Abstract
Broad-spectrum analysis for pathogens in patients with respiratory tract infections is becoming more relevant as the number of potential infectious agents is still increasing. Here we describe the new multiparameter RespiFinder assay, which is based on the multiplex ligation-dependent probe amplification (MLPA) technology. This assay detects 15 respiratory viruses in one reaction. The MLPA reaction is preceded by a preamplification step which ensures the detection of both RNA and DNA viruses with the same specificity and sensitivity as individual monoplex real-time reverse transcription-PCRs. The RespiFinder assay was validated with 144 clinical samples, and the results of the assay were compared to those of cell culture and a respiratory syncytial virus (RSV)-specific immunochromatography assay (ICA). Compared to the cell culture results, the RespiFinder assay showed specificities and sensitivities of 98.2% and 100%, respectively, for adenovirus; 96.4% and 100%, respectively, for human metapneumovirus; 98.2% and 100%, respectively, for influenza A virus (InfA); 99.1% and 100%, respectively, for parainfluenza virus type 1 (PIV-1); 99.1% and 80%, respectively, for PIV-3; 90.1% and 100%, respectively, for rhinovirus; and 94.6% and 100%, respectively, for RSV. Compared to the results of the RSV-specific ICA, the RespiFinder assay gave a specificity and a sensitivity of 82.4% and 80%, respectively. PIV-2, PIV-4, influenza B virus, InfA H5N1, and coronavirus 229E were not detected in the clinical specimens tested. The use of the RespiFinder assay resulted in an increase in the diagnostic yield compared to that obtained by cell culture (diagnostic yields, 60% and 35.5%, respectively). In conclusion, the RespiFinder assay provides a user-friendly and high-throughput tool for the simultaneous detection of 15 respiratory viruses with excellent overall performance statistics.
Acute respiratory tract infections (RTIs) are the most widespread types of infections in adults and children and are responsible for considerable morbidity and mortality worldwide (18). Unfortunately, the etiology remains undetermined in more than 50% of cases (9). Among the pathogens responsible for undiagnosed infections, respiratory viruses are thought to contribute to a substantial number of RTIs. Influenza viruses, respiratory syncytial viruses (RSVs), and parainfluenza viruses (PIVs) have been identified as important pathogens in community-acquired pneumoniae. These viruses are also a significant cause of disease in immunocompromised patients (11). Currently, the clinical diagnosis of RTIs is mostly restricted to the detection and identification of these few pathogens. However, evidence is accumulating that other viruses, such as coronaviruses 229E, OC43, and NL63 (Cor-OC43, Cor-229E, and Cor-NL63, respectively) (35) and human metapneumovirus (hMPV) (20, 33), are frequently associated with infected lower respiratory tracts. Even an unexpectedly high prevalence of rhinovirus in lower RTIs has been demonstrated in children as well as adults (8). The clinical presentation of patients with RTIs is generally not indicative of a specific pathogen, so a rapid diagnosis could be helpful in therapeutic decision making. In addition, there is an increasing threat of uncommon yet significant respiratory viruses, such as the severe acute respiratory syndrome (SARS) coronavirus and influenza A virus (InfA) H5N1 (28). As a result, it is becoming more important to detect a broad panel of respiratory viruses.
Cell culture is still the “gold standard” for the laboratory detection of respiratory viruses. However, cell culture is slow and has a low sensitivity. Therefore, its implementation for routine virus detection is suboptimal. Although rapid antigen detection tests are available for some of the respiratory viruses, these tests have been shown to be less sensitive and less specific than cell culture-based approaches (5, 29). Nucleic acid amplification tests have proven to be rapid, very sensitive, and specific alternatives and can be used in either a monoplex or a multiplex format. Moreover, multiplex assays allow the coamplification of more than one target, thus providing insight into the significance of mixed infections for the prognosis and recrudescence of the respiratory disease. In addition, the incorporation of the ability to detect viruses such as the SARS coronavirus and InfA H5N1 in a multiplex assay would allow monitoring of these viruses and could act as an early built-in detection system (28).
An all-embracing multiparameter test is a prerequisite to reducing the costs involved with such a comprehensive monitoring system (19). Currently, several assays with multiplex formats detect up to nine respiratory viruses in one reaction. For instance, several real-time multiplex assays allow the real-time detection of up to four targets in a reaction, depending on the number of channels available in the real-time PCR machines (13, 30, 34). Based on the Roche LightCycler480 instrument, a real-time TaqMan PCR that detects five different targets simultaneously has been developed (21). Existing multiplex PCR assays that use agarose gel electrophoresis or capillary electrophoresis as the detection system detect five to eight targets per reaction (3, 7, 24). Multiplex PCR assays combined with an enzyme-linked immunosorbent assay currently detect up to nine targets simultaneously (12, 26).
Recently, two multiplex assays that detect respiratory viruses have been evaluated with 360 clinical samples (17). The NGEN respiratory virus analyte-specific assay (Nanogen, San Diego, CA) detects InfA, InfB, PIV type 1 (PIV-1), PIV-2, PIV-3, and RSV on a NanoChip 400 electronic microarray. The ResPlex II assay (Genaco Biomedical Products, Inc., Huntsville, AL) detects InfA, InfB, PIV-1 to PIV-4, RSV, hMPV, rhinovirus, enterovirus, and the SARS coronavirus on a Luminex 100 instrument (Luminex, Austin, TX). Both assays exhibited specificities comparable to those of cell culture and the monoplex real-time reverse transcription (RT)-PCR. However, the sensitivities of both assays were lower than the sensitivity of the monoplex real-time RT-PCR.
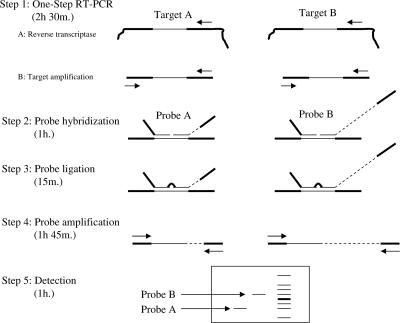
The multiplex ligation-dependent probe amplification (MLPA) technology (27) uses a simple-to-perform, multiplex PCR method which is able to amplify up to 45 different targets simultaneously. MLPA is based on the oligonucleotide ligation assay, and identification is accomplished by size by using gel electrophoresis methods. MLPA has three steps: first, an annealing step hybridizes the probes to their target regions; second, a ligation step links the two probes; and third, the final PCR exponentially amplifies the ligated probes with only two primers (Fig. 1). So far, MLPA has mainly been used to detect changes in the copy numbers of specific chromosomal regions (27), in expression profiling studies (6), for the detection of CpG methylation of genes (22), and for the detection of recombination events (16). We developed a multiparameter test (the RespiFinder assay) that is based on the MLPA technology and that can be used clinically to detect infectious disease agents.
FIG. 1.
