Abstract
In the last 2 decades, a variety of different molecular typing methods have been developed to differentiate strains of Mycobacterium avium subsp. paratuberculosis. The most successful techniques are based on insertion sequences, repetitive loci, comparative genomics, or single nucleotide polymorphisms. In the present study, we chose to examine whether a single M. avium subsp. paratuberculosis gene could serve as a means of differentiation of a variety of isolates. The MAP1506 gene locus encodes a member of the polymorphic PPE protein family that has putative roles relevant to M. avium subsp. paratuberculosis pathogenicity. The MAP1506 locus was sequenced from a collection of 58 M. avium subsp. paratuberculosis isolates from different sources, hosts, and typing profiles. Following sequence alignment and analysis, it was found that bovine (type II) strains of M. avium subsp. paratuberculosis consistently differed from ovine (type I) and intermediate (type III) strains in seven and eight nucleotides, respectively. Polymorphic regions of the MAP1506 locus were selected for analysis by denaturing gradient gel electrophoresis, allowing visual discrimination of the three subtypes of M. avium subsp. paratuberculosis isolates. This is the first report describing the use of PCR and denaturing gradient gel electrophoresis on a single gene as a method to distinguish types I, II, and III of M. avium subsp. paratuberculosis.
Mycobacterium avium subsp. paratuberculosis is the causative organism of Johne's disease (or paratuberculosis), a debilitating chronic gastroenteritis in ruminants (50). Animals typically become infected by the fecal-oral route in the first few months of life. The chronic wasting and profuse diarrhea that characterize clinical paratuberculosis are not usually observed until three or more years following infection (12). Paratuberculosis is prevalent in domestic animals worldwide and has a significant impact on the global economy, including the Canadian economy (10, 38). M. avium subsp. paratuberculosis has also been implicated in Crohn's disease in humans (22).
Molecular techniques for strain typing based on mobile genetic elements, repetitive elements, and single nucleotide polymorphisms (SNPs) of M. avium subsp. paratuberculosis have been well explored in the last decade (reviewed by Motiwala et al. [39]). The most widely used method to type M. avium subsp. paratuberculosis isolates is restriction fragment length polymorphism (RFLP) analysis, with detection of polymorphisms by hybridization to an IS900 probe (IS900 RFLP) (49). This technique is able to distinguish bovine (type II), ovine (type I), and intermediate (type III) isolates (42) but is slow, technically demanding, and applicable only to cultivable strains of M. avium subsp. paratuberculosis, as it requires a considerable amount of genomic DNA. Moreover, IS900 RFLP requires analysis of complex banding patterns and has limited discriminatory power (39). Another technique for typing M. avium subsp. paratuberculosis is pulsed-field gel electrophoresis of digested genomic DNA (21, 48, 49); however, this technique suffers from many of the same limitations associated with IS900 RFLP. IS1311 PCR-restriction endonuclease analysis (REA), described by Marsh et al. (32), is also used to type M. avium subsp. paratuberculosis. While this technique has the benefit of employing a PCR step to reduce the need for a large quantity of starting DNA, it suffers in its low discriminatory power, as only the bovine (C) and ovine (S) types can be differentiated. Thus, rapid molecular typing methods with discriminatory power greater than that seen for methods in current use need to be assessed as alternatives when studying the genetic diversity in M. avium subsp. paratuberculosis.
Mycobacteria contain two unique polymorphic protein families, the PE and PPE proteins, which are unknown for any other species. These families are particularly expanded in the pathogenic mycobacterial species. The names PE and PPE are derived from the motifs Pro-Glu and Pro-Pro-Glu, respectively, found in conserved domains near the N termini of these proteins. Although no precise function is known for any member of these families, some members in M. tuberculosis have been found to associate with the cell wall (7, 19) and to influence interactions with other cells (7). It has been suggested that some PE/PPE proteins play a role in immune evasion and antigenic variation (4, 6, 13, 18). Members of the PE and PPE families have also been linked to virulence (31, 44), and some PPE proteins have been found to be immunodominant antigens (11). There are 10 PE and 37 PPE genes in the M. avium subsp. paratuberculosis genome (comprising 1% of the genome) (26, 30), but no information on their putative roles exists.
