Abstract
Norovirus infections were detected in 114 of 762 children with acute gastroenteritis in South Korea from November 2005 to November 2006. Seasonality peaks in December, March, and October were also assessed in this study. We identified seven noroviral genotypes (GI-6, GII-2, GII-3, GII-4, GII-5, GII-6, and GII-8) and a C1-120 strain showing low identity (79.3%) with GII-13 and GII-17.
Norovirus (NoV) is one of the most important viruses that cause nonbacterial acute gastroenteritis in humans. The mortality of acute gastroenteritis was estimated to be 2.1 million in the year 2000, and mortality due to gastroenteritis in children was higher in developing countries than in the developed countries (2). Recently, NoVs have been recognized as novel emerging pathogens.
NoV is a member of the family Caliciviridae and harbors a positive-sense, single-stranded RNA (7.6 kb). The NoVs are classified into five genogroups (genogroup I [GI] to GV), and human NoV is divided into three genogroups, GI, GII, and GIV, which are further classified into 8, 17, and 1 genotypes, respectively (15). Previous studies have demonstrated that the GII-4 genotype is the dominant circulating genotype worldwide (3, 8, 14).
In South Korea, a study was conducted in an effort to characterize the molecular epidemiology of gastroenteritis outbreaks induced by human NoV infection (9); however, no study has yet addressed the molecular epidemiology of human NoVs from various districts of South Korea.
Stool specimens were collected from children under 5 years of age suffering from diarrheal disease from eight domestic hospitals (Our Lady of Mercy Hospital in Kangnam, St. Mary's Hospital, St. Vincent Hospital, Severance Hospital, Wonju Christian Hospital, Jeonju Jesus Hospital, Changwon Fatima Hospital, and Chungnam National University Hospital) in South Korea from November 2005 to November 2006.
The viral genomic RNA was extracted from 140 μl of 10% fecal suspensions via the application of the Qiaamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) was conducted using a One Step RT-PCR kit (Qiagen, Hilden, Germany) for the NoV genotypes. We amplified a 330-bp fragment (positions 5342 to 5671 of NoV M87661) of the capsid gene (GI) with the consensus forward primer NV-GIF1 (5′-CTG CCC GAA TTY GTA AAT GAT CAT-3′) and the reverse primer NV-GIR1 (5′-CCA ACC CAR CCA TTR TAC ATY TG-3′) (9). We also amplified a 341-bp (positions 5061 to 5401 of Lordsdale virus X86557) fragment of the capsid gene (GII) harboring the consensus forward primer NV-GIIF1 (5′-GGG AGG GCG ATC GCA ATC T-3′) and the reverse primer NV-GIIR1 (5′-CCR CCI GCA TRI CCR TTR TAC AT-3′) (9). The reaction was conducted with an initial RT step at 50°C for 30 min, followed by PCR activation at 95°C for 15 min and then 35 cycles of amplification (45 s at 94°C, 50 s at 58°C, and 45 s at 72°C) and a final extension step of 10 min at 72°C in a PCR System Px2 thermal cycler (Thermo Hybaid, Middlesex, United Kingdom). The PCR products were run on 1.5% agarose gels, stained with ethidium bromide, and visualized under UV light. The products were extracted using a Qiaquick PCR purification kit (Qiagen, Hilden, Germany) and were sequenced by Genotech (Daejeon, South Korea).
The phylogenetic analyses were conducted using the DNAStar version 5.07 software package. The DNA sequences were aligned by the Clustal W method. The dendrograms were constructed by the neighbor-joining method.
Among the 762 stool specimens, 114 (15.0%) fecal samples were identified as infected by NoVs by RT-PCR and nucleotide sequence analysis. Twelve (10.5%) of the 114 specimens were determined to belong to GI strains, and 102 (89.5%) of the 114 specimens belonged to GII strains.