Overview of the RespiFinder technology. A one-step RT-PCR is performed with specific primers for all targets (steps 1A and 1B). Subsequently, an MLPA reaction is performed with MLPA probes specific for all targets (steps 2, 3, and 4). An MLPA probe consists of two oligonucleotides: one synthetic oligonucleotide and one M13-derived oligonucleotide. The synthetic oligonucleotide contains a universal forward priming site, and the M13-derived oligonucleotide contains a universal reverse priming site. In addition, the M13-derived oligonucleotide contains a unique stuffer sequence. The length of this stuffer sequence is specific for each probe and varies between the different probes. The length of the MLPA probe is the combined length of both oligonucleotides. This length is unique for each probe due to the specific stuffer sequence. The two oligonucleotides hybridize specifically to the target adjacent to each other. Subsequently, the two oligonucleotides are joined by ligation. The ligated oligonucleotides are amplified by one universal primer set. After amplification, the MLPA reaction is analyzed by electrophoresis (step 5). Each MLPA probe can be discerned due to its specific length. Recently, the protocol has been further optimized by combining the probe ligation (step 3) and probe amplification (step 4) in one reaction.
The RespiFinder assay allows the detection and differentiation of 15 respiratory viruses and one internal amplification control (IAC) (2, 14). The test includes 14 viruses involved in RTIs. These are InfA, influenza virus B (InfB), PIV-1 to PIV-4, respiratory syncytial virus A (RSVA), respiratory syncytial virus B (RSVB), rhinovirus, Cor-229E, Cor-OC43, Cor-NL63, hMPV, and adenovirus. Additionally, the test includes a probe for InfA H5N1.
The RespiFinder assay was validated with 144 clinical samples, and the results were compared with those of cell culture and an RSV-specific immunochromatography assay (ICA). We demonstrate that the RespiFinder assay provides an excellent tool for the simultaneous detection of 15 respiratory viruses.
MATERIALS AND METHODS
Viral cultures and monoplex real-time RT-PCR.
Serial dilutions of cultures of RSVA, RSVB, InfA, InfB, rhinovirus, Cor-OC-43, Cor-229E, Cor-NL63, PIV-1, PIV-2, PIV-3, PIV-4, and hMPV and the results of the monoplex real-time RT-PCR obtained with the dilutions were kindly provided by H. G. Niesters, Department of Virology, Erasmus MC, Rotterdam, The Netherlands.
Clinical samples.
A total of 144 nasal wash specimens (from both adults and children) were submitted for the routine diagnosis of viral infection; of these, 110 samples were tested by cell culture on four different cell lines (Virology Department, University Hospital St. Radboud, Nijmegen. The Netherlands). This included adenovirus, hMPV, InfA, InfB, PIV-1 to PIV-4, rhinovirus, and RSV. The samples were not routinely checked for the presence of coronaviruses. A rapid ICA was performed for the detection of RSV in 34 samples from children by using the RSV Respi-Strip from Coris BioConcept (Gembloux, Belgium). All clinical samples were stored at −80°C until further analysis by the RespiFinder assay.
Nucleic acid extraction.
Viral RNA or DNA was isolated by using the automated MagNA Pure LC system (Roche Diagnostics, Almere, The Netherlands). The MagNA Pure LC total nucleic acid isolation kit and the total nucleic acid lysis extraction MagNA Pure protocol were applied. Extraction was performed according to the manufacturer's instructions. Briefly, 200 μl of starting material was used, and the purified nucleic acid was eluted in a final volume of 100 μl. Before the start of extraction, 5 μl of the IAC, which contained an encephalomyocarditis virus (EMC) RNA transcript, was added to the lysed samples.
In vitro transcripts of viral targets.
Synthetic viral target genes, which contained both the PCR primer regions and the MLPA probe sequence, were constructed by using the software package of DNA Works (version 3.1). The DNA Works software calculates the composition and the number of oligonucleotides required to synthesize the viral target. The oligonucleotides generated are characterized by highly homogeneous melting temperatures and a minimized tendency for hairpin formation. By using a two-step PCR protocol, the synthetic viral target gene can easily be synthesized and amplified (15).
The amplification products were purified with an MSB Spin PCRapace kit (Invitek, Berlin, Germany) and were subsequently cloned in pGEM-T Easy vector system II (Promega, Leiden, The Netherlands). The nucleotide sequences of the cloned viral target genes were sequenced on an ABI 3100 DNA capillary sequencer. All cloned viral targets were linearized and transcribed in vitro from the SP6 promoter by using the Riboprobe in vitro transcription kit (Promega, Leiden, The Netherlands). Residual plasmid DNA was removed by DNase treatment (TURBO DNA-free; Ambion, Huntingdon, United Kingdom), according to the manufacturer's instructions. The concentration of the SP6 RNA transcripts was determined by measuring the absorbance at 260 nM (1 unit of the optical density at 260 nm is equal to 40 μg/ml of RNA), and the absence of residual plasmid DNA was checked by performing a real-time PCR without an RT step. Aliquots of the RNA transcripts were stored at −20°C, and dilutions were made in Tris-EDTA (pH 7.6) containing 1 μg/ml of carrier DNA (herring sperm DNA; Fermentas, St. Leon-Rot, Germany).
IAC.
The IAC was a SP6 RNA transcript of the polyproteon gene from EMC. The IAC was produced as described above. Twofold serial dilutions of the IAC were spiked into a clinical sample which previously tested virus negative by the RespiFinder assay. Subsequently, the spiked clinical samples were analyzed by the RespiFinder assay. The optimal spiked concentration was defined as two times the lowest detectable concentration.
Preamplification.
A one-step RT-PCR protocol was performed with a OneStep RT-PCR kit (Qiagen, Hilden, Germany). Combined RT and multiplex target amplification PCR were performed in a 25-μl reaction mixture, according to the manufacturer's instructions, in the presence of all preamplification primers (Table 1). The mixture contained 0.2 μM of each primer and 10 μl of the RNA or DNA sample. Amplification was performed on a T1 thermocycler (Biometra, Göttingen, Germany), as follows: an initial cycle of 30 min at 50°C and 15 min at 95°C, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C and, finally, a 10-min extended elongation.
TABLE 1.