The aim of this study was to examine the polymorphic variability of a PPE protein family gene locus, namely, the MAP1506 locus, in a collection of M. avium subsp. paratuberculosis isolates from different hosts and geographic locations and of different IS900 RFLP profiles. The MAP1506 locus was chosen, as it is one of the few PPE gene loci that was previously discovered to be expressed in vitro and has its protein product surface exposed on the cell wall of M. avium subsp. paratuberculosis (J. De Buck, unpublished results). We hypothesized that SNPs would be present in the MAP1506 gene locus and allow differentiation of M. avium subsp. paratuberculosis isolates. A secondary aim of this study was to apply denaturing gradient gel electrophoresis (DGGE), a widely used method for mutation analysis (25) and studies of microbial diversity in multispecies communities (41), for the visualization of SNPs in the MAP1506 locus.
MATERIALS AND METHODS
M. avium subsp. paratuberculosis isolates.
A set of seven type II, three type I, and three type III M. avium subsp. paratuberculosis strains (15, 42) (Table 1) with different IS900 RFLP profiles (M. Behr [McGill University, Canada] and D. Collins [AgResearch, New Zealand], personal communication) was provided by D. Collins. Another 45 isolates from 11 countries on 3 continents (Table 1) and from a variety of hosts were kindly provided by S. Sreevatsan (Veterinary Population Medicine Department, College of Veterinary Medicine, University of Minnesota), S. Naser (Department of Molecular Biology and Microbiology, University of Central Florida), S. Nielsen (The Royal Veterinary and Agricultural University, Denmark), R. Juste (Animal Health and Production Department, Neiker Tecnalia, Spain), K. Stevenson (Moredun Research Institute, Scotland), M. Behr (McGill University, Canada), M. Ngeleka (Prairie Diagnostics Services, Canada), and D. Collins (AgResearch, New Zealand).
TABLE 1.
SNPs in the MAP1506 gene locus of 58 M. avium subsp. paratuberculosis isolates, including three type I, seven type II, and three type III isolates with different IS900 RFLP profiles
| Isolate | Host animal | Country | IS1311 PCR-REA typea | IS900 RFLP typeb | IS900 RFLP minor pattern differencesc | SNP at:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bp 293 | bp 328 | bp 344 | bp 411 | bp 542 | bp 944 | bp 945 | bp 946 | ||||||
| TC1613 | Bovine | United States | C | II | b1 | T | G | G | T | T | T | G | G |
| 6772 | Caprine | New Zealand | C | II | b2 | T | G | G | T | T | T | G | G |
| 5979 | Cervine | New Zealand | C | II | b3 | T | G | G | T | T | T | G | G |
| 7428 | Bovine | New Zealand | C | II | b4 | T | G | G | T | T | T | G | G |
| 87/8880 | Bovine | Australia | C | II | b5 | T | G | G | T | T | T | G | G |
| 6601 | Bovine | Australia | C | II | b6 | T | G | G | T | T | T | G | G |
| 316F | Bovine | United Kingdom | C | II | b7 | T | G | G | T | T | T | G | G |
| 6756 | Ovine | New Zealand | S | I | s1 | A | T | G | G | C | -d | - | - |
| 86-45 | Ovine | Canada | S | III | s2 | A | T | A | G | C | - | - | - |
| P133/79 | Ovine | Faeroe Islands | S | I | s3 | A | T | G | G | C | - | - | - |
| P465 | Ovine | Iceland | S | III | s4 | A | T | A | G | C | - | - | - |
| 6759 | Ovine | New Zealand | S | I | s5 | A | T | G | G | C | - | - | - |
| 85/14 | Ovine | Canada | S | III | s6 | A | T | A | G | C | - | - | - |
| LN20 | Porcine | Canada | S | III | s2 | A | T | A | G | C | - | - | - |
| 269ov | Ovine | Spain | S | NDe | ND | A | T | A | G | C | - | - | - |
| 11G | Ovine | Spain | S | ND | ND | A | T | A | G | C | - | - | - |
| Ovicap18 | Caprine | Spain | S | ND | ND | A | T | A | G | C | - | - | - |
| 311 | Caprine | Spain | S | ND | ND | A | T | A | G | C | - | - | - |
| M173/04 | Cervine | Netherlands | S | ND | ND | A | T | A | G | C | - | - | - |
| M214/04 | Cervine | Czech Republic | S | ND | ND | A | T | A | G | C | - | - | - |
| 571 | Leporine | Scotland | C | ND | ND | T | G | G | T | T | T | G | G |
| 834 | Bovine | Spain | C | ND | ND | T | G | G | T | T | T | G | G |
| 791 | Bovine | Spain | C | ND | ND | T | G | G | T | T | T | G | G |
| 808 | Bovine | Spain | C | ND | ND | T | G | G | T | T | T | G | G |
| Bison10.3 | Bison | United States | Bf | ND | ND | T | G | G | T | T | T | G | G |
