Abstract
Coronavirus (CoV) transcription includes a discontinuous mechanism during the synthesis of sub-genome-length minus-strand RNAs leading to a collection of mRNAs in which the 5′ terminal leader sequence is fused to contiguous genome sequences. It has been previously shown that transcription-regulating sequences (TRSs) preceding each gene regulate transcription. Base pairing between the leader TRS (TRS-L) and the complement of the body TRS (cTRS-B) in the nascent RNA is a determinant factor during CoV transcription. In fact, in transmissible gastroenteritis CoV, a good correlation has been observed between subgenomic mRNA (sg mRNA) levels and the free energy (ΔG) of TRS-L and cTRS-B duplex formation. The only exception was sg mRNA N, the most abundant sg mRNA during viral infection in spite of its minimum ΔG associated with duplex formation. We postulated that additional factors should regulate transcription of sg mRNA N. In this report, we have described a novel transcription regulation mechanism operating in CoV by which a 9-nucleotide (nt) sequence located 449 nt upstream of the N gene TRS core sequence (CS-N) interacts with a complementary sequence just upstream of CS-N, specifically increasing the accumulation of sg mRNA N. Alteration of this complementarity in mutant replicon genomes showed a correlation between the predicted stability of the base pairing between 9-nt sequences and the accumulation of sg mRNA N. This interaction is exclusively conserved in group 1a CoVs, the only CoV subgroup in which the N gene is not the most 3′ gene in the viral genome. This is the first time that a long-distance RNA-RNA interaction regulating transcriptional activity specifically enhancing the transcription of one gene has been described to occur in CoVs.
Transmissible gastroenteritis virus (TGEV) is a member of the Coronaviridae family, included in the Nidovirales order (6), which is characterized by a common gene expression strategy based on the generation of a nested set of subgenomic (sg) mRNAs, despite significant differences in their genome sizes. Coronaviruses (CoVs) have been classified in groups 1, 2, and 3. The most representative members of group 1 are TGEV and human CoV (HCoV) 229E, which belong to subgroups 1a and 1b, respectively. Group 2 has been divided into subgroups 2a, including mouse hepatitis virus (MHV), and 2b, including the severe acute respiratory syndrome CoV (SARS-CoV). Group 3 includes the infectious bronchitis virus (IBV) as a prototype (6, 9). TGEV has a single-stranded, plus-sense 28.5-kb RNA genome (17). About two-thirds of the entire RNA from the 5′ end comprises open reading frames 1a and 1ab, which encode the replicase. The 3′ third of the genome includes the genes encoding structural and nonstructural proteins (5′-S-3a-3b-E-M-N-7-3′). Engineering of the TGEV genome for the study of fundamental viral processes, such as transcription, has been possible by the construction of TGEV infectious cDNA clones (2, 25) and TGEV-derived replicons (1). CoV transcription is an RNA-dependent RNA synthesis that includes a discontinuous RNA synthesis step. According to current experimental data, this process occurs during the production of sub-genome-length minus-strand RNAs (19, 27) and involves the fusion of a copy of the genomic 5′ terminal leader sequence to the 3′ end of each of the nascent RNAs complementary to the coding (body) sequences. The resulting chimeric sg RNAs of minus polarity serve as templates to yield sg mRNAs that share both 5′ and 3′ terminal sequences with the genome RNA. Genes expressed through sg mRNAs are preceded by conserved transcription-regulating sequences (TRSs) that include the conserved core sequence (CS; 5′-CUAAAC-3′) identical in all TGEV genes and the 5′ and 3′ flanking sequences (5′ TRS and 3′ TRS, respectively).
According to this working model, the TRS preceding each gene (the body TRS [TRS-B]) would act as an attenuation and dissociation signal for the transcription complex during the synthesis of the RNA minus strand in order to promote a strand transfer to the leader region to resume the synthesis of minus-strand sg RNA. This process is named discontinuous extension of the minus strand and can be considered a variant of similarity-assisted template switching that operates during viral RNA recombination (5). Transcription in CoVs is probably regulated by several factors: (i) base pairing between the leader TRS (TRS-L) and the complement of the TRS-B (cTRS-B) in the nascent minus strand, (ii) RNA-protein and protein-protein interactions involving TRSs and viral and cellular proteins, (iii) the relative position in the genome of TRS-B, and (iv) the TRS-B secondary structure. The proximity of TRSs to the 3′ end of the genome probably influences the relative amount of sg mRNA, because the RNA-dependent RNA polymerase finds a smaller number of attenuation and dissociation TRS-B signals during the synthesis of the smaller minus-strand sg RNAs, favoring the synthesis of these sg RNAs. This is the case for other viruses that produce multiple sg RNAs (15) and, in general, for CoVs. Nevertheless, a perfect correlation between mRNA abundance and the relative position of TRS in the genome could not be strictly established (17, 23).
Previous studies on the TGEV transcription mechanism have shown that base pairing between the TRS-L and cTRS-B in the nascent RNA is a key factor during CoV transcription. In fact, the requirement for base pairing between the CS at the 3′ end of the leader and the complement of each CS-B in the nascent minus strand during transcription has been formally demonstrated in arteriviruses (16, 24) and CoVs (27). It was also shown that the synthesis of sg mRNAs proceeds only when a minimum complementarity between TRS-L and cTRS-B is reached. Moreover, a good correlation between sg mRNA levels and the free energy (ΔG) of duplex formation between TRS-L and cTRS-B was observed in TRS-B mutants that extended complementarity with TRS-L (22), indicating that base pairing is a main determinant factor during CoV transcription.
In this report, the relevance of the ΔG of duplex formation between TRS-L and cTRS-B for the sg mRNAs synthesis has been reinforced by demonstrating a good correlation between the levels of viral sg mRNA during TGEV infection and the ΔG of TRS-L and cTRS-B duplex formation. However, sg mRNA N represented a clear exception to this observation, being the most abundant sg mRNA during viral infection in spite of its minimum ΔG. As a consequence, we postulated that there must be additional factors regulating sg mRNA N transcription. The distal 5′ TRS-N region included a 9-nucleotide (nt) sequence (nt −449 relative to the N gene CS [CS-N]) unique throughout the whole TGEV genome, complementary to another 9-nt segment localized immediately upstream of the CS-N. Within this paper, we show that complementarity between both 9-nt elements in TGEV is functionally relevant in the transcriptional activation of the N gene. Moreover, a positive correlation between the predicted stability of base-pairing interaction and accumulation levels of sg mRNA N has been shown. The sequence within the distal 5′ TRS-N region has an enhancer activity similar to that described for other plus-stranded RNA viruses, such as tombusviruses (26). Although there is experimental evidence for the importance of long-distance RNA-RNA interactions in sg mRNA synthesis in other viral systems (12), this is the first time that a long-distance RNA-RNA interaction associated with transcription enhancement has been described to occur in the Nidovirales order as a transcription-regulating mechanism.