Sequences of the preamplification primers
| Virus | Primer | Sequence (5′-3′) |
|---|---|---|
| InfA | Forward | CAAGACCAATCCTGTCACCTCT |
| Reverse | ATCGATGGCGCATGCAACTGGCAAG | |
| InfB | Forward | ATGTCGCTGTTTGGAGACACAATTG |
| Reverse | GCATCTTTTGTTTTTTATCCATTC | |
| InfA/H5N1 | Forward | TGCATGGGTCTCATATACAACA |
| Reverse | TCCATAGCCTTAGCTGTAGTGC | |
| RSVA | Forward | TCCCATAATATACAAGTATGATCTCAA |
| Reverse | AACCCAGTGAATTTATGATTAGCA | |
| RSVB | Forward | TGTGGTATGCTATTAATCACTGAAGA |
| Reverse | GGAGCCACTTCTCCCATCTC | |
| PIV-1 | Forward | TTCTGGAGATGTCCCGTAGG |
| Reverse | TCCTGTTGTCGTTGATGTCATA | |
| PIV-2 | Forward | TGGGGATAATACAACAATCTGC |
| Reverse | AGAAAGCAAGTCTCAGTTCAGC | |
| PIV-3 | Forward | TCAATCTCAACAACAAGATTTAAGAA |
| Reverse | GACACCCAGTTGTGTTGCAG | |
| PIV-4 | Forward | TACAGGCCACATCAATGCAG |
| Reverse | GCATGTTCTGCATCTCTGGA | |
| Rhinovirus | Forward | GCCTGCGTGGCTGCC |
| Forward | CCTGCGTGGCGGCC | |
| Reverse | GAAACACGGACACCCAAAGTAGT | |
| Cor-229E | Forward | CGCAGCCGGTGGAAAAGACAGCCTAA |
| Reverse | AGCATAGCAGCTGTAGACGGCACAAGC | |
| Cor-OC43 | Forward | AAGAGCTCAACCCAAGCAAA |
| Reverse | ATACCATCGTGGCAGCAGTT | |
| Cor-NL63 | Forward | GGCTGCGTTACTTTGGCTTTAAAGAACTTAGGTTTTG |
| Reverse | GACGCTCCAACGAGGTTTCTTCAACTG | |
| hMPV | Forward | CAAAGAGGCAAGAAAAACAATGG |
| Reverse | GCCTGGCTCTTCTGACTGTGGTCTC | |
| Adenovirus | Forward | GCCCATGCGCTGGACATGACTTTTGAGGTGGATCCCATGGA |
| Forward | GACATGACTTTTGAGGTGGATCCCATGGA | |
| Reverse | TTATGTGGTGGCGTTGCCGGC | |
| Reverse | AAGAAGCAAGAGGCTTCTTATGTGGTGGCGTTACCGGC | |
| EMC (IAC) | Forward | ACATGTAACCGCCCCCATT |
| Reverse | TCCACGCACGCACTACTATG |
Primer and probe design.
The primers and probes were designed against conserved genes within each virus. The sequences from these genes were obtained from GenBank and from the Los Alamos influenza virus database (http://www.flu.lanl.gov/). The sequences were aligned by using the Clustal X program (version 1.81) to identify highly conserved regions within each gene. PCR primers and MLPA probes were designed on the basis of the sequences of these highly conserved regions. The primers were designed with Primer3 software (version 0.2) (http://primer3.sourceforge.net/). These primers were used for both specific RT and target amplification. The primers flanked the target region of the MLPA probe. The MLPA probes were designed as described earlier (27). Subsequently, all primers and probes were evaluated by performing a BLAST analysis against the sequences in the NCBI database. The primers and probes were approved when no mismatches within the critical regions of the probes (e.g., no mismatch within 5 nucleotides from the ligation site) and primers (e.g., no mismatch at the 3′ end of a primer) were found. The primers and the probes were synthesized by Biolegio (Malden, The Netherlands) and are shown in Tables 1 and 2.
TABLE 2.
Target genes and sequences of virus specific regions of the MLPA probes
| Virus | Gene | MLPA oligonucleotide | Sequence of virus-specific regions of MLPA probe (5′-3′) |
|---|---|---|---|
| InfA | Matrix protein gene (M1) | Synthetic | CCATGCACGCTCACCGTGCCCAGTGAGCGAGG |
| Long | ACTGCAGCGTAGACGCTTTGTCCAAAATGCCCTCAATGGGAATG | ||
| InfB | Matrix protein gene (M1) | Synthetic | GACAGAAGATGGAGAAGGCAAAGCAGA |
| Long | ACTAGCAGAAAAATTACACTGTTGGTTCGGTGGGAAAGAA | ||
| InfA H5N1 | Matrix protein gene (M1) | Synthetic | CIGTIACTACIGAAGTGGCTTTTGGCCTA |
| Long | GTGTGTGCCACTTGTGAGCAGATTGCAGATTCACAGCA | ||
| RSVA | Major nucleocapsid protein gene (N) | Synthetic | GGCTCTTAGCAAAGTCAAGTTGAATGATACACTC |
| Long | AACAAAGATCAACTTCTGTCATCCAGCAAATACACCATCCAACGGA | ||
| RSVB | Major nucleocapsid protein gene (N) | Synthetic | GTCCAGGTTAGGAAGGGAAGACACTATAAAGATACTT |
| Long | AAAGATGCTGGATATCATGTTAAAGCTAATGGAGTAGATATAACAA | ||
| PIV-1 | Hemagglutinn-neuraminidase gene (HN) | Synthetic | CAGGATGTGTTAGACTACCTTCATTATCAATTGGT |
| Long | GATGCAATATATGCGTATTCATCAAACTTAATCACTCAAGGATGTGC | ||
| PIV-2 | Hemagglutinn-neuraminidase gene (HN) | Synthetic | CAGGACTATGAAAACCATTTACCTAAGTGATGG |
| Long | AATCAATCGCAAAAGCTGTTCAGTCACTGCTATACCAGGAGGTTGT | ||
| PIV-3 | Hemagglutinn-neuraminidase gene (HN) | Synthetic | CCATCTGTTGGACCAGGGATATACTACAAA |
| Long | GGCAAAATAATATTTCTCGGGTATGGAGGTCTTGAACATCCAA | ||
| PIV-4 | Hemagglutinn-neuraminidase gene (HN) | Synthetic | GATTATACATTAACTATTAATCCAAGATCTGGCTGGGATG |
| Long | ACATCAAGATCAGAGCTTATAGAGCATTATCCAGAGATTTGC | ||
| Rhinovirus | 5′ untranslated region polyprotein gene (PP) | Synthetic | GACAIGGTGTGAAGAGCCCCGTGTGCTIIIITTGAITCCTCC |
| Long | GGCCCCTGAATGTGGCTAACCTTAACCC | ||
| Cor-229E | Nucleocapsid protein gene (NP) | Synthetic | CCCAGAGACCTTGACCACAACTTTGGAAGT |
| Long | GCAGGTGTTGTGGCCAATGGTGTTAAAGCTAAAGGCTATC | ||
| Cor-OC43 | Nucleocapsid protein gene (NP) | Synthetic | CTATTGCACCAGGAGTCCCAGCTACTGAA |
| Long | GCTAAGGGGTACTGGTACAGACACAACAGACGTTCTTTTAAAAC | ||
| Cor-NL63 | Nucleocapsid protein gene (NP) | Synthetic | CCAGTCGAAGTCACCTAGTTCTTCTGGTA |
| Long | CTTCCACTCCTAAGAAACCTAATAAGCCTCTTTCTCAACCCAGGGCT | ||
| hMPV | Nucleocapsid protein gene (NP) | Synthetic | GCTCATGCATCCCACAAAATCAGAGGCCTTCAGCACCAG |
| Long | ACACACCAATAATTTTATTATGTGTAGGTGCCTTAATATTCACTAAACTAGCATCAA | ||
| Adenovirus | Hexon gene (H) | Synthetic | GAAGTTTTCGACGTGGTCCGAGTGCACCAGCCGCACCGC |
| Synthetic | GAAGTTTTCGACGTGGTCAGTGTGCATCAGCCACACCGC | ||
| Long | GGCGTCATCGAGGCCGTCTACCTGCGCACACCGTTCTC | ||
| EMC (IAC) | Polyprotein gene (PP) | Synthetic | GCAGTCAGGTGAAGCACCCAGACTTGCCTCCTTGT |
| Long | GAGAGGCCGCACCTTGGTAGTAAATAGACACATGGCCGAGT |
MLPA reaction.
