrometry-based technique for the rapid high-resolution identification of Campylobacter jejuni strain types. This method readily distinguishes C. jejuni from C. coli, has a resolving power comparable to that of multilocus sequence typing (MLST), is applicable to mixtures, and is highly automated. The strain typing approach is based on high-performance mass spectrometry, which “weighs” PCR amplicons with enough mass accuracy to unambiguously determine the base composition of each amplicon (i.e., the numbers of A's, G's, C's, and T's). Amplicons are derived from PCR primers which amplify short (<140-bp) regions of the housekeeping genes used by conventional MLST strategies. The results obtained with a challenge panel that comprised 25 strain types of C. jejuni and 25 strain types of C. coli are presented. These samples were parsed and resolved with demonstrated sensitivity down to 10 genomes/PCR from pure isolates.
Food-borne pathogens are estimated to cause 76 million illnesses annually in the United States alone (19), and considering the impact on society from health care costs and lost productivity, the time and effort expended to understand the epidemiology of the underlying agents are well justified. Campylobacter has been identified as one of the leading causative bacterial agents associated with food-borne illnesses in the United States as well as worldwide (1, 3, 9, 10, 18, 21, 29). It is believed that many of these cases of illness are due to improperly handled poultry products, as Campylobacter contamination rates have been reported to be as high as 71% in commercial chicken products (31). The underlying etiology of 90% of the cases of campylobacteriosis is associated with Campylobacter jejuni, and the majority of the remaining cases are related to Campylobacter coli (11). Although they are not generally considered severe for healthy individuals, septicemia, meningitis, abortions, Guillian-Barré syndrome, and deaths have occurred (2, 23).
The routine identification of infections or contamination typically relies on culture-based protocols (16). Culturing of these gram-negative, microaerophilic organisms is difficult and often requires several days to weeks; moreover, biochemical strain typing is not always accurate (24, 25). Alternative testing methods can include matrix-assisted laser desorption ionization-time of flight mass spectrometry biomarker analysis (8, 17), pulsed-field gel electrophoresis (PFGE), amplified fragment length polymorphism (AFLP) analysis, phage typing, resistotyping, serotyping, and PCR and genomic analysis (21).
Campylobacter jejuni and C. coli PCR-multilocus sequence typing (MLST) has been at the forefront of recent research in this arena and is based on the characterization of genomic heterogeneity in highly polymorphic regions of seven housekeeping genes (4, 20, 22). MLST applies a phylogenetic ordering of strains that is nearly unambiguous as a means of identification, and a detailed database is being assembled to define strain sequence types (STs) (14). The disadvantages of conventional MLST-based approaches include the analysis time, the cost per sample, and the requirement that samples be derived from pure isolates.
Here we report on an alternative technique for the rapid high-resolution differentiation of Campylobacter strains that maintains a resolving power comparable to that of the MLST approach, can function with mixtures, and is automated from end to end such that hundreds of samples can be analyzed on a single instrument each day. The strain typing approach is based on technology originally developed for the identification of a diverse collection of pathogens by PCR amplification of broadly conserved housekeeping genes and analysis of the resulting amplicons by mass spectrometry (12). As the exact molecular weight of each nucleotide is known with very high precision, one can use the measured molecular masses of the forward and reverse strands of the amplicons (along with Watson-Crick base-pairing constraints) to unambiguously determine the base composition of each PCR product (12). The approach has previously been used to detect and type the strains of bacterial pathogens associated with respiratory pneumonia (6), adenoviruses (26), human coronaviruses (27), Acinetobacter baumannii (7), and more recently, a diverse collection of human and avian influenza viruses (28). This approach uses a unique mass spectrometry-based platform to identify organisms across broad biological kingdoms by targeting broadly conserved genes (e.g., DNA encoding 16S or 23S ribosomal subunits in bacteria) (12).
More relevant to this work, by targeting genes that are highly conserved among a given genus or species, the same platform can be used to parse closely related bacterial or viral species with the appropriate primer panel. As demonstrated below, closely related strain variants of C. jejuni are readily distinguished from one another on the basis of subtle base composition differences in conserved genes. The base compositions from eight C. jejuni amplicons are adequate to provide high-resolution ST identification. As each PCR is analyzed in just under a minute, the integrated platform (known as the Ibis T5000 biosensor system) is capable of analyzing over 1,400 PCRs in 24 h or 175 samples a day (8 PCRs/sample). Additional enhancements in throughput will be achieved by PCR multiplexing and enhanced automation.
