Abstract
During lyssavirus surveillance, 1,221 bats of at least 30 species were collected from 25 locations in Kenya. One isolate of Lagos bat virus (LBV) was obtained from a dead Eidolon helvum fruit bat. The virus was most similar phylogenetically to LBV isolates from Senegal (1985) and from France (imported from Togo or Egypt; 1999), sharing with these viruses 100% nucleoprotein identity and 99.8 to 100% glycoprotein identity. This genome conservancy across space and time suggests that LBV is well adapted to its natural host species and that populations of reservoir hosts in eastern and western Africa have sufficient interactions to share pathogens. High virus concentrations, in addition to being detected in the brain, were detected in the salivary glands and tongue and in an oral swab, suggesting that LBV is transmitted in the saliva. In other extraneural organs, the virus was generally associated with innervations and ganglia. The presence of infectious virus in the reproductive tract and in a vaginal swab implies an alternative opportunity for transmission. The isolate was pathogenic for laboratory mice by the intracerebral and intramuscular routes. Serologic screening demonstrated the presence of LBV-neutralizing antibodies in E. helvum and Rousettus aegyptiacus fruit bats. In different colonies the seroprevalence ranged from 40 to 67% and 29 to 46% for E. helvum and R. aegyptiacus, respectively. Nested reverse transcription-PCR did not reveal the presence of viral RNA in oral swabs of bats in the absence of brain infection. Several large bat roosts were identified in areas of dense human populations, raising public health concerns for the potential of lyssavirus infection.
Lagos bat virus (LBV) is a species in the Lyssavirus genus (family Rhabdoviridae, order Mononegavirales). It was first isolated from a pool of brains of Eidolon helvum fruit bats at Lagos Island, Nigeria, in 1956 (4). Relatedness between LBV and classical Rabies virus (RABV) was not established for 14 years. An electron microscopy study undertaken from 1969 to 1970 demonstrated that LBV and Mokola virus (MOKV) were rhabdoviruses. Additional studies revealed serologic cross-reactivity of these viruses to each other and to RABV, and the concept of rabies-related viruses (subsequently classified into genus Lyssavirus) was established (53).
At present, seven species are recognized within the Lyssavirus genus. Besides RABV, LBV, and MOKV, these include Duvenhage virus (DUVV), European bat lyssavirus type 1 (EBLV-1), EBLV-2, and Australian bat lyssavirus (ABLV) (61). Four other lyssaviruses have been incorporated into the genus as putative species: Aravan virus (ARAV), Khujand virus (KHUV), Irkut virus (IRKV), and west Caucasian bat virus (WCBV) (32, 34). One other putative species, Rochambeau virus, is currently listed within the genus but was shown recently to have no significant phylogenetic relatedness to lyssaviruses (33).
Four lyssavirus species have been documented in Africa. Of these, RABV occurs worldwide but LBV, MOKV, and DUVV have not been naturally encountered outside of Africa. Although RABV infection of bats is well known in the Americas, this viral species has been associated only with infections of terrestrial mammals in Africa. To date, MOKV has been isolated exclusively from terrestrial mammals as well, whereas LBV and DUVV are bat lyssaviruses, with only occasional isolation from other mammals (47). In total, 28 cases of LBV infection were reported from several African countries, but only 16 isolates were obtained (39, 40). After its first isolation in Nigeria, LBV was isolated in 1974 in the Central African Republic from the fruit bat Micropteropus pusillus (57). From 1980 to 1982 and in 1990 several isolations were made in South Africa from Epomophorus wahlbergi fruit bats and from a cat (58). In 1985, LBV isolation was reported from Senegal, where the virus was obtained from the brain of an E. helvum bat, and from Guinea, where it was isolated from the insectivorous bat Nycteris gambiensis (39). In addition, LBV was isolated from a cat in Zimbabwe (1986) and from a dog in Ethiopia (between 1989 and 1990) (43, 58). In 1999, LBV was isolated from a fruit bat of the species Rousettus aegyptiacus that was imported to Belgium from Africa (presumably from Togo or Egypt) and that later died in France (2, 49). Enhanced surveillance in the KwaZulu-Natal Province of South Africa resulted in several LBV isolates obtained between 2003 and 2006. The majority of these originated from a single species of fruit bats (E. wahlbergi); however, the virus was also isolated from a dog and a mongoose (39, 40, 41).
Recent studies have demonstrated the complex phylogeny of LBV (39, 45). The original isolate (Nigeria; 1956) is genetically distant from other LBV isolates encountered to date. The two viruses originating from Senegal (1985) and found in France (having been introduced via Togo or Egypt; 1999) are similar to each other and constitute another phylogenetic lineage. A third lineage is formed by isolates from Ethiopia, the Central African Republic, Zimbabwe, and South Africa, identified from 1974 to 2006. Genetic distances between these lineages are greater than those described for other lyssavirus species (39). However, the limited number of isolates and lack of surveillance data do not allow conclusive assessment of distribution, host specificity, and circulation patterns of LBV across the African continent.
Given the emergence of new viruses associated with bats, additional surveillance is needed to appreciate the zoonotic importance of these agents. In the present study we report information obtained after initiation of the first bat lyssavirus surveillance in Kenya.
MATERIALS AND METHODS
Bat sampling and identification.
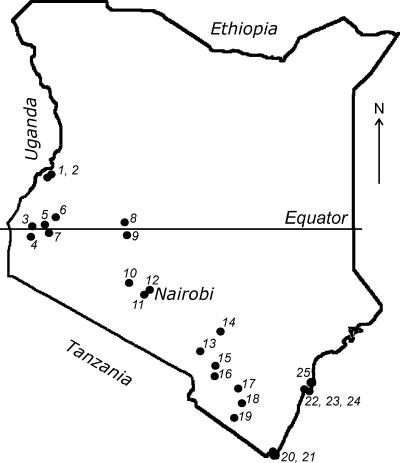
The study was performed in the framework of the Global Disease Detection Program, dedicated to the detection of emerging infectious agents in Kenyan bats. A pilot survey was conducted from July to August 2006 across the southern part of Kenya (Fig. 1). Bats (n = 290) were collected from 17 locations. Selection of sampling sites was based on the available information about bat roosts and on field observations of flying and foraging bats. Whenever possible, 10 to 20 animals of each species present were collected from each roost. Based on the results obtained in 2006, additional bat sampling (n = 931) was performed from June to July 2007 from 14 locations, including new sites and sites that had been sampled in 2006 (Table 1). The focus was given to the species that demonstrated the presence of lyssavirus-neutralizing antibodies and to those reported previously as reservoirs of emerging pathogens (58). The number of samples and the collection protocol were justified and approved by the National Museums of Kenya and the Kenyan Wildlife Service.
FIG. 1.
Map of Kenya, with the locations of the bat collections indicated.
TABLE 1.