MLPA analysis was performed essentially as described earlier (27), with minor modifications. The preamplification reaction mixture was diluted five times with sterile water. Two microliters of the diluted preamplification mixture was mixed with 3 μl of sterile water, 1.5 μl probe mixture, and 1.5 μl MLPA buffer (1.5 M KCl, 300 mM Tris-HCl, pH 8.5, 1 mM EDTA); and denaturation was performed for 5 min at 98°C in a thermocycler. After denaturation, the mixture was incubated for 1 h at 60°C and was subsequently diluted to 40 μl with ligation buffer (2.6 mM MgCl2, 5 mM Tris-HCl, pH 8.5, 0.013% nonionic detergents, 0.2 mM NAD). Ligation was performed by adding 1 U of the heat-labile ligase 65 enzyme and incubating at 54°C for 15 min, followed by ligase inactivation at 98°C for 5 min. Subsequently, 4 μl ligation mixture was used in a 20-μl reaction volume containing 50 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, deoxynucleoside triphosphates (0.2 mM each; Fermentas), 1 U Taq polymerase, and 0.2 μM of the two PCR primers (forward primer, 5′-GGGTTCCCTAAGGGTTGGA-3′; reverse primer, 5′-GTGCCAGCAAGATCCAATCTAGA-3′). Amplification was performed on a Biometra T1 thermocycler, as follows: an initial cycle of 2 min at 95°C, followed by 33 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C and, finally, a 10-min extended elongation. All buffers and enzymes for MLPA were obtained from MRC-Holland (Amsterdam, The Netherlands).
Analysis of PCR products.
The amplified MLPA products can be analyzed on different detection platforms, e.g., by electrophoresis in 2.5% agarose gels, by use of the HT DNA 5000 SE55 kit on a LabChip 90 system (Caliper LifeSciences, Teralfene, Belgium), in 6.5% acrylamide slab gels in a 4300 DNA analyzer (LI-COR Biosciences, Lincoln, NE), by capillary electrophoresis on a MegaBACE DNA analysis system (GE Healthcare Europe GmbH, Diegem, Belgium), and/or in an ABI 3100 genetic analyzer (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands).
For detection on the 4300 DNA analyzer, an IR-700-labeled reverse primer was used in the final PCR. The final PCR mixture was diluted 30 times and was analyzed on the 4300 DNA analyzer according to the manufacturer's instruction. When analysis was performed on a MegaBACE DNA analysis system or an ABI 3100 genetic analyzer, a 6-carboxyfluorescein-labeled forward primer was used in the final PCR. The final PCR mixture was diluted 10 times and was subsequently treated according to the manufacturers' instructions. All clinical samples were analyzed by capillary electrophoresis on a MegaBACE DNA analysis system or an ABI 3100 genetic analyzer.
RESULTS
MLPA reaction.
The MLPA technology allows highly complex analyses of up to 45 targets in a single reaction. However, a standard MLPA reaction requires at least 6,000 copies of target DNA (27). We made some modifications to the MLPA technology to obtain the sensitivity required for the effective detection of infectious pathogens. An RT step was introduced for the conversion of RNA into DNA. This was followed by a PCR to amplify the target DNA, which was then present at levels above the detection level of MLPA. Subsequently, an MLPA reaction was performed and comprised a probe hybridization step, a probe ligation step, and a probe amplification step. A schematic representation of the RespiFinder assay is shown in Fig. 1.
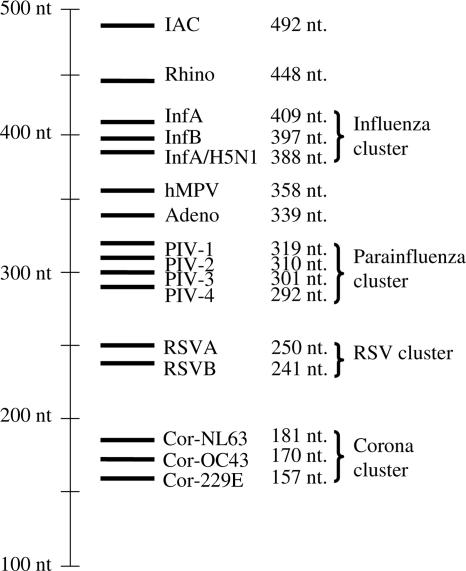
An IAC is included in the assay to discriminate between samples with true-negative results and samples with false-negative results due to PCR failure. This added up to a total of 16 probes. The total lengths of the probes varied from 157 to 492 nucleotides. The sizes of the probes of the related viruses were clustered. Four clusters are distinguished: an influenza virus cluster of about 400 nucleotides, a PIV cluster of about 300 nucleotides, an RSV cluster of about 250 nucleotides, and a coronavirus cluster of about 170 nucleotides. An overview of all probes is presented in Fig. 2.
FIG. 2.
Overview of the RespiFinder probes and their length. nt, nucleotides.
MLPA probes.
An MLPA probe consists of one short synthetic oligonucleotide and one phage M13-derived, long oligonucleotide probe (27). Each oligonucleotide contains a target-specific hybridizing part and one universal priming site. In addition, the long oligonucleotide probe contains a stuffer sequence to facilitate size-based separation. The most conserved region within each viral family was selected to design the two hybridizing parts of the MLPA probe.
The adenovirus group is a very large and heterogeneous group, including 51 different serotypes divided into six subgenera. To cover all six subgenera with one MLPA probe, we combined one long oligonucleotide probe with two synthetic oligonucleotides. This MLPA probe was tested with 15 different adenovirus serotypes covering the six subgenera. Included were serotypes 2, 4, 5, and 7, which are generally more often associated with RTIs (25), and serotypes 8, 9, 12, 19, 30, 31, 35, 37, 40, 48, and 49. All 15 serotypes could be detected by use of this probe (data not shown).
Specificity.