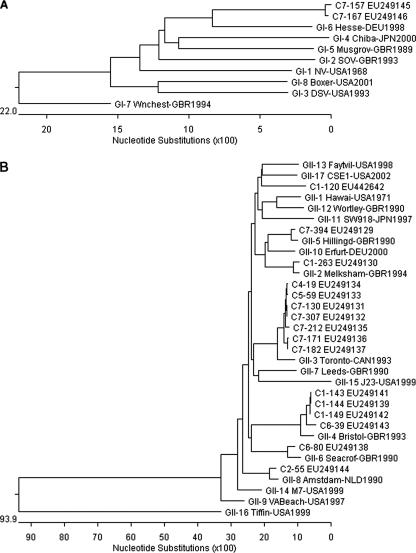
Whereas 12 GI NoVs among the total 114 NoVs were classified further into only one genotype, GI-6, accounting for 10.5% (12 of 114), 102 GII NoVs were classified further into GII-2, GII-3, GII-4, GII-5, GII-6, and GII-8, as well as a C1-120 strain, accounting for 0.9% (1 of 114), 7.9% (9 of 114), 71.9% (82 of 114), 5.3% (6 of 114), 1.8% (2 of 114), 0.9% (1 of 114), and 0.9% (1 of 114), respectively. The NoVs identified in this study were constructed via the phylogenetic analyses of nucleotide sequences on the basis of the GI (314-bp) and GII (305-bp) capsid regions (Fig. 1A and 1B).
FIG. 1.
Phylogenetic analysis of identified NoVs based on GI (314-bp region) (A) and GII (305-bp region) (B) of the NoV capsid gene. The compared strains were GI-1NV-USA1968 (Norwalk, M87661), GI-2SOV-GBR1993 (Southampton, L07418), GI-3DSV-USA1993 (Desert Shield, U04469), GI-4Chiba-JPN2000 (AB042808), GI-5Musgrov-GBR1989 (Musgrove, AJ277614), GI-6Hesse-DEU1998 (AF093797), GI-7Wnchest-GBR1994 (Winchester, AJ277609), GI-8Boxer-USA2001 (AF538679), GII-1Hawaii-USA1971 (U07611), GII-2Melksham-GBR1994 (X81879), GII-3Toronto-CAN1993 (U02030), GII-4Bristol-GBR1993 (X76716), GII-5Hillingd-GBR1990 (Hillingdon, AJ277607), GII-6Seacrof-GBR1990 (Seacroft, AJ277620), GII-7Leeds-GBR1990 (AJ277608), GII-8Amstdam-NLD1990 (Amsterdam, AF195848), GII-9VABeach-USA1997 (AY038599), GII-10Erfurt-DEU2000 (AF427118), GII-11SW918-JPN1997 (AB074893), GII-12Wortley-GBR1990 (AJ277618), GII-13Faytvil-USA1998 (Fayetteville, AY113106), GII-14M7-USA1999 (AY130761), GII-15J23-USA1999 (AY130762), GII-16Tiffin-USA1999 (AY502010), GII-17CSE1-USA2002 (AY502009), and C1-120 (EU442642).
The difference in the NoV infection rates between males and females was not significant (50.9% and 49.1%, respectively) (data not shown). The distribution of NoV GI and GII prevalence in children with acute gastroenteritis by age was as follows: 6 GI and 43 GII (0 to 1 year), 4 GI and 32 GII (1 to 2 years), 1 GI and 20 GII (2 to 3 years), 1 GI and 6 GII (3 to 4 years), and 1 GI and 0 GII (4 to 5 years). Although NoV infections were detected in all age groups, NoV infections were found most frequently in the <1-year-old group (43.0%; 49 of 114). It was also determined that most of the NoV infections occurred in children <2 years of age (74.6%; 85 of 114).