MATERIALS AND METHODS
RNA metabolic labeling.
TGEV strain PUR46-MAD (18) was used to infect swine testis (ST) cells (14) grown in 35-mm-diameter plates with a multiplicity of infection of 10 PFU per cell. Infection was performed for 1 h. Then, the inoculum was replaced with phosphate-deficient medium containing 2% dialyzed fetal calf serum (FCS). Cells were incubated at 37°C for 2 h, actinomycin D was added to give a final concentration of 2.5 μg/ml, and cells were incubated for an additional 3 h at 37°C. Then, [33P]H3PO4 was added to give a final concentration of 100 μCi/ml and viral mRNAs were labeled for 3 h at 37°C. At 9 h postinfection (h p.i.), cytoplasmic RNA was extracted using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. RNAs were separated in denaturing 1% agarose-2.2 M formaldehyde gels. The gel was dried under vacuum onto Whatman 3MM paper without relevant loss of radioactivity during the process, and RNA products were visualized and quantitated by PhosphorImager analysis (Molecular Dynamics). The amounts of different sg mRNAs were corrected by taking into account the length of mRNA and the background in each lane. The relative levels of viral mRNAs were normalized to those of the mRNA of the S gene.
Cells and transfection.
Baby hamster kidney (BHK) cells stably transformed with the porcine amino peptidase N gene (4) and with the Sindbis virus replicon pSINrep21 (8), expressing TGEV N protein (BHK-N), were grown in Dulbecco's modified medium supplemented with 5% FCS and G418 (1.5 mg/ml) as a selection agent. BHK-N cells were grown to 95% confluence on 35-mm-diameter plates and transfected with 4 μg of each TGEV replicon, representing on average 100 molecules per cell, by using 12 μg of Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications.
Plasmid constructs.
cDNAs of TGEV-derived replicons including mutated sequences of TRS-N and the coding sequence of the TGEV N gene were generated by PCR-directed mutagenesis. Mutants TRS-N and E-TRS-N were generated by PCR using the plasmid pBAC-TGEV (2), containing the TGEV genome (GenBank accession no. AJ271965), as a template, with the same reverse primer and specific forward oligonucleotides (Table 1). PCR fragments including AscI restriction sites were cloned into the plasmid pBAC-REP-1 (1), previously digested with MluI and AscI enzymes, generating compatible ends. To generate replicon E-TRS-GFP, two overlapping PCR fragments were obtained using pcDNA3-GFP and pBAC-TGEV as templates and specific oligonucleotides given in Table 1. The final amplification product (2,068 bp), which includes the TRS from the N gene fused to the green fluorescent protein (GFP)-coding sequence, was generated with outer oligonucleotides E-800 AvrII VS and 3′AscI GFP RS, digested with AvrII and AscI, and cloned into the same restriction sites of the plasmid pBAC-REP-1. To obtain mutant E-TRS3a-N, a PCR fragment was amplified using pBAC-TGEV as a template and oligonucleotides E-800 AvrII VS and E-120 AvrII RS. The amplification fragment (680 bp) was digested by AvrII and cloned into the same restriction sites of intermediate plasmid pBAC-REP-TRS3a-N (J. L. Moreno, I. Sola, and L. Enjuanes, unpublished).
TABLE 1.
Oligonucleotides used for site-directed mutagenesis
| Mutant | Oligonucleotide | Oligonucleotide sequence (5′ → 3′)a |
|---|---|---|
| E-TRS-N | N-800 AscI VS | TTGGCGCGCCACTCCTTGCTTGAACTAAACAAAATG |
| 3′-AscI RS | TTGGCGCGCCTTAGTTCGTTACCTCATCAATTATC | |
| TRS-N | N-120 AscI VS | TTGGCGCGCCTTGAAAGCAAGTAGTGCGACTGG |
| 3′-AscI RS | ||
| E-TRS-GFP | E-800 AvrII VS | TTCCTAGGACTCCTTGCTTGAACTAAACAAAATG |
| GFP 5′ RS | CAGCTCCTCGCCCTTGCTCACCATTTAGAAGTTTAGTTA | |
| GFP 5′ VS | ACATATGGTATAACTAAACTTCTAAATGGTGAGCAAGGGC | |
| 3′AscI GFP RS | AAGGCGCGCCGGAAGAAAGCGAAAGGAGCGGGCGC | |
| E-TRS3a-N | E-800 AvrII VS | |
| E-120 AvrII RS | AACCTAGGCTTCTTGCCAACAAGTGTGTAGACAATAGTC | |
| E1-TRS-N | E-800 AvrII VS | |
| E-460 AvrII RS | AACCTAGGGACACTTGGTATTCCGAGTATGCATTAAAAATC | |
| E2-TRS-N | E-740 AvrII VS | TTCCTAGGTGGAACTTCAGCTGGTCTATAATATTGATC |
| E-160 AvrII RS | AACCTAGGCATAGCTTCTTCCTAATGCACTAACGCAAAG | |
| E3-TRS-N | E-460 AvrII VS | TTCCTAGGCAGATATGTAATGTTCGGCTTTAGTATTGC |
| E-120 AvrII RS | ||
| M1-TRS-N | Rep Mut 2.1 VS | GTCCAGATTACATATGTTCGGCTTTAGTAT |
| Rep Mut 2.1 RS | CCGAACATATGTAATCTGGACACTTGGTAT | |
| M2-TRS-N | Rep Mut 2.2 VS | AAAATTATATGTAATGGTATAACTAAACTT |
| Rep Mut 2.2 RS | TATACCATTACATATAATTTTTCTTGCTCA | |
| M4-TRS-N | Rep Mut 2 A1G VS | GTGTCCAGGTATGTAATGTTCGGCTTTAGTA |
| Rep Mut 2 A1G RS | AGCCGAACATTACATACCTGGACACTTGGTATTC | |
| M5-TRS-N | Rep Mut 2 A1C VS | GTGTCCAGCTATGTAATGTTCGGCTTTAGTA |
| Rep Mut 2 A1C RS | AGCCGAACATTACATAGCTGGACACTTGGTATTC | |
| M6-TRS-N | Rep Mut 2 A1C-U9C VS | GTGTCCAGCTATGTAACGTTCGGCTTTAGTA |
| Rep Mut 2 A1C-U9C RS | AGCCGAACGTTACATAGCTGGACACTTGGTATTC | |
| M7-TRS-N | Rep Mut 2 CC-U9C VS | GTGTCCCCATACGTAATGTTCGGCTTTAGTA |
| Rep Mut 2 CC-U9C RS | AGCCGAACATTACGTATGGGGACACTTGGTATTC |
The mutated nucleotides are shown in boldface. The restriction endonuclease sites used for cloning are underlined (AvrII, CCTAGG; AscI, GGCGCGCC).