To asses the specificity of the primers and the probes, all PCR primers and MLPA probes were tested in either a monoplex or a multiplex MLPA reaction with different samples. No cross-reactivity among the 15 viral probes and the IAC probe was observed when they were tested with culture samples of individual viruses. In addition, clinical samples which tested positive for other pathogens by routine diagnostic analysis by pathogen-specific PCR assays were analyzed by the RespiFinder assay. These samples contained enterovirus, echovirus, coxsackievirus, and herpes simplex virus type 1, as well as the bacterial pathogens Mycoplasma pneumoniae, Chlamydia pneumoniae, Legionella pneumophila, Bordetella pertussis, Staphylococcus warneri, Staphylococcus intermedius, Staphylococcus haemolyticus, Staphylococcus epidermidis, Enterococcus faecalis, Pseudomonas spp., Haemophilus influenzae, Haemophilus parainfluenzae, and Streptococcus spp. No sample showed any cross-reactivity with the RespiFinder assay probes (data not shown).
Internal control.
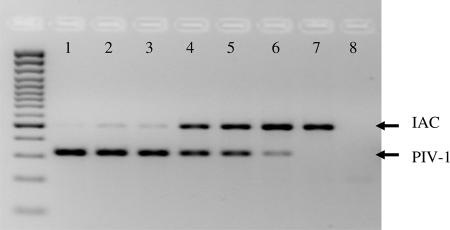
The use of an IAC allows discrimination between a true-negative result and a false-negative result due to a PCR/MLPA failure. However, since the MLPA reaction uses only two primers in the final PCR, the amplification of any respiratory virus-specific MLPA probe competes with the amplification of the IAC MLPA probe. Figure 3 shows the results of the RespiFinder assay with fivefold serial dilutions of a PIV-1 culture, all of which were spiked with an equal amount of IAC. Only a weak IAC signal was detected in the presence of the highest PIV-1 concentration. As the PIV-1 concentration decreased, the IAC signal increased. This demonstrates competition between simultaneously amplified MLPA probes.
FIG. 3.
Detection of the IAC in spiked serial virus dilutions on a 2.5% agarose gel. RespiFinder assay analysis was performed with fivefold serial dilutions of the supernatant of a PIV-1 culture. Lanes: 1, undiluted; 2, diluted 51; 3, diluted 52; 4, diluted 53; 5, diluted 54; 6, diluted 55; 7, diluted 56; 8, blank. Each dilution except the blank was spiked with the IAC.
Analytical sensitivity: RNA transcripts and serial virus dilutions.
First, to determine the analytical sensitivity of the RespiFinder assay, serial dilutions of viral RNA transcripts of all RNA viruses and serial dilutions of a plasmid with the adenovirus target sequence were analyzed by the RespiFinder assay, and the reactions were analyzed on a LI-COR 4300 DNA analyzer. The limit of detection of the 15 viral RNA transcripts did not vary significantly, and for each viral probe an analytical detection limit of 10 to 20 copies was determined.
Second, the sensitivity of the RespiFinder assay was compared to that of the monoplex real-time PCR. Nucleic acid preparations from fivefold serial dilutions of culture supernatants of 13 viruses (no InfA H5N1 or adenovirus cultures were available) were tested. The results are shown in Table 3. In the monoplex real-time PCR, 20 μl of an RNA or a DNA sample was used, whereas 10 μl of an RNA or a DNA sample was used in the preamplification step of the RespiFinder assay. The results indicate that the sensitivity of the RespiFinder assay was comparable to that of the monoplex real-time RT-PCR.
TABLE 3.
Sensitivity of RespiFinder assay compared to that of monoplex real-time RT-PCRa
| Virus | Highest viral dilution detected by:
|
|
|---|---|---|
| RespiFinder assay | Real-time RT-PCR | |
| RSVA | 4 | 4 |
| RSVB | 5 | 5 |
| InfA | 6 | 6 |
| InfB | 7 | 7 |
| Rhino | 7 | 7 |
| Cor-OC43 | 6 | 5 |
| Cor-229E | 5 | 6 |
| Cor-NL63 | 6 | 6 |
| PIV-1 | 6 | 7 |
| PIV-2 | 7 | 7 |
| PIV-3 | 6 | 6 |
| PIV-4 | 6 | 7 |
| hMPV | 5 | 4 |
Fivefold serial dilutions of all indicated viruses were analyzed.
Validation with respiratory specimens.
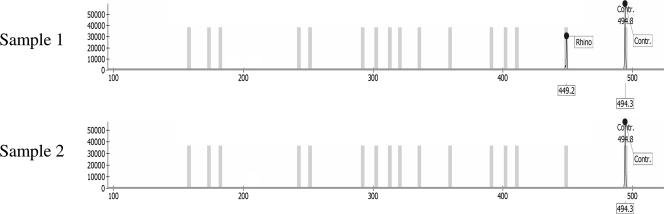
To validate the performance of the RespiFinder assay with clinical samples, 110 nasal lavage fluid specimens from patients with symptoms of RTIs were analyzed retrospectively. All clinical samples were analyzed by capillary electrophoresis on a MegaBACE DNA analysis system or an ABI 3100 genetic analyzer. Typical results of this RespiFinder assay screening and analyses by capillary electrophoresis are shown in Fig. 4. The results of the validation assay are shown in Tables 4 and 5. The RespiFinder assay identified the same virus as that identified by cell culture in 37 of 39 samples (94.9%). The two aberrant results were for samples in which an untypeable influenza virus was detected by cell culture and an InfA was detected by RespiFinder assay and a sample in which PIV-3 was detected by cell culture and hMPV was detected by the RespiFinder assay. The eight adenovirus infections detected were caused by adenovirus type 2 (subgenera C) and adenovirus types 3 and 7 (subgenera B). By cell culture, 71 clinical samples were found to be negative. In 22 (31%) of these culture-negative samples, a virus was identified by the RespiFinder assay. These included nine rhinovirus infections, five RSVA infections, one RSVB infection, two hMPV infections, two InfA infections, one PIV-3 infection, one adenovirus infection, and a rhinovirus and PIV-1 coinfection. In addition, one Cor-NL63 infection and two Cor-OC43 infections were detected by the RespiFinder assay. The coronaviruses were not included in the cell culture screening. Therefore, the coronavirus results of the RespiFinder assay are reported but were not used in the validation of RespiFinder assay. In total, five coinfections were detected by RespiFinder assay. These involved three coinfections of rhinovirus with PIV-1, PIV-3, and hMPV, respectively, and two coinfections of RSVA with adenovirus and RSVB, respectively. No coinfection was detected by cell culture. Overall, the RespiFinder assay detected 66 (60%) viruses in the 110 samples. All viruses involved in the coinfections were counted individually, and the three coronavirus infections were not included. Cell culture detected 39 (35.5%) viruses. This is an increase in diagnostic yield of 24.5%. With two samples, no signal was observed by the RespiFinder assay in the initial screening. Upon retesting, one sample again failed to show any signal.
