Abstract
Escherichia coli strains of serotype O51:H40 were studied with regard to the presence of several virulence properties and their genetic diversity and enteropathogenicity in rabbit ileal loops. This serotype encompasses potential enteropathogenic strains mostly classified as being atypical enteropathogenic E. coli (EPEC) strains, which are genetically closer to enterohemorrhagic E. coli than to typical EPEC strains.
Enterohemorrhagic Escherichia coli (EHEC) and enteropathogenic E. coli (EPEC) comprise two of the six diarrheagenic E. coli pathotypes (16). While EHEC secretes Shiga toxin(s), EPEC adheres to HeLa/HEp-2 cells, forming compact clusters that distinguish the localized pattern of adherence (LA) (16). The LA is mediated by the bundle-forming pilus encoded by the EPEC adherence factor plasmid (pEAF) (16).
EHEC and EPEC share the ability to promote attaching-effacing (AE) lesions in intestinal cells (11) characterized by intimate bacterial adherence, localized microvillus effacement, and accumulation of polymerized actin and other cytoskeleton elements, resulting in pedestal-like structures underneath adherent bacteria (11). The AE lesion-associated genes reside in the locus of enterocyte effacement (LEE) (20), a pathogenicity island encoding a type III secretion system, regulators, chaperones, and effectors that interfere with diverse cell signaling processes (revised in reference 11). The LEE also encodes the outer membrane adhesive protein intimin (15) and its translocated receptor, Tir (18).
EPEC is currently subgrouped into typical EPEC (tEPEC) and atypical EPEC (aEPEC), where pEAF is present only in the former group (17, 34). Most tEPEC and aEPEC strains belong to defined (traditional) EPEC serogroups (34). Nevertheless, we previously identified and characterized several E. coli strains of non-EPEC serogroups isolated in three cities in Brazil carrying the intimin-encoding gene (eae) and lacking Shiga toxin-encoding genes (12). That study emphasized the large diversity of such strains but allowed no identification of the virulence potential of individual serotypes. Moreover, as both diarrheic and nondiarrheic patients may carry aEPEC strains (34, 36), it is important to evaluate the virulence potential of selected strains to identify those strains that are truly enteropathogenic.
In this study, we examined all 10 strains of serotype O51:H40, the most frequently found serotype in our previous study (12), and an additional uncharacterized O51:H40 strain (7) for novel E. coli virulence properties, their enteropathogenic potentials in vivo, genetic diversity, and genetic relatedness to tEPEC and EHEC. These results were compiled with those of our previous study (12) to attain an overview of the virulence potential of serotype O51:H40.
Table 1 presents the origins and the clinical and microbiological data of the patients carrying the 11 O51:H40 strains studied (7, 12, 13). Their adherence patterns in HeLa and differentiated intestinal Caco-2 cells were determined previously (12, 36), and in this study, strains were further tested in differentiated T84 cells as described previously (1). In HeLa cells, strain 0151-1 expressed LA after 3 h of infection, whereas seven strains presented an LA-like (LAL) pattern (27), and three strains showed a noncharacteristic pattern in prolonged (6-h) assays (12, 36). In Caco-2 cells, all strains, except strain 21242, presented LAL patterns, whereas in T84 cells, eight strains (including 0151-1) presented LAL patterns, and three (strains 21323, 21242, and 21075) showed an aggregative adherence (AA) pattern (8) (Table 1). The potential to cause AE lesions was examined indirectly in the same cell lineages using a fluorescent actin staining (FAS) assay that detects actin accumulation (19). The divergent results among the three cell lineages could be due to different environmental factors (e.g., medium composition or the presence of specific receptors) or the induction of the expression of different bacterial adhesins. T84 was the most sensitive lineage, detecting eight (72.7%) FAS-positive (FAS+) strains (Table 1), whereas only three (27.3%) and seven (63.6%) strains were FAS+ in HeLa and Caco-2 cells, respectively (not shown). In T84 cells, all FAS+ strains and one FAS-negative (FAS−) strain promoted the tyrosine phosphorylation of Tir (28) (Table 1), thus behaving as tEPEC strain E2348/69, in contrast to EHEC strain EDL933 (9).
TABLE 1.