MATERIALS AND METHODS
Primers for genotyping.
Housekeeping genes provide a good basis for species identification (15); thus, in this work primer targets were established on the same housekeeping genes used by the Campylobacter MLST project (4, 14): aspA, glnA, gltA, glyA, pgm, tkt, and uncA. By using a combined set of 1,208 C. jejuni strain types and 256 C. coli strain types, sequence alignments for each of the genes were concatenated to yield a total of 3,309 nucleotide positions. The primer pairs were selected for maximal diversity by using previously described criteria (6, 12), with 16 initial primer pairs used for C. jejuni. On the basis of the strain resolution ability, amplification specificity, and amplification sensitivity with laboratory strains and sample throughput considerations, the C. jejuni-specific primer pairs were competitively narrowed to a panel of eight primer pairs (Table 1). As part of the initial primer selection process, all primer candidates were tested in silico in simulated PCRs against bacterial databases to minimize the possibility of cross-genus amplification, to avoid confounding of the results for future research aimed at detection directly from complex bacterial backgrounds.
TABLE 1.
Panel of primers used for MLST on the T5000 platform for C. jejuni strain typing by base composition analysis
| Primer code | Primer location | Forward primer | Reverse primer |
|---|---|---|---|
| 2588 | TKT_NC002163-1569415-1569903_107_236 | TCGCTACAGGCCCTTTAGGACAAG | TCCCCATCTCCGCAAAGACAATAAA |
| 2590 | GLYA_NC002163-367572-368095_214_340 | TGCCTATCTTTTTGCTGATATAGCACATATTGC | TCCTCTTGGGCCACGCAAAGTTTT |
| 2592 | PGM_NC002163_21_142 | TCCTAATGGACTTAATATCAATGAAAATTGTGGA | TCAAACGATCCGCATCACCATCAAAAG |
| 2593 | PGM_NC002163_149_277 | TAGATGAAAAAGGCGAAGTGGCTAATGG | TCCCCTTTAAAGCACCATTACTCATTATAGT |
| 2594 | GLNA_NC002163-658085-657609_79_179 | TGTCCAAGAAGCATAGCAAAAAAAGCAA | TCAAAAACAAAGAATTCATTTTCTGGTCCAAA |
| 2595 | ASPA_NC002163-96685-97196_367_497 | TCCTGTTATTCCTGAAGTAGTTAATCAAGTTTGTTA | TCAAGCTATATGCTACAACTGGTTCAAAAAC |
| 2598 | GLTA_NC002163-1604931-1604529_165_261 | TGGCAGCTAGAATAGTAGCTAAAATCCCTAC | TCACGATCTAAATTTGGATAAGCCATAGGAAA |
| 2599 | GLTA_NC002163-1604931-1604529_252_381 | TGGGTCGTGGTTTTACAGAAAATTTCTTATATATG | TTTTGCTCATGATCTGCATGAAGCATAAA |
Primer sensitivity with isolates.
Studies performed with quantified, isolated DNA demonstrated that the assay is sensitive down to at least 10 genome equivalents per 50-μl PCR mixture by use of the purified genome of C. jejuni RM3211, as quantified by UV measurement at 260 nm.
Test genomes.
Fifty genomes from isolates were obtained from the Campylobacter collection of the USDA, ARS, WRRC, PSMRU. Of this set, 45 samples possessed unique STs by MLST, 2 C. jejuni samples (strains RM4275 and RM4279) had the same ST, and 3 samples (strains RM3210, RM4190 and RM4280) had STs yet to be determined.
PCR amplification.