To generate replicons including different sequences from the distal 5′ TRS-N region, PCR fragments of the same size (360 bp) were amplified with specific oligonucleotides containing AvrII restriction sequences (Table 1) and pBAC-TGEV as a template. PCR products were digested with AvrII and cloned into the same restriction sites of plasmid TRS-N, leading to plasmids E1-TRS-N, E2-TRS-N, and E3-TRS-N. To obtain mutations in the complementary 9-nt sequences, overlapping PCR fragments were amplified by using oligonucleotides given in Table 1 and pBAC-TGEV as a template. The final PCR product (340 bp), amplified with outer oligonucleotides E-740 AvrII VS and E-160 AvrII RS, was digested with AvrII and cloned into the same restriction site of the plasmid TRS-N, leading to the mutants M1-TRS-N, M4-TRS-N, M5-TRS-N, M6-TRS-N, and M7-TRS-N. The mutant M2-TRS-N was engineered using TRS-N as a template. Overlapping PCR fragments including mutations were amplified by using specific oligonucleotides (Table 1). The final amplification product (1,645 nt), generated with outer oligonucleotides E-740 AvrII VS and 3′AscI RS, was digested with AvrII and AscI and cloned into the same restriction sites of the plasmid pBAC-REP-1. To generate mutant M3-TRS-N, the AvrII digestion product (340 bp) from plasmid E2-TRS-N was cloned into the same restriction sites of the plasmid M2-TRS-N. All cloning steps were checked by sequencing the PCR-amplified fragments and cloning junctions.
RNA analysis by real-time RT-PCR.
Total intracellular RNA was extracted at 24 h posttransfection (h p.t.) from transfected BHK-pAPN-N cells or at 12 h p.i. from infected ST cells as previously described. cDNAs were synthesized at 50°C for 1.5 h with avian myeloblastosis virus reverse transcriptase (ThermoScript; Invitrogen) and the reverse primers given in Table 2. The cDNAs generated were used as templates for specific PCR amplification. Real-time reverse transcription-PCR (RT-PCR) was used for quantitative analysis of genome RNA (gRNA; used as an endogenous standard) and sg mRNAs from TGEV and TGEV-derived replicons. Oligonucleotides used for RT and PCRs (Table 2) were designed by Primer Express software. Sybr green PCR master mix (Applied Biosystems) was used in the PCR step, according to the manufacturer's specifications. Detection was performed with an ABI PRISM 7000 sequence detection system (Applied Biosystems). Data were analyzed with ABI PRISM 7000 SDS version 1.2.3 software.
TABLE 2.
Oligonucleotides used for real-time RT-PCR analysis
| Amplicon | Forward primer
|
Reverse primer
|
||
|---|---|---|---|---|
| Name | Sequence (5′ → 3′) | Name | Sequence (5′ → 3′) | |
| gRNA | RT-REP-VS | TTCTTTTGACAAAACATACGGTGAA | RT-REP-RS | CTAGGCAACTGGTTTGTAACATCTTT |
| mRNA-S1 | L-CS1 VS | CCAACTCGAACTAAACTTTGGTAACC | L-CS1 RS | TCAATGGCATTACGACCAAAAC |
| mRNA-3a.1 | rt-3aVS2 | CGGACACCAACTCGAACTAAACTTAC | rt3a-RS | ATCAAGTTCGTCAAGTACAGCATCTAC |
| mRNA-E | Ldrt-VS | CGTGGCTATATCTCTTCTTTTACTTTAACTAG | mRNAE-RS | CAATGCCCTAGGAAACGTCATAG |
| mRNA-M | Ldrt-VS | CGTGGCTATATCTCTTCTTTTACTTTAACTAG | mRNAM-RS | GCATGCAATCACACACGCTAA |
| mRNA-N | Ldrt-VS | CGTGGCTATATCTCTTCTTTTACTTTAACTAG | N82RS | TCTTCCGACCACGGGAATT |
| mRNA-7 | Ldrt-VS | CGTGGCTATATCTCTTCTTTTACTTTAACTAG | 7(38)RS | AAAACTGTAATAAATACAGCATGGAGGAA |
| mRNA-GFP | Ldrt-VS | CGTGGCTATATCTCTTCTTTTACTTTAACTAG | RT-GFP RS | CGCCCTTGCTCACCATTTAG |
Pulse and chase assay.
Viral RNAs were metabolically labeled as described above. At 9 h p.i. cells were washed with fresh medium containing unlabeled phosphate and 2% dialyzed FCS. Cells were incubated at 37°C, and the total intracellular RNA was isolated and quantitated at different chase hours as described above.
In silico analysis.
Potential base-pairing score calculations were done as previously described (27). ΔG calculations were performed using the two-state hybridization server (http://www.bioinfo.rpi.edu/applications/hybrid/twostate.php) (13). The analysis of sequences implicated in sg mRNA N synthesis was performed using DNASTAR Lasergene software 7.0.
RESULTS
Quantification of TGEV sg mRNAs.