Clinical, phenotypic, and genotypic characteristics of the O51:H40 strains studied
| Strain | Age of patient | Other clinical information | Other pathogen(s) | Adherence patterng | FASg | TPg,h | AE lesions in rabbitsi | Virulence gene(s)j | Antibiotic resistance(s)k | ERIC type |
|---|---|---|---|---|---|---|---|---|---|---|
| 1757/01b | 27 yr | Nonee | Campylobacter sp. | LAL | + | + | Yes | eae shf | None | IA1 |
| 21075c | 8 mo | None | Rotavirus, DAEC | AA | + | + | Yes | eae shf | Amp Amc Sut Tet Azi | IA1 |
| 21323c | 1 yr | Nonef | None | AA | + | + | Yes | eae shf | Amp Amc Sut Tet | IA2 |
| 1711-4a | 4 yr 5 mo | Vomit, fever | DAEC | LAL | + | + | NT | eae shf | None | IB |
| 1931-2a | 2 yr 4 mo | Vomit, fever | Rotavirus | LAL | + | + | NT | eae | None | IIA1 |
| 3062-1a | 2 yr 5 mo | Bronchitisd | None | LAL | − | − | No | eae | None | IIA1 |
| 3102-1a | 1 yr 7 mo | Measlesd | None | LAL | − | − | No | eae | Amp Amc Sut Tet | IIA1 |
| 2022/01b | 1 yr | None | None | LAL | + | + | Yes | eae | None | IIA1 |
| 21242c | 1 yr | Blood in feces | Rotavirus, DAEC | AA | − | + | Yes | eae | None | IIA1 |
| 4361-2a | 1 yr 10 mo | Vomit | None | LAL | + | + | NT | eae | Amp Amc Sut Crx | IIA2 |
| 0151-1a | 2 mo | Vomit, mucous in feces | None | LAL | + | + | NT | eae EAF bfpA perA toxB | None | IIB |
Universidade Federal de Sao Paulo.
Instituto Adolfo Lutz.
Universidade Federal do Rio de Janeiro.
Nondiarrheic patient.
Patient with AIDS.
Patient with persistent diarrhea.
Tests performed in T84 cells for 6 h.
TP, tyrosine phosphorylation of Tir.
NT, not tested.
Strains were reported previously to lack homology with gene probes for E. coli virulence factors STh, STp, LTI, INV, Stx1, Stx2, CDT, CNF, E-Hly, diffuse adherence, Afa, Sfa, Pap, enteroaggregative E. coli plasmid, EAST-1, aggregative adherence fimbriae I, II, and III, Hly, aggR, irp-2, pic, aap, and pet (12, 13, 36). All strains were positive for intimin subtype θ.
As tested by the disc diffusion assay for 22 antibiotics. Amp, ampicillin; Amc, amikacin; Sut, sulfazotrim; Crx, cefuroxime; Tet, tetracycline; Azi, azithromycin.
To verify the enteropathogenic potential in vivo, whole cultures of seven O51:H40 strains (three FAS− and four FAS+) were inoculated in rabbit ileal loops and examined by transmission electron microscopy (33). One FAS− and all FAS+ strains tested promoted AE lesions in this model (Table 1), confirming that at least five O51:H40 strains are potentially enteropathogenic. Interestingly, the two FAS− strains (3062-1 and 3102-1) that were unable to promote AE lesions were isolated from nondiarrheic children. It is likely that mutations within the LEE (or their regulatory elements) rendered these strains nonpathogenic.
The O51:H40 strains were previously tested by colony blot or PCR for 29 diarrheagenic E. coli and extraintestinal pathogenic E. coli virulence genes (12, 13, 36). In this study, they were further tested for novel virulence genes (efa1, saa, paa, lpfO113, iha, toxB, ldaG, and agg3C) by PCR (3, 5, 10, 21, 23, 29, 31, 32). Besides eae, four strains carried shf, and strain 0151-1 carried EPEC adherence factor, bfpA, and perA (which comprise pEAF sequences) as well as toxB (encoding an EHEC adhesin). Although the role of Shf in pathogenesis is unknown, the shf sequence has been detected in Shigella flexneri, EHEC O157:H7, and enteroaggregative E. coli strains (8). Its closest homolog is IcaB, a protein implicated in the intercellular adhesion of Staphylococcus epidermidis (14).
The plasmid profiles and antibiotic resistance patterns of the O51:H40 strains were analyzed by alkaline extraction (6) and disc diffusion agar (4), respectively. Although no common profile was found, a ∼45-MDa plasmid band was observed in the three strains (not shown) that were resistant to ampicillin, amikacin, sulfazotrim, and tetracycline (Table 1), suggesting that it comprises a common resistance plasmid. Moreover, strain 0151-1 presented a 59-MDa band that hybridized with two pEAF sequences (EAF and bfpA) (2) as well as toxB (not shown). Since strain 0151-1 carried pEAF and expressed LA in HeLa cells, it was classified as being a tEPEC strain, whereas the remaining strains lacking pEAF and the stx genes and showing a LAL pattern were classified as being aEPEC strains.
LEE insertion sites vary according to the clonal origins of the E. coli strains (37). In tEPEC and EHEC strains, it is generally located adjacent to selC or pheU (26, 30, 37). However, as in the O51:H40 strains, these sites were intact, whereas when pheV was disrupted, the LEE is apparently located in pheV in these strains (12).