PCR was performed in 40-μl reaction mixtures consisting of 10× PCR buffer, deoxynucleoside triphosphates (dNTPs), primers, genomic sample, and Fast Start polymerase (2.4 U per reaction mixture). A mass-modified dGTP containing 10 13C atoms per base was employed in place of standard dGTP to enhance the mass separation of base compositions that would otherwise be very similar (S. A. Hofstadler, Y. Jiang, T. A. Hall, C. Massire, K. Sannes-Lowery, R. Sampath, L. Blyn, and D. E. Ecker, unpublished data). The reactions were performed in 96-well plates (Bio-Rad, Hercules, CA) with an Eppendorf thermal cycler (Westbury NY). The following PCR conditions were used to amplify the sequences used for PCR/electrospray ionization (ESI)-mass spectrometry analysis: 95°C for 10 min, followed by eight cycles of 95°C for 30 s, 48°C for 30 s, and 72°C for 30 s, with the 48°C annealing temperature increasing 0.9°C each cycle. The PCR was then continued for 37 additional cycles of 95°C for 15 s, 56°C for 20 s, and 72°C for 20 s.
Sample preparation and mass spectrometry base composition determination.
Following PCR amplification, 96-well plates containing the amplicon mixtures were rigorously desalted by use of a protocol based on the weak anion-exchange method published elsewhere (13). As part of the T5000 platform, a custom eight-channel, fixed-needle liquid-handling robot based on a mini autosampler (LEAP Technologies, Carrboro, NC) was used to dispense and aspirate the rinse buffers and transfer sample aliquots. Initially, aliquots of PCR solutions were transferred to the desalting plate, where the amplicons were bound to a weak anion-exchange resin. Unconsumed dNTPs, salts, polymers, and other low-molecular-weight species that might interfere with subsequent ESI-mass spectrometry analysis were removed by rinsing the resin with solutions containing volatile salts and organic solvents. Elution of the final purified/desalted PCR products was accomplished by rinsing the resin with an aliquot (typically 25 μl) of a high-pH buffer. For optimal calibration of the mass spectrometer, internal mass standards, which bracket the m/z range of the PCR products' charge state envelope, were added to the elution buffer to allow internal calibration of each mass spectrum. ESI-time of flight (TOF) data were obtained on the Ibis T5000 platform, which utilizes a Bruker Daltonics MicrOTOF apparatus, with each 75-μs ESI-TOF scan (a 37.5-μs delay followed by a 37.5-μs digitization event at 2 GHz) comprising 75,000 data points. For each spectrum, 660,000 scans were coadded. All aspects of data acquisition were controlled by the Bruker MicrOTOF software package (version 1.0). Sample aliquots of 20 μl were automatically introduced into the mass spectrometer on the T5000 platform, which utilizes a CTC HTS PAL autosampler (LEAP Technologies). Analyte solutions were electrosprayed at 1.3 μl/min against a heated countercurrent bath gas made up of dry air. Spectral calibration and data processing are automatically initiated as the next sample is injected into the mass spectrometer. Base compositions are derived by the use of an algorithm constrained by Watson and Crick base pairing and acceptable mass error limits. For MLST analysis on the T5000 platform, where the amplicon size is typically less than 130 bp and mass errors are routinely less than 20 ppm, unique base compositions are obtained. The use of [13C]guanosine increases the mass and base compositional separation, providing further assurance that a unique base composition is obtained while permitting the option of analyzing larger amplicons or the use of greater mass errors, if such an option is required. For example, when normal dNTPs are used, the base composition A47, G19, C32, and T39 (42,031.18 Da) and the base composition A48, G18, C31, and T40 (42,030.19 Da) differ by a G→A base and a C→T base, resulting in a mass difference of 1 Da (24 ppm). By using the modified 13C-enriched dGTP during PCR, the mass difference is 10 Da (237 ppm) for A47, G19, C32, and T39 (42,197.81 Da) and A48, G18, C31, and T40 (42,187.02 Da) and is easily resolved (Hofstadler et al., unpublished). The automation scheme and the associated software of the Ibis T5000 platform are described in detail elsewhere (5).
Strain typing and clonal complex association.
For each Campylobacter strain examined, the base compositions for all eight primer pair regions were compared to a base composition database derived from the sequence information for the 2,927 STs in the Campylobacter MLST database. All ST matches for each amplified primer pair region were compiled to find the highest degree of correlation among the STs. Less defined samples, novel base compositions, and multiallelic loci can be added to the base composition database as they are encountered for future analysis. Thus, enhancement of the database can be effected by new sequence information or newly derived base composition signatures.