Samples of bats collected in Kenya from 2006 to 2007 and subjected to lyssavirus diagnosis and LBV-neutralizing antibody detection
| Location | Species | No. of positive samples/no. tested from:
|
||
|---|---|---|---|---|
| Brains by DFA or MITa | Oral swabs by nRT-PCR | Serum by RFFITb | ||
| 1 | Miniopterus sp. | 0/155 | 0/110 | 0/132 |
| Rhinolophus sp. | 0/31 | 0/24 | 0/16 | |
| Rousettus aegyptiacus | 0/76 | 0/75 | 33/76 | |
| 2 | Hipposideros ruber | 0/4 | —c | 0/3 |
| Rhinolophus sp. | 0/1 | 0/1 | 0/1 | |
| Rousettus aegyptiacus | 0/56 | 0/54 | 25/54 | |
| 3 | Chaerephon pumila | 0/8 | 0/8 | 0/8 |
| Eidolon helvum | 1/18d | 1/17d | 12/18e | |
| Epomophorus labiatus | 0/6 | 0/6 | 0/5 | |
| 4 | Chaerephon pumila | 0/3 | — | 0/4 |
| Chaerephon sp. | 0/8 | — | 0/8 | |
| 5 | Eidolon helvum | 0/86 | 0/86 | 41/79 |
| 6 | Hipposideros ruber | 0/2 | — | 0/2 |
| Rousettus angolensis | 0/10 | — | 0/11 | |
| Miniopterus inflatus | 0/12 | — | 0/12 | |
| 7 | Chaerephon sp. | 0/17 | — | 0/16 |
| Eptesicus tenuipinnis | 0/4 | — | 0/4 | |
| 8 | Miniopterus sp. | 0/47 | 0/50 | 0/46 |
| Rhinolophus hildebrandti | 0/1 | 0/1 | 0/1 | |
| Rhinolophus landeri | 0/6 | 0/6 | 0/4 | |
| Rhinolophus sp. | 0/1 | 0/1 | 0/1 | |
| 9 | Rhinolophus landeri | 0/9 | 0/8 | 0/6 |
| 10 | Otomops martiensseni | 0/19 | — | 0/19 |
| 11 | Pipistrellus sp. | 0/1 | — | 0/1 |
| Rhinolophus sp. | 0/6 | — | 0/6 | |
| 12 | Epomophorus wahlbergi | 0/3 | — | 0/3 |
| Epomophorus labiatus | 0/1 | — | — | |
| 13 | Miniopterus africanus | 0/29 | 0/31 | 0/30 |
| Rhinolophus huldebrandti | 0/21 | 0/16 | 0/19 | |
| Rhinolophus landeri | 0/2 | 0/2 | 0/2 | |
| Rhinolophus sp. | 0/4 | 0/4 | 0/4 | |
| 14 | Pipistrellus sp. | 0/1 | 0/1 | 0/1 |
| 15 | Chaerephon pumila | 0/13 | 0/13 | 0/12 |
| Epomophorus wahlbergi | 0/2 | 0/2 | 0/2 | |
| Nycteris sp. | 0/1 | — | 0/1 | |
| Neoromicia sp. | 0/2 | 0/2 | 0/2 | |
| 16 | Coleura afra | 0/12 | 0/12 | 0/13 |
| Rhinolophus landeri | 0/1 | 0/1 | 0/1 | |
| Rhinolophus sp. | 0/15 | 0/13 | 0/14 | |
| 17 | Epomophorus wahlbergi | 0/7 | 0/8 | 0/8 |
| Nycteris sp. | 0/1 | 0/1 | 0/1 | |
| Pipistrellus sp. | 0/2 | 0/2 | 0/1 | |
| 18 | Chaerephon pumila | 0/6 | — | 0/6 |
| Coleura afra | 0/18 | — | 0/2 | |
| Taphozous sp. | 0/2 | — | 0/2 | |
| 19 | Cardioderma cor | 0/12 | — | 0/11 |
| Species unidentified | 0/4 | — | 0/4 | |
| 20 | Coleura afra | 0/5 | 0/2 | 0/1 |
| Hipposideros commersoni | 0/6 | 0/4 | 0/6 | |
| Miniopterus minor | 0/134 | 0/120 | 0/111 | |
| Nycteris hispida | 0/4 | 0/4 | 0/4 | |
| Rhinolophus sp. | 0/1 | — | — | |
| Rousettus aegyptiacus | 0/107 | 0/106 | 30/93 | |
| Triaenops persicus | 0/16 | 0/18 | 0/12 | |
| 21 | Coleura afra | 0/1 | — | 0/1 |
| Hipposideros commersoni | 0/10 | — | 0/10 | |
| Rhinolophus sp. | 0/2 | — | 0/2 | |
| Taphozous hildegardeae | 0/3 | — | 0/2 | |
| 22 | Cardioderma cor | 0/14 | — | 13 |
| 23 | Pipistrellus sp. | 0/1 | — | 0/1 |
| Rousettus aegyptiacus | 0/106 | 0/117 | 34/116 | |
| Scotophilus sp. | 0/1 | — | 0/1 | |
| 24 | Eidolon helvum | 0/5 | 0/5 | 2/5 |
| 25 | Chaerephon sp. | 0/20 | — | 0/19 |
| Total | 1/1,182d | 1/931d | 177/1,069e | |
Both the DFA test and the MIT were implemented for 397 samples, whereas the DFA test only was implemented for the remaining 787 samples.
Test for LBV-neutralizing antibody.
—, no samples tested.
The single positive record indicates the dead E. helvum bat, from which the KE131 virus was isolated.
Including the dead E. helvum bat, from which the KE131 virus was isolated.
Locations 1, 2, 6, 8 to 11, 13, 16, and 19 to 23 were caves; locations 3, 5, and 24 were tree roosts of E. helvum (also including several bats of other species mistnetted under these roosts at night); locations 4, 7, 18, and 25 were buildings; and locations 12, 14, 15, and 17 were sites of nocturnal foraging of several bat species. Locations 3 to 5, 7, 12, 17, 18, 21, and 23 to 25 were situated within or in immediate proximity to human settlements; locations 1, 2, 6, 13 to 16, and 20 were often visited by local people and by tourists; and locations 8, 9, 10, 11, 13, 19, and 22 were visited by the public only infrequently.
Bats were collected by hand nets or manually in the caves and human dwellings and mistnetted around roosts or in locations of nocturnal foraging. Both adult and subadult animals (based on body size) were randomly collected in 2006, whereas in 2007 the preference was given to adults. Captured bats were anesthetized by an intramuscular injection of ketamine hydrochloride (0.05 to 0.1 mg/g body weight) and euthanized under sedation in compliance with the field protocol, approved by the Animal Institute Care and Use Committee of the Centers for Disease Control and Prevention. The bats were measured, sexed, and identified to species. If species determination in the field was not possible, DNA specimens (pieces of liver in ethanol or tissue impressions on FTA (Flinders Technology Associates) cards (Whatman, Florham Park, NJ) were submitted for identification to Guelph University (Ontario, Canada), where partial sequences of the cytochrome oxidase gene were generated and compared to those available from the database of the Barcode of Life Data Systems (http://www.boldsystems.org). For virological studies brains and pooled organs (spleen, liver, and lung) were collected in sterile plastic tubes. Oral swabs were placed in tubes containing minimum essential medium supplemented with 10% fetal calf serum (MEM-10; Invitrogen, Grand Island, NY) for further virus isolation or TRIzol (Invitrogen, Carlsbad, CA) for RNA extraction. For a subset of animals, fecal and nasal swabs were also collected in sterile dry tubes. Serum was separated from blood clots by centrifugation. When sick or dead bats were encountered, additional tissues (salivary glands, tongue, reproductive organs, adrenal glands, kidneys, stomach, intestine, bladder, and heart) and vaginal swabs were collected. All samples were transported on dry ice and stored at −80°C until use.
Lyssavirus antigen detection.
Bat brains (n = 1,182) were subjected to the direct fluorescent antibody (DFA) test as described elsewhere (13) using monoclonal (Fujirebio Diagnostics Inc., Malvern, PA) or polyclonal (Chemicon Int., Temecula, CA) fluorescein isothiocyanate-labeled anti-rabies virus antibodies. The same test was applied to brains of mice that developed clinical signs of disease during virus isolation and titration and to the mouse neuroblastoma (MNA) cell culture used for the same purposes.
In addition, the frozen-section DFA test was implemented for the tissues of the LBV-positive bat. Representative tissue samples (adrenal glands, bladder, heart, intestine, kidney, liver, lung, reproductive tract, salivary glands, spleen, stomach, and tongue; approximately 0.1 to 0.2 g of each) were embedded in Tris-buffered saline tissue freezing medium (Triangle Biomedical Sciences, Durham, NC). Serial sections of 8 μm each were cut on a cryostat (Microm, HM 505N; Richard Allen Scientific, Kalamazoo, MI) at −22°C. Sections (50 to 75 from each tissue) were collected on glass slides (precleaned Gold Seal slides; Gold Seal Products, Portsmouth, NH), air dried, and fixed in acetone (EMD Chemicals Inc., Gibbstown, NJ) at −20°C for 30 min. Tissues of another E. helvum bat that did not demonstrate the presence of lyssavirus antigen in the brain were used as negative controls. Representative slides containing cuts from different layers of the embedded-tissue block were selected for DFA staining. Stained slides were rinsed twice in phosphate-buffered saline, and coverslips were applied using 10% glycerol-phosphate-buffered saline solution. All slides were examined for the presence of lyssavirus antigen using an Axioplan 2 imaging microscope (Carl Zeiss, Germany) at 200× magnification.
Virus isolation and titration.
Bat brains collected in 2006 (n = 277) were homogenized and tested in the intracerebral mouse inoculation test (MIT) as described elsewhere (30) using 3-week-old outbred ICR mice. For the specimens collected in 2007, the isolation was attempted in 2-day-old suckling mice. However, this was done for a subset of brains only (n = 120), including the specimens from all sick and dead bats (n = 11). For the bat that demonstrated the presence of lyssavirus antigen in the brain, the titers of the virus in homogenates of the brain and salivary glands were determined by intracerebral and intramuscular inoculation of 3-week-old ICR mice. The 50% mouse lethal dose (MLD50) was calculated using the Spearman-Karber method (1). In addition, for this bat the isolation in MNA cells was attempted from a number of tissues and swabs, as described previously (65). The test was performed in 25-cm2 plastic flasks (Corning Inc., Cambridge, MA), with the control of inoculation in LabTek slides (Nalge Nunc Int., Naperville, IL). If no lyssavirus antigen was detected in the MNA cells placed in LabTek slides 72 h postinoculation, the cells from the flask were subjected to two subpassages at 72-h intervals. Absence of viral antigen in the cells after the last passage was considered a negative result.
Detection of viral RNA by nested reverse transcription-PCR (nRT-PCR).