| ATCC19698g | Bovine | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| K10 | Bovine | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| D060162 | Bovine | Canada | C | ND | ND | T | G | G | T | T | T | G | G |
| D0616257 | Bovine | Canada | C | ND | ND | T | G | G | T | T | T | G | G |
| D0624010 | Bovine | Canada | C | ND | ND | T | G | G | T | T | T | G | G |
| D0635848 | Bovine | Canada | C | ND | ND | T | G | G | T | T | T | G | G |
| R0629132 | Bovine | Canada | C | ND | ND | T | G | G | T | T | T | G | G |
| R0632107 | Bovine | Canada | C | ND | ND | T | G | G | T | T | T | G | G |
| R0636342 | Bovine | Canada | C | ND | ND | T | G | G | T | T | T | G | G |
| F76 | Ovine | Scotland | C | ND | ND | T | G | G | T | T | T | G | G |
| JD143 | Ovine | Scotland | C | ND | ND | T | G | G | T | T | T | G | G |
| JD146 | Ovine | Scotland | C | ND | ND | T | G | G | T | T | T | G | G |
| JD18 | Bovine | Scotland | C | ND | ND | T | G | G | T | T | T | G | G |
| JD29 | Ovine | Scotland | C | ND | ND | T | G | G | T | T | T | G | G |
| M48/04 | Ovine | Scotland | C | ND | ND | T | G | G | T | T | T | G | G |
| M212/04 | Cervine | Czech Republic | C | ND | ND | T | G | G | T | T | T | G | G |
| M153/C | Caprine | Scotland | C | ND | ND | T | G | G | T | T | T | G | G |
| Map99 | Bovine | Denmark | C | ND | ND | T | G | G | T | T | T | G | G |
| Map103 | Bovine | Denmark | C | ND | ND | T | G | G | T | T | T | G | G |
| V20683474 | Bovine | Denmark | C | ND | ND | T | G | G | T | T | T | G | G |
| V20683587 | Bovine | Denmark | C | ND | ND | T | G | G | T | T | T | G | G |
| 9319 | Bovine | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| 9346 | Bovine | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| 7300 | Bovine | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| 9286 | Bovine | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| 9287 | Bovine | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| 9354 | Bovine | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| Ben | Human | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| Map3 | Human | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| Linda | Human | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| Map4 | Human | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| Map5 | Human | United States | C | ND | ND | T | G | G | T | T | T | G | G |
| 04-4531 | Bovine | Canada | C | II | ND | T | G | G | T | T | T | G | G |
IS1311 PCR-REA according to the method of Motiwala et al. (40).
Discrimination based on IS900 RFLP minor pattern differences (M. Behr, McGill University, Canada, and D. Collins, AgResearch, New Zealand, personal communication).
-, nucleotide deletion compared to the full genome sequence of M. avium subsp. paratuberculosis K10 (30).
ND, not determined.
IS1311 type B (48).
American Type Culture Collection (www.atcc.org).
Genomic DNA extraction and MAP1506 locus PCR.
M. avium subsp. paratuberculosis cultures were grown in Middlebrook 7H9 broth (Becton Dickinson, Oakville, ON, Canada) supplemented with 10% oleic acid albumin dextrose complex (Becton Dickinson), 0.5% glycerol, and 2 mg/liter mycobactin J (Allied Monitor, MO). Lysis was performed by boiling for 30 min in 10 mM Tris-HCl (pH 7.4) containing 1 mM EDTA and 1% Triton X-100. Genomic DNA was purified with the DNeasy blood and tissue kit (Qiagen, Mississauga, Ontario, Canada). MAP1506 gene loci (1,224 bp) were amplified by PCR from all isolates (Map1506F, 5′-GAGTCAATGATGTTGGATTATGG-3′; Map1506R, 5′-CAATTCCGGATGACACTGG-3′). PCRs (50-μl volumes) were performed with high-fidelity platinum Taq polymerase (Invitrogen) and contained 2 μl template DNA, each deoxynucleoside triphosphate at a concentration of 200 μM, 20 pmol of each primer, 5 μl of the manufacturer's PCR buffer containing MgCl2 (final concentration of MgCl2, 1.5 mM), and 1.75 U of Taq polymerase. PCR conditions were denaturation at 94°C for 5 min followed by 30 cycles of PCR with denaturation at 94°C for 45 s, annealing at 55°C for 1 min, and extension at 72°C for 2 min. The final extension time was 10 min at 72°C. The PCR products were sequenced and aligned with the MAP1506 gene locus sequence of the K10 strain (nucleotides 1653138 to 1654361; NCBI accession number AE016958.1) by use of DNAMAN 5.2.9 (Lynnon Bio-Soft, Quebec, Canada).