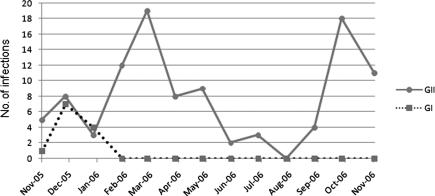
NoVs were continuously detected throughout the year, but the principal peaks of detection in South Korea were in December, March, and October (Fig. 2). GI NoV infections exhibited a peak in December, corresponding to the winter season, whereas GII NoV infections evidenced two peaks, in March and October, corresponding to the spring and fall, respectively. The peaks for GI NoV infections preceded those for GII NoV infections by at least 3 months.
FIG. 2.
Seasonality of GI and GII NoV infections in South Korea, November 2005 to November 2006.
NoVs, which cause acute gastroenteritis, have emerged as an increasingly urgent public health problem in societies and hospitals. Due to the absence of a cell culture system and an experimental-animal model for NoVs, the development of sensitive diagnostic methods for the detection of these viruses is required. With the introduction of RT-PCR and enzyme-linked immunosorbent assay as diagnostic methods, NoV has come to be understood as an important etiologic agent of acute gastroenteritis worldwide (4). Although a previous study of NoV infections in South Korea was conducted in which outbreaks of NoV in Jeju Island, South Korea, were described (9), it remains an open question whether many varied genotypes of NoVs were actually detected in these outbreaks.
Our results demonstrated that the detected NoVs belonged to two distinct genogroups, namely, GI and GII, and these represented 10.5% and 89.5%, respectively. These findings were concordant with previous epidemiological studies conducted worldwide; NoV GI was consistently present at low incidence in fecal specimens compared to NoV GII (11, 12). Among the NoV GII isolates, we detected one strain, C1-120, which showed low identity (79.3%) with the closest references, NoV GII-13 and GII-17, on the basis of a 305-bp region (positions 5085 to 5389 of Lordsdale virus X86557) of the NoV capsid gene (Fig. 1B).
The overall frequency (15.0%) of NoV detected in our study is consistent with previous reports regarding the molecular epidemiology of NoV infection worldwide, in which the prevalence fell within a range of 6% to 19% (1, 5, 6). The highest incidence of NoV infection was in the 1-year-old group, and the incidence decreased with increasing age over 2 years.
In many countries, NoV infections prevail during the winter months (10, 13), though several studies evidenced a peak of seasonal distribution (7, 13). In our study, the principal peaks of NoV infection were in December (for GI) and in March and October (for GII). Thus, the NoV GI and GII infections evidenced very different seasonality characteristics. Obviously, there are three peaks of NoV infections that cause acute gastroenteritis in South Korea. However, the seasonality pattern observed in this study must be analyzed with caution, as our study involved only a 1-year detection period; longer periods will be required in order to determine with more accuracy the possible pattern of seasonality.
Our study is the first large-scale epidemiological study in South Korea showing diverse NoV genogroups and a potential novel strain from a large number of samples from eight hospitals located in a variety of provinces. Nevertheless, continuous epidemiological studies and monitoring of NoV infections in South Korea will be necessary to effectively and efficiently address and solve public health problems in South Korean communities.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this article have been deposited at GenBank (accession numbers EU249129 to EU249146 and EU442642).
