Abstract
Subgenomic (sg) mRNAs are small viral messages that are synthesized by polycistronic positive-strand RNA viruses to allow for the translation of certain viral proteins. Tombusviruses synthesize two such sg mRNAs via a premature termination mechanism. This transcriptional process involves the viral RNA-dependent RNA polymerase terminating minus-strand RNA synthesis prematurely at internal RNA signals during copying of the viral genome. The 3′-truncated minus-strand RNAs generated by the termination events then serve as templates for sg mRNA transcription. A higher-order RNA structure in the viral genome, located just upstream from the termination site, is a critical component of the RNA-based polymerase attenuation signal. Here, we have analyzed the role of this RNA structure in mediating efficient sg mRNA2 transcription. Our results include the following: (i) we define the minimum overall thermodynamic stability required for an operational higher-order RNA attenuation structure; (ii) we show that the distribution of stability within an attenuation structure affects its function; (iii) we establish that an RNA quadruplex structure can act as an effective attenuation structure; (iv) we prove that the higher-order RNA structure forms and functions in the plus strand; (v) we provide evidence that a specific attenuation structure-binding protein factor is not required for transcription; (vi) we demonstrate that sg mRNA transcription can be controlled artificially through small-molecule activation using RNA aptamer technology. These findings provide important new insights into the premature termination mechanism and present a novel approach to regulate the transcriptional process.
Subgenomic (sg) mRNAs are transcribed by most polycistronic positive-strand RNA viruses to allow for the expression of viral proteins that are encoded internally or 3′-proximally in these viral genomes (12). Within sg mRNAs, such translationally silent open reading frames in the viral genome are repositioned to 5′-proximal positions, where scanning eukaryotic ribosomes can readily access their initiation codons. These smaller messages are transcribed from viral genomes during infections by their encoded RNA-dependent RNA polymerases (RdRps) (12).
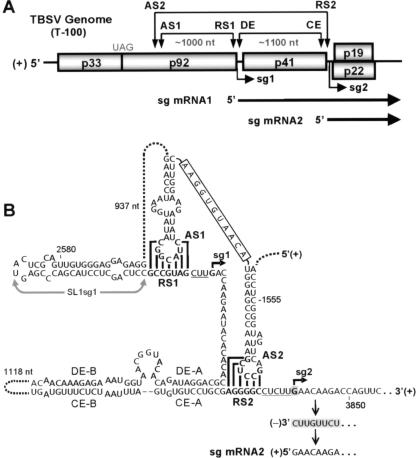
Tomato bushy stunt virus (TBSV), the type member of the genus Tombusvirus, transcribes two sg mRNAs (5). sg mRNA1 templates the translation of coat protein (p41), and its smaller counterpart sg mRNA2 mediates expression of two overlapping open reading frames that encode the movement (p22) and gene silencing suppressor (p19) proteins (19) (Fig. 1). The mechanism of transcription in TBSV has been studied extensively, and the results point to it using a premature termination (PT) mechanism for the synthesis of both of its sg mRNAs (2, 3, 8, 9, 22, 23). In this model, the viral replicase, which includes the p92 RdRp and its accessory protein, p33, is stalled at an internal site while copying the viral genome (18, 22). The 3′-truncated minus strands generated are then used as templates for transcription of sg mRNAs (Fig. 1). The replicase termination step is mediated by a multicomponent RNA attenuation signal that includes a long distance RNA-RNA interaction, a spacer element, and downstream sequences (2, 8, 9). The long distance interactions, modeled in the positive strand, involve cRNA sequences located immediately 5′ to the sites of transcriptional initiation (called receptor sequences [RSs]) and partner segments positioned hundreds of nucleotides upstream (called activator sequences [ASs]) (2, 8) (Fig. 1). The higher-order RNA structures formed by AS-RS interactions are recognized as part of the attenuation signal by the viral replicase, and this causes it to stall and terminate synthesis, thereby generating the minus-strand RNA templates used for sg mRNA transcription (18). In Red clover necrotic mosaic virus (RCNMV), which is also thought to utilize a PT mechanism, an AS-RS-like interaction is required for sg mRNA transcription from RNA-1; however, this interaction forms in trans between the two genomic RNA segments, RNA-1 and RNA-2 (16). For RCNMV, it has been suggested that a protein factor may bind to and stabilize the bimolecular RNA-RNA interaction (16). Similarly, it is possible that the cis-forming AS/RS structures in TBSV could be bound by a protein factor that stabilizes them, thereby facilitating the termination step. However, this prospect has not been explored experimentally for either TBSV or RCNMV.