IS1311 PCR-REA.
Genomic DNA (prepared as described above) was used as the template in the PCR amplification of a 1,259-bp fragment of the IS1311 insertion sequence (40, 48). The PCR product was digested with the restriction endonuclease HinfI (Invitrogen) and separated on a 1.5% agarose gel as previously described (40, 48). Some copies of IS1311 from C strains carry a point mutation creating a recognition site for the restriction endonuclease HinfI, resulting in an extra band on the gel (32).
DGGE.
Two pairs of primers were used to amplify different regions of the MAP1506 locus by PCR. The template for these reactions was the entire MAP1506 gene locus, which had been previously amplified using a high-fidelity Taq polymerase. A 288-bp fragment of the MAP1506 gene locus (comprising nucleotides 1653357 to 1653644 of the M. avium subsp. paratuberculosis K10 strain; NCBI accession number AE016958.1) was amplified with a GC-clamp (shown in bold) on the forward primer (DGGE1F, 5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGGTGGGGTGGATGAGTACGAC-3′; DGGE1R, 5′-CTGAAACGGCGTCACCTTAC-3′). A 259-bp fragment of the MAP1506 gene locus (comprising nucleotides 1653482 to 1653740 of the M. avium subsp. paratuberculosis K10 strain; NCBI accession number AE016958.1) was amplified with a GC-clamp (shown in bold) on the reverse primer (DGGE2F, 5′-GATGCAGGTCTCGCAGTTG-3′; DGGE2R, 5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGGCTTGGTGGTGTTGGTCGAG-3′). PCRs (50-μl volumes) were performed with the Roche Expand high-fidelity PCR system and contained 2 μl template DNA, each deoxynucleoside triphosphate at a concentration of 200 μM, 15 pmol of each primer, 4% (vol/vol) dimethyl sulfoxide, 5 μl of the manufacturer's PCR buffer containing MgCl2 (final concentration of MgCl2, 1.5 mM), and 1.75 U of Taq polymerase. PCR conditions were denaturation at 94°C for 4 min followed by 30 cycles of PCR with denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min. The final extension time was 10 min at 72°C. The products were separated by DGGE using a DCode universal mutation detection system (Bio-Rad, Ontario, Canada) and 16- by 16-cm by 1-mm gels. Eight-percent polyacrylamide gels were prepared and electrophoresed with 1× Tris-acetate-EDTA buffer. The gels contained a 55 to 65% (DGGE1 product) or a 50 to 65% (DGGE2 product and DGGE1/2 products combined) denaturing gradient of urea and formamide that increased in the direction of electrophoresis. A 100% denaturing solution contained 40% (vol/vol) formamide and 7.0 M urea. Electrophoresis was performed at a constant 75 V for 19 h at 60°C. Gels were stained in 0.1% ethidium bromide for 30 min and visualized with UV light.
Nucleotide sequence accession numbers.
MAP1506 gene locus sequences for types I and III were deposited in GenBank under accession numbers EU200348 and EU200350, respectively.
RESULTS
IS1311 PCR-REA and sequence analysis of MAP1506 gene loci from various isolates.
IS1311 PCR-REA was performed on all M. avium subsp. paratuberculosis isolates (Table 1) to confirm C and S subtypes by use of a validated methodology.
Sequencing of the MAP1506 gene loci from the first 13 isolates shown in Table 1 (7 type II, 3 type I, and 3 type III) revealed a number of consistent nucleotide substitutions (nucleotides 293, 328, 411, and 542) between the C and S subtypes (Table 1). The transition of nucleotide 344 was not present in all S strains but was restricted to those with IS900 RFLP profiles s2, s4, and s6 (Table 1), corresponding to IS900 RFLP type III. A complete codon comprising nucleotides 944 to 946 was absent from all S isolates. All SNPs were nonsynonymous, leading to conservative amino acid substitutions. The MAP1506 gene locus was sequenced in an additional collection of 45 M. avium subsp. paratuberculosis isolates from different origins, comprising 37 C types, 1 B type (47), and 7 S types (according to IS1311 PCR-REA typing). These sequencing results reliably confirmed the presence of the SNPs that differentiate between type I, type II, and type III isolates.
DGGE.