RESULTS
MLST strain typing on the T5000 platform and clonal complex association for primer selection.
The assay for Campylobacter on the T5000 platform incorporates two primer pairs that unambiguously distinguish C. coli and C. jejuni, determining whether one or both species are present in a sample, and then incorporates a panel of eight C. coli-specific and/or eight C. jejuni-specific primers to determine the STs and the ST-clonal complex association. In this work we focus the discussion on the C. jejuni panel. Initial primer selection was computationally performed with a database of 1,208 campylobacters to produce concatenated sequences from the seven housekeeping genes, from which 16 primer pairs were chosen for maximum ST diversity on the basis of the base composition. A panel of eight primer pairs (Table 1) was selected to provide a high resolution among the vast majority of STs in the database (i.e., the ability to resolve any database strain from the remainder of the databases strains). While an expanded primer panel would provide additional resolution, practical constraints associated with the use of a large number of primers kept the panel relatively small. Nonetheless, the worst-case resolution with the eight selected C. jejuni-specific primers and the 1,208 campylobacters is 94%, but on average, the resolution is greater than 99.8%.
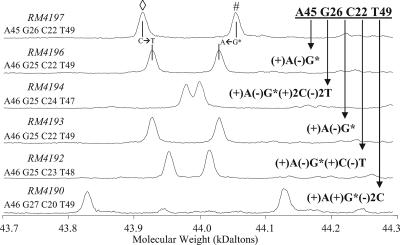
The base composition differentiation obtained with a single primer pair is illustrated in Fig. 1, which shows the deconvoluted mass spectrum for both strands of the tkt gene amplicon for strain RM4197, along with the identical regions for five other C. jejuni strains. Strain identification and the T5000 software-derived base compositions are listed to the left of the spectra. By using as a reference strain RM4197, which has a base composition of A45, G26, C22, and T49 in the targeted region of the tkt gene, it is possible to visualize the change in signal associated with the single-nucleotide polymorphism corresponding to the [13C]guanosine-to-adenosine transition (−26 Da) on one strand along with the complementary transition of cytidine to thymidine (+15 Da) on the complementary strand. Although differences in the spectra are readily visible, the T5000 software automatically converts the raw mass spectra to forward/reverse strand molecular masses, which are in turn converted to base compositions. Collections of base compositions obtained with multiple primers are then used to strain type the organism of interest, thus eliminating the need to manually interpret the mass spectra.
FIG. 1.
Deconvoluted, ESI-TOF mass spectra of PCR amplicons derived from the tkt housekeeping genes from six different C. jejuni strains. Both the forward (⋄) and the reverse (#) strands of the PCR amplicons from each strain are clearly evident in the spectra (e.g., for strain RM4197, the forward strand is A49, G22, C26, and T45 and the reverse strand is A45, G26, C22, and T49). As can be observed in the stacked spectra, differences due to variations in the sequence (and, thus, the base composition) are readily discernible. Note that any mass differences resulting from changes in the number of guanosines are enhanced by the use of [13C]guanosine (G*). The T5000 software automatically determines the base composition of each strain and provides an ST association by using a set of eight primer pairs.
Diversity in Campylobacter isolates.
Base composition allelic calls were produced for all 50 Campylobacter isolates by using the eight C. jejuni-specific primer pairs and two of the primers pairs that differentiate C. jejuni and C. coli. With the C. jejuni-specific primers, a high degree of allelic diversity among the C. jejuni isolates was expected, while on the basis of the bioinformatic criteria used to choose the primers, the C. coli isolates were expected to be less well differentiated from one another but readily distinguished from C. jejuni.