FIG. 1.
TBSV genome, sg mRNAs and transcriptional regulatory elements. (A) Linear representation of the TBSV RNA genome and its coding organization. Boxes indicate encoded proteins. The p92 RdRp is translated directly from the viral genome by readthrough of the p33 stop codon (UAG). Proteins encoded downstream are translated from two sg mRNAs that are transcribed during infections. The relative positions (arrowheads) of interacting RNA elements that are involved in sg mRNA transcription are shown above the viral genome. Initiation sites for sg mRNA transcription are labeled sg1 and sg2 and bold arrows below represent corresponding structures of the two sg mRNAs. (B) RNA elements that regulate sg mRNA transcription in TBSV. Relevant sequences of the genome are shown with corresponding coordinates provided. The AS1/RS1 base-pairing interaction is essential for sg mRNA1 transcription, whereas the AS2/RS2 (and DE/CE) interaction is essential for transcription of sg mRNA2. Stem-loop structures containing AS1 and AS2 are connected by an 11 nt long sequence (boxed). Initiation sites for the two sg mRNAs are indicated by small arrows and are separated from their respective AS/RS interactions by spacer elements (underlined). The minus-strand template shown is generated by a PT mechanism and contains a sg mRNA promoter sequence (shaded) at its 3′ end that is used to transcribe sg mRNA2.
In this study we have examined the nature of the higher-order RNA component of the attenuation signal required for TBSV sg mRNA2 transcription, with the following specific goals in mind: (i) defining the minimum overall thermodynamic stability required for an operational higher-order RNA component of the attenuation signal; (ii) investigating whether the distribution of stability within an attenuation structure affects its function; (iii) determining if a double-stranded helical RNA structure is essential for attenuator activity; (iv) providing indisputable evidence for the higher-order RNA structure forming and functioning in the plus strand; (v) examining the likelihood that a specific structure-binding protein factor is required to facilitate the attenuation process; (vi) testing if sg mRNA transcription can be regulated through small-molecule activation. Our results successfully address each of these objectives and offer novel insights into the PT transcriptional mechanism and a new strategy to control this process.
MATERIALS AND METHODS
Construction of plasmids.
Constructs described previously that were used in this study include the following: T100, the wild-type (wt) TBSV genome construct (5); RS2m1, a modified TBSV genome containing substitutions in RS2 that prevent the AS2-RS2 interaction (8). RS2m1 was the base construct used to generate the stem-loop (SL)-, aptamer-, and quadruplex-containing mutants. The detailed structures of the modifications introduced into these different constructs are presented in the respective figures, the exception being SW88-myc, into which an additional sequence (5′-GAGCAGAAACTCATCTCTGAAGAGGATCTG-3′) corresponding to the c-Myc epitope tag was inserted at the N terminus of the p19 coding region. All modifications were made using PCR-based mutagenesis and standard cloning techniques (15). The PCR-derived regions introduced into the constructs were sequenced completely to ensure that only the intended modifications were present.
In vitro transcription and protoplast inoculation.
In vitro RNA transcripts of genomic RNAs were generated using T7 RNA polymerase as described previously (20). Preparation and inoculation of cucumber protoplasts were carried out as described elsewhere (3). Briefly, isolated cucumber protoplasts (∼3 × 105) were prepared from cucumber cotyledons and polyethylene glycol transfected with 3 μg genomic RNA or water (mock control). Transfected protoplasts were incubated at 22°C or 25°C for 22 h under constant lighting. For the aptamer-containing mutants, transfected protoplasts were incubated with the indicated concentrations of theophylline or caffeine (both from Sigma).
Viral RNA analysis.