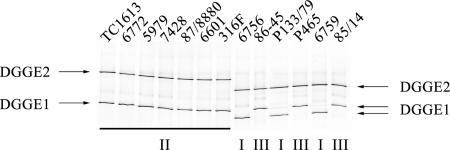
The sequencing data were used to select polymorphic regions of the MAP1506 locus that would be suitable for subsequent PCR and DGGE analysis. A region of the MAP1506 locus encompassing the SNPs at nucleotide positions 293, 328, 344, and 411 (DGGE1 amplicon) was first selected for analysis but did not possess the discriminatory power to distinguish all C and S subtypes. A second region of the MAP1506 locus encompassing T→G (nucleotide 411) and T→C (nucleotide 542) transitions (DGGE2 amplicon) allowed discrimination of the C and S subtypes. DGGE1 and DGGE2 PCR amplicons were combined and electrophoresed on a single DGGE to demonstrate the discrimination of C isolates from two subtypes of S isolates (Fig. 1), corresponding to IS900 RFLP types II, I, and III, respectively.
FIG. 1.
DGGE1 and DGGE2 PCR products separated by DGGE. Using this technique, types I, II, and III of M. avium subsp. paratuberculosis can be differentiated. Isolate names are shown and identified as type I, II, or III.
DISCUSSION
Genomic differences between C and S subtypes of M. avium subsp. paratuberculosis have been previously demonstrated. Large-scale genomic deletions have been described for S strains (33, 34) and three smaller deletions totaling 978 bp were found in C strains (20). In a recent study comparing a C and an S isolate of M. avium subsp. paratuberculosis, 11 SNPs were discovered in a region covering 12 kb (35). Another study examined polymorphisms in the gyrA (2,233 bp) and gyrB (1,838 bp) genes in multiple type I, II, and III strains and found four and five SNPs, respectively (9). In the current study, we found four SNPs and one codon deletion in a 1,224-bp region of the M. avium subsp. paratuberculosis PPE gene, the MAP1506 locus, consistently differing between all C (n = 45) and S (n = 13) isolates that were examined. The results from our study and the other recent studies (17, 35) comparing the C and S strains of M. avium subsp. paratuberculosis suggest that SNPs may be quite common between the two subtypes. Clearly, a more accurate understanding of the number of SNPs between strains or subspecies will be achieved when larger regions or whole genomes are compared.
All of the conserved MAP1506 locus polymorphisms that were found in this study in the ovine subtype S are also present in two MAP1506 locus homologs (MAV_2924 and MAV_2926) in the genetically similar (3) M. avium subsp. avium. This finding is in agreement with a previous suggestion that the S strain is an evolutionary intermediate between M. avium subsp. avium and the C strain of M. avium subsp. paratuberculosis based on SNPs in IS1311 (52) and by the discovery of three polymorphic regions unique to the S strain of M. avium subsp. paratuberculosis (20). Several studies (35, 51) show that SNPs are a major source of genotypic variation within the M. avium complex, and as demonstrated by Semret et al. (47), SNPs can be used in conjunction with large sequence polymorphisms to identify possible evolutionary paths within the M. avium complex. Further detailed investigations of a larger number of M. avium subsp. avium isolates are required to understand the phylogenetic and ancestral relationship of the M. avium complex.
Previously, a correlation of SNPs with phenotypic diversity has been demonstrated in some mycobacterial species, thereby encouraging its use for bacterial strain differentiation (1, 8, 23, 35). Moreover, SNPs are frequently used in epidemiological and evolutionary studies to differentiate between closely related species, subspecies, and strains of bacteria without knowledge of what effect the SNP may have on gene function or protein activity (1, 24, 27, 28, 49). However, SNPs in the rpoV and mma3 genes of M. bovis have a marked impact on virulence and cellular functions, and SNPs in M. tuberculosis are thought to be responsible for altered phenotypes (5, 16). Targeting functional coding genes for strain typing purposes, as recently done with the gyrA and gyrB genes of M. avium subsp. paratuberculosis (9), might have advantages over targeting noncoding insertion sequences in M. avium subsp. paratuberculosis DNA to discover differences that are linked with host specificity.
We reasoned that selecting a surface-exposed PPE protein would increase our chances of locating SNPs. Although it has not been proven that SNPs occur more frequently in genes coding for surface-exposed proteins, it is tempting to hypothesize that selection pressures occurring specifically at the cell surface, e.g., host immune reactions (2), environmental interactions, and bacteriophage binding (43), favor certain mutations. As demonstrated in this study, sequence polymorphisms in the MAP1506 gene locus are detectable by DGGE and allow discrimination of M. avium subsp. paratuberculosis isolates corresponding to known IS900 RFLP subtypes I, II, and III. Discrimination of C and S subtypes is possible by PCR (14), IS1311 PCR-REA (32), or specific-locus PCR (20), but the additional differentiation of subtypes I and III within the S isolates illustrates the usefulness of DGGE as a new method to subtype M. avium subsp. paratuberculosis isolates. Moreover, DGGE, being a PCR-based technique, will be of great advantage in the subtyping of difficult-to-culture or nonculturable M. avium subsp. paratuberculosis organisms as well as M. avium subsp. paratuberculosis DNA extracted from tissues, blood, or milk (both human and veterinary origin). In short, MAP1506 locus DGGE is a valuable tool to characterize isolates when there is not enough bacterial growth to perform pulsed-field gel electrophoresis or IS900 RFLP.