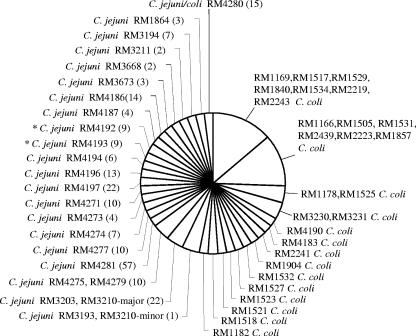
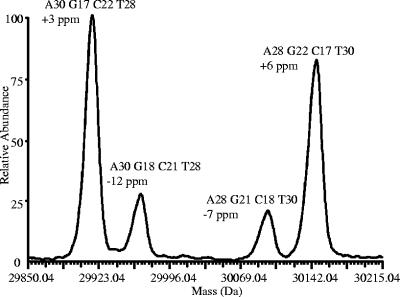
The pie chart in Fig. 2 shows the breakdown of the 50 strain isolates plus the addition of 1 strain found to be a component within a mixed sample. The C. jejuni-specific primer set was able to resolve the strains identified as C. jejuni into 21 sets (22 sets if the two primers used to differentiate C. jejuni and C. coli are added to separate strain RM4192 from strain RM4193), with the largest set containing two C. jejuni isolates. The clustered set of RM4275 and RM4279 proved identical by both T5000 software base composition analysis and conventional MLST. The other two-member set included RM3203 and RM3210 (to date, RM3210 has no defined profile by MLST); however, RM3210, intended to be a pure isolate, was observed to be a mixture by T5000 software base compositional analysis, with the minor strain exhibiting ca. 20% the amplitude of the major strain in the mass spectra. The base compositions of two strains in the mixture differed at five of the eight C. jejuni-specific primer pair amplicons. The deconvoluted mass spectrum in Fig. 3 illustrates the detection of the mixture for RM3210 by the use of primer pair 2598. Four peaks are observed in the spectrum: the two peaks of the greatest amplitude define the molecular masses for the complementary strands derived from the most abundant strain (strain RM3210-major) in the mixture, and the two minor peaks are for the least abundant strain (strain RM3210-minor) in the mixture. Above the peaks are the base compositions determined with the T5000 software and the ppm mass errors for the molecular masses. RM3210-major had a base composition of A30, G17, C22, and T28 for the forward strand; and RM3210-minor had a base composition of A30, G18, C21, and T28, which represents a G/C single-nucleotide polymorphism. By using the profile obtained with the eight C. jejuni-specific primer pairs, RM3210-major matched 22 of the 2,987 C. jejuni STs in the database, thus excluding 99.3% of the known STs within the database. The clonal complex for these 22 STs included ST-45 and ST-283. This same profile was found for RM3203. Assuming the absence of null alleles for the three primer pairs producing only a single amplicon, RM3210-minor had a base compositional profile that matched a single ST, ST-362, from the clonal complex of ST-362. The profile for the RM3210-minor was identical to that for sample RM3193. Verification of sample RM3210 as a mixture was accomplished independently by MLST and by PCR and sequencing of the lipooligosaccharide locus.
FIG. 2.
Pie chart illustrating the ability of the T5000 platform to parse the C. coli strains from the C. jejuni strains and provide high-resolution typing of C. jejuni strains The largest C. jejuni clusters consisted of two-member sets. RM4275 and RM4279 also proved to be identical by conventional MLST. The other two-member sets included the mixed sample of RM3210, of which the major strain in the mix had a profile identical to that of RM3203 and the minor strain had a profile identical to that of RM3193. A conventional MLST profile has yet to be defined for RM3210. Also, samples RM4190 and RM4183 (originally classified as C. jejuni) were parsed with C. coli by the base compositional assay with the T5000 platform. RM4183 was later confirmed to be C. coli by conventional MLST analysis, while an MLST profile for RM4190 has not yet been established. The number of matching STs for each C. jejuni samples is provided in parentheses. *, RM4192 and RM4193 required the differentiating primer pairs to fully separate the strains.
FIG. 3.
Detection of two strains in sample RM3210 is clearly illustrated in the deconvoluted mass spectrum of the amplicons produced with primer pair 2598. The two peaks of greatest amplitude (peak A30, G17, C22, and T28 and peak A28, G22, C17, T30) result from the most abundant strain in the mixed sample, while the peaks at a ca. 20% relative amplitude (peak A30, G18, C21, and T28 and peak A28, G21, C18, and T30) are from the least-abundant strain in the mix. Theoretical molecular mass accuracies are listed below the respective base compositions.