In animal models and in human intestinal biopsies, tEPEC preferentially colonizes the small intestines, whereas EHEC is restricted to the large intestine (24). Moreover, tropism to different colonization sites apparently depends on the intimin subtype (38). All O51:H40 strains carried the less common intimin subtype theta (Int-θ), as identified by PCR (12). Although the preferential adherence site of Int-θ has yet to be identified, it could comprise the small intestine since most O51:H40 strains tested promoted AE lesions in the rabbit ileum.
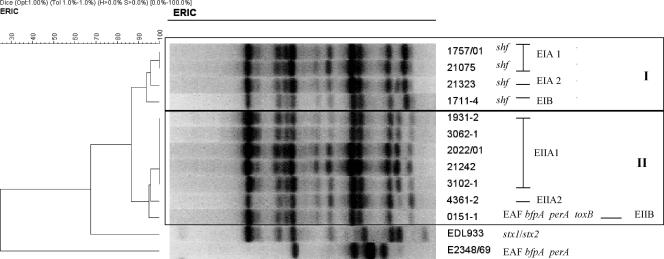
The genetic diversity of the O51:H40 strains and their relatedness to tEPEC and EHEC were investigated. Ribotyping (25) performed with BglI showed a single banding pattern, as expected by the fact that the strains belonged to a single serotype (22). However, enterobacterial repetitive intergenic consensus PCR (ERIC-PCR) (35) generated six distinct electrophoretic types (E patterns) distributed in two main clonal groups, EI and EII, with a similarity of 87%, as calculated by the Dice unweighted-pair group method with arithmetic mean (Table 1 and Fig. 1). The genome differences found could reflect the horizontal acquisition of virulence factors (e.g., shf, toxB, and pEAF) or deletions. Interestingly, EI comprised four strictly correlated strains (all FAS and shf positive) despite their distinct geographical origins. Notwithstanding some heterogeneous characteristics within EII, five strains were identical by this technique. Note that ERIC-PCR showed a much closer relationship between the O51:H40 strains and EHEC strain EDL933 (68% similarity) than to tEPEC strain E2348/69 (<30% similarity). Although these findings corroborate a previous study showing that aEPEC strains are more related to EHEC than to tEPEC strains (35), they should be confirmed with a larger group of strains of both pathotypes.
FIG. 1.
Dendrogram of ERIC-PCR patterns of E. coli strains of serotype O51:H40 carrying eae. The analysis of similarity of the normalized images was accomplished using the unweighted-pair group method with arithmetic mean with the Dice optimized coefficient. Control strains were EPEC prototype strain E2348/69 and EHEC prototype strain EDL 933 (serotype O157:H7). Two different ERIC patterns (EI and EII) were found among the O51:H40 strains showing a similarity index of 87%.
In conclusion, serotype O51:H40 encompasses potential enteropathogenic strains mostly classified as being aEPEC strains, which are genetically more related to EHEC than to tEPEC strains.
Acknowledgments
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-Edital Universal no. 478243/2003-0). F.C.M. received a scholarship from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 04/14713-0).
We thank Sylvia P. C. Leão for the construction and analysis of the dendrograms and Jorge Luiz Mello Sampaio for assistance with ERIC-PCR.
Footnotes
Published ahead of print on 6 February 2008.
REFERENCES
- 1.Abe, C. M., S. Knutton, M. Z. Pedroso, E. Freymuller, and T. A. T. Gomes. 2001. An aggregative Escherichia coli strain of serotype O111:H12 damages and invades cultured T84 cells and human colonic mucosa. FEMS Microbiol. Lett. 203199-205. [DOI] [PubMed] [Google Scholar]
- 2.Baldini, M. M., J. B. Kaper, M. M. Levine, and H. W. Moon. 1983. Plasmid-mediated adhesion of enteropathogenic E. coli. J. Pediatr. Gastroenterol. 2534-538. [DOI] [PubMed] [Google Scholar]
- 3.Batisson, I., M. Guimond, F. Girard, H. A. C. Zhu, E. Oswald, J. M. Fairbrother, M. Jacques, and J. Harel. 2003. Characterization of novel factor PAA involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect. Immun. 714516-4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45493-496. [PubMed] [Google Scholar]
- 5.Bernier, C., P. Gounon, and C. Le Bouguenec. 2002. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 704302-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 71513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake, P. A., S. R. T. S. Ramos, K. L. MacDonald, V. Rassi, T. A. T. Gomes, C. Ivey, N. H. Bean, and L. R. Trabulsi. 1993. Pathogen-specific risk factors and protective factors for acute diahrreal disease in urban Brazilian infants. J. Infect. Dis. 167627-632. [DOI] [PubMed] [Google Scholar]
- 8.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 672692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeVinney, R., M. Stein, D. Reinscheid, A. Abe, S. Ruschkowsi, and B. B. Finlay. 1999. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cells membrane but is not tyrosine phosphorylated. Infect. Immun. 2389-2398. [DOI] [PMC free article] [PubMed]