Total RNA was extracted from the oral swabs that had been collected in TRIzol (n = 785) according to the manufacturer's instructions. For certain bats (n = 146) the swabs collected in MEM-10 were the only ones available. For these, 200 μl of swab medium was mixed with 1 ml of TRIzol and subjected to RNA extraction. For the bat that demonstrated the presence of lyssavirus antigen in the brain, RNA was extracted from all available tissues and swabs. Primers were designed within the coding region of the nucleoprotein (N) gene based on the alignment of available gene sequences of LBV, MOKV, and WCBV. The initial reaction was performed with sense primer N1F, ATGGAKTCWGAMAASATTGT (positions 71 to 90), which was also used for reverse transcription, and antisense primer N550B, GTRCTCCARTTAGCRCACAT (positions 647 to 666). The nested reaction was performed with sense primer N70F, GAYCAATATGARTATAARTA (positions 140 to 159), and antisense primer N490B, TCCATYCTRTCTGCWACATT (positions 560 to 579; all positions are given according to the Street Alabama Dufferin RABV strain genome sequence [GenBank accession number M31046]). The reactions were performed as described elsewhere (26). No housekeeping gene was used as a control for the presence of the host RNA in swab samples, as we dealt with many species of bat species from different families for which no genetic information was available. All positive results were confirmed by nucleotide sequencing, performed on an ABI Prism 377 DNA sequencer (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions.
Sequencing of the complete LBV genome and sequence analysis.
Total RNA was extracted from the bat brain that demonstrated the presence of lyssavirus antigen using TRIzol and subjected to RT-PCR. The primer pairs described above were used initially, and sequencing of the RT-PCR product demonstrated that the virus belongs to LBV. Design of primers for amplification of the remaining part of the N, phosphoprotein (P), matrix protein (M), and glycoprotein (G) genes was based on the alignment of several LBV sequences determined earlier (39). For amplification of the polymerase (L) gene, specific sense primers were constructed close to the 5′ end of the previously generated sequence and degenerate antisense primers within the L gene were constructed based on the alignment of the L gene sequences of RABV, MOKV, and ABLV, available from GenBank. Overlapping segments of 1.0 to 1.5 kb were amplified and sequenced at each step.
Further, as extremities of all previously described lyssaviruses were similar, we used the common lyssavirus forward primer LYS001F (ACGCTTAACGAMAAA), starting in the beginning of the lyssavirus genome, to amplify a significant part of the leader RNA. Similarly, the reverse primer LYSEND (ACGCTTAACAAAWAAA), which is complementary to the 5′ terminus of the lyssavirus genome (and reversely complementary to the 3′ terminus) was used to amplify a part of the trailer RNA. Indeed, the 3′ and 5′ extremities of the genome (the annealing regions of LYS001F and LYSEND primers) remained unknown when this method was used.
For determination of the 3′ and 5′ genome extremities, circularization of the RNA by ligation, with subsequent amplification of the ligated extremities by nRT-PCR, cloning of the nRT-PCR product, and sequencing of the clones, was performed. In brief, 13 μl of RNA solution (concentration, 0.5 to 1.0 μg/ml) was mixed with 2 μl of T4 RNA ligase (20 U), 4 μl of 10× ligation buffer (Promega; supplied with the ligation kit), 20 μl of 40% water solution of polyethylene glycol 8000, and 1 μl (40 U) of RNase inhibitor (Roche Diagnostics, Mannheim, Germany) in a total volume of 40 μl. The mixture was incubated at 37°C for 30 min. Thereafter the samples were subjected to ethanol precipitation twice and resuspended in 13 μl of diethyl pyrocarbonate-treated water. The ligated RNA was subjected to nRT-PCR with sense primers located within the 5′ end of the viral L gene and antisense primers located within the 3′ end of the N gene (a fragment of 450 nucleotides [nt] was amplified in the primary RT-PCR, and a fragment of 300 nt was amplified in the nested reaction). As the ligated genome termini are truncated frequently, cloning was the obligatory prerequisite for sequence determination. The nRT-PCR products were purified with the Wizard PCR Preps DNA purification system (Promega, Madison, WI), inserted into the pGEM-T Easy vector (Promega), and cloned in Escherichia coli JM109 competent cells (Promega). Fifteen randomly selected clones were subjected to sequencing. Both DNA strands of a given PCR product were sequenced at least twice.
The sequence assembly, alignment, and consensus sequence generation, as well as DNA translation and estimation of identities, were performed with BioEdit software (22). Phylogenetic comparison with other LBV representatives was performed by the neighbor-joining method with the Kimura-2 distance estimation, implemented in the MEGA program, version 2.1 (31). The entire N gene sequences were compared, and branching support was determined for 1,000 bootstrap replicates.
RFFIT.
The virus-neutralizing antibodies (VNA) in bat sera were determined by a modification of the rapid fluorescent focus inhibition test (RFFIT) (54) using four-well (6-mm) Teflon-coated glass slides (Cel-Line; Erie Scientific, Portsmouth, NH). Initially all serum samples were screened in dilutions of 1:10 and 1:25. In brief, 3.5 μl of serum was mixed in a well with 14 μl of MEM-10. Further, 5 μl of this mixture was transferred to another well and mixed with 7.5 μl of MEM-10 (final volume in each well, 12.5 μl). Thereafter 12.5 μl of viral inoculum was added to each well (virus dose, 28 to 100 focus-forming units, as determined by titration on a control slide with each set of sera), and the slides were incubated in a humidity chamber for 90 min at 37°C in the presence of 5% CO2. After the incubation, MNA cells (25 μl of 2 × 106 cells/ml) were added into each well, and slides were incubated at the same conditions for 20 to 44 h (depending on the virus used) before acetone fixation and staining. At microscopy, 10 separate fields were counted for each well. If a reduction or absence of fluorescence was observed, the serum sample was subjected to additional titration, in dilutions 1:10 to 1:1,250. The 50% end point neutralizing titers were calculated by the method of Reed and Muench (54). Only the samples that had a 50% end point neutralizing titer greater than 1 log10 (e.g., less than five fields contained infected cells at a serum dilution of 1:10) were considered positive. Previous trials for RABV VNA demonstrated that results obtained by this micromethod are comparable to those obtained by the classical test with chamber slides (54).
For samples collected in 2006, the neutralizing activity against representatives of three known phylogenetic lineages of LBV (LBVAFR1999, LBVSA1982, and LBVNIG1956; see the Fig. 3 legend), MOKV (isolated in South Africa in 1997), DUVV (isolated in South Africa in 1970), and RABV (laboratory strain CVS-11) was determined. For samples collected in 2007, the neutralizing activity against the LBV isolate LBVAFR1999 only was determined.
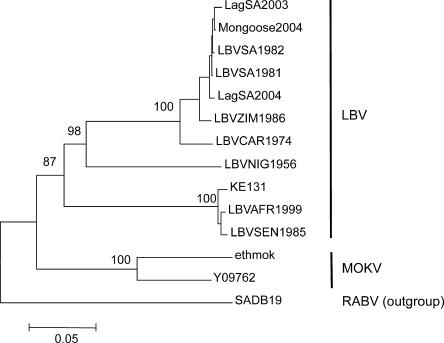
FIG. 3.
Phylogenetic position of the KE131 isolate among other LBV sequences, based on the entire N gene (1,350 nt). The tree was obtained by the neighbor-joining method. Bootstrap values are presented for key nodes, and branch lengths are drawn to scale. The LBV sequences, location and date of isolation, and species are as follows: LagSA2003 (EF547451), South Africa, 2003, Epomophorus wahlbergi; Mongoose2004 (EF547453), South Africa, 2004, water mongoose; LBVSA1982 (EF547455), South Africa, 1982, E.wahlbergi; LBVSA1981 (EF547457), South Africa, 1980 to 1981, E. wahlbergi; LagSA2004 (EF547458), South Africa, 2004, E. wahlbergi; LBVZIM1986 (EF547450), Zimbabwe, 1986, cat; LBVCAR1974 (EF547449), Central African Republic, 1974, Micropteropus pusillus; LBVNIG1956 (EF547459), Nigeria, 1956, Eidolon helvum; LBVSEN1985 (EF547448), Senegal, 1985, E. helvum; and LBVAFR1999 (EF547447), France via Togo or Egypt, Rousettus aegyptiacus.
Statistical analysis.
The 95% confidence intervals for virus titers, indicated in the text, were calculated by Neoprobit method (1). Seroprevalence values for different demographic groups of E. helvum and R. aegyptiacus were compared using the chi-square test. Antibody titers between males and females of these bat species were compared by the two-sided Student t test for independent samples, since distribution of the log10 titers in each group was close to normal, and variances in the groups were assumed to be equal. P values less than 0.05 were considered statistically significant.
RESULTS
Bat sightings and detection of LBV.
Most bats observed and collected during our field trials appeared healthy. No fresh bat carcasses, which could be suitable for virological testing, were encountered in July and August 2006. Only one sick bat, a male Taphozous hildegardeae bat, was found in location 21. During June and July 2007, 11 fresh bat carcasses were collected, including three E. helvum bats (locations 3, 4 and 24), six Coleura afra bats (location 20), and two R. aegyptiacus bats (location 20). One sick Hipposideros commersoni bat was found in location 20. According to the information provided by representatives of the local public, people encounter sick or dead bats infrequently, except at location 3, where numerous large bats (presumably E. helvum) were seen dead on the ground in February 2007.