Also, two samples in the PSMRU strain collection, strains RM4183 and RM4190, were originally classified as C. jejuni, but base composition analysis of RM4183 revealed a closer association to STs from C. coli isolates of clonal complex ST-828. Subsequent sequence analysis of the MLST housekeeping genes confirmed that RM4183 was properly assigned to C. coli by the mass spectrometry-derived base compositions on the T5000 platform. Sample RM4190's highest degree of correlation was to seven strains of C. coli that matched five of the eight primer pairs' amplicons. MLST data for RM4190 are not available at this time. Both RM4183 and RM4190 were also identified as C. coli by the use of the two sets of primer pairs that differentiate C. jejuni and C. coli, but a more accurate association for both of these C. coli samples would require the panel of C. coli-specific primers.
Also to be noted in Fig. 2 is isolate RM4280, which matched 14 C. jejuni isolates and 1 C. coli isolate. This C. coli isolate, which is ST-1993, was the only C. coli isolate in the set of 39 STs for clonal complex ST-574 in the combined C. jejuni-C. coli database.
By using eight primer pair regions of less than 140 bp, it is expected that the resolving power will be slightly less than that of conventional MLST, resulting in the correlation of some base composition-characterized strains to more than one ST, as illustrated above for RM3203 and RM3210-major. The number of matching STs for each C. jejuni isolate identified is listed in parentheses in Fig. 2. The resolving power of the panel of C. jejuni-specific primer pairs on the T5000 platform provided a range of ST matches from 1 for RM3193 and RM3210-minor to 57 for RM4281, but even with 57 matching STs, the exclusionary percentage is 98.1%.
Overall, the eight sets of C. jejuni-specific primer pairs were able separate the 23 C. jejuni isolates identified into 21 sets, reassign two C. jejuni isolates to C. coli, and parse the remaining 25 C. coli isolates into 12 sets.
DISCUSSION
Of the alternative Campylobacter typing methods, MLST, PFGE, and AFLP analysis are generally thought to provide the best discriminatory power and reproducibility. Both PFGE and AFLP analysis have been reviewed previously for their abilities to type Campylobacter species (30): neither method provided the discriminatory power of MLST. Furthermore, the performance of these methods can be intricate and time-consuming (i.e., 2 to 3 days for analysis). MLST provides the highest degree of resolving power for typing and subtyping; its main disadvantages are the time and cost for sequencing and the requirement for pure cultured isolates.
The T5000 platform proved to be highly effective in differentiating the C. jejuni isolates in the panel of 50 Campylobacter isolates, and it also determined the correct classification of RM4183 as C. coli instead of C. jejuni, placed RM4190 as a C. coli isolate, and identified and characterized both components of a mixed sample, RM3210. Importantly, the T5000 platform provides high throughput and a fully automated means of analysis of PCR amplicons at a rate of one well per minute. Thus, the analyses described above with a panel of eight primer pairs can be performed at a rate of one sample every 8 min or 175 samples a day. While the eight-primer-pair panel described above was designed to resolve the majority of the most clinically relevant strain types and represents a high-throughput, low-cost approach to the study of C. jejuni isolates associated with campylobacteriosis, additional efforts are under way to both increase the resolution and improve the throughput of this assay by going to a four-well triplex format (12 primer pairs in four wells). This multiplexed format, in conjunction with the use of a novel dual-head electrospray source under development, which will facilitate ESI-TOF analysis at a rate of 30 s/well, will allow the analysis of over 700 samples/day on a single instrument with a resolution exceeding that described above.
Acknowledgments
We thank R. Meinersmann, A. Lastovica, D. Woodward, W. Johnson, I. Wesley, M. Englen, and L. Stanker for the contribution of strains.