Acknowledgments
This work was supported by the National Institute of Environment Research (Ministry of Environment) and The Office of Waterworks (Seoul Metropolitan) Fund, MOEHRD (KRF-2006-E00010), and the BK21 project team for Biomedical Science.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Buesa, J., B. Collado, P. Lopez-Andjar, R. Abu-Mallouh, J. Rodrgues-Daz, A. Garca-Daz, J. Prat, S. Guix, T. Llovet, G. Prats, and A. Bosh. 2002. Molecular epidemiology of calicivirus causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J. Clin. Microbiol. 402854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dey, S. K., T. A. Nguyen, T. G. Phan, O. Nishio, A. F. M. Salim, M. Rahman, F. Yagyu, S. Okitsu, and H. Ushijima. 2007. Molecular and epidemiological trend of norovirus associated gastroenteritis in Dhaka City, Bangladesh. J. Clin. Virol. 13908-911. [DOI] [PubMed] [Google Scholar]
- 3.Dove, W., N. A. Cunliffe, J. S. Gondwe, R. L. Broadhead, M. E. Molyneux, O. Nakagomi, and C. A. Hart. 2005. Detection and characterization of human caliciviruses in hospitalized children with acute gastroenteritis in Blantyre, Malawi. J. Med. Virol. 77522-527. [DOI] [PubMed] [Google Scholar]
- 4.Estes, M. K., B. V. Prasad, and R. L. Atmar. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19467-474. [DOI] [PubMed] [Google Scholar]
- 5.Farkas, T., X. Jiang, M. L. Guerrero, W. Zhong, N. Wilton, T. Berk, D. O. Matson, L. K. Pickering, and G. Ruiz-Palacios. 2000. Prevalence and genetic diversity of human caliciviruses (HuCVs) in Mexican children. J. Med. Virol. 62217-223. [PubMed] [Google Scholar]
- 6.Foley, B., J. O'Mahony, S. M. Morgan, C. Hill, and J. G. Morgan. 2000. Detection of sporadic cases of Norwalk-like virus (NLV) and astrovirus infection in a single Irish hospital from 1996 to 1998. J. Clin. Virol. 17109-117. [DOI] [PubMed] [Google Scholar]
- 7.Hansman, S., L. T. P. Doan, T. A. Nguyen, S. Okitsu, K. Katayama, S. Ogawa, K. Natori, N. Takeda, Y. Kato, O. Nishio, M. Noda, and H. Ushijima. 2007. Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. J. Med. Virol. 79582-590. [DOI] [PubMed] [Google Scholar]
- 8.Kageyama, T., M. Shinohara, K. Uchida, S. Fukushi, F. B. Hoshino, S. Kojima, R. Takai, T. Oka, N. Takeda, and K. Katayama. 2004. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J. Clin. Microbiol. 422988-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, S. H., D. S. Cheon, J. H. Kim, D. H. Lee, W. H. Jheong, Y. J. Heo, H. M. Chung, Y. Jee, and J. S. Lee. 2005. Outbreaks of gastroenteritis that occurred during school excursions in Korea were associated with several waterborne strains of norovirus. J. Clin. Microbiol. 434836-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koopmans, M., J. Vinje, M. de Wit, I. Leenen, W. van der Poel, and Y. van Duynhoven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181262-269. [DOI] [PubMed] [Google Scholar]
- 11.Oh, D. Y., G. Gaedicke, and E. Schreier. 2003. Viral agents of acute gastroenteritis in German children: prevalence and molecular diversity. J. Med. Virol. 7182-93. [DOI] [PubMed] [Google Scholar]
- 12.Phan, T. G., M. Okame, T. A. Nguyen, N. Maneekarn, O. Nishio, S. Okitsu, and H. Ushijima. 2004. Human astrovirus, norovirus (GI, GII), and sapovirus infections in Pakistani children with diarrhea. J. Med. Virol. 73256-261. [DOI] [PubMed] [Google Scholar]
- 13.Phan, T. G., S. Takanashi, K. Kaneshi, Y. Ueda, S. Nakaya, S. Nishimura, K. Sugita, T. Nishimura, A. Yamamoto, F. Yagyu, S. Okitsu, N. Maneekarn, and H. Ushijima. 2006. Detectiion and genetic characterization of norovirus strains circulating among infants and children with acute gastroenteritis in Japan during 2004-2005. Clin. Lab. 52519-525. [PubMed] [Google Scholar]
- 14.Radford, A. D., R. M. Gaskell, and C. A. Hart. 2004. Human norovirus infection and the lessons from animal caliciviruses. Curr. Opin. Infect. Dis. 17471-478. [DOI] [PubMed] [Google Scholar]
- 15.Zheng, D.-P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]