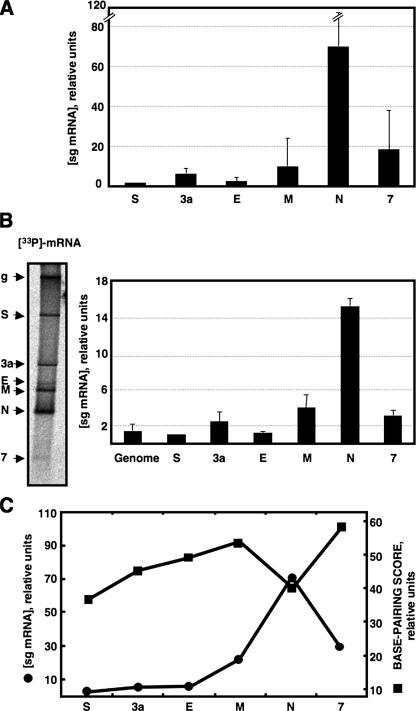
It was previously shown that potential base pairing between TRS-L and the cTRS-B in the nascent minus-strand RNA was a determinant factor for discontinuous transcription in TGEV (22, 27). To study the correlation between TGEV viral sg mRNA levels and base pairing between TRS-L and cTRS-Bs, the amount of viral sg mRNAs was quantified by two different approaches. In the first one, viral sg mRNA accumulation during infection was quantified by real-time RT-PCR. The relative amount of each sg mRNA was expressed in relation to that of sg mRNA S, the least abundant sg mRNA during TGEV infection. Viral sg mRNAs were expressed at different levels, and interestingly, sg mRNA N was the most abundant sg mRNA during TGEV infection, around 70-fold more abundant than sg mRNA S and 4.5-fold more abundant than sg mRNA 7, the second most abundant sg mRNA during infection (Fig. 1A). In the second approach, sg mRNA levels were quantified by mRNA metabolic labeling with [33P]orthophosphate. These experiments confirmed the results obtained by real-time RT-PCR, showing that sg mRNA N was significantly more abundant than the other sg mRNAs and around 4.5-fold more abundant than sg mRNA 7 (Fig. 1B). In general, as expected from previous results (22), a good correlation was observed between the relative amounts of TGEV sg mRNAs and the ΔG of the TRS-L and cTRS-B duplex formation, with the noticeable exception of sg mRNA N, the most abundant sg mRNA during TGEV infection in spite of its minimum ΔG value (Fig. 1C). Therefore, an additional regulation mechanism might be involved in transcriptional activation of sg mRNA N.
FIG. 1.
Relative levels of TGEV sg mRNAs and correlation with the base-pairing scores of leader and nascent RNAs of minus polarity complementary to TRS-Bs. (A) Quantification of TGEV sg mRNA accumulation at 12 h p.i. by real-time RT-PCR. The data presented correspond to the average of six independent experiments performed in triplicate in each case. (B) Quantification of viral mRNAs by metabolic labeling with [33P]orthophosphate. TGEV-infected ST cells were labeled with [33P]orthophosphate from 7 to 10 h p.i. Purified total cytoplasmic RNA was separated by electrophoresis through 1% agarose containing formaldehyde. The labeled RNA was visualized (left) and quantified by PhosphorImager analysis (right). Viral mRNAs are indicated on the left side of the figure. The RNA blot is representative of results from three experiments. Error bars represent standard deviations. (C) Relation between viral mRNA relative levels (circles) and the base-pairing scores of TRS-L and cTRS-Bs (squares).
Study of TGEV sg mRNA stability.
To analyze whether a higher stability of sg mRNA N might explain its high levels during TGEV infection, pulse and chase experiments were performed. Viral RNAs were metabolically labeled with [33P]orthophosphate, and RNA from infected cells was isolated at different chase times. RNAs were separated by electrophoresis, and labeled sg mRNAs were quantified by gel densitometry (Fig. 2 and data not shown). As observed from autoradiography and quantification, the stability of sg mRNA N was not higher than that of the other viral sg mRNAs. Therefore, the exceptional high levels of mRNA N during transcription must be caused by another regulatory mechanism.
FIG. 2.
TGEV sg mRNA stability as determined by pulse-chase experiments. Viral sg mRNAs were metabolically labeled with [33P]orthophosphate. At 9 h p.i., cells were incubated with fresh medium containing unlabeled phosphate and the total intracellular RNA was harvested and analyzed by electrophoresis in a denaturing gel at the indicated times.
Localization of signals responsible for transcriptional activation of sg mRNA N.
The increase in transcription levels over those predicted by the ΔG value of base pairing between TRS-L and cTRS-B was observed only for the N gene; therefore, we decided to investigate potential regulatory sequences near this gene. The presence of signals responsible for the specific transcriptional activation of sg mRNA N was analyzed within the N gene coding sequence and within the distal and proximal 5′ TRSs of the N gene, comprising the M gene, by using a TGEV-derived replicon (REP-1). The cDNA of this replicon includes the N gene in the same sequence context and at the same distance from the 3′ end as in the whole viral genome (1). In addition, since efficient RNA synthesis activity of this replicon is associated with the presence of the nucleoprotein, and the expression of this protein is prevented in some designed mutants, the analysis of their functionality was performed with BHK cells expressing in trans TGEV N protein. A set of three mutants was engineered using the TGEV-derived replicon (Fig. 3A). The E-TRS-N mutant contained the coding sequence of the N gene and 800 nt from the 5′ TRS of the N gene, representing the complete M gene. This sequence context was identical to that of the N gene in the wild-type (wt) virus. In the E-TRS-GFP mutant, the N gene coding sequence from E-TRS-N was replaced by the GFP coding sequence (600 nt) plus a nonrelated 500-nt region to mimic the length of original N gene (1,100 bp). To study the effect of the proximal 5′ TRS of the N gene in the levels of sg mRNA N, a replicon mutant TRS-N that includes only the N gene coding sequence and 120 nt of the 5′ TRS was engineered. BHK-N cells were transfected with the cDNAs encoding mutant replicons, and the levels of intracellular sg mRNA N were analyzed by real-time RT-PCR. The amount of sg mRNA N was expressed in relation to that of sg mRNA 7. The relative level of sg mRNA N in the E-TRS-N mutant was similar to that obtained during TGEV infection (Fig. 3B), indicating that similar transcription-regulating mechanisms most likely were operating in the replicon and in the whole virus and validating the study of transcriptional activation of the N gene in the replicon system. The transcriptional activity of E-TRS-GFP was similar to that of E-TRS-N, including the same regulating sequences and the N gene coding sequence, indicating that the signals responsible for transcriptional activation of the N gene were not included within the N gene coding sequence and were mostly located within the 5′ TRS. In contrast, the amount of sg mRNA N from mutant TRS-N was significantly reduced, indicating that the proximal 5′ TRSs comprising 120 nt immediately upstream of CS-N were not sufficient to promote transcription of the N gene up to the levels observed in a standard virus infection and that the distal 5′ TRS-N sequences, including nt −800 to −120 upstream of the CS-N, were necessary for transcriptional activation of the N gene.
FIG. 3.