A portion of this work was funded by USDA-ARS CRIS project 5325-42000-045-00D.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 528-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser, M. J., G. P. Perez, P. F. Smith, C. Patton, F. C. Tenover, A. J. Lastovica, and W. I. Wang. 1986. Extraintestinal Campylobacter jejuni and Campylobacter coli infections: host factors and strain characteristics. J. Infect. Dis. 153552-559. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2004. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—selected sites, United States, 2003. MMWR Morb. Mortal. Wkly. Rep. 53338-343. [PubMed] [Google Scholar]
- 4.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 3914-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ecker, D. J., J. Drader, J. Gutierrez, A. Gutierrez, J. Hannis, A. Schink, R. Sampath, J. A. Ecker, L. B. Blyn, M. W. Eshoo, T. A. Hall, M. Tobarmosquera, Y. Jiang, K. Sannes-Lowery, L. Cummins, B. Libby, D. J. Walcott, C. Massire, R. Ranken, S. M. Manalili, C. Ivy, R. Melton, H. Levene, V. Harpin, F. Li, N. White, M. Pear, V. Samant, D. Knize, D. Robbins, K. Rudnick, F. Hajjar, and S. A. Hofstadler. 2006. The Ibis T5000 universal biosensor—an automated platform for pathogen identification and strain typing. J. Assoc. Lab. Automation 11341-351. [Google Scholar]
- 6.Ecker, D. J., R. Sampath, L. B. Blyn, M. W. Eshoo, C. Ivy, J. A. Ecker, B. Libby, V. Samant, K. Sannes-Lowery, R. E. Melton, K. Russell, N. Freed, C. Barrozo, J. Wu, K. Rudnick, A. Desai, E. Moradi, D. J. Knize, D. W. Robbins, J. C. Hannis, P. M. Harrell, C. Massire, T. A. Hall, Y. Jiang, R. Ranken, J. J. Drader, N. White, J. A. McNeil, S. T. Crooke, and S. A. Hofstadler. 2005. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl. Acad. Sci. USA 1028012-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ecker, J. A., C. Massire, T. A. Hall, R. Ranken, T. T. Pennella, C. Agasino Ivy, L. B. Blyn, S. A. Hofstadler, T. P. Endy, P. T. Scott, L. Lindler, T. Hamilton, C. Gaddy, K. Snow, M. Pe, J. Fishbain, D. Craft, G. Deye, S. Riddell, E. Milstrey, B. Petruccelli, S. Brisse, V. Harpin, A. Schink, D. J. Ecker, R. Sampath, and M. W. Eshoo. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 442921-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagerquist, C. K., W. G. Miller, L. A. Harden, A. H. Bates, W. H. Vensel, G. Wang, and R. E. Mandrell. 2005. Genomic and proteomic identification of a DNA-binding protein used in the “fingerprinting” of Campylobacter species and strains by MALDI-TOF-MS protein biomarker analysis. Anal. Chem. 774897-4907. [DOI] [PubMed] [Google Scholar]
- 9.Friedman, C. R., J. Niemann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. American Society for Microbiology, Washington, DC.
- 10.Frost, J. A., I. A. Gillespie, and S. J. O'Brien. 2002. Public health implications of Campylobacter outbreaks in England and Wales, 1995-9: epidemiological and microbiological investigations. Epidemiol. Infect. 128111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillespie, I. A., S. J. O'Brien, J. A. Frost, G. K. Adak, P. Horby, A. V. Swan, M. J. Painter, and K. R. Neal. 2002. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg. Infect. Dis. 8937-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofstadler, S. A., R. Sampath, L. B. Blyn, M. W. Eshoo, T. A. Hall, Y. Jiang, J. J. Drader, J. C. Hannis, K. A. Sannes-Lowery, L. L. Cummins, B. Libby, D. J. Walcott, A. Schink, C. Massire, R. Ranken, N. White, V. Samant, J. A. McNeil, D. Knize, D. Robbins, K. Rudnik, A. Desai, E. Moradi, and D. J. Ecker. 2005. TIGER: the universal biosensor. Int. J. Mass Spectrom. 24223-41. [Google Scholar]
- 13.Jiang, Y., and S. A. Hofstadler. 2003. A highly efficient and automated method of purifying and desalting PCR products for analysis by electrospray ionization mass spectrometry. Anal. Biochem. 31650-57. [DOI] [PubMed] [Google Scholar]
- 14.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinform. 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiratisin, P., L. Li, P. R. Murray, and S. H. Fischer. 2005. Use of housekeeping gene sequencing for species identification of viridans streptococci. Diagn. Microbiol. Infect. Dis. 51297-301. [DOI] [PubMed] [Google Scholar]
- 16.Kramer, J. M., J. A. Frost, F. J. Bolton, and D. R. Wareing. 2000. Campylobacter contamination of raw meat and poultry at retail sale: identification of multiple types and comparison with isolates from human infection. J. Food Prot. 631654-1659. [DOI] [PubMed] [Google Scholar]
- 17.Mandrell, R. E., L. A. Harden, A. Bates, W. G. Miller, W. F. Haddon, and C. K. Fagerquist. 2005. Speciation of Campylobacter coli, C. jejuni, C. helveticus, C. lari, C. sputorum, and C. upsaliensis by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 716292-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandrell, R. E., and W. G. Miller. 2006. Campylobacter, p. 476-521. In Y. Motarjemi and M. D. Adams (ed.), Emerging Campylobacter species. CRC Press, Boca Raton, FL, and Woodhead Publishing Limited, Cambridge, United Kingdom.