In this study, the resolving power of DGGE was used to separate and differentiate amplicons of a gene containing SNPs. While DGGE is usually used to characterize multispecies bacterial communities (41), single-band DGGE has previously only been used with Mycobacterium sp. to analyze and detect polymorphisms in genes associated with antibiotic resistance in M. tuberculosis (36, 37, 45, 46) or in the 16S rRNA gene (29). The DGGE technique developed in this study has added discriminatory power because it can discriminate two types within the S type, while IS1311 PCR-REA cannot discriminate any subtypes within the S type. In this respect, this technique complements other rapid molecular techniques and may have future use as additional informative SNPs are discovered in other M. avium subsp. paratuberculosis genes that have significance in host specificity.
Acknowledgments
This work was supported by the Faculty of Veterinary Medicine, University of Calgary, and in part by a grant from the Crohn's and Colitis Foundation of Canada.
We thank Victoria Newton for excellent technical assistance.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbon, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 1853392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrzejewska, J., S. K. Lee, P. Olbermann, N. Lotzing, E. Katzowitsch, B. Linz, M. Achtman, C. I. Kado, S. Suerbaum, and C. Josenhans. 2006. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J. Bacteriol. 1885865-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., Q. Zhang, L. L. Li, and V. Kapur. 2003. Genomic homogeneity between Mycobacterium avium subsp avium and Mycobacterium avium subsp paratuberculosis belies their divergent growth rates. BMC Microbiol. 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banu, S., N. Honore, B. Saint-Joanis, D. Philpott, M. C. Prevost, and S. T. Cole. 2002. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 449-19. [DOI] [PubMed] [Google Scholar]
- 5.Behr, M. A., B. G. Schroeder, J. N. Brinkman, R. A. Slayden, and C. E. Barry. 2000. A point mutation in the mma3 gene is responsible for impaired methoxymycolic acid production in Mycobacterium bovis BCG strains obtained after 1927. J. Bacteriol. 1823394-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, M. J., and G. Delogu. 2002. The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol. 10246-249. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, M. J., G. Delogu, Y. P. Chen, S. Bardarov, J. Kriakov, M. Alavi, and W. R. Jacobs. 2001. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect. Immun. 697326-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 993684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castellanos, E., A. Aranaz, B. Romero, L. De Juan, G. R. Alvarez, J. Bezos, S. Rodriguez, K. Stevenson, A. Mateos, and L. Dominguez. 2007. Polymorphisms in gyrA and gyrB genes among Mycobacterium avium subsp. paratuberculosis types I, II, and III. J. Clin. Microbiol. 453439-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi, J., J. A. VanLeeuwen, A. Weersink, and G. P. Keefe. 2002. Direct production losses and treatment costs from bovine viral diarrhoea virus, bovine leukosis virus, Mycobacterium avium subspecies paratuberculosis, and Neospora caninum. Prev. Vet. Med. 55137-153. [DOI] [PubMed] [Google Scholar]
- 11.Choudhary, R. K., S. Mukhopadhyay, P. Chakhaiyar, N. Sharma, K. J. R. Murthy, V. M. Katoch, and S. E. Hasnain. 2003. PPE antigen Rv2430c of Mycobacterium tuberculosis induces a strong B-cell response. Infect. Immun. 716338-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, C. J. 1997. The pathology and pathogenesis of paratuberculosis in ruminants and other species. J. Comp. Pathol. 116217-261. [DOI] [PubMed] [Google Scholar]
- 13.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 14.Collins, D. M., M. De Zoete, and S. M. Cavaignac. 2002. Mycobacterium avium subsp. paratuberculosis strains from cattle and sheep can be distinguished by a PCR test based on a novel DNA sequence difference. J. Clin. Microbiol. 404760-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins, D. M., D. M. Gabric, and G. W. deLisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 281591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins, D. M., R. P. Kawakami, G. W. Delisle, L. Pascopella, B. R. Bloom, and W. R. Jacobs. 1995. Mutation of the principal sigma-factor causes loss of virulence in a strain of the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 928036-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Juan, L., J. Alvarez, B. Romero, J. Bezos, E. Castellanos, A. Aranaz, A. Mateos, and L. Dominguez. 