Localization of signals responsible for transcriptional activation of mRNA N. (A) Scheme showing the genetic structure of TGEV-derived replicons. The TGEV genome and the genetic structure of TGEV-derived replicon REP-1 are shown in the first and second lines, respectively. TGEV genes are represented by letters above the boxes. The scheme of different mutant replicons generated to localize the signals responsible for transcriptional activation of sg mRNA N is shown at the bottom, and their names are indicated to the left. Numbers indicate distances in nucleotides upstream of CS-N. Distal 5′ TRS-N includes nt −800 to −120 upstream of the CS of the N gene (larger black box). Deletion mutants from this region are represented with smaller black boxes. 5′ TRS-N represents the first 120 nt upstream of the CS of the N gene (dark gray boxes). 3′ TRS-N includes the 3′ TRS of gene N. N indicates the N gene coding sequence (light gray boxes). 5′ TRS-3a includes the first 120 nt upstream of the CS of gene 3a. 3′ TRS-3a includes the 3′ TRS of gene 3a. UTR, untranslated region. (B) Quantification of sg mRNA N by real-time RT-PCR. The amounts of sg mRNA N expressed from each mutant replicon are shown relative to that of sg mRNA 7. The data are the averages of four independent experiments performed in triplicate in each case. Error bars represent standard deviations.
To localize the signals responsible for sg mRNA N transcription activity within the distal 5′ TRS-N region, three deletion mutants were engineered (Fig. 3A). These mutants, E1-TRS-N, E2-TRS-N, and E3-TRS-N, included overlapping sequences from the distal 5′ TRS-N region, nt −800 to −460, nt −630 to −290, and nt −460 to −120, respectively, preventing the interruption of potential elements responsible for transcriptional activation of the N gene at the level of primary sequence or secondary structure. These sequences were fused to the proximal region included in the TRS-N mutant, comprising 120 nt immediately upstream of the CS-N and 3′ TRS-N. To study whether the effect in transcriptional activation of the distal 5′ TRS-N region was independent of the proximal 5′ TRS-N region, these sequences were replaced in mutant E-TRS-N by the proximal 5′ TRS from gene 3a (TRS-3a), generating the E-TRS3a-N mutant. BHK-N cells were transfected with cDNAs encoding the mutant replicons, and the levels of sg mRNA N were analyzed by real-time RT-PCR. The levels of sg mRNA N in the E2-TRS-N mutant were even higher than those in the replicon containing the complete 5′ TRS-N regulating sequences, probably as a consequence of approaching the distal and proximal 5′ TRS-N domains containing the elements involved in the transcriptional activation of the N gene. In contrast, the amounts of sg mRNA N in mutants E1-TRS-N and E3-TRS-N were significantly lower than those in E-TRS-N and TGEV (Fig. 3B). Therefore, the sequences from the distal 5′ TRS-N region included in the E2-TRS-N mutant were required for high-level transcription of the N gene. Nevertheless, the sg mRNA N levels in the E-TRS3a-N mutant, containing the complete distal 5′ TRS-N region fused to the proximal 5′ TRS from gene 3a, were lower than those in E-TRS-N and TGEV, indicating that the distal 5′ TRS-N regions were not able to enhance the transcription of the N gene when associated with other proximal 5′ TRSs (Fig. 3B). All together, these results indicated that transcriptional activation of gene N requires the presence of sequences located within the distal 5′ TRS-N region (nt −630 to −290) in addition to sequences located just upstream of the CS of gene N (the first 120 nt).
Long-distance RNA-RNA interaction is required for transcriptional activation of gene N.
Sequence analysis of the distal 5′ TRS-N region included in the E2-TRS-N mutant and the proximal 5′ TRS-N region revealed the presence of a 9-nt sequence (5′-AUAUGUAAU-3′) within the distal 5′ TRS-N region, 449 nt upstream of CS-N, unique along the whole genome of the TGEV-PUR46-MAD strain, which was complementary to another 9-nt segment (5′-AUUACAUAU-3′) located 7 nucleotides upstream of CS-N (Fig. 4A). To test whether the transcriptional activation of the N gene was regulated by the putative base-paring interaction between the distal and proximal 9-nt sequences, a set of mutants in which the predicted interaction was disrupted was constructed (Fig. 4A). The distal 9-nt sequence is located into a transmembrane domain of the M protein, according to previous publications (7). In the M1-TRS-N mutant, the 9-nt sequence in the distal 5′ TRS-N region was replaced by a mutant sequence (5′-AUUACAUAU-3′) that disrupted the potential base-pairing interaction in 5 out of 9 nt, whereas in the M2-TRS-N mutant, the 9-nt sequence located in the proximal 5′ TRS-N region was replaced by another sequence (5′-AUAUGUAAU-3′) that similarly disrupted the predicted base-pairing interaction. Mutant M3-TRS-N included complementarity 9-nt mutant sequences at distal and proximal locations that restored potential base pairing. BHK-N cells were transfected with the cDNA of mutant replicons, and sg mRNA N levels were determined by real-time RT-PCR (Fig. 3B). The sg mRNA N levels in the M1-TRS-N and M2-TRS-N mutants were significantly lower than those in the original E2-TRS-N replicon. In contrast, the amount of sg mRNA N in the M3-TRS-N mutant increased almost to levels similar to those observed in the E2-TRS-N replicon. The remaining activities of these mutants were probably due to the retention of partial base pairing between the mutant sequence and its complementary element. These results reinforce the functional relevance of the proposed long-distance base pairing between proximal and distal 9-nt elements for N gene transcriptional activation.
FIG. 4.
Long-distance RNA-RNA interaction required for transcriptional activation of the N gene. (A) Scheme showing the genetic structure of TGEV-derived replicons. The 9-nt sequence segments are represented by small white boxes within the proximal and distal 5′ TRS-N regions. The scheme of different mutant replicons generated to analyze the functional relevance of the putative RNA-RNA interaction is shown at the bottom, and their names are indicated to the left. The sequences of the 9-nt motif mutants located within the proximal and distal 5′ TRS-N regions are represented in smaller black boxes. Asterisks represent nucleotides identical to those in E2-TRS-N. Potential base pairing between proximal and distal sequences is shown in the right column by vertical lines. See the legend to Fig. 3 for more details. UTR, untranslated region. (B) Quantification of sg mRNA N by real-time RT-PCR. The amounts of sg mRNA N expressed by each mutant replicon are shown relative to that of sg mRNA 7. The data are the averages of four independent experiments performed in triplicate in each case. Error bars represent standard deviations.
Influence of ΔG duplex formation between the distal and proximal 9-nt elements on sg mRNA N synthesis.