Total nucleic acid preparations isolated from virus-transfected protoplasts were subjected to Northern blot analysis for detection of viral RNAs as described previously (3). Nucleic acids were separated in 1.4% agarose gels, and equal loading of lanes was confirmed prior to transfer via staining the gels with ethidium bromide. Following electrophoretic transfer to nylon membranes, viral RNAs were detected using 32P-labeled DNA oligonucleotide probes, and relative isotopic levels of detected bands were determined using a PharosFx-Plus Molecular Imager (Bio-Rad).
Viral protein analysis.
After a 22-h incubation, protoplasts were vortexed in 50 μl 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer for 30 s and then heated at 100°C for 5 min. Proteins were separated by 13% acrylamide sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After electrotransfer to nitrocellulose membrane, Myc-tagged p19 was detected using a monoclonal antibody against the c-Myc tag. Horseradish peroxidase-conjugated secondary antibody and Amersham ECL Plus Western blotting detection reagents (RPN 2132; GE Healthcare) were used for further processing of the blots.
Prediction of RNA structures and free energy.
RNA structures were predicted and free energy changes calculated at 22°C using mfold version 2.3 (11, 25).
RESULTS AND DISCUSSION
Structural properties of RNA stem-loops important for sg mRNA transcription.
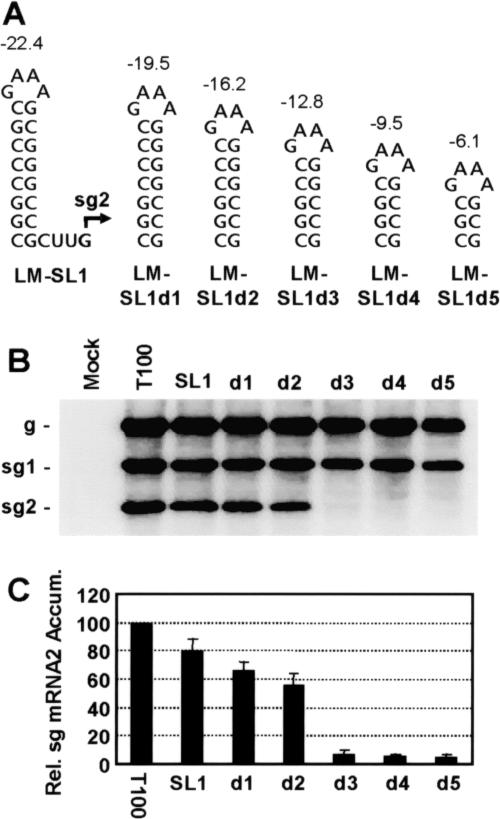
Previous analysis demonstrated that a local stem-loop (SL) RNA structure can functionally replace the wt long-range tertiary interaction formed by the AS2 and RS2 elements (8). Here, we sought to define the minimum thermodynamic stability of the SL structure required for readily detectable sg mRNA2 transcription. In an attempt to delineate this lower limit, SL structures with predicted progressive reductions in stability (i.e., ΔΔGs predicted in the range of 2.9 to 3.4 kcal/mol) were tested for their ability to mediate sg mRNA2 transcription (Fig. 2A). Mutant TBSV genomes containing SL structures with predicted stabilities of −22.4 to −6.1 kcal/mol (11, 25) were transfected into plant protoplasts, and viral RNA accumulation was monitored by Northern blotting 22 h posttransfection (Fig. 2B). Progressive and roughly proportional decreases in sg mRNA2 levels were observed for mutants LM-SL1 through LM-SL1d2 (Fig. 2C). However, further reductions in predicted stability below that of LM-SL1d2 (i.e., ΔG of −16.2 kcal/mol) resulted in only minor levels of accumulation (i.e., <10% of wt) (Fig. 2C).
FIG. 2.