FIG. 4.
Results of the RespiFinder assay with two clinical samples which were analyzed by capillary electrophoresis. Sample 1 represents a clinical sample with a rhinovirus (Rhino) infection. The electropherogram shows a signal at the position of the rhinovirus probe and the IAC. Sample 2 represents a clinical sample with no viral infection. Only the IAC signal was detected in the electropherograms. Contr, internal amplification control.
TABLE 4.
Analysis of concordant results between RespiFinder assay and cell culture for 110 clinical samplesa
| Cell culture and RespiFinder assay result | No. (%) of samples | Remarks |
|---|---|---|
| Adenovirus | 8 | Including serotypes 2, 3, and 7 |
| hMPV | 2 | |
| InfA | 4 | |
| PIV-1 | 1 | |
| PIV-3 | 4 | 1 CIb with rhinovirus detected by RespiFinder assay |
| Rhinovirus | 5 | 1 CI with hMPV detected by RespiFinder assay |
| RSV | 13 | 7 RSVA (1 CI with adenovirus), 5 RSVB, and 1 CI of RSVA + RSVB detected by RespiFinder assay |
| Negative | 45 | |
| Total | 82 (74.5) |
The results were considered consistent if the virus identified by cell culture was the same virus as that identified by the RespiFinder assay. Additional viruses identified by the RespiFinder assay are indicated. An RSV-positive result by cell culture was considered consistent with the RespiFinder assay result if either RSVA or RSVB was identified by the RespiFinder assay.
CI, coinfection.
TABLE 5.
Analysis of nonconsistent results between RespiFinder assay and cell culturea
| Cell culture result (no. of samples) | RespiFinder assay result | No. (%) of samples |
|---|---|---|
| Untypeable inflenza virus (1) | InfA | 1 |
| PIV-3 (1) | hMPV | 1 |
| Negative (26) | Rhinovirus | 9 |
| Cor-NL63 | 1 | |
| Cor-OC43 | 2 | |
| RSVA | 5 | |
| RSVB | 1 | |
| hMPV | 2 | |
| InfA | 2 | |
| Adenovirus | 1 | |
| PIV-3 | 1 | |
| Rhinovirus + PIV-1 | 1 | |
| Failureb | ||
| Total | 28 (25.5) |
The results were considered nonconsistent if the virus identified by cell culture was not identified by the RespiFinder assay.
No IAC signal.
In addition, 34 clinical samples which were routinely tested by the RSV-specific ICA were retrospectively analyzed by the RespiFinder assay (Table 6). Among the 15 samples RSV positive by the RSV-specific ICA, 12 samples were also RSV positive by the RespiFinder assay. The RespiFinder assay detected InfA in a sample which scored weakly positive by the RSV-specific ICA. The other two samples positive by the RSV-specific ICA were negative and failed to amplify the internal control in the RespiFinder assay. Retesting of the last sample again showed no amplification of the internal control. Three (15.8%) of the 19 samples which tested negative by ICA were also negative by the RespiFinder assay. In six (31.6%) samples, an RSV infection was detected by the RespiFinder assay, and these comprised four RSVA infections and two RSVB infections. In the remaining 10 (52.6%) samples, additional virus infections were detected by the RespiFinder assay. These included four InfA infections, four rhinovirus infections, one Cor-OC43 infection, and a Cor-OC43 and Cor-NL63 coinfection.
TABLE 6.
Comparison of RespiFinder assay and RSV-specific ICA results for 34 clinical samples
| RSV-specific ICA score (no. of samples) | RespiFinder assay result (no. of samples)
|
|
|---|---|---|
| RSV score | Additional virus(es) detected | |
| Positive (15) | Positive (12) | RSVA (6) |
| RSVB (5) | ||
| RSVB + rhinovirus (1) | ||
| Negative (2) | InfA (1) | |
| Negative (1) | ||
| Failurea (1) | ||
| Negative (19) | Negative (13) | Negative (3) |
| InfA (4) | ||
| Rhinovirus (4) | ||
| Cor-OC43 (1) | ||
| Cor-OC43 + Cor-NL63 (1) | ||
| Positive (6) | RSVA (4) | |
| RSVB (2) | ||
No IAC signal.
The sensitivity and specificity of the RespiFinder assay were assessed in relation to the results of cell culture and ICA (Table 7). In agreement with the cell culture results, the RespiFinder assay identified 10 samples with adenovirus infections (9.1%), 6 with hMPV infections (5.5%), 7 with InfA infections (6.4%), 2 with PIV-1 infections (1.8%), 5 with PIV-3 infections (4.5%), 16 with rhinovirus infections (14.5%), and 19 with RSV infections (17.3%). In addition, the RespiFinder assay detected 27 additional positive samples, including 2 positive for adenovirus, 4 for hMPV, 2 for InfA, 1 for PIV-1, 1 for PIV-3, 11 for rhinovirus, and 6 for RSV. One sample with a false-negative result for PIV-3 was observed by the RespiFinder assay. Compared with the results of ICA, the RespiFinder assay detected 18 samples positive for RSV (52.9%), corresponding to a specificity of 82.4% and a sensitivity of 80%. Six additional RSV-positive samples were observed by the RespiFinder assay. The results for three samples which scored RSV positive by ICA were not confirmed by the RespiFinder assay.
TABLE 7.
Sensitivity and specificity of RespiFinder assay compared with those of cell culture and an RSV-specific ICAa
| Virus | No. of samples with the following CC or CC/ICA result and RF resultb:
|
Specificity (%) | Sensitivity (%) | |||
|---|---|---|---|---|---|---|
| CC+ or CC+/ICA+ and RF+ | CC− or CC−/ICA− and RF+ | CC+ or CC+/ICA+ and RF− | CC− or CC−/ICA− and RF− | |||
| Adenovirus | 8 | 2 | 0 | 101 | 98.2 | 100 |
| hMPV | 2 | 4 | 0 | 105 | 96.4 | 100 |
| InfA | 5 | 2 | 0 | 104 | 98.2 | 100 |
| PIV-1 | 1 | 1 | 0 | 109 | 99.1 | 100 |
| PIV-3 | 4 | 1 | 1 | 105 | 99.1 | 80 |
| Rhinovirus | 5 | 11 | 0 | 95 | 90.1 | 100 |
| RSV | 13/12 | 6/6 | 0/2 | 92/13 | 94.6/81.8c | 100/85.7c |
The RespiFinder assay results for coronaviruses were not validated.
CC, cell culture; RF, RespiFinder assay; +, positive result; −, negative result.
Data represent the results by cell culture/ICA.