To study the effect of duplex stability between the distal and proximal 9-nt sequences in the sg mRNA N synthesis, four mutants with nucleotide substitutions within the distal 9-nt sequence were generated (Fig. 5A). In the M4-TRS-N and M5-TRS-N mutants, the most 5′ nucleotide at the distal 9-nt sequence was mutated to a G and a C, respectively, decreasing the stability, measured as ΔG of duplex between the distal and proximal 9-nt elements (Fig. 5A). The ΔG value of duplex formation in the M4-TRS-N mutant was higher than that in the M5-TRS-N mutant because a non-Watson-Crick base pair could be established between the mutated G and the U in the proximal sequence. In the M6-TRS-N mutant, two point mutations at the 5′ and 3′ ends of the distal sequence severely decreased the ΔG. In the M7-TRS-N mutant, in addition to an internal mutation that disrupts continuous complementarity, two nucleotide substitutions that increased the complementarity of the distal 9-nt element with the proximal element up to a ΔG value similar to that of the wt sequence were introduced just upstream of the distal 9-nt element. BHK-N cells were transfected with cDNAs encoding the mutant replicons, and the levels of sg mRNA N were analyzed by real-time RT-PCR (Fig. 5B). The amounts of sg mRNA N in replicons M4, M5, and M6, including mutations that decreased duplex stability between proximal and distal elements, were lower than that in the E2-TRS-N replicon. In contrast, the level of sg mRNA N in the M7-TRS-N mutant, with a ΔG for duplex formation similar to that of the E2-TRS-N replicon, was similar to that observed in the wt replicon, in spite of the internal mutation that disrupts complementarity between sequence elements. A good correlation was observed between the amount of sg mRNA N and duplex formation ΔG in the distal and proximal 9-nt elements, indicating that the stability of the duplex between these elements may be a decisive factor for transcriptional activation of the TGEV N gene.
FIG. 5.
Influence of ΔG duplex formation between the distal and proximal elements on sg mRNA N synthesis. (A) Scheme showing the genetic structures of REP-1, E-TRS-N, and E2-TRS-N TGEV-derived replicons. The names of mutant replicons are indicated in the left column. Sequences of the proximal and distal 9-nt motifs in mutant replicons are shown in the central columns. Asterisks represent nucleotides identical to those in replicon E2-TRS-N. Letters within the light gray box in mutant M7-TRS-N represent nucleotides increasing the complementarity with the proximal 9-nt element. Potential base pairing between proximal and distal sequences are indicated in the right column. Solid vertical bars represent Watson-Crick base pairing. Dotted vertical bars represent non-Watson-Crick interactions. See the legend to Fig. 4 for more details. UTR, untranslated region. (B) Quantification of sg mRNA N by real-time RT-PCR. The mutants are represented in decreasing order of ΔG duplex formation between the distal and proximal 9-nt sequences. The data are the averages of four independent experiments performed in triplicate in each case. Error bars represent standard deviations.
Conservation of long-distance RNA-RNA interaction in CoVs.
To determine whether the 9-nt sequence elements were present in other TGEV strains, a sequence analysis was performed using several TGEV strains (Fig. 6). The two 9-nt sequences, located at the distal and proximal 5′ TRS-N regions, were conserved in most analyzed TGEV strains. However, in recent TGEV isolates, point mutations were observed in the first nucleotide of the distal 9-nt sequence (A-to-G substitution in ENG96 and TAW83), which leads no amino acid change in the M protein, and in the eighth nucleotide of the proximal sequence (A-to-U substitution in HN2002) (Fig. 6). In two porcine respiratory viruses (PRCV-RM4 and PRCV-ISU-I), TGEV-derived mutants with differences in tropism and pathogenicity, the same point mutation at the first nucleotide of the distal 9-nt sequence was found. Interestingly, in the PRCV-ISU-I strain, an additional point mutation (C-to-U substitution) was observed at position 175 upstream of CS-N, generating a new 9-nt sequence domain with a perfect complementarity to the proximal 9-nt segment. This intermediate domain (Fig. 6) includes in the other TGEV isolates an internal mutation (5′-AUACGUAAU-3′ [the mutated nucleotide is underlined]) that disrupts the potential complementarity to the proximal 9-nt sequence. This intermediate sequence domain, found in most TGEV isolates, was not able to promote transcriptional activation of the N gene, as shown in the E3-TRS-N mutant, which includes a region containing this motif (Fig. 3B). These observations reinforce the relevance of the extent of complementarity between the distal and proximal 5′ TRS-N regions to enhance the transcriptional activation of sg mRNA N.
FIG. 6.
Alignment of TRSs from different group 1a CoVs in the region involved in transcriptional activation of the TGEV N gene. In the first line, TGEV-derived replicon REP-1 is shown. TGEV genes are represented by letters above the boxes. In the second line, a scheme including 9-nt sequences involved in sg mRNA N synthesis is shown (not to scale). Numbers indicate distances in nucleotides upstream of CS-N. Distal, intermediate, and proximal 9-nt motifs are shown within light gray rectangles. The nucleotide inside the white box disrupts the base pairing with the proximal 9-nt element. In virus names, the specific strain and virus isolate is followed by three letters referring to either a historical name or a geographical area where the virus was isolated and a number indicating the year of isolation where known. The GenBank accession numbers of the analyzed viruses are shown in the right column. Proximal and distal 9-nt sequences and the 9-nt motif 170 nt upstream of CS-N are represented by bold letters. The asterisks represent identity between analyzed sequences. UTR, untranslated region.
The presence of primary sequences that may establish long-distance RNA-RNA interactions in the context of 5′ TRS-N, similar to those involved in transcriptional activation of the TGEV N gene, was also analyzed in other CoVs and arteriviruses. In the feline infectious peritonitis virus (FIPV) and canine enteric CoV (CCoV), the two 9-nt elements were also found at the same relative positions within the 5′ TRS-N region. Therefore, these sequence elements were conserved in members of group 1a CoV. Moreover, in these viruses the N gene has also a minimum value of ΔG for the duplex formation between TRS-L and cTRS-B and most likely its levels of sg mRNA N are also the most abundant during infection, as described in general for other CoVs. In contrast, these sequence elements were not present in phylogenetically more-distant viruses, such as HCoV 229E and porcine epidemic diarrhea virus (group 1b CoV), MHV, SARS-CoV (group 2), IBV (group 3), and equine arteritis arterivirus (Table 3). Therefore, the presence in the Nidovirales order of signals that may establish long-distance RNA-RNA interactions in the 5′ TRS-N region, similar to those increasing the transcriptional activity of the TGEV N gene, seems restricted to group 1a CoV.
TABLE 3.