Effect of SL stability on sg mRNA2 transcription. (A) Depiction of RNA SLs introduced into TBSV genomes. Free energy changes (ΔG) for formation at 22°C are indicated in kcal/mol above each structure in this and other figures. (B) Northern blot analysis of TBSV viral RNAs. Wild-type (T100) and mutant TBSV genomes were transfected into protoplasts and viral RNAs were analyzed following a 22 h incubation at 22°C. The identities of the viral genomes used in the transfections are indicated at the top. The positions of the viral genome (g) and sg mRNAs (sg1 and sg2) are indicated to the left. (C) In this and all other figures, corresponding relative levels of accumulation of sg mRNA2 were determined from radio-analytical scanning of blots. The relative sg mRNA2 accumulation levels correspond to means with standard deviations from three independent experiments and represent the ratios of sg mRNA2 levels to their corresponding genomic RNA levels, all normalized to that for T100, set at 100.
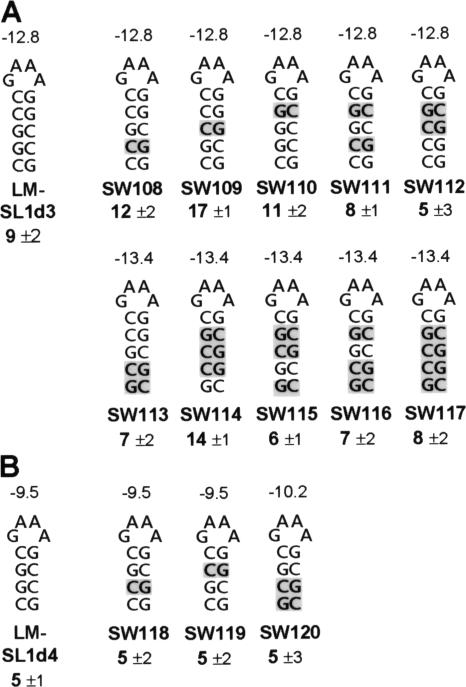
Two possibilities could account for this effect. The first is that the SLs did form but their stabilities were not sufficient for effective function. Alternatively, the SLs may not have formed, making them incapable of conferring structure-dependent function. In an attempt to distinguish between these two options, additional sets of SL structures were tested under the premise that formation of the SLs may be context dependent. That is, flanking or other RNA sequences may be pairing with sections of the SLs, thereby preventing their formation. To reduce the likelihood of this occurring consistently, numerous SLs with identical predicted stability but different primary sequences were designed and tested (Fig. 3). TBSV genomes containing three different sets of SLs corresponding to stabilities of −12.8 (as for LM-SL1d3), −13.4, or −9.5 kcal/mol (as for LM-SL1d4) were generated (Fig. 3A and B). Analysis of sg mRNA accumulation from transfections involving these mutant genomes revealed that the highest relative levels of accumulation from the −12.8 and −13.4 series were only ∼17% and ∼14% of wt, respectively (Fig. 3A). In contrast, extrapolation of the accumulation levels seen for LM-SL1 through LM-SL1d2 in Fig. 2C suggests that a hairpin with predicted stability of −12.8 kcal/mol should accumulate to ∼43% that of wt. Thus, the highest experimental value (17%) was ≥2.5-fold lower than the predicted value (43%). Similarly, an extrapolated value for a hairpin with predicted stability of −9.5 kcal/mol would be ∼30%; however, the experimentally observed levels did not exceed ∼5%, which is sixfold lower (Fig. 3B). In the latter case it could be argued that such small hairpins may not be able to form in the context of this viral genome; however, SL2-RIV (5′-CCGGACAACCGG; the loop sequence is underlined), which is located internally in the 3′ untranslated region of the TBSV genome, has a lower predicted stability of −5.6 kcal/mol yet forms efficiently in vivo (4, 13). Therefore, considering the variety of hairpins tested and the functionality of another with notably lower stability, our data suggest that the higher-order RNA component of the attenuation signal has a minimum required stability for moderate function (i.e., ≥50% of wt) of approximately −16.2 kcal/mol (at 22°C) and that further decreases in stability dramatically and disproportionately reduce this activity (as illustrated in Fig. 2C and 3A, −16.2 series). Thus, there appears to be a reasonably well-defined cutoff value for the overall stability of a functional higher-order RNA component.
FIG. 3.
Analysis of SLs with alternative sequences but equivalent stabilities. Depiction of RNA SLs introduced into TBSV genomes with corresponding ΔGs above each. Substituted nucleotides are shaded. Relative levels of sg mRNA2 accumulation (compared to that for wt T100) are indicated below each structure. Structures and activities of (A) SLs with ΔGs of −12.8 or −13.4 and (B) −9.5 kcal/mol. Transfected protoplasts were incubated at 22°C for 22 h.