No lyssavirus antigen was detected in the brains of bats collected in 2006, and no neurotropic agents were isolated from these brains in MIT (n = 277). In 2007, lyssavirus antigen was detected in one sample, the brain of an adult female E. helvum bat found dead under the roost in location 3. The estimated time between bat death and sample collection was several hours. The body was in rigor mortis, all tissues at necropsy were in a good condition, and serum was successfully separated from the blood. All mice inoculated intracerebrally with 10% suspensions of the bat brain and salivary glands developed signs of encephalitis with incubation periods of 6 to 8 days. The isolate was named KE131. The mouse intracerebral titer of the virus in the bat brain was 4.9 ± 0.53 log10 MLD50/0.03 ml, and in the bat salivary glands it was 3.3 ± 1.35 log10 MLD50/0.03 ml. In addition, the brain suspension was pathogenic for mice when given intramuscularly, with a titer of 1.3 ± 0.49 log10 MLD50/0.05 ml, whereas salivary gland suspension did not kill mice by this route.
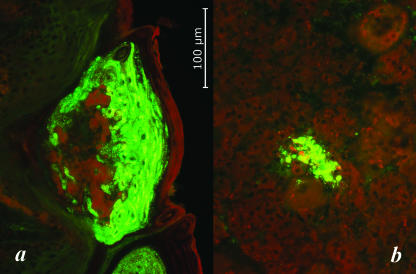
Results of virus isolation from various tissues of the infected bat, in comparison with RNA and antigen detection, are presented in Table 2. Only samples of brain, salivary glands, and tongue demonstrated the presence of viral antigen in MNA cells 72 h after the first inoculation. However, additional subpassages revealed the presence of the virus in several additional tissues, as well as in oral, nasal, and vaginal swabs. Viral RNA was detected in all examined tissues except spleen and intestine. Various distributions of viral antigen in tissue cryosections were observed. The greatest amount of the antigen was detected in the tongue and salivary glands. Positive areas of the tongue papillae included epithelial cells and associated connective tissue ganglia. In addition, numerous positive foci within the muscular layer nerves and nerve bundles were registered (Fig. 2a). Viral antigen in the submandibular salivary glands was observed in ganglion, mucous, and serous acini (Fig. 2b).
TABLE 2.
Results of virus isolation in MNA cells, detection of viral RNA by nRT-PCR, and detection of viral antigen by DFA test in the tissues and swabs from an LBV-infected E. helvum bat
| Specimen source | Virus isolation | RNA detection | Antigen detection |
|---|---|---|---|
| Brain | + | + | NTa |
| Salivary gland | + | + | + |
| Tongue | + | + | + |
| Oral swab | + | + | NT |
| Bladder | + | + | +b |
| Nasal swab | + | + | NT |
| Intestine | − | − | +b |
| Lung | − | + | ±b,c |
| Stomach | + | + | +b |
| Adrenal | − | + | +b |
| Liver | − | + | ±b |
| Heart | − | + | +b |
| Ovaryd | + | + | +b |
| Kidney | − | + | ±b |
| Vaginal swab | + | + | NT |
| Spleen | − | − | ±b |
NT, not tested.
Viral antigen associated with neural tissue and ganglia only.
±, limited presence of viral antigen in a few cryosections only.
Complete longitudinal sections of the reproductive system were examined for the presence of viral antigen.
FIG. 2.
Detection of viral antigen by the DFA test of frozen tissue sections of the LBV-positive E. helvum bat. Shown is viral antigen within papillae on the dorsal surface of the tongue (a) and in acinar cells of the submandibular salivary glands (b). Total magnification, ×200. The photo was by Michael Niezgoda.
The antigen was also detected in all sections of adrenal glands, heart, intestine, reproductive tract, and stomach. Tissue morphology in cryosections was less than ideal; however, the majority of focal antigen in these organs was clearly associated with connective tissue nerves and ganglia. For example, in the adrenal glands viral antigen was identified within ganglia of the medulla. No positive muscle was identified in the heart; however, small antigen foci were detected in associated nerves.
Molecular characterization of the KE131 LBV isolate.
Sequencing of the initial RT-PCR product (fragment of the N gene), obtained from the bat brain, and subsequent comparison of this sequence with those of other lyssaviruses demonstrated that the virus belongs to LBV. Further genome fragments were amplified using specific primers, designed for the alignment of LBV gene sequences (39), and common degenerate primers for the lyssavirus L gene. The use of primers LYS001 and LYSEND provided amplification and sequencing of major parts of the leader and trailer regions. Finally, the genome extremities were successfully determined from the RNA ligation product, amplified by nRT-PCR, and cloned. Of 15 clones sequenced, 9 contained the nontruncated leader region and 5 contained the nontruncated trailer region.
The length of the KE131 genome (GenBank accession number EU259198) was 12,017 nt. The genome consisted of five structural genes, found in all lyssaviruses: the N (1,350 nt coding for 450 amino acids [aa]), P (915 nt coding for 305 aa), M (606 nt coding for 202 aa), G (1,566 nt coding for 522 aa), and L (6,381 nt coding for 2,127 aa) genes. The major gene characteristics were similar to those of other lyssaviruses described previously (3, 5, 21, 33, 34, 42, 45, 46, 50, 64). The B-cell epitope NI (aa 374 to 383) (14) of the KE131 nucleoprotein is shared with isolates LBVAFR1999 and LBVSEN1985, whereas other LBV nucleoproteins have substitution R/K376, similar to MOKV. The NIII epitope (aa 313 to 337) is almost invariant in all LBV and MOKV nucleoproteins, as well as the TH site (aa 410 to 413) (16). Among described T-cell epitopes (12, 19) significant conservation was observed, and KE131 shared maximum identity of these regions with the LBVSEN1985 and LBVAFR1999 isolates. The critical position of the binding site for the cytoplasmic light chain of dynein LC8 within the phosphoprotein (aa 143 to 148) (51) is conserved: N(Q/R)QTQT is found in all LBV representatives as well as in other lyssavirus species except MOKV, where it consists of S(I/V)QIQT, and WCBV, where it is apparently absent (32). Among antigenic sites I to III of the glycoprotein, which are not well conserved between lyssaviruses (3, 34), the KE131 sequences share maximum identity with other LBV sequences (and among these, particularly with LBVSEN1985 and LBVAFR1999 sequences) and to a lesser extent with MOKV sequences. The R(K)/D333 substitution in the glycoprotein ectodomain, which is thought to be responsible for the limited peripheral pathogenicity of certain lyssavirus strains (3, 15), is present in the KE131 glycoprotein as well as in glycoproteins of all LBV and MOKV isolates sequenced to date. Functional blocks described previously for the polymerase proteins of lyssaviruses and other Mononegavirales (50) are well conserved in the KE131 polymerase. The 3′ and 5′ extremities of the KE131 genome are complementary to each other along the 10 terminal nucleotides.
No complete LBV genomes are present in the GenBank to date. Among other complete lyssavirus genomes available for comparison, the noncoding regions of KE131 were most similar to those of the MOKV sequence (GenBank accession no. Y09762). The N-P intergenic regions of both these viruses consisted of 3 nt (in RABV, EBLV-1, EBLV-2, ABLV, ARAV, KHUV, and IRKV genomes there are 2 nt, and in the WCBV genome there are 4 nt), and the M-G intergenic regions consisted of 16 nt (in RABV, EBLV-1, EBLV-2, ABLV, ARAV, KHUV, and IRKV genomes there are 5 nt, and in the WCBV genome there are 39 nt).
Phylogenetic analysis implemented for the entire N gene (Fig. 3) demonstrated that the KE131 isolate was most similar to two viruses originating from Senegal (LBVSEN1985) and from France via Togo or Egypt (LBVAFR1999). The N gene sequences shared 98.5 to 98.8% nucleotide identity, and the associated amino acid sequences shared 100% amino acid identity. We also compared the G gene and deduced glycoprotein sequences, because the G is responsible for VNA production, which was important for the assessment of specificity and sensitivity of our serologic assay. For the G, the KE131 isolate shared with isolates LBVAFR1999 and LBVSEN1985 99.1% and 99.6% nucleotide identity and 99.8% and 100% amino acid identity, respectively (only a single amino acid substitution, G/E518, was detected in the glycoproteins of KE131 and LBVSEN1985 isolates compared to the LBVAFR1999 isolate).
Detection of LBV RNA in oral swabs.
All collected oral swabs (n = 931) were negative in the nRT-PCR except the one obtained from the bat from which the KE131 virus was isolated.
Serologic evidence of LBV circulation in bats.
Anti-LBV VNA were detected in a substantial proportion of serum samples collected from E. helvum and R. aegyptiacus bats and were not detected in any other bat species (Table 1). To assess the specificity of the RFFIT, all serum samples collected in 2006 (n = 269) were tested against representatives of three LBV lineages, MOKV, DUVV, RABV, and WCBV (Table 3). Most of the samples that neutralized the LBVAFR1999 isolate also neutralized the LBVSA1982 and LBVNIG1956 isolates, indicating significant cross-reactivity between LBVs. Several samples had a greater neutralizing titer against LBVNIG1956 than against other LBV representatives. However, this distinction might be caused by operational differences in RFFIT procedures. The LBVNIG1956 replicates in MNA cells slowly and never reaches high titers.