Conservation of enhancer sequences in Nidovirales
| Virus | Coronavirus group | Enhancer elementa | Most 3′ geneb |
|---|---|---|---|
| TGEV | 1a | + | 7 |
| FIPV | 1a | + | 7 |
| CCoV | 1a | + | 7 |
| HCoV 229E | 1b | − | N |
| Porcine epidemic diarrhea virus | 1b | − | N |
| MHV | 2a | − | N |
| SARS-CoV | 2b | − | N |
| IBV | 3a | − | N |
| Equine arteritis arterivirus | Arteriviridae | − | N |
The presence of an enhancer element, a distal sequence complementary to a proximal sequence segment, is indicated by the symbol +.
For each virus, the most 3′ gene in the genome is indicated.
DISCUSSION
Although multiple factors seem to regulate the transcription process (19, 27), the base pairing between the TRS-L and the cTRS-B in the nascent RNA is a crucial regulating factor controlling CoV transcription (22). In this report, a good correlation was observed between sg mRNA levels and the ΔG of TRS-L and cTRS-B duplex formation, with the only exception being sg mRNA N, the most abundant mRNA during viral infection in spite of its minimum ΔG. Viral sg mRNA N was quantified by two different approaches, real-time RT-PCR and RNA metabolic labeling, with similar results, showing that sg mRNA N was significantly the most abundant sg mRNA during TGEV infection. To analyze whether a higher stability of sg mRNA N might explain its high levels during TGEV infection, pulse-chase experiments showing that the stability of sg mRNA N was not higher than that of the other viral sg mRNAs were performed. Therefore, we postulated the existence of an additional regulation mechanism controlling the exceptionally high levels of sg mRNA N transcribed during infection that acts on top of transcriptional regulation by ΔG of duplex formation between TRS-L and cTRS-B. In this report, we have described a mechanism for a second level of transcription regulation in the Coronaviridae family in which complementary sequences at distal and proximal locations upstream of CS-N enhance the transcriptional activity of the TGEV N gene.
The presence of sequences responsible for transcriptional regulation of sg mRNA N was analyzed by reverse genetics using a replicon derived from the TGEV genome (1). The cDNA encoding this replicon includes the N gene in the same sequence context and at the same distance from the 3′ end as in the whole viral genome. Moreover, the relative amount of sg mRNA N transcribed by replicon E-TRS-N, including the coding sequence of the N gene and 800 nt from the 5′ TRS of the N gene, was similar to that observed in TGEV infection. Therefore, more likely similar regulating mechanisms were operating in the replicon and the whole virus, validating the study of transcriptional activation of the N gene in the replicon system.
Transcriptional activation of the N gene required the presence of sequences located within the distal 5′ TRS, in addition to sequences just upstream of the N gene CS. The distal sequences mapped within a sequence domain located between nt −630 and −290 relative to the N gene CS. This distal region included a 9-nt sequence unique throughout the whole TGEV genome that was complementary to another 9-nt segment localized immediately upstream of CS-N. We have demonstrated that putative base pairing between the distal 9-nt sequence and the 9-nt element located immediately upstream of CS-N regulates the transcriptional activity of the N gene. Moreover, it has been shown that the stability of the duplex between the distal and proximal 9-nt sequences was functionally relevant for transcriptional activation of the TGEV N gene, as a good correlation was observed between sg mRNA N levels and ΔG associated with duplex formation. Accordingly, the sequence within the distal 5′ TRS-N region has an enhancer activity similar to that described for other plus-stranded RNA viruses, such as Tombusvirus (26), in which this enhancer sequence (i) acts to stimulate the accumulation of an mRNA, (ii) interacts with elements near the site of mRNA synthesis initiation, (iii) functions from a distal location, and (iv) maintains its activity when it is repositioned in the genome. This is the first time that an enhancer sequence has been described to occur within the Nidovirales order. Nevertheless, the functional relevance of the base pairing between long-distance RNA-RNA sequences for sg mRNA synthesis has also been demonstrated in other RNA viruses, such as Dianthovirus (21), Potexvirus (10), and Tombusvirus (26), and also in DNA viruses, such as Hepadnavirus (11). In the context of TGEV discontinuous transcription, we propose that this long-distance RNA-RNA interaction in the plus-strand RNA would act as a stop signal for the transcription complex during the synthesis of the minus-strand RNA. This interruption of minus-strand-RNA synthesis would specifically increase the frequency of the template switch to the leader during sg mRNA N synthesis (Fig. 7). A similar model has been proposed for Tombusvirus to explain regulation of RNA synthesis in a premature termination transcription (26). In Dianthovirus, with a segmented genome composed of two single-stranded RNA molecules, a model in which direct binding of the two RNA components trans-activates sg RNA genome synthesis is proposed (21).
FIG. 7.
Working model for sg mRNA N transcription regulation. The first line is a scheme of the TGEV genome (gRNA) showing the elements involved in the regulation of sg mRNA N transcription (not to scale). The leader sequence (L) is represented in black, and the CSs are in gray. TRSs flanking the CS are shown in lighter gray. The distances between elements involved in sg mRNA N synthesis are shown by numbers below the arrows. The long-distance RNA-RNA interaction in the plus-strand RNA that would act as a stop signal for the transcription complex during the synthesis of the minus-strand RNA is represented at the bottom. The curved thick arrow indicates the template switch step during transcription. Nascent minus-strand RNA is shown in a lighter shade than the plus-strand RNA. cL, complement of the leader sequence; cCS-N, complement of CS-N; cTRS-N, complement of TRS-N; ENH, distal 9-nt enhancer sequence; cENH, proximal 9-nt sequence complementary to ENH.
Although the relevance of duplex formation between the distal and proximal 9-nt elements has been demonstrated in the transcriptional activation of the N gene, the involvement in this process of other factors, such as higher-order RNA structure or protein factor(s), cannot be excluded. In fact, in the Tombusvirus enhancer system, it has been described that for optimal sg mRNA transcription, the coordinated activity of several distinct and highly integrated RNA elements that form a multicomponent RNA-based control system was required (12).