- 10.Doughty, S., J. Sloan, V. Bennett-Wood, M. Robertson, R. M. Robins-Browne, and E. L. Hartland. 2002. Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 706761-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 732573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes, T. A. T., K. Irino, D. M. Girão, V. B. C. Girão, B. E. C. Guth, T. M. I. Vaz, F. C. Moreira, S. H. Chinarelli, and M. A. M. Vieira. 2004. Emerging Escherichia coli strains? Emerg. Infect. Dis. 101851-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes, T. A. T., M. A. M. Vieira, C. M. Abe, D. Rodrigues, P. M. Griffin, and S. R. T. S. Ramos. 1998. Adherence patterns and adherence-related DNA sequences in Escherichia coli isolates from children with and without diarrhea in São Paulo City, Brazil. J. Clin. Microbiol. 363609-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 201083-1091. [DOI] [PubMed] [Google Scholar]
- 15.Jerse, A. E., J. U. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesion on tissue culture cells. Proc. Natl. Acad. Sci. USA 877839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2123-140. [DOI] [PubMed] [Google Scholar]
- 17.Kaper, J. P. 1996. Defining EPEC. Rev. Microbiol. 27130-133. [Google Scholar]
- 18.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic Escherichia coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91511-520. [DOI] [PubMed] [Google Scholar]
- 19.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 571290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDaniel, T. K., K. G. Jarvis, M. S. Donnemberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens Proc. Natl. Acad. Sci. USA 921664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35275-288. [DOI] [PubMed] [Google Scholar]
- 22.Orskov, F., T. S. Whittam, A. Cravioto, and I. Orskov. 1990. Clonal relationships among classic enteropathogenic Escherichia coli (EPEC) belong to different O groups. J. Infect. Dis. 16276-81. [DOI] [PubMed] [Google Scholar]
- 23.Paton, A. W., P. Srimanot, M. C. Woodrow, and J. C. Paton. 2001. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 696999-7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips, A. D., and G. Frankel. 2000. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 1811496-1500. [DOI] [PubMed] [Google Scholar]
- 25.Popovic, T., C. A. Bopp, O. Olsvik, and A. Kiehlbauch. 1993. Ribotyping in molecular epidemiology, p. 573-583. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.). Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, DC.
- 26.Reid, R. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 40664-67. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigues, J., I. C. A. Scaletsky, L. C. Campos, T. A. T. Gomes, T. S. Whittam, and L. R. Trabulsi. 1996. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect. Immun. 642680-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenshine, I., M. S. Donnemberg, J. B. Kaper, and B. B. Finlay. 1992. Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 113551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scaletsky, I. C. A., J. Michalski, A. G. Torres, M. V. Dulguer, and J. B. Kaper. 2005. Identification and characterization of the locus for diffuse adherence, which encodes a novel afimbrial adhesin found in atypical enteropathogenic Escherichia coli. Infect. Immun. 734753-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarr, C. L., and T. S. Whittam. 2002. Molecular evolution of the intimin gene in O111 clones of the pathogenic Escherichia coli. J. Bacteriol. 184479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarr, P. I., S. S. Bilge, C. James, J. R. Varu, S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 681400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 696660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trabulsi, L. R. 1964. Revelação de colibacilos associados às diarréias infantis pelo método da infecção experimental da alça ligada do intestino de coelho. Rev. Inst. Med. Trop. São Paulo 6197-203. [PubMed] [Google Scholar]
- 34.Trabulsi, L. R., R. Keller, and T. A. T. Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli (EPEC). Emerg. Infect. Dis. 8508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 196823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vieira, M. A. M., J. R. C. Andrade, L. R. Trabulsi, A. C. P. Rosa, A. M. G. Dias, S. R. T. S. Ramos, G. Frankel, and T. A. T. Gomes. 2001. Phenotypic and genotypic characteristics of Escherichia coli strains of non-enteropathogenic E. coli (EPEC) serogroups that carry eae and lack the EPEC adherence factor and Shiga toxin DNA probe sequences. J. Infect. Dis. 183762-772. [DOI] [PubMed] [Google Scholar]
- 37.Wieler, L. H., T. K. McDaniel, T. S. Whittam, and J. B. Kaper. 1997. Insertion site of the locus of enterocyte effacement in enteropathogenic and enterohemorrhagic Escherichia coli differs in relation to the clonal phylogeny of the strains. FEMS Microbiol. Lett. 15649-53. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, W. L., B. Köhler, E. Oswald, L. Beutin, H. Karch, S. Morabito, A. Caprioli, S. Seurbaum, and H. Schmidt. 2002. Genetic diversity of intimin genes of attaching and effacing Escherichia coli strains. J. Clin. Microbiol. 404486-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]