DISCUSSION
The number of possible viral agents involved in RTIs is high, and new viruses are still emerging. In recent years, five new viruses, including human bocavirus (1), the SARS coronavirus (4), hMPV (31), and coronaviruses NL63 (32) and HKU1 (36), were identified to play a role in RTIs. Although the gold standard for the laboratory detection of respiratory viruses is viral culture, monitoring of this increasing number of respiratory viruses solely by viral culture is not possible for practical reasons. PCR is a fast and sensitive alternative. However, the use of conventional PCR techniques is not practical in a study for the detection of a broad spectrum of pathogens due to the handling time and the high reagent costs (19).
Here we present a new multiparameter test that is based on the MLPA technology and that is able to detect 15 respiratory viruses simultaneously. In order to obtain the required sensitivity, the MLPA technology was adapted by incorporating a preamplification step. This preamplification step involves an RT step and a 30-cycle target amplification step. Due to the introduction of the preamplification step, the hybridization time in the MLPA reaction could be reduced to 1 h, while the original MLPA protocol required a hybridization time of 16 h. As a result, the whole RespiFinder assay could be completed within a working day.
In the initial experiments, the RespiFinder assay products were analyzed on 2.5% agarose gels. However, due to the complexity of the RespiFinder assay, the resolution of the agarose gel was insufficient to distinguish all products amplified by MLPA. The RespiFinder assay reactions were also analyzed on a LabChip 90 system from Caliper LifeSciences. The resolution of this system was also not sufficient for the nonambiguous identification of all individual products amplified by MLPA. However, due to the clustering of probes of more related viruses, use of the LabChip 90 system allowed the detection of the individual probes for rhinovirus, adenovirus, and hMPV and the four probe clusters for influenza virus, PIV, RSV, and coronavirus. On high-resolution electrophoresis systems, such as the LI-COR 4300 DNA analyzer, the MegaBACE DNA analysis system, and the ABI 3100 genetic analyzer, all individual probes could be clearly distinguished.
Well-designed MLPA probes have the ability to discriminate between single-nucleotide polymorphisms (27). The viral MLPA probes require a more generic design, as viruses, especially RNA viruses, show a great deal of genetic variability. Therefore, a major concern in the design of the MLPA probes was the compatibility of virus-specific probes with as many known viral sequences as possible, including those of the serotypes common among humans. However, this generic design should not compromise the specificity of the probe. The probes were positioned in well-conserved regions. No mismatch within 5 nucleotides from the ligation site was tolerated. With the adenovirus-specific probe two short synthetic oligonucleotides were combined with one M13-derived long oligonucleotide probe to enclose all 51 serotypes of the heterogeneous adenovirus group. No cross-reactivity was detected with any of the probes.
The sensitivity and the specificity of the RespiFinder assay were assessed by comparison of the results of the RespiFinder assay to those of cell culture obtained with 110 clinical samples. All samples were spiked with the IAC. The spiking was done after addition of the lysis buffer in order to prevent degradation of the IAC. For one (1%) sample, the RespiFinder assay failed to produce a result. The RespiFinder assay detected 27 (24.5%) viruses which were missed by cell culture. These included 2 adenoviruses, 4 hMPVs, 2 InfAs, 1 PIV-1, 1 PIV-3, 11 rhinoviruses, and 6 RSVs. This increase in diagnostic yield is in accordance with the findings of other studies that used molecular methods (10, 23). One sample with a false-negative result for PIV-3 was observed by the RespiFinder assay. Among the eight adenoviruses detected, three were of different serotypes (serotypes 2, 3, and 7) representing two different subgenera (subgenera B and C). This illustrates the ability of the generic adenovirus probe to detect different serotypes in clinical samples. The RespiFinder assay found an incidence of coinfection of 7.8%, whereas cell culture did not detect any double infections. The percentage of mixed infections that we detected is in agreement with the percentages detected in other studies, which showed incidences of coinfections of 3.4% (34), 6.1% (21), and 18.6% (3).
Compared to the results of a commercial ICA-based rapid assay for RSV with 34 clinical samples, the RespiFinder assay detected 18 (52.9%) RSV infections, corresponding to a specificity of 82.4% and a sensitivity of 80%. The results for two RSV-positive samples were not confirmed. This included one sample which scored weakly positive by the rapid assay for RSV. The RespiFinder assay detected an InfA infection in this sample. In addition, the RespiFinder assay detected six additional RSV-positive samples. Additional viruses were detected in 52.6% of the ICA RSV-negative samples, including InfA, rhinovirus, Cor-OC43, and Cor-NL63.
In conclusion, we have developed a new multiparameter respiratory assay based on the MLPA technique for the detection of 15 respiratory viruses plus an ICA. The analytical sensitivity of the RespiFinder assay matches the sensitivity of the monoplex real-time RT-PCR. The validation with clinical samples shows that the RespiFinder assay gives an excellent overall performance. The assay does not need specialized equipment but makes use of the PCR and capillary electrophoresis systems generally available in molecular laboratories. The assay can be performed within one working day and is suitable for implementation in a diagnostic setting. Implementation of this assay for routine diagnostics allows the fast analysis of samples for a broad spectrum of pathogens at a reduced cost compared to the costs of individual monoplex real-time RT-PCR assays.
At present, the full analytical power of the RespiFinder assay can be used only when the results are analyzed on a high-resolution system like a capillary electrophoresis system. However, by adapting the design of the MLPA probes, it will also be possible to identify most probes on LabChip systems, like the Caliper LabChip 90 system, the Experion system (Bio-Rad Laboratories, Hercules, CA), or the 2100 bioanalyzer (Agilent Technologies, Foster City, CA). The MLPA technology allows the highly complex analysis of up to 45 fragments. This gives room for future expansion of the number of probes in the RespiFinder assay. Recently, four bacterial probes, including probes for Mycoplasma pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and Bordetella pertussis, have been added to the RespiFinder assay. This allows the detection in a clinical sample of 19 pathogens and one IAC in one reaction. In addition, for better implementation in a diagnostic setting, the RespiFinder assay protocol was recently adapted by combining the probe ligation step and the probe amplification step in a single reaction. As a result, the RespiFinder assay protocol consists at present of four steps, i.e., preamplification (one-step RT-PCR), probe hybridization, probe ligation/amplification, and probe detection.
Acknowledgments
We thank Bert Niesters (Erasmus Medical Centre, Rotterdam) for the serial dilutions of viral RNA and performing the monoplex real-time PCR analysis. We thank Eric Claas (Leiden University Medical Centre) for providing us with the DNA of the different adenovirus serotypes. Sandra Bokkers (Canisius Wilhelmina Hospital Nijmegen) is acknowledged for technical assistance. In addition, we thank Lothar Kruska and André Frontzek (Stein + Kollegen, Mönchengladbach, Germany) for assisting us with the cross-reactivity testing.
The research described here has been facilitated by a grant provided by the Dutch Ministry of Economic Affairs (grant TSGE3074).
Footnotes
Published ahead of print on 6 February 2008.