2006. Comparison of four different culture media for isolation and growth of type II and type I/III Mycobacterium avium subsp. paratuberculosis strains isolated from cattle and goats. Appl. Environ. Microbiol. 725927-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delogu, G., and M. J. Brennan. 2001. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect. Immun. 695606-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delogu, G., C. Pusceddu, A. Bua, G. Fadda, M. J. Brennan, and S. Zanetti. 2004. Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 52725-733. [DOI] [PubMed] [Google Scholar]
- 20.Dohmann, K., B. Strommenger, K. Stevenson, L. de Juan, J. Stratmann, V. Kapur, T. J. Bulls, and G. F. Gerlach. 2003. Characterization of genetic differences between Mycobacterium avium subsp. paratuberculosis type I and type II isolates. J. Clin. Microbiol. 415215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feizabadi, M. M., I. D. Robertson, A. Hope, D. V. Cousins, and D. J. Hampson. 1997. Differentiation of Australian isolates of Mycobacterium paratuberculosis using pulsed field gel electrophoresis. Aust. Vet. J. 75887-889. [DOI] [PubMed] [Google Scholar]
- 22.Feller, M., K. Huwiler, R. Stephan, E. Altpeter, A. Shang, H. Furrer, G. E. Pfyffer, T. Jemmi, A. Baumgartner, and M. Egger. 2007. Mycobacterium avium subspecies paratuberculosis and Crohn's disease: a systematic review and meta-analysis. Lancet Infect. Dis. 7607-613. [DOI] [PubMed] [Google Scholar]
- 23.Filliol, I., A. S. Motiwala, M. Cavatore, W. H. Qi, M. H. Hazbon, M. B. del Valle, J. Fyfe, L. Garcia-Garcia, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. Leon, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendon, J. Sifuentes-Osornio, A. P. de Leon, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 1883162-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleischmann, R. D., D. Alland, J. A. Eisen, L. Carpenter, O. White, J. Peterson, R. Deboy, R. Dodson, M. Gwinn, D. Haft, E. Hickey, J. F. Kolonay, W. C. Nelson, L. A. Umayam, M. Ermolaeva, S. L. Salzberg, A. Delcher, T. Utterback, J. Weidman, H. Khouri, J. Gill, A. Mikula, W. Bishai, W. R. Jacobs, J. C. Venter, and C. M. Fraser. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 1845479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fodde, R., and M. Losekoot. 1994. Mutation detection by denaturing gradient electrophoresis (DGGE). Hum. Mutat. 383-94. [DOI] [PubMed] [Google Scholar]
- 26.Gey van Pittius, N. C., S. L. Sampson, H. Lee, Y. Kim, P. D. van Helden, and R. M. Warren. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol. Biol. 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutacker, M. M., J. C. Smoot, C. A. L. Migliaccio, S. M. Ricklefs, S. Hua, D. V. Cousins, E. A. Graviss, E. Shashkina, B. N. Kreiswirth, and J. M. Musser. 2002. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 1621533-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes, A. L., R. Friedman, and M. Murray. 2002. Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerg. Infect. Dis. 81342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leys, N. M., A. Ryngaert, L. Bastiaens, P. Wattiau, E. M. Top, W. Verstraete, and D. Springael. 2005. Occurrence and community composition of fast-growing Mycobacterium in soils contaminated with polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 51375-388. [DOI] [PubMed] [Google Scholar]
- 30.Li, L. L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 10212344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y. J., E. Miltner, M. Wu, M. Petrofsky, and L. E. Bermudez. 2005. A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell. Microbiol. 7539-548. [DOI] [PubMed] [Google Scholar]
- 32.Marsh, I., R. Whittington, and D. Cousins. 1999. PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311. Mol. Cell. Probes 13115-126. [DOI] [PubMed] [Google Scholar]
- 33.Marsh, I. B., J. P. Bannantine, M. L. Paustian, M. L. Tizard, V. Kapur, and R. J. Whittington. 2006. Genomic comparison of Mycobacterium avium subsp. paratuberculosis sheep and cattle strains by microarray hybridization. J. Bacteriol. 1882290-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marsh, I. B., and R. J. Whittington. 2005. Deletion of an mmpL gene and multiple associated genes from the genome of the S strain of Mycobacterium avium subsp paratuberculosis identified by representational difference analysis and in silico analysis. Mol. Cell. Probes 19371-384. [DOI] [PubMed] [Google Scholar]