The presence of primary sequences that might establish long-distance RNA-RNA interactions in the context of 5′ TRS-N, similar to those involved in transcriptional activation of the TGEV N gene, was analyzed in CoVs and arteriviruses, both included in the Nidovirales order. Only group 1a CoVs (TGEV, FIPV, and CCoV) conserve the 9-nt sequences responsible for transcriptional activation of the TGEV N gene by a long-distance RNA-RNA interaction. In contrast, neither the described 9-nt sequences nor alternative analog complementary sequences were found in other CoVs or arteriviruses. Group 1a CoVs include the group-specific gene 7 downstream of the N gene at the 3′ end of the genome, whereas in the other CoV groups and in arteriviruses, the N gene is the most 3′ gene in the genome (Table 3) (5, 9). In these viruses, the proximity of TRS-N to the 3′ end of the genome probably favors sg mRNA N synthesis, because the RNA-dependent RNA polymerase finds no attenuating and dissociating TRS-B signals preceding its synthesis. Moreover, in group 2 CoVs (MHV and SARS-CoV), the ΔG of duplex formation between TRS-L and cTRS-N has a maximum value, thus also increasing the sg mRNA N synthesis. This is not the case for group 1a CoVs, in which the transcription of sg mRNA N is not promoted either by its genome position or by the ΔG of duplex formation between the TRS-L and cTRS-N (Fig. 1). In this context, we postulate that group 1a CoVs have evolved to include a new regulating mechanism enhancing N gene expression levels, since N protein is essential in different viral processes, such as morphogenesis (7) and RNA synthesis (1, 3, 20).
The presence of the 9-nt sequence elements in other TGEV strains was studied by a comparative sequence analysis (Fig. 6) showing that 9-nt sequences at the distal and proximal 5′ TRS-N regions were conserved in most analyzed TGEV strains. However, in recent isolates of TGEV (ENG96 and TAW83) a point mutation that disrupts perfect complementarity between distal and proximal 9-nt elements was identified. This mutation in the first nucleotide of the distal 9-nt sequence (A-to-G substitution) was introduced in mutant M4-TRS-N, showing that in the TGEV-derived replicons 70% of N gene transcriptional activation was maintained, suggesting that the long-distance RNA-RNA interaction is functional in these TGEV strains, increasing the transcriptional activation of sg mRNA N in spite of a mismatch in the duplex. Interestingly, the conservation of perfect long-distance base pairing between distal and proximal 9-nt elements in 5′ TRS-N was also observed in the PRCV-ISU-I. In this strain, in addition to the described point mutation affecting this first nucleotide of the distal 9-nt sequence, another point mutation (C-to-U substitution) was observed at an intermediate position, 175 nt upstream of CS-N (Fig. 6). This additional mutation generates a new 9-nt sequence with a perfect complementarity to the proximal 9-nt segment, probably restoring the sg mRNA N transcriptional activation. Altogether, the phylogenetic conservation of these sequence elements supports the relevance of the described long-distance RNA-RNA interaction as a regulating mechanism during CoV transcription.
In this report, we have described a novel regulatory mechanism for discontinuous transcription in the Nidovirales order. The presence of functional long-distance RNA-RNA interactions associated with transcription enhancement has not been previously described to occur in nidoviruses, although it has been studied for other nonrelated viral systems, suggesting that this mechanism represents a general strategy for regulating viral RNA synthesis.
Acknowledgments
We thank J. C. Oliveros for the in silico analysis and M. González for technical assistance.
This work was supported by grants from the European Union (Frame VI, DISSECT PROJECT, SP22-CT-2004-511060, and RiViGene PROJECT, SSPE-CT-2005-022639) and a grant (CPE03-021-C4-3) from the National Institute of Agronomics Research (INIA), Spain. J.L.M., S.Z., and I.S.G. received fellowships from Consejo Superior de Investigaciones Científicas (CSIC).
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Almazán, F., C. Galán, and L. Enjuanes. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 7812683-12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almazán, F., J. M. González, Z. Pénzes, A. Izeta, E. Calvo, J. Plana-Durán, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 975516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., G. W. Nelson, J. O. Fleming, R. J. Deans, J. G. Keck, N. Casteel, and S. A. Stohlman. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 624280-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delmas, B., J. Gelfi, H. Sjöström, O. Noren, and H. Laude. 1993. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342293-298. [DOI] [PubMed] [Google Scholar]
- 5.Enjuanes, L., A. E. Gorbalenya, R. J. de Groot, J. A. Cowley, J. Ziebuhr, and E. J. Snijder. The Nidovirales. In B. W. J. Mahy, M. Van Regenmortel, P. Walker, and D. Majumder-Russell (ed.), Encyclopedia of virology, 3rd ed., in press. Elsevier Ltd., Oxford, United Kingdom.
- 6.Enjuanes, L., W. Spaan, E. Snijder, and D. Cavanagh. 2000. Nidovirales, p. 827-834. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the international committee on taxonomy of viruses. Academic Press, New York, NY.
- 7.Escors, D., E. Camafeita, J. Ortego, H. Laude, and L. Enjuanes. 2001. Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core. J. Virol. 7512228-12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolov, I., T. A. Hoffman, B. M. Prágai, S. A. Dryga, H. V. Huang, S. Schlesinger, and C. M. Rice. 1996. Alphavirus-based expression vectors: strategies and applications. Proc. Natl. Acad. Sci. USA 9311371-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 11717-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, K.-H., and C. L. Hemenway. 1999. Long-distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA 5636-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewellyn, E. B., and D. D. Loeb. 2007. Base pairing between cis-acting sequences contributes to template switching during plus-strand DNA synthesis in human hepatitis B virus. J. Virol. 816207-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin, H.-X., W. Xu, and K. A. White. 2007. A multicomponent RNA-based control system regulates subgenomic mRNA transcription in a tombusvirus. J. Virol. 812429-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 14.McClurkin, A. W., and J. O. Norman. 1966. Studies on transmissible gastroenteritis of swine. II. Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can. J. Comp. Med. Vet. Sci. 30190-198. [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA virus. Virology 2731-8. [DOI] [PubMed] [Google Scholar]
- 16.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 207220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penzes, Z., J. M. González, E. Calvo, A. Izeta, C. Smerdou, A. Mendez, C. M. Sánchez, I. Sola, F. Almazán, and L. Enjuanes. 2001. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes 23105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez, C. M., G. Jiménez, M. D. Laviada, I. Correa, C. Suñe, M. J. Bullido, F. Gebauer, C. Smerdou, P. Callebaut, J. M. Escribano, and L. Enjuanes. 1990. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology 174410-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 8120-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schelle, B., N. Karl, B. Ludewig, S. G. Siddell, and V. Thiel. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 796620-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281829-832. [DOI] [PubMed] [Google Scholar]
- 22.Sola, I., J. L. Moreno, S. Zuñiga, S. Alonso, and L. Enjuanes. 2005. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 792506-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Most, R. G., and W. J. M. Spaan. 1995. Coronavirus replication, transcription, and RNA recombination, p. 11-31. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, NY.
- 24.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 9612056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: the transmissible gastroenteritis virus model. J. Virol. 7410600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, G., V. Slowinski, and K. A. White. 1999. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA 5550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zúñiga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]