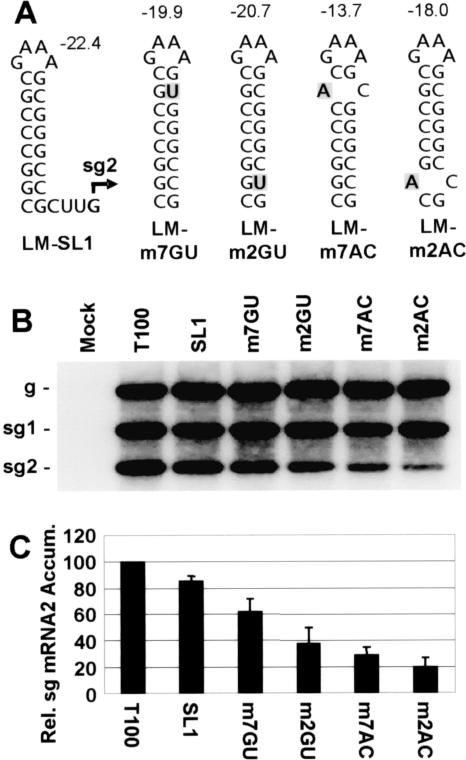
Having established the overall stability requirements for activity of the SL component, we next asked whether the distribution of stability within the SL was important for function. To address this question, modifications were introduced into the SL in LM-SL1 that were predicted to destabilize the stem at either the top or the bottom of the structure. This was accomplished using substitutions that converted upper and lower penultimate GC base pairs in the stem to either GU wobble pairs or AC mismatches (Fig. 4A). When tested, the GU substitution located near the bottom of the stem was found to be more inhibitory to transcription than the same substitution introduced near the top of the stem (Fig. 4B, C), and similar results were also observed for the two AC mismatches (Fig. 4B and C). Thus, the same trend was seen with two different sets of mutants, even though in both cases the modifications at the bottom of the stem were predicted to form SLs that were more stable (Fig. 4A, compare LM-m7GU with LM-m2GU and LM-m7AC with LM-m2AC). These results suggest that although the overall stability of an SL is critical to activity, the distribution of stability within the SL is also an important determinant of function. The fact that stability at the bottom of the SL was more critical to activity is consistent with the important role played by the spacer element (9, 22), i.e., both act to define the distance between the transcriptional initiation site and the closest stable portion of the higher-order RNA structure (8). The greater activities seen for the GU-containing mutants versus their AC-containing counterparts at both the upper and lower positions in the SL are consistent with the proposed plus-strand activity of the SL structure (8), as the GU pairs would be more stable in the plus strand. However, this type of evidence is not indisputable, as it is possible that noncanonical AC base pairs form (10) and generate functional complementary structures in the minus strand.
FIG. 4.
Effect of distribution of SL stability on sg mRNA2 transcription. (A) Depiction of RNA SLs introduced into TBSV genomes with corresponding ΔGs above each. Substituted nucleotides are shaded. (B) Northern blot analysis of TBSV viral RNAs. (C) Relative levels of accumulation of sg mRNA2. Transfected protoplasts were incubated at 22°C for 22 h.
Small-molecule induction of sg mRNA transcription.
In a previous study, we showed that the activity of a viral SL structure, termed SL5 (representing an essential viral RNA replication element) could be modulated exogenously by replacing it with a small-molecule-binding RNA aptamer (17). This was accomplished by replacing SL5 (a perfect RNA hairpin), located in the 5′ untranslated region of the TBSV genome, with a modified theophylline-binding aptamer, which is an imperfect RNA hairpin containing internal loops. In this viral context, the unbound aptamer was not stable enough to structurally mimic the form and function of SL5. Accordingly, viral genomes containing the aptamer in place of SL5 were highly defective when tested in protoplasts. However, when the ligand for this aptamer was added to the protoplast medium, the stabilized ligand-bound aptamer structure was able to efficiently activate viral genome replication (17).