- 35.Marsh, I. B., and R. J. Whittington. 2007. Genomic diversity in Mycobacterium avium: single nucleotide polymorphisms between the S and C strains of M. avium subsp paratuberculosis and with M. a. avium. Mol. Cell. Probes 2166-75. [DOI] [PubMed] [Google Scholar]
- 36.McCammon, M. T., J. S. Gillette, D. P. Thomas, S. V. Ramaswamy, E. A. Graviss, B. N. Kreiswirth, J. Vijg, and T. N. Quitugua. 2005. Detection of rpoB mutations associated with rifampin resistance in Mycobacterium tuberculosis using denaturing gradient gel electrophoresis. Antimicrob. Agents Chemother. 492200-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCammon, M. T., J. S. Gillette, D. P. Thomas, S. V. Ramaswamy, I. I. Rosas, E. A. Graviss, J. Vijg, and T. N. Quitugua. 2005. Detection by denaturing gradient gel electrophoresis of pncA mutations associated with pyrazinamide resistance in Mycobacterium tuberculosis isolates from the United States-Mexico border region. Antimicrob. Agents Chemother. 492210-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna, S. L. B., G. P. Keefe, A. Twari, J. VanLeeuwen, and H. W. Barkema. 2006. Johne's disease in Canada. Part II. Disease impacts, risk factors, and control programs for dairy producers. Can. Vet. J. 471089-1099. [PMC free article] [PubMed] [Google Scholar]
- 39.Motiwala, A. S., L. L. Li, V. Kapur, and S. Sreevatsan. 2006. Current understanding of the genetic diversity of Mycobacterium avium subsp paratuberculosis. Microbes Infect. 81406-1418. [DOI] [PubMed] [Google Scholar]
- 40.Motiwala, A. S., M. Strother, A. Amonsin, B. Byrum, S. A. Naser, J. R. Stabel, W. P. Shulaw, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 412015-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek 73127-141. [DOI] [PubMed] [Google Scholar]
- 42.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, R. du Maine, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38155-167. [DOI] [PubMed] [Google Scholar]
- 43.Power, M. L., B. C. Ferrari, J. Littlefield-Wyer, D. M. Gordon, M. B. Slade, and D. A. Veal. 2006. A naturally occurring novel allele of Escherichia coli outer membrane protein A reduces sensitivity to bacteriophage. Appl. Environ. Microbiol. 727930-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 2881436-1439. [DOI] [PubMed] [Google Scholar]
- 45.Scarpellini, P., S. Braglia, P. Carrera, M. Cedri, P. Cichero, A. Colombo, R. Crucianelli, A. Gori, M. Ferrari, and A. Lazzarin. 1999. Detection of rifampin resistance in Mycobacterium tuberculosis by double gradient-denaturing gradient gel electrophoresis. Antimicrob. Agents Chemother. 432550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scarpellini, P., P. Carrera, P. Cichero, C. Gelfi, A. Gori, M. Ferrari, A. Zingale, and A. Lazzarin. 2003. Detection of resistance to isoniazid by denaturing gradient-gel electrophoresis and DNA sequencing in Mycobacterium tuberculosis clinical isolates. New Microbiol. 26345-351. [PubMed] [Google Scholar]
- 47.Semret, M., C. Y. Turenne, P. de Haas, D. M. Collins, and M. A. Behr. 2006. Differentiating host-associated variants of Mycobacterium avium by PCR for detection of large sequence polymorphisms. J. Clin. Microbiol. 44881-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sevilla, I., S. V. Singh, J. M. Garrido, G. Aduriz, S. Rodriguez, M. V. Geijo, R. J. Whittington, V. Saunders, R. H. Whitlock, and R. A. Juste. 2005. Molecular typing of Mycobacterium avium subspecies paratuberculosis strains from different hosts and regions. Rev. Sci. Tech. 241061-1066. [PubMed] [Google Scholar]
- 49.Stevenson, K., V. M. Hughes, L. de Juan, N. F. Inglis, F. Wright, and J. M. Sharp. 2002. Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 401798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sweeney, R. W. 1996. Transmission of paratuberculosis. Vet. Clin. N. Am. Food Anim. Pract. 12305-312. [DOI] [PubMed] [Google Scholar]
- 51.Turenne, C. Y., M. Semret, D. V. Cousins, D. M. Collins, and M. A. Behr. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J. Clin. Microbiol. 44433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whittington, R., I. Marsh, E. Choy, and D. Cousins. 1998. Polymorphisms in IS1311, an insertion sequence common to Mycobacterium avium and M. avium subsp. paratuberculosis, can be used to distinguish between and within these species. Mol. Cell. Probes 12349-358. [DOI] [PubMed] [Google Scholar]