REFERENCES
- 1.Allander, T., M. T. Tammi, M. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 10212891-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beld, M., R. Minnaar, J. Weel, C. Sol, M. Damen, H. van der Avoort, P. Wertheim-van Dillen, A. van Breda, and R. Boom. 2004. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J. Clin. Microbiol. 423059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coiras, M. T., J. C. Aguilar, M. L. Garcia, I. Casas, and P. Perez-Brena. 2004. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested-PCR assays. J. Med. Virol. 72484-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 3481967-1976. [DOI] [PubMed] [Google Scholar]
- 5.Effler, P. V., M. C. Ieong, T. Tom, and M. Nakata. 2002. Enhancing public health surveillance for influenza virus by incorporating newly available rapid diagnostic tests. Emerg. Infect. Dis. 823-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eldering, E., C. A. Spek, H. L. Aberson, A. Grummels, I. A. Derks, A. F. de Vos, C. J. McElgunn, and J. P. Schouten. 2003. Expression profiling via novel multiplex assay allows rapid assessment of gene regulation in defined signalling pathways. Nucleic Acids Res. 31e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erdman, D. D., G. A. Weinberg, K. M. Edwards, F. J. Walker, B. C. Anderson, J. Winter, M. Gonzalez, and L. J. Anderson. 2003. GeneScan reverse transcription-PCR assay for detection of six common respiratory viruses in young children hospitalized with acute respiratory illness. J. Clin. Microbiol. 414298-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falsey, A. R., E. E. Walsh, and F. G. Hayden. 2002. Rhinovirus and coronavirus infection-associated hospitalizations among older adults. J. Infect. Dis. 1851338-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.File, T. M. 2003. Community-acquired pneumonia. Lancet 3621991-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freymuth, F., A. Vabret, D. Cuvillon-Nimal, S. Simon, J. Dina, L. Legrand, S. Gouarin, J. Petitjean, P. Eckart, and J. Brouard. 2006. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J. Med. Virol. 781498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenberg, S. B. 2002. Respiratory viral infections in adults. Curr. Opin. Pulm. Med. 8201-208. [DOI] [PubMed] [Google Scholar]
- 12.Grondahl, B., W. Puppe, A. Hoppe, I. Kuhne, J. A. Weigl, and H. J. Schmitt. 1999. Rapid identification of nine microorganisms causing acute respiratory tract infections by single-tube multiplex reverse transcription-PCR: feasibility study. J. Clin. Microbiol. 371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunson, R. N., T. C. Collins, and W. F. Carman. 2005. Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions. J. Clin. Virol. 33341-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoorfar, J., B. Malorny, A. Abdulmawjood, N. Cook, M. Wagner, and P. Fach. 2004. Practical considerations in design of internal amplification controls for diagnostic PCR assays. J. Clin. Microbiol. 421863-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoover, D. M., and J. Lubkowski. 2002. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 30e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langerak, P., A. O. Nygren, J. P. Schouten, and H. Jacobs. 2005. Rapid and quantitative detection of homologous and non-homologous recombination events using three oligonucleotide MLPA. Nucleic Acids Res. 33e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, H., M. A. McCormac, R. W. Estes, S. E. Sefers, R. K. Dare, J. D. Chappell, D. D. Erdman, P. F. Wright, and Y. W. Tang. 2007. Simultaneous detection and high-throughput identification of a panel of RNA viruses causing respiratory tract infections. J. Clin. Microbiol. 452105-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez, A. D., C. D. Mathers, M. Ezzati, D. T. Jamison, and C. J. Murray. 2006. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 3671747-1757. [DOI] [PubMed] [Google Scholar]
- 19.Luna, L. K., M. Panning, K. Grywna, S. Pfefferle, and C. Drosten. 2007. Spectrum of viruses and atypical bacteria in intercontinental air travelers with symptoms of acute respiratory infection. J. Infect. Dis. 195675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maertzdorf, J., C. K. Wang, J. B. Brown, J. D. Quinto, M. Chu, M. de Graaf, B. G. van den Hoogen, R. Spaete, A. D. Osterhaus, and R. A. Fouchier. 2004. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J. Clin. Microbiol. 42981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molenkamp, R., A. van der Ham, J. Schinkel, and M. Beld. 2007. Simultaneous detection of five different DNA targets by real-time TaqMan PCR using the Roche LightCycler480: application in viral molecular diagnostics. J. Virol. Methods 141205-211. [DOI] [PubMed] [Google Scholar]
- 22.Nygren, A. O., N. Ameziane, H. M. Duarte, R. N. Vijzelaar, Q. Waisfisz, C. J. Hess, J. P. Schouten, and A. Errami. 2005. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 33e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oosterheert, J. J., A. M. van Loon, R. Schuurman, A. I. Hoepelman, E. Hak, S. Thijsen, G. Nossent, M. M. Schneider, W. M. Hustinx, and M. J. Bonten. 2005. Impact of rapid detection of viral and atypical bacterial pathogens by real-time polymerase chain reaction for patients with lower respiratory tract infection. Clin. Infect. Dis. 411438-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osiowy, C. 1998. Direct detection of respiratory syncytial virus, parainfluenza virus, and adenovirus in clinical respiratory specimens by a multiplex reverse transcription-PCR assay. J. Clin. Microbiol. 363149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pehler-Harrington, K., M. Khanna, C. R. Waters, and K. J. Henrickson. 2004. Rapid detection and identification of human adenovirus species by Adenoplex, a multiplex PCR-enzyme hybridization assay. J. Clin. Microbiol. 424072-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Puppe, W., J. A. Weigl, G. Aron, B. Grondahl, H. J. Schmitt, H. G. Niesters, and J. Groen. 2004. Evaluation of a multiplex reverse transcriptase PCR ELISA for the detection of nine respiratory tract pathogens. J. Clin. Virol. 30165-174. [DOI] [PubMed] [Google Scholar]
- 27.Schouten, J. P., C. J. McElgunn, R. Waaijer, D. Zwijnenburg, F. Diepvens, and G. Pals. 2002. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speers, D. J. 2006. Clinical applications of molecular biology for infectious diseases. Clin. Biochem. Rev. 2739-51. [PMC free article] [PubMed] [Google Scholar]
- 29.Storch, G. A. 2003. Rapid diagnostic tests for influenza. Curr. Opin. Pediatr. 1577-84. [DOI] [PubMed] [Google Scholar]
- 30.Templeton, K. E., S. A. Scheltinga, M. F. Beersma, A. C. Kroes, and E. C. Claas. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 421564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, R. de Groot, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viazov, S., F. Ratjen, R. Scheidhauer, M. Fiedler, and M. Roggendorf. 2003. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J. Clin. Microbiol. 413043-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vicente, D., G. Cilla, M. Montes, and E. Perez-Trallero. 2003. Human metapneumovirus and community-acquired respiratory illness in children. Emerg. Infect. Dis. 9602-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijgen, L., E. Keyaerts, E. Moes, P. Maes, G. Duson, and M. Van Ranst. 2005. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 435452-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]