- 19.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. Wang, L. Pittenger-Alley, R. M. Siletz, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152245-255. [DOI] [PubMed] [Google Scholar]
- 21.Miller, W. G., and R. E. Mandrell. 2005. Prevalence of Campylobacter in the food and water supply: incidence, outbreaks, isolation and detection, p. 101-163. In J. Ketley and M. E. Konkel (ed.), Campylobacter jejuni: new perspectives in molecular and cellular biology. Horizon Scientific Press, Norfolk, United Kingdom.
- 22.Miller, W. G., S. L. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 432315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nachamkin, I., M. J. Blaser, and L. S. Tompkins. 1992. Campylobacter jejuni: current status and future trends. American Society for Microbiology Press, Washington, DC.
- 24.Nicholson, M. A., and C. M. Patton. 1993. Application of Lior biotyping by use of genetically identified Campylobacter strains. J. Clin. Microbiol. 313348-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roop, R. M., II, R. M. Smibert, J. L. Johnson, and N. R. Krieg. 1984. Differential characteristics of catalase-positive Campylobacters correlated with DNA homology groups. Can. J. Microbiol. 30938-951. [DOI] [PubMed] [Google Scholar]
- 26.Russell, K. L., M. P. Broderick, S. E. Franklin, L. B. Blyn, N. E. Freed, E. Moradi, D. J. Ecker, P. E. Kammerer, M. A. Osuna, A. E. Kajon, C. B. Morn, and M. A. Ryan. 2006. Transmission dynamics and prospective environmental sampling of adenovirus in a military recruit setting. J. Infect. Dis. 194877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampath, R., S. A. Hofstadler, L. Blyn, M. Eshoo, T. Hall, C. Massire, H. Levene, J. Hannis, P. M. Harrell, B. Neuman, M. J. Buchmeier, Y. Jiang, R. Ranken, J. Drader, V. Samant, R. H. Griffey, J. A. McNeil, S. T. Crooke, and D. J. Ecker. 2005. Rapid identification of emerging pathogens: coronavirus. Emerg. Infect. Dis. 11373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampath, R., K. L. Russell, C. Massire, M. W. Eshoo, V. Harpin, L. B. Blyn, R. Melton, C. Ivy, T. T. Pennella, F. Li, H. Levene, T. Hall, B. Libby, N. Fan, D. J. Walcott, R. Ranken, M. Pear, A. Schink, J. Gutierrez, J. Drader, D. Moore, D. Metzgar, L. Addington, R. Rothman, C. A. Gaydos, S. Yang, K. St. George, M. E. Fuschino, A. B. Dean, D. Stallknecht, G. Goekjian, S. Yingst, M. Monteville, M. D. Saad, C. A. Whitehouse, C. Baldwin, K. H. Rudnick, S. A. Hofstadler, S. M. Lemon, and D. J. Ecker. 2007. Global surveillance of emerging influenza virus genotypes by mass spectrometry. PLoS ONE 2e489. doi: 10.1371/journal.pone.0000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savill, M., A. Hudson, M. Devane, N. Garrett, B. Gilpin, and A. Ball. 2003. Elucidation of potential transmission routes of Campylobacter in New Zealand. Water Sci. Technol. 4733-38. [PubMed] [Google Scholar]
- 30.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao, C., B. Ge, J. De Villena, R. Sudler, E. Yeh, S. Zhao, D. G. White, D. Wagner, and J. Meng. 2001. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the greater Washington, D.C., area. Appl. Environ. Microbiol. 675431-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]