TABLE 3.
Neutralizing activity of samples, collected in 2006, against a panel of lyssavirusesa
| Bat no. | Species | Neutralization activityb againstc:
|
||||
|---|---|---|---|---|---|---|
| LBVAFR1999 | LBVSA1982 | LBVNIG1956 | MOKV | RABV | ||
| 284 | R. aegyptiacus | 1.56 ± 0.23 | 1.56 ± 0.23 | 1.64 ± 0.33 | neg | neg |
| 286 | R. aegyptiacus | 1.34 ± 0.29 | 1.61 ± 0.22 | 1.91 ± 0.23 | neg | neg |
| 289 | R. aegyptiacus | 2.31 ± 0.33 | 2.32 ± 0.27 | 1.54 ± 0.19 | 1.59 ± 0.26 | neg |
| 290 | R. aegyptiacus | 2.19 ± 0.31 | 1.79 ± 0.17 | 2.37 ± 0.38 | 2.36 ± 0.28 | 1.53 ± 0.30 (0.25 IUd) |
| 291 | R. aegyptiacus | 1.56 ± 0.18 | 1.69 ± 0.34 | 1.06 ± 0.27 | neg | neg |
| 300 | R. aegyptiacus | 1.20 ± 0.23 | 1.28 ± 0.16 | 1.17 ± 0.27 | neg | neg |
| 304 | R. aegyptiacus | 1.61 ± 0.22 | neg | 1.32 ± 0.24 | neg | neg |
| 307 | R. aegyptiacus | 1.33 ± 0.36 | 1.24 ± 0.16 | 1.57 ± 0.18 | neg | 1.26 ± 0.16 (0.20 IUd) |
| 308 | R. aegyptiacus | 1.68 ± 0.78 | 1.48 ± 0.20 | neg | neg | neg |
| 269 | E. helvum | 2.24 ± 0.43 | 2.26 ± 0.38 | 1.91 ± 0.31 | 1.79 ± 0.30 | neg |
| 274 | E. helvum | 1.68 ± 0.31 | 1.49 ± 0.19 | 1.64 ± 0.28 | neg | neg |
| 275 | E. helvum | 1.49 ± 0.23 | neg | neg | neg | neg |
| 279 | E. helvum | 1.04 ± 0.44 | 1.12 ± 0.25 | 1.73 ± 0.26 | neg | neg |
| 198 | R. aegyptiacus | 2.35 ± 0.21 | 1.88 ± 0.27 | 1.54 ± 0.19 | 1.14 ± 0.14 | neg |
| 206 | R. aegyptiacus | 1.65 ± 0.23 | 1.61 ± 0.22 | neg | 1.49 ± 0.16 | neg |
| 216 | R. aegyptiacus | 1.68 ± 0.34 | 1.49 ± 0.20 | neg | neg | neg |
| 227 | R. aegyptiacus | 1.56 ± 0.20 | 1.02 ± 0.21 | neg | neg | neg |
| 228 | R. aegyptiacus | 1.13 ± 0.17 | 1.81 ± 0.26 | 2.13 ± 0.16 | 2.19 ± 0.30 | neg |
| 232 | R. aegyptiacus | 1.67 ± 0.33 | 1.61 ± 0.22 | 2.05 ± 0.27 | 1.09 ± 0.24 | neg |
| 233 | R. aegyptiacus | 1.04 ± 0.36 | 1.69 ± 0.34 | 1.33 ± 0.16 | 1.56 ± 0.22 | neg |
| 222B | R. aegyptiacus | 2.34 ± 0.29 | 1.56 ± 0.23 | 1.16 ± 0.25 | neg | neg |
Only the samples that neutralized LBV are included.
The log10 50% end point neutralizing titers ± 95% confidence intervals are indicated. Samples were considered negative (neg) if the 50% end point neutralizing titer at a serum dilution of 1:10 was 1 log10 or less (e.g., 50% or more observed fields contained the infected cells). None of the samples neutralized DUVV (the isolate from South Africa, human, 1970) and WCBV (the isolate from Russia, Miniopterus schreibersi, 2002).
MOKV, the MOKV isolate from South Africa (1997; cat); RABV, laboratory strain CVS-11.
Expressed in IU based on a comparison with the activity of a standard anti-rabies virus immunoglobulin, 2 IU/ml (NIH, Bethesda, MD).
In addition, 38% of specimens that neutralized LBV also neutralized MOKV, and only two of them demonstrated limited neutralizing activity against RABV. None of the samples that neutralized LBV demonstrated any activity against DUVV and WCBV. Considering the detected cross-reactivity between different LBV isolates and the observation that glycoproteins of KE131 and LBVAFR1999 isolates are very similar to each other, we used only the latter virus for screening of serum samples collected in 2007 (n = 813).
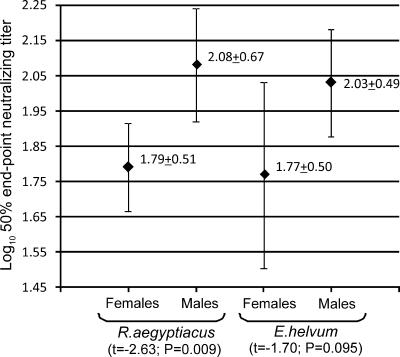
Seropositive R. aegyptiacus (n = 339; seroprevalence range by roost location, 29 to 46%) and E. helvum (n = 102; seroprevalence range by roost location, 40 to 67%) bats were detected in each roost where these species were present. No significant differences in seroprevalence were observed between different roosts, and no variations between 2006 and 2007 were detected for each roost. Seroprevalence in males was greater than in females, although statistically insignificant (55% of males [n = 55] and 41% of females [n = 14] for E. helvum [χ2 = 1.72, P = 0.19]; 35% of males [n = 65] and 32% of females [n = 55] for R. aegyptiacus [χ2 = 0.45, P = 0.50]). In addition, for both bat species antibody titers in males were greater than in females (Fig. 4). Comparison of seroprevalence in adult versus subadult R. aegyptiacus bats was available for the animals collected in 2006. The seroprevalence in adults (60% [n = 20]) was greater than that in subadults (31% [n = 16]), while statistically insignificant (χ2 = 2.95, P = 0.086). Based on this observation, we did not collect subadult bats in 2007, and further comparison between age groups was unavailable. The dead bat which was the source of KE131 isolation was seropositive, with a 50% end point log10 neutralizing titer of 2.86 ± 0.27.
FIG. 4.
Titers of anti-LBV VNA in the sera of male and female Eidolon helvum and Rousettus aegyptiacus bats (means ± standard deviations are indicated; whiskers show 1.96 times the standard errors).
DISCUSSION
We performed the first bat lyssavirus surveillance in eastern Africa. This study resulted in isolation of LBV from an E. helvum fruit bat. The LBV isolate KE131, obtained in our study, was related phylogenetically to the virus LBVSEN1985, isolated from E. helvum in Senegal (∼7,800 km away) 22 years ago, and to the virus LBVAFR1999, translocated to France from Togo or Egypt in 1999 by a sick R. aegyptiacus bat. This genetic stability across time and space suggests that a given LBV variant is well adapted to its primary host and that host populations in western and eastern Africa have sufficient interactions for pathogen exchange. If one considers Egypt as the potential origin of the LBVAFR1999 isolate, we should include north Africa and the Mediterranean region (distribution area of R. aegyptiacus) in this range as well. Unfortunately, the available three isolates do not allow a conclusion as to whether the primary host of this LBV variant is E. helvum or R. aegyptiacus. Interestingly, the initial isolate of LBV collected in Nigeria in 1956 from an E. helvum bat (LBVNIG1956) differs significantly from all other LBV representatives (Fig. 3). We detected evidence of LBV circulation in Kenya in both E. helvum and R. aegyptiacus. However, we do not know whether these species maintain circulation of only a single LBV variant or whether there are additional LBV variants circulating as well, as only the single isolate has been available to date. E. helvum roosts on high trees, whereas R. aegyptiacus roosts in caves (Fig. 5). These species may interact and exchange pathogens during nocturnal foraging. Meanwhile, as is well established in the Americas, different bat species maintain circulation of specific RABV variants (18, 46, 52). Therefore we may expect circulation of distinct LBV variants in different Old World bat species as well.
FIG. 5.
A roosting group of Eidolon helvum bats on a tree (a) and a colony of Rousettus aegyptiacus bats in a cave (b). The photos were by Ivan V. Kuzmin.
The isolate KE131 demonstrated genomic organization typical for all lyssaviruses. Among other complete lyssavirus genomes available for comparison from GenBank (no LBV genomes are available for a comparison yet), the KE131 genome is most similar to the genome of MOKV. This similarity is evident not only in the genetic distances and structure of crucial functional elements but also in intergenic regions. Together with serologic cross-reactivity, this observation supports the assumption that MOKV and LBV are members of one phylogroup (3).
We sequenced the complete lyssavirus genome from the brain of a naturally infected bat. The most challenging part of this procedure was to determine the genome extremities via RNA circularization, followed by amplification and cloning of the circularized extremities. Circularization was described previously for viral genomic RNA only (6, 38, 44). However, we demonstrated that this approach may be implemented for total RNA extracted from an infected animal brain. The representation of nontruncated genome extremities in the sequenced clones was quite efficient, despite the fact that several hours separated the animal's death from the sampling (ambient temperature around the roost was approximately 20°C). Furthermore, the harvested brain was subjected to freeze-thaw cycles at least twice prior to RNA extraction, and the extracted RNA was frozen and thawed prior to the ligation procedure and nRT-PCR. This method should facilitate the generation of a greater number of the complete lyssavirus genome sequences from field specimens. At present, we do not know the speed of accumulated mutations in lyssavirus genomes during passages in laboratory animals or cell cultures or their functional significance. Extensive passaging may alter virulence dramatically, leading to adaptation to a new replication model and attenuation for other models. Therefore, it is preferable to generate complete viral genomes from field samples. Furthermore, as more complete genomes are generated, there will be greater insight into virus phylogeny and evolution.
We did not detect LBV or serologic reactivity against this virus in other bat species, including Epomophorus bats, which are the presumed hosts of LBV in South Africa (39, 40). One plausible explanation is that this LBV variant is not present in Kenya (although it was isolated not only in South Africa and Zimbabwe but also in countries neighboring Kenya, such as the Central African Republic and Ethiopia), or that our collection of Epomophorus bats was too limited (n = 19) and we missed positive bats from a spatiotemporal or collection bias.
In general, the infection prevalence among all collected bats was low (1 of 1,182 brains tested, or about 0.1%), and only 0.2% if calculated considering only E. helvum and R. aegyptiacus (n = 441). Among all sick and dead bats (n = 12) the infection prevalence was 9%. In contrast, the seroprevalence within various roosts ranged from 40 to 67% and 29 to 46% for E. helvum and R. aegyptiacus, respectively. Similar results were published for colonial North American bats that maintain circulation of RABV: the infection rate among randomly collected bats was usually less than 1%, whereas among moribund and dead bats it was 4 to 14%; seroprevalence in colonies of Tadarida brasiliensis was sometimes over 70% (9, 11, 56). This may suggest similar circulation patterns of RABV and LBV in gregarious bat species, which have high conspecific exposure rates. Perhaps, due to limited susceptibility, possibly resulting from coevolution, a majority of exposures lead to the development of immunity, attributed to peripheral virus activity rather than to central nervous system infection. We can speculate that immunocompromised, sick, or stressed bats (for example, as a result of superinfection, physical depletion caused by migrations, breeding behavior, limited food supply, etc.) have a greater probability to develop disease. While statistically insignificant, the greater seroprevalence and higher VNA titers in males may suggest that certain behavioral aspects of the sexes are important for LBV exposure. In foxes, which maintain circulation of RABV in Europe, rabies is diagnosed more frequently in males than in females. This is attributed to their territorial behavior and increased aggression during the mating season (60). In contrast, in North American insectivorous bat species, rabies was detected evenly in males and females (11, 20, 24) or the occurrence of infection among females was greater than among males (7, 8). The latter was also true for EBLV-1-infected insectivorous bats in The Netherlands (63). It is interesting that in Australia, where fruit bats maintain circulation of ABLV, a seroprevalence survey of a mixture of sick and apparently healthy bats demonstrated the presence of anti-ABLV VNA in 16% of samples (25).
In addition, seroprevalence in adult R. aegyptiacus bats was greater than in subadults. Studies of T. brasiliensis demonstrated that seroprevalence in juvenile bats and fetuses was similar to that in adult females, suggesting the possibility of prenatal VNA transfer. In August, seroprevalence in young T. brasiliensis bats was limited, suggesting that by that season young bats have already lost maternal antibodies (11, 56). We did not test juvenile bats or fetuses from Kenya. Even if prenatal VNA transport occurred, those passively acquired antibodies should have been eliminated from the blood of the subadult bats that we collected. At the same time, subadult bats have a rather limited chance to obtain active immunity. Their relatively short life history and minimal opportunity for frequent contacts with more aggressive adults (in colonies subadults most often roost together, segregated from adults) may potentially reduce their chance of exposure.
Interestingly, the bat from which the LBV was isolated was seropositive. According to several reports from North America, sera of rabid bats rarely demonstrated virus-neutralizing activity (11, 62). The relatively high neutralizing titer of the serum of the infected bat may suggest that the animal was ill for a considerable time (several days) and developed a serologic response. Detection of the infectious virus, viral RNA, and antigen in various tissues contributes to this assumption. The lack of virus isolation from several tissues that demonstrated the presence of viral RNA and antigen might be caused not only by limited virus load but also by neutralization of the infectious virus by VNA detected in the serum. High virus load in mucous and serous acini of salivary glands, as well as in the tongue epithelium cells and in the oral swab, suggests that LBV infection may be transmitted by saliva. Detection of virus in a nasal swab is not indicative. The nasal cavity might be contaminated by the infectious saliva during the clinical period of the disease (as the result of altered swallowing) or after death (as the result of passive leaking). However, at least one communication has reported the presence of RABV in the nasal mucosa of naturally infected T. brasiliensis bats (10). We did not test cryosections of nasal mucosa for the presence of viral antigen. The presence of infectious virus in reproductive organs and in a vaginal swab may suggest alternative routes of LBV transmission. Detection of infectious virus in gastric and bladder tissues is not indicative of virus excretion. As demonstrated by the DFA test, viral antigen in these and other extraneural tissues, except the salivary glands and tongue, was associated with peripheral neural innervation of tissues and ganglia.
Significant serologic cross-reactivity between LBV and MOKV and very limited cross-reactivity of these viruses with RABV were reported frequently from the initial recognition of LBV and MOKV as rabies-related viruses (3, 23, 27, 53). We have no substantive reason to consider the possibility of MOKV circulation in fruit bats based on our cumulative serologic results. Only 38% of LBV-neutralizing samples additionally neutralized MOKV. Historically, LBV has repeatedly been isolated from fruit bats in different areas of Africa, whereas MOKV has never been identified in these animals.
No suggestions for virus shedding in saliva, in the absence of brain infection, were obtained in our study, as all oral swabs (except the one obtained from the rabid bat) were negative.
Both E. helvum and R. aegyptiacus are abundant fruit bat species throughout major parts of the African continent. E. helvum is distributed in sub-Saharan Africa only. While this species is abundant and forms vast colonies in those areas where there is a yearlong abundance of fruit, in less favorable areas it forms smaller colonies or occurs only as a visitor during seasonal migrations (28). The migratory activity of E. helvum is broadly recognized; however, the predominant driving forces, routes, and distances of the migrations are largely unknown (17). R. aegyptiacus is distributed broadly in sub-Saharan Africa and also in Cyprus and along the eastern part of the Mediterranean coast (Turkey, Syria, Jordan, Israel, and Egypt). No information about migratory patterns of R. aegyptiacus is available, and we do not know whether the Sahara is a significant natural barrier between northern and southern populations. In addition, a very closely related species, Rousettus leschenaulti is distributed broadly in southern Asia, and data on bat lyssaviruses from that area are very limited (35, 36, 48, 55).
Most of the roosts of E. helvum and R. aegyptiacus encountered in Kenya were situated within or in close proximity to human settlements. Caves inhabited by R. aegyptiacus are frequently visited by tourists. Usually bats avoid contacts with people and fly away when disturbed. However, contacts of people with sick bats that are unable to fly may occur. We do not know the reason for mass mortality of bats in location 3 in February 2007, as no samples were available for testing.
To date, LBV has not been reported as a cause of human disease. Reduced pathogenicity in the mouse model was demonstrated for LBV and MOKV. This was attributed largely to the R(K)/D333 substitution in their glycoprotein ectodomains (3). However, the initial pathogenicity studies of LBV were performed on the prototype isolate (Nigeria; 1956) only. Mice and dogs did not present a productive infection after intramuscular administration of this virus, even with doses of 6.5 to 7.5 log10 MLD50. Nevertheless, one of six monkeys inoculated intramuscularly with 6 log10 MLD50 developed bilateral paresis on day 22 but recovered on day 86, and no virus was isolated from the animal after euthanasia on day 108 (59). When various LBV representatives were compared in the mouse model, isolates closely related to KE131 (LBVSEN1985 and LBVAFR1999) demonstrated the same peripheral pathogenicity as RABV (39). In our study, isolate KE131 was also pathogenic peripherally for mice, although a high virus dose was needed to produce the disease. Therefore, the previous assumption that LBV is lacking peripheral pathogenicity was incorrect.
In Kenya, as in many other African countries, rabies surveillance is lacking (29). The majority of lyssavirus isolates are not identified, and the actual significance of LBV and other lyssaviruses for public and veterinary health is unknown. A recent study in Malawi demonstrated that 11.5% of human cases of cerebral malaria were actually misdiagnosed rabies cases (37). Public awareness and education must be increased, and additional surveillance is needed for a better understanding of the epizootic situation, circulation patterns, and threat of lyssavirus emergence in Kenya and other African countries.
Acknowledgments
We thank Evelyne Mulama, Heather Burke, Dorine Bonyo, Edwin Danga, Leonard Nderitu, and Solomon Gikundi (CDC, Nairobi, Kenya) for excellent logistical support and for providing the laboratory facilities during our field trials. We appreciate the exceptional technical support that Lydia Kigo (National Museum of Kenya, Nairobi) and Nadia Stegeman (Tufts University, Boston, MA) provided during bat sampling. We are grateful to Alex Borisenko, Natalia Ivanova, and other staff of Guelph University (Ontario, Canada), as well as to Sergey Kruskop (Moscow State University, Russia), for assistance in bat species identification.
The study was supported in part by the Global Disease Detection program (CDC, Atlanta, GA). J.C.B. and N.S. were funded by the O. C. Hubert Fellowship (CDC, Atlanta, GA), and W.M. was funded by the National Research Foundation (South Africa).
Use of trade names and commercial sources are for identification only and do not imply endorsement by the U.S. Department of Health and Human Services. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
Footnotes
Published ahead of print on 27 February 2008.
REFERENCES
- 1.Aubert, M. F. A. 1996. Methods of the calculation of titers, p. 445-456. In F.-X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. WHO, Geneva, Switzerland.
- 2.Aubert, M. F. A. 1999. Rabies in individual countries. France. Rabies Bull. Eur. 236. [Google Scholar]
- 3.Badrane, H., C. Bahloul, P. Perrin, and N. Tordo. 2001. Evidence of two lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 753268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulger, L. R., and J. S. Porterfield. 1958. Isolation of a virus from Nigerian fruit bats. Trans. R. Soc. Trop. Med. Hyg. 52421-424. [DOI] [PubMed] [Google Scholar]
- 5.Bourhy, H., B. Kissi, and N. Tordo. 1993. Molecular diversity of the Lyssavirus genus. Virology 19470-81. [DOI] [PubMed] [Google Scholar]
- 6.Briese, T., A. Schneemann, A. J. Lewis, Y. S. Park, S. Kim, H. Ludwig, and W. I. Lipkin. 1994. Genomic organization of Borna disease virus. Proc. Natl. Acad. Sci. USA 914362-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnett, C. D. 1989. Bat rabies in Illinois: 1965 to 1986. J. Wildl. Dis. 2510-19. [DOI] [PubMed] [Google Scholar]
- 8.Childs, J. E., C. V. Trimarchi, and J. W. Krebs. 1994. The epidemiology of bat rabies in New York State, 1988-92. Epidemiol. Infect. 113501-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constantine, D. G. 1967. Bat rabies in the southwestern United States. Public Health Rep. 82867-888. [PMC free article] [PubMed] [Google Scholar]
- 10.Constantine, D. G. 1972. Rabies virus in nasal mucosa of naturally infected bats. Science 1751255-1256. [DOI] [PubMed] [Google Scholar]
- 11.Constantine, D. G., E. S. Tierkel, M. D. Kleckner, and D. M. Hawkins. 1968. Rabies in New Mexico cavern bats. Public Health Rep. 83303-316. [PMC free article] [PubMed] [Google Scholar]
- 12.da Cruz, F. W., A. J. A. McBride, F. R. Conceição, J. W. Dale, J. McFadden, and O. A. Dellagostin. 2001. Expression of the B-cell and T-cell epitopes of the rabies virus nucleoprotein in Mycobacterium bovis BCG and induction of an humoral response in mice. Vaccine 20731-736. [DOI] [PubMed] [Google Scholar]
- 13.Dean, D. J., M. K. Abelseth, and P. Atanasiu. 1996. The fluorescent antibody test, p. 88-93. In F.-X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. WHO, Geneva, Switzerland.
- 14.Dietzschold, B., M. Lafon, H. Wang, L. J. Otvos, E. Celis, W. H. Wunner, and H. Koprowski. 1987. Localization and immunological characterization of antigenic domains of the rabies virus internal N and NS proteins. Virus Res. 8103-125. [DOI] [PubMed] [Google Scholar]
- 15.Dietzschold, B., W. H. Wunner, T. J. Wiktor, A. D. Lopes, M. Lafon, C. L. Smith, and H. Koprowski. 1983. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. USA 8070-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ertl, H., B. Dietzchold, and L. Otvos. 1991. T helper cell epitope of rabies virus nucleoprotein defined by tri- and tetrapeptides. Eur. J. Immunol. 211-10. [DOI] [PubMed] [Google Scholar]
- 17.Fleming, T. H., and P. Eby. 2003. Ecology of bat migration, p. 156-208. In T. H. Kunz, and M. B. Fenton (ed.), Bat ecology. The University of Chicago Press, Chicago, IL.
- 18.Franka, R., D. G. Constantine, I. V. Kuzmin, A. Velasco-Villa, A. Wong, and C. E. Rupprecht. 2006. New phylogenetic lineage of rabies virus associated with the western pipistrelle bat (Pipistrellus hesperus). J. Gen. Virol. 872309-2321. [DOI] [PubMed] [Google Scholar]
- 19.Fu, Z. F., W. H. Wunner, and B. Dietzschold. 1994. Immunoprotection by rabies virus nucleoprotein, p. 207-218. In C. E. Rupprecht, B. Dietzschold, and H. Koprowski (ed.), Lyssaviruses. Springer, Berlin, Germany.
- 20.Girard, K. F., H. B. Hitchcock, G. Edsall, and R. A. Maccready. 1965. Rabies in bats in southern New England. N. Engl. J. Med. 27275-80. [DOI] [PubMed] [Google Scholar]
- 21.Gould, A. R., J. A. Kattenbelt, S. G. Gumley, and R. A. Lunt. 2002. Characterisation of an Australian bat lyssavirus variant isolated from an insectivorous bat. Virus Res. 891-28. [DOI] [PubMed] [Google Scholar]
- 22.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 4195-98. [Google Scholar]
- 23.Hanlon, C. A., I. V. Kuzmin, J. D. Blanton, W. C. Weldon, J. S. Manangan, and C. E. Rupprecht. 2005. Efficacy of rabies biologics against new lyssaviruses from Eurasia. Virus Res. 11144-54. [DOI] [PubMed] [Google Scholar]
- 24.Hester, L. C., T. L. Best, and M. K. Hudson. 2007. Rabies in bats from Alabama. J. Wildl. Dis. 43291-299. [DOI] [PubMed] [Google Scholar]
- 25.Hooper, P. T., R. A. Lunt, A. R. Gould, H. Samaratunga, A. D. Hyatt, L. J. Gleeson, B. J. Rodwell, C. E. Rupprecht, J. S. Smith, and P. K. Murray. 1997. A new lyssavirus—the first endemic rabies-related virus recognized in Australia. Bull. Inst. Pasteur 95209-218. [Google Scholar]
- 26.Hughes, G. J., I. V. Kuzmin, A. Schmitz, J. Blanton, J. Manangan, S. Murphy, and C. E. Rupprecht. 2006. Experimental infection of big brown bats (Eptesicus fuscus) with Eurasian bat lyssaviruses Aravan, Khujand, and Irkut virus. Arch. Virol. 1512021-2035. [DOI] [PubMed] [Google Scholar]
- 27.Jallet, C., Y. Jacob, C. Bahloul, A. Drings, E. Desmezieres, N. Tordo, and P. Perrin. 1999. Chimeric lyssavirus glycoproteins with increased immunological potential. J. Virol. 73225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingdon, J. 1974. East African mammals. An atlas of evolution in Africa, vol. II, part A. Academic Press, London, United Kingdom.
- 29.Kitala, P. M., J. J. McDermott, M. N. Kyule, and J. M. Gathuma. 2000. Community-based active surveillance for rabies in Machakos District, Kenya. Prev. Vet. Med. 4473-85. [DOI] [PubMed] [Google Scholar]
- 30.Koprowski, H. 1996. The mouse inoculation test, p. 80-86. In F.-X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. WHO, Geneva, Switzerland.
- 31.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 171244-1245. [DOI] [PubMed] [Google Scholar]
- 32.Kuzmin, I. V., G. J. Hughes, A. D. Botvinkin, L. A. Orciari, and C. E. Rupprecht. 2005. Phylogenetic relationships of Irkut and west Caucasian bat viruses within the Lyssavirus genus and suggested quantitative criteria based on the N gene sequence for lyssavirus genotype definition. Virus Res. 11128-43. [DOI] [PubMed] [Google Scholar]
- 33.Kuzmin, I. V., G. J. Hughes, and C. E. Rupprecht. 2006. Phylogenetic relationships of seven previously unclassified viruses within the family Rhabdoviridae using partial nucleoprotein gene sequences. J. Gen. Virol. 872323-2331. [DOI] [PubMed] [Google Scholar]
- 34.Kuzmin, I. V., L. A. Orciari, Y. T. Arai, J. S. Smith, C. A. Hanlon, Y. Kameoka, and C. E. Rupprecht. 2003. Bat lyssaviruses (Aravan and Khujand) from central Asia: phylogenetic relationships according to N, P and G gene sequences. Virus Res. 9765-79. [DOI] [PubMed] [Google Scholar]
- 35.Kuzmin, I. V., M. Niezgoda, D. S. Carroll, N. Keeler, M. J. Hossain, R. F. Breiman, T. G. Ksiazek, and C. E. Rupprecht. 2006. Lyssavirus surveillance in bats, Bangladesh. Emerg. Infect. Dis. 12486-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lumlertdacha, B., K. Boongird, S. Wanghongsa, S. Wacharapluesadee, L. Chanhome, P. Khawplod, T. Hemachudha, I. Kuzmin, and C. E. Rupprecht. 2005. Survey for bat lyssaviruses, Thailand. Emerg. Infect. Dis. 11232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallewa, M., A. R. Fooks, D. Banda, P. Chikungwa, L. Mankhambo, E. Molyneux, M. E. Molyneux, and T. Solomon. 2007. Rabies encephalitis in malaria-endemic area, Malawi, Africa. Emerg. Infect. Dis. 13136-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandl, C. W., F. X. Heinz, E. Puchhammer-Stockl, and C. Kunz. 1991. Sequencing the termini of capped viral RNA by 5′-3′ ligation and PCR. BioTechniques 101485-486. [PubMed] [Google Scholar]
- 39.Markotter, W. 2007. Molecular epidemiology and pathogenesis of Lagos bat virus, a rabies-related virus specific to Africa. Ph.D. dissertation, University of Pretoria, Pretoria, South Africa.
- 40.Markotter, W., I. Kuzmin, C. E. Rupprecht, J. Randles, C. T. Sabeta, A. I. Wandeler, and L. H. Nel. 2006. Isolation of Lagos bat virus from water mongoose. Emerg. Infect. Dis. 121913-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markotter, W., J. Randles, C. E. Rupprecht, C. T. Sabeta, A. I. Wandeler, P. J. Taylor, and L. H. Nel. 2006. Lagos bat virus, South Africa. Emerg. Infect. Dis. 12504-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marston, D. A., L. M. McElhinney, N. Johnson, T. Muller, K. K. Conzelmann, N. Tordo, and A. R. Fooks. 2007. Comparative analysis of the full genome sequence of European bat lyssavirus type 1 and type 2 with other lyssaviruses and evidence for a conserved transcription termination and polyadenylation motif in the G-L 3′ non-translated region. J. Gen. Virol. 881302-1314. [DOI] [PubMed] [Google Scholar]
- 43.Mebatsion, T., J. H. Cox, and J. W. Frost. 1992. Isolation and characterization of 115 street rabies virus isolates from Ethiopia by using monoclonal antibodies: identification of 2 isolates as Mokola and Lagos bat viruses. J. Infect. Dis. 166972-977. [DOI] [PubMed] [Google Scholar]
- 44.Morzunov, S. P., J. R. Winton, and S. T. Nichol. 1995. The complete genome structure and phylogenetic relationship of infectious hematopoietic necrosis virus. Virus Res. 38175-192. [DOI] [PubMed] [Google Scholar]
- 45.Nadin-Davis, S. A., M. Abdel-Malik, J. Armstrong, and A. Wandeler. 2002. Lyssavirus P gene characterization provides insight into the phylogeny of the genus and identities structural similarities and diversity within the encoded phosphoprotein. Virology 298286-305. [DOI] [PubMed] [Google Scholar]
- 46.Nadin-Davis, S. A., W. Huang, J. Armstrong, G. A. Casey, C. Bahloul, N. Tordo, and A. I. Wandeler. 2001. Antigenic and genetic divergence of rabies viruses from bat species indigenous to Canada. Virus Res. 74139-156. [DOI] [PubMed] [Google Scholar]
- 47.Nel, L. H., and C. E. Rupprecht. 2007. Emergence of lyssaviruses in the Old World: the case of Africa. Curr. Top. Microbiol. Immunol. 315161-393. [DOI] [PubMed] [Google Scholar]
- 48.Pal, S. R., B. M. Arora, P. N. Chuttani, S. Broer, S. Choudhury, and R. M. Joshi. 1980. Rabies virus infection of a flying fox bat, Pteropus poliocephalus, in Chandigarh, northern India. Trop. Geogr. Med. 32265-267. [PubMed] [Google Scholar]
- 49.Picard-Meyer, E., J. Barrat, M. Wasniewski, A. Wandeler, S. Nadin-Davis, J. P. Lowings, A. R. Fooks, L. McElhinney, V. Bruyère, and F. Cliquet. 2004. Epidemiology of rabid bats in France, 1989 to 2002. Vet. Rec. 155774-777. [PubMed] [Google Scholar]
- 50.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 711153-1162. [DOI] [PubMed] [Google Scholar]
- 51.Poisson, N., E. Real, Y. Gaudin, M. C. Vaney, S. King, Y. Jacob, N. Tordo, and D. Blondel. 2001. Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8: dissociation of dynein-binding properties and transcriptional functionality of P. J. Gen. Virol. 822691-2696. [DOI] [PubMed] [Google Scholar]
- 52.Shankar, V., L. A. Orciari, C. De Mattos, I. Kuzmin, W. J. Pape, T. J. O'Shea, and C. E. Rupprecht. 2005. Genetic divergence of rabies viruses from bat species of Colorado, USA. Vector Borne Zoonot. Dis. 5330-341. [DOI] [PubMed] [Google Scholar]
- 53.Shope, R. E., F. A. Murphy, A. K. Harrison, O. R. Causey, G. E. Kemp, D. L. Simpson, and D. L. Moore. 1970. Two African viruses serologically and morphologically related to rabies virus. J. Virol. 6690-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, J. S., P. A. Yager, and G. M. Baer. 1996. A rapid fluorescent focus inhibition test (RFFIT) for determining rabies virus-neutralizing antibody, p. 181-192. In F.-X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. WHO, Geneva, Switzerland.
- 55.Smith, P. C., K. Lawhaswasdi, W. E. Vick, and J. S. Stanton. 1967. Isolation of rabies virus from fruit bats in Thailand. Nature 216384. [DOI] [PubMed] [Google Scholar]
- 56.Steece, R., and J. S. Altenbach. 1989. Prevalence of rabies specific antibodies in the Mexican free-tailed bat (Tadarida brasiliensis mexicana) at Lava Cave, New Mexico. J. Wildl. Dis. 25490-496. [DOI] [PubMed] [Google Scholar]
- 57.Sureau, P., M. Germain, J. P. Herve, B. Geoffrey, J. P. Cornet, G. Heme, and Y. Robin. 1977. Isolement du virus Lagos bat en Empire Centrafricain. Bull. Exotic Pathol. Soc. 70467-470. [PubMed] [Google Scholar]
- 58.Swanepoel, R. 2004. Rabies, p. 1123-1182. In J. A. W. Coetzer and R. C. Tustin (ed.), Infectious diseases of livestock: with special reference to southern Africa, 2nd ed. Oxford University Press, Cape Town, South Africa.
- 59.Tignor, G. H., R. E. Shope, P. N. Bhatt, and D. H. Percy. 1973. Experimental infection of dogs and monkeys with two rabies serogroup viruses, Lagos bat and Mokola (IbAn 27377): clinical, serologic, virologic and fluorescent-antibody studies. J. Infect. Dis. 128471-478. [DOI] [PubMed] [Google Scholar]
- 60.Toma, B., and L. Andral. 1977. Epidemiology of fox rabies. Adv. Virus Res. 211-36. [DOI] [PubMed] [Google Scholar]
- 61.Tordo, N., A. Benmansour, C. Calisher, R. G. Dietzgen, R. X. Fang, A. O. Jackson, G. Kurath, S. Nadin-Davis, R. B. Tesh, and P. J. Walker. 2005. Rhabdoviridae, p. 623-644. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). Virus taxonomy, VIIIth report of the ICTV. Elsevier, London, United Kingdom.
- 62.Trimarchi, C. V., and J. G. Debbie. 1977. Naturally occurring rabies virus and neutralizing antibody in two species of insectivorous bats of New York state. J. Wildl. Dis. 13366-369. [DOI] [PubMed] [Google Scholar]
- 63.Van der Poel, W. H. M., R. Van der Heide, E. R. A. M. Verstraten, K. Takumi, P. H. C. Lina, and J. A. Kramps. 2005. European bat lyssaviruses, The Netherlands. Emerg. Infect. Dis. 111854-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warrilow, D., I. L. Smith, B. Harrower, and G. A. Smith. 2002. Sequence analysis of an isolate from a fatal human infection of Australian bat lyssavirus. Virology 297109-119. [DOI] [PubMed] [Google Scholar]
- 65.Webster, W. A., and G. A. Casey. 1996. Virus isolation in neuroblastoma cell culture, p. 96-104. In F.-X. Meslin, M. M. Kaplan, and H. Koprowski (ed.), Laboratory techniques in rabies, 4th ed. WHO, Geneva, Switzerland.