Abstract
Whole blood has been found to be a reliable matrix for the detection and quantitation of cytomegalovirus (CMV) DNA. In this study, the performance of the artus CMV LightCycler (LC) PCR kit in conjunction with automated sample preparation on a BioRobot EZ1 workstation was evaluated. The accuracy, linearity, analytical sensitivity, and inter- and intra-assay variations were determined. A total of 102 clinical EDTA whole-blood samples were investigated, and results were compared with those obtained with the in vitro diagnostics (IVD)/Conformité Européene (CE)-labeled CMV HHV6,7,8 R-gene quantification kit. When the accuracy of the new kit was tested, seven of eight results were found to be within ±0.5 log10 unit of the expected panel results. Determination of linearity resulted in a quasilinear curve over more than 5 log units. The lower limit of detection of the assay was determined to be 139 copies/ml in EDTA whole blood. The interassay variation ranged from 15 to 58%, and the intra-assay variation ranged from 7 to 35%. When clinical samples were tested and the results were compared with those of the routinely used IVD/CE-labeled assay, 53 samples tested positive and 13 samples tested negative by both of the assays. One sample was found to be positive with the artus CMV LC PCR kit only, and 35 samples tested positive with the routinely used assay only. The majority of discrepant results were found with low-titer samples. In conclusion, use of the artus CMV LC PCR kit in conjunction with automated sample preparation on the BioRobot EZ1 workstation may be suitable for the detection and quantitation of CMV DNA in EDTA whole blood in the routine low-throughput laboratory; however, low-positive results may be missed by this assay.
Human cytomegalovirus (CMV) has the ability to establish lifelong persistent and latent infection following primary exposure. Under certain conditions, CMV can reactivate, resulting in asymptomatic viral shedding or the development of disease (10). While in the immunocompetent individual the infection is held in check by the host's immune response, CMV disease is generally restricted to the immunocompromised or immunologically immature host (15).
To avoid a lethal outcome of CMV disease, the start of treatment at the earliest stage is of extreme significance (3, 8). The level of CMV DNA has been found to be an important prognostic marker for the ongoing disease (1, 2, 4). Today, laboratories use different sample materials for the detection and quantitation of CMV DNA. Although whole blood has been found to be superior to blood cells or plasma, current guidelines do not recommend a specific kind of sample (5, 7, 8, 9, 11, 12, 16).
Recently, the artus CMV LightCycler (LC) PCR kit (Qiagen, Hilden, Germany) for the quantitative detection of CMV DNA has been introduced. This molecular assay is designed and in vitro diagnostics (IVD)/Conformité Européene (CE) labeled for the amplification and detection of CMV DNA after the manual extraction of CMV DNA from human plasma and serum samples.
The aim of this study was to evaluate the performance of the artus CMV LC PCR kit in conjunction with the automated extraction of EDTA whole blood on a BioRobot EZ1 workstation. Accuracy was tested with a reference material. Linearity was analyzed by use of a dilution series of a high-titer sample, and the lower limit of detection was determined by probit analysis. Both interassay and intra-assay variations were tested. The clinical performance of the artus CMV LC PCR kit in the routine diagnostic laboratory was evaluated with routine clinical EDTA whole-blood samples, and the results were compared to those obtained with the IVD/CE-labeled CMV HHV6,7,8 R-gene quantification kit (Argene SA, Varilhes, France).
MATERIALS AND METHODS
Sample preparation and molecular assays.
Prior to amplification and detection with the artus CMV LC PCR kit, CMV DNA was extracted with the EZ1 virus mini kit (Qiagen) on a BioRobot EZ1 workstation (Qiagen). When the CMV HHV6,7,8 R-gene quantification kit was used, samples were prepared with MagNa Pure compact nucleic acid isolation kit 1 (Roche) on a MagNa Pure compact instrument (Roche). For both assays, the LC (version 2.0) instrument was used for real-time PCR and detection. According to the manufacturers' package inserts, the lower limit of detection is 78.9 copies/ml for the artus CMV LC PCR kit with plasma as the sample matrix and 150 copies/ml for the CMV HHV6,7,8 R-gene quantification kit with EDTA or citrate whole blood as the sample matrix.
Both the BioRobot EZ1 and the MagNA Pure compact instruments use magnetic particle technology to capture nucleic acids. For extraction with the BioRobot EZ1 workstation, a mixture of 75 μl protease, 2.5 μl carrier RNA, and 7.5 μl internal control (IC) was manually prepared for each sample and was loaded on the instrument together with the reagent cartridges and the EDTA whole-blood samples. The sample input volume was 200 μl, and the elution volume was 75 μl. For extraction with the MagNA Pure compact instrument, the ready-to-use reagents were placed on the instrument. The sample input volume was 200 μl, and a total of 5 μl of IC was automatically added to each sample. The elution volume was 50 μl.
For real-time PCR with the artus CMV LC PCR kit and the CMV HHV6,7,8 R-gene quantification kit, 15 μl of the master mixture and 10 μl of the extracted sample were pipetted into LC capillaries and loaded on the LC instrument. Both of the kits allow quantitation of target nucleic acids on the basis of a standard curve prepared with known concentrations of the same target (homologous external standards). The four standard samples included in each of the kits are amplified in separate capillaries but within the same run. The LC software calculates the validity of the standard curve by taking several variables, including the slope and the correlation coefficient, into consideration. When the same lot of the kit is used, the standard curve which was generated in a previous run and stored may be used; however, at least one quantitation standard must be included in each run as a calibrator for the imported standard curve. In this study, LC software (version 4.05.415) was used to analyze fluorescence curves, with channel 530 used for the target fluorescence signal of both assays, channel 705/back 530 used for the IC fluorescence signal of the artus CMV LC PCR kit, and channel 560/back 530 used for the IC fluorescence signal of the CMV HHV6,7,8 R-gene quantification kit.
Study design.
Determinations of the accuracy, linearity, and inter-and intra-assay variations and testing of routine samples were done in an International Standards Organization (ISO9001, 2000)-certified laboratory, the Molecular Diagnostics Laboratory, Institute of Hygiene.
The accuracy of the artus CMV LC PCR kit was determined with the Quality Control for Molecular Diagnostics (QCMD) 2006 human CMV proficiency panel (www.qcmd.org). The panel consisted of eight plasma samples. CMV DNA was extracted by using the identical extraction protocol used for EDTA whole blood throughout this study. The panel contained various concentrations of CMV DNA (range, 2.0 × 102 to 2.5 × 104 copies/ml) and two samples negative for CMV DNA.
The linearity of the artus CMV LC PCR kit was tested with a high-titer routine clinical EDTA whole-blood sample. A dilution series (0.5-log steps, i.e., 1:3.16 dilutions) was prepared by using CMV-negative EDTA whole blood. Each dilution was analyzed three times, and the mean CMV DNA titer of each sample was determined.
The lower limit of detection for EDTA whole-blood samples with the artus CMV LC PCR kit in conjunction with the LC (version 2.0) instrument after sample preparation with the BioRobot EZ1 workstation in conjunction with the EZ1 virus mini kit was determined by use of serial dilutions of prequantified CMV (Toledo strain) from the cell culture supernatant (0.5-log steps in Buffer AE [Qiagen]) spiked into human EDTA whole-blood samples to create CMV concentrations ranging from 316 to 0.316 copies/ml. Analyses were performed on 3 days with six replicates per dilution per day. The results were subjected to probit analysis.
The interassay variation of the artus CMV LC PCR kit was determined by using eight routine clinical samples within the linear range of the assay. The samples contained different concentrations of CMV DNA, ranging from 7.5 × 102 to 4.9 × 107 copies/ml, and were tested five times on five different days. The intra-assay variation of the new assay was tested by using four routine clinical samples. The samples contained different concentrations of CMV DNA, ranging from 9.9 × 102 to 5.7 × 106 copies/ml. Aliquots were analyzed five times each in a single run.
The performance of the artus CMV LC PCR kit in the routine diagnostic laboratory was evaluated by testing 102 clinical EDTA whole-blood samples and comparing the results with those obtained with the IVD/CE-labeled CMV HHV6,7,8 R-gene quantification kit, which was used as the reference assay.
RESULTS
When 10 samples of the QCMD 2006 human CMV proficiency panel containing various concentrations of CMV DNA were tested with the artus CMV LC PCR kit, seven of eight samples with positive results were found to have results within ±0.5 log10 unit of the expected panel results (Table 1). One sample with an expected CMV DNA concentration of 2.0 × 102 copies/ml tested negative. Both of the samples without CMV DNA were found to be negative.
TABLE 1.
Results obtained by the new molecular assay in comparison with those obtained by reference laboratories with samples from the QCMD 2006 human CMV proficiency program panela
| Sample no. | Result (no. of copies of CMV DNA/ml) obtained by:
|
Log10 unit difference | |
|---|---|---|---|
| artus CMV LC PCR kit | Reference laboratories | ||
| 1 | 6.8 × 104 | 2.5 × 104 | 0.44 |
| 2 | 5.9 × 103 | 5.0 × 103 | 0.08 |
| 3 | 8.3 × 102 | 1.0 × 103 | 0.08 |
| 4 | 4.5 × 102 | 2.0 × 102 | 0.36 |
| 5 | 3.0 × 104 | 2.5 × 104 | 0.08 |
| 6 | 4.7 × 103 | 5.0 × 103 | 0.02 |
| 7 | 7.0 × 102 | 1.0 × 103 | 0.15 |
| 8 | TNDb | 2.0 × 102 | |
One replicate of each sample was tested.
TND, target not detected.
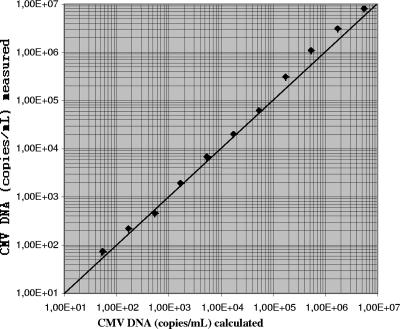
Linearity was tested with a dilution series of a high-titer routine clinical sample. A quasilinear curve was observed up to the original concentration of 5.3 × 106 copies/ml (Fig. 1). CMV DNA was inconsistently detected in dilutions containing less than 2.2 × 102 copies/ml.
FIG. 1.
Linearity of the results for a 0.5-log-unit dilution series of a high-titer routine clinical sample obtained by the new molecular assay. Diagonal line, line of identity.
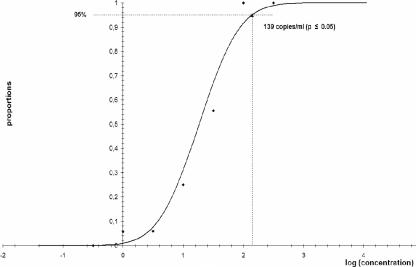
The lower limit of detection of the artus CMV LC PCR kit in combination with the LC (version 2.0) instrument for the amplification and detection of CMV DNA in human EDTA whole-blood samples after sample preparation with the BioRobot EZ1 workstation was determined to be 139 copies/ml (P ≤ 0.05), with a confidence interval of 74 to 406 copies/ml (Fig. 2).
FIG. 2.
Results of probit analysis for determination of the lower limit of detection of the artus CMV LC PCR kit for the detection of CMV DNA in EDTA whole-blood samples after sample preparation with the BioRobot EZ1 workstation.
For the determination of interassay variation, eight clinical EDTA whole-blood samples were analyzed five times on different days. The coefficients of variation were found to be between 15 and 58% (Table 2). The intra-assay variation was determined by analyzing four routine clinical samples five times each in a single run. The coefficients of variation were found to be between 7 and 35% (Table 3).
TABLE 2.
Results of interassay testing
| Sample no. | No. of CMV DNA copies/ml detecteda
|
Coefficient of variation (%) | |
|---|---|---|---|
| Mean | SD | ||
| 1 | 5.0 × 107 | 7.7 × 106 | 15 |
| 2 | 5.0 × 107 | 8.0 × 106 | 16 |
| 3 | 1.9 × 107 | 3.6 × 106 | 19 |
| 4 | 2.1 × 106 | 4.1 × 105 | 20 |
| 5 | 1.6 × 105 | 3.7 × 104 | 24 |
| 6 | 4.2 × 104 | 8.7 × 103 | 21 |
| 7 | 1.1 × 104 | 3.6 × 103 | 33 |
| 8 | 7.5 × 102 | 4.3 × 102 | 58 |
The samples were tested five times on five different days.
TABLE 3.
Results of intra-assay testing
| Sample no. | No. of CMV DNA copies/ml detecteda
|
Coefficient of variation (%) | |
|---|---|---|---|
| Mean | SD | ||
| 1 | 5.8 × 106 | 3.9 × 105 | 7 |
| 2 | 2.0 × 105 | 2.3 × 104 | 11 |
| 3 | 1.3 × 104 | 2.9 × 103 | 22 |
| 4 | 9.9 × 102 | 3.5 × 102 | 35 |
The samples were tested five times each in a single run.
Of 102 clinical EDTA whole-blood samples, 53 tested positive by both of the assays and 13 were found to be negative by both of the assays (Table 4). One sample was found to be positive with the artus CMV LC PCR kit only. The viral load in this sample was found to be less than 139 copies/ml. A total of 35 samples were found to be positive with the CMV HHV6,7,8 R-gene quantification kit only. The viral loads in those samples ranged from less than 150 copies/ml (under the lower limit of detection) to 2.3 × 103 copies/ml, with 24 of 35 samples (69%) having viral loads of less than 150 copies/ml. In order to clarify whether those negative results were caused by either the automated sample preparation protocol or the artus CMV LC PCR kit, 20 of them were retested by both of the assays but with the alternative sample preparation instrument. There was no sample material available for the remaining 15 samples. When samples were prepared on the BioRobot EZ1 workstation and amplified and detected with the CMV HHV6,7,8 R-gene quantification kit, 5 of 20 samples gave positive results, with the viral loads ranging from 2.6 × 102 to 3.2 × 103 copies/ml (Table 5). Additionally, the weakly positive sample of the QCMD proficiency panel which had given a negative result with the artus CMV LC PCR kit tested positive (6.8 × 102 copies/ml) with the CMV HHV6,7,8 R-gene quantification kit. When samples were prepared on the MagNa Pure compact instrument and amplified and detected with the artus CMV LC PCR kit, 4 of 20 samples gave positive results, with the viral loads ranging from less than 139 copies/ml up to 330 copies/ml (Table 5).
TABLE 4.
Results obtained with the artus CMV LC PCR kit and the routinely used CMV HHV6,7,8 R-gene quantification kit
| Result with artus CMV LC PCR kit | No. of samples with the following result with CMV HHV6,7,8 R-gene quantification kit:
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 53 | 1 | 54 |
| Negative | 35 | 13 | 48 |
| Total | 88 | 14 | 102 |
TABLE 5.
Comparison of results obtained with different test systems for quantitation of CMV DNA
| Sample no. | No. of CMV DNA copies/ml obtained with:
|
|||
|---|---|---|---|---|
| EZ1/artusa | EZ1/R-geneb | MPC/artusc | MPC/R-gened | |
| 1 | TNDe | TND | TND | <150f |
| 2 | TND | TND | TND | <150 |
| 3 | TND | TND | TND | <150 |
| 4 | TND | TND | TND | 200 |
| 5 | TND | TND | TND | <150 |
| 6 | TND | TND | <139f | <150 |
| 7 | TND | TND | TND | 460 |
| 8 | TND | 260 | TND | 765 |
| 9 | TND | 950 | TND | <150 |
| 10 | TND | TND | <139 | <150 |
| 11 | TND | TND | TND | 400 |
| 12 | TND | TND | TND | 200 |
| 13 | TND | TND | <139 | 2,300 |
| 14 | TND | TND | TND | <150 |
| 15 | TND | TND | 330 | 300 |
| 16 | TND | TND | TND | 740 |
| 17 | TND | 3,180 | TND | 600 |
| 18 | TND | TND | TND | 300 |
| 19 | TND | 2,200 | TND | 1,740 |
| 20 | TND | 540 | TND | <150 |
EZ1/artus, BioRobot EZ1 workstation/artus CMV LC PCR kit.
EZ1/R-gene, BioRobot EZ1 workstation/CMV HHV6,7,8 R-gene quantification kit.
MPC/artus, MagNa Pure compact instrument/artus CMV LC PCR kit.
MPC/R-gene, MagNa Pure compact instrument/CMV HHV6,7,8 R-gene quantification kit.
TND, target not detected.
The result is under the lower limit of quantification, i.e., weakly positive.
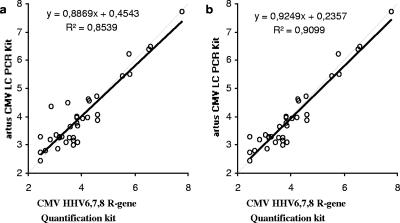
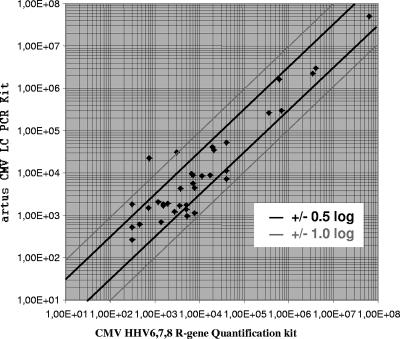
When the results obtained for the 37 samples with viral loads above the lower limit of detection by both of the assays were compared, a correlation (R2) of 0.8593 was observed (Fig. 3a). Of those results, 28 were found to be within ±0.5 log10 units and 7 were found to be between ±0.5 and ±1.0 log10 units. The viral loads in the two remaining samples showed a difference of more than ±1.0 log10 unit (Fig. 4). Without both of them, an R2 value of 0.9099 was found (Fig. 3b).
FIG. 3.
Correlation between the results (copies/ml) obtained with the artus CMV LC PCR kit and the CMV HHV6,7,8 R-gene quantification kit. Black solid lines, regression curve; gray dashed lines, identity lines. (a) Correlation of all quantified samples; (b) correlation of all quantified samples within ±1.0 log unit.
FIG. 4.
Log deviation between the results (copies/ml) obtained with the artus CMV LC PCR kit and the CMV HHV6,7,8 R-gene quantification kit.
DISCUSSION
For both the prophylactic and the preemptive therapy of CMV disease, quantitation of CMV DNA in whole blood has been reported to be an important marker (6, 7, 11, 13, 16). In this study, use of the quantitative artus CMV LC PCR kit in conjunction with automated sample preparation on the BioRobot EZ1 workstation was evaluated. The quantitative results for 102 EDTA whole-blood samples obtained by the new molecular assay were compared to those obtained by the CMV HHV6,7,8 R-gene quantification kit.
When samples from the QCMD 2006 human CMV proficiency panel were tested, 9 of 10 samples gave correct results. One sample with an expected viral load of 200 copies/ml tested negative. The lower limit of detection for EDTA whole-blood samples was analyzed by probit analysis and was found to be 139 copies/ml (P ≤ 0.05) for the artus CMV LC PCR kit in conjunction with automated sample preparation. This is slightly higher than the detection limit for plasma (78.9 copies/ml), as stated in the package insert.
The linear range of the artus CMV LC PCR kit was determined by analysis of dilutions of an EDTA whole-blood sample with a high titer of CMV DNA. The new assay revealed a sufficient linearity up to 5.3 × 106 copies/ml. CMV DNA was inconsistently detected in dilutions containing less than 2.2 × 102 copies/ml. The interassay variation ranged from 15 to 58% and the intra-assay variation ranged from 7 to 35%, with a trend toward higher deviations for samples with lower viral loads. These results are in concordance with those reported for other molecular assays based on automated sample preparation and real-time PCR (14).
When clinical samples were tested by the new assay, discrepant results were found particularly for samples with lower viral loads. The reason for these discrepancies remains unclear, but the possibility of an incompatibility of the buffer systems cannot be excluded. The highest number of positive results was obtained when the samples were extracted on the MagNa Pure compact instrument, followed by amplification and detection with the CMV HHV6,7,8 R-gene quantification kit. However, the CMV DNA concentrations in the majority of those samples were found to be weakly positive or within half a log unit above the lower limit of detection. In routine diagnostic laboratories, CMV levels this low are usually not considered significant.
In conclusion, use of the artus CMV LC PCR kit in conjunction with automated sample preparation on the BioRobot EZ1 workstation may be suitable for the detection and quantitation of CMV DNA in EDTA whole blood in the routine low-throughput laboratory. The detection of CMV DNA in low-positive samples may be improved by the use of an alternative test system.
Acknowledgments
We gratefully acknowledge Philippe Bourgeois for stimulating discussions.
This study was supported in part by the Austrian-Croatian Scientific and Education Cooperation Action Program (11/2006) and by Qiagen.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Allice, T., M. Enrietto, F. Pittaluga, S. Varetto. A. Franchello, G. Marchiaro, and V. Ghisetti. 2006. Quantitation of cytomegalovirus DNA by real-time polymerase chain reaction in peripheral blood specimens of patients with solid organ transplants: comparison with end-point PCR and pp65 antigen test. J. Med. Virol. 78915-922. [DOI] [PubMed] [Google Scholar]
- 2.Boeckh, M., M. Huang, J. Ferrenberg, T. Stevens-Ayers, L. Stensland, W. G. Nichols, and L. Corey. 2004. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 421142-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boriskin, Y. S., K. Fuller, R. L. Powles, I. B. Vipond, P. S. Rice, J. C. Booth, E. O. Caul, and P. D. Butcher. 2002. Early detection of cytomegalovirus (CMV) infection in bone marrow transplant patients by reverse transcription-PCR for CMV spliced late gene UL21.5: a two site evaluation. J. Clin. Virol. 2413-23. [DOI] [PubMed] [Google Scholar]
- 4.Caliendo, A. M., J. Ingersoll, A. M. Fox-Canale, S. Pargman, T. Bythwood, M. K. Hayden, J. W. Bremer, and N. S. Lurain. 2007. Evaluation of real-time PCR laboratory-developed tests using analyte-specific reagents for cytomegalovirus quantification. J. Clin. Microbiol. 451723-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrigue, I., S. Boucher, L. Couzi, A. Caumont, C. Dromer, M. Neau-Cransac, R. Tabrizi, M.-H. Schrive, H. Fleury, and M.-E. Lafon. 2006. Whole blood real-time quantitative PCR for cytomegalovirus infection follow-up in transplant recipients. J. Clin. Virol. 3672-75. [DOI] [PubMed] [Google Scholar]
- 6.Khoury, J. A., G. A. Storch, D. L. Bohl, R. M. Schuessler, S. M. Torrence, M. Lockwood, M. Gaudreault-Keener, M. J. Koch, B. W. Miller, K. L. Hardinger, M. A. Schnitzler, and D. C. Brennan. 2006. Prophylactic versus preemptive oral valganciclovir for the management of cytomegalovirus infection in adult renal transplant recipients. Am. J. Transplant. 62134-2143. [DOI] [PubMed] [Google Scholar]
- 7.Ljungman, P., P. Reusser, R. de la Camara, H. Einsele, D. Engelhard, P. Ribaud, K. Ward, and C. Cordonnier. 2004. Management of CMV infections: recommendations from the infectious diseases working party of the EBMT. Bone Marrow Transplant. 331075-1081. [DOI] [PubMed] [Google Scholar]
- 8.Meijer, E., G. J. Boland, and L. F. Verdonck. 2003. Prevention of cytomegalovirus disease in recipients of allogeneic stem cell transplants. Clin. Microbiol. Rev. 16647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mengelle, C., K. Sandres-Sauné, C. Pasquier, L. Rostaing, J.-M. Mansuy, M. Marty, I. Da Silva, M. Attal, P. Massip, and J. Izopet. 2003. Automated extraction and quantification of human cytomegalovirus DNA in whole blood by real-time PCR assay. J. Clin. Microbiol. 413840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mocarski, E. S. 1996. Cytomegaloviruses and their replication, p. 2447-2492. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 11.Razonable, R. R., and V. C. Emery. 2004. Management of CMV Infection and disease in transplant patients. Herpes 1177-86. [PubMed] [Google Scholar]
- 12.Sia, I. G., J. A. Wilson, M. J. Espy, C. V. Paya, and T. F. Smith. 2000. Evaluation of the COBAS AMPLICOR CMV MONITOR test for detection of viral DNA in specimens taken from patients after liver transplantation. J. Clin. Microbiol. 38600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skapova, D., Z. Racil, D. Dvorakova, D. Minarikova, and J. Mayer. 2005. Significance of qualitative PCR detection methods for preemptive therapy of cytomegalovirus infection in patients after allogeneic hematopoietic stem cell transplantation—single-center experience. Neoplasma 52137-142. [PubMed] [Google Scholar]
- 14.Stelzl, E., Z. Muller, E. Marth, and H. H. Kessler. 2004. Rapid quantification of hepatitis B virus DNA by automated sample preparation and real-time PCR. J. Clin. Microbiol. 422445-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vancikova, Z., and P. Dvorak. 2001. Cytomegalovirus infection in immunocompetent and immunocompromised individuals—a review. Curr. Drug Targets Immune Endocr. Metab. Disord. 1179-187. [PubMed] [Google Scholar]
- 16.Weinberg, A., D. Schissel, and R. Giller. 2002. Molecular methods for cytomegalovirus surveillance in bone marrow transplant recipients. J. Clin. Microbiol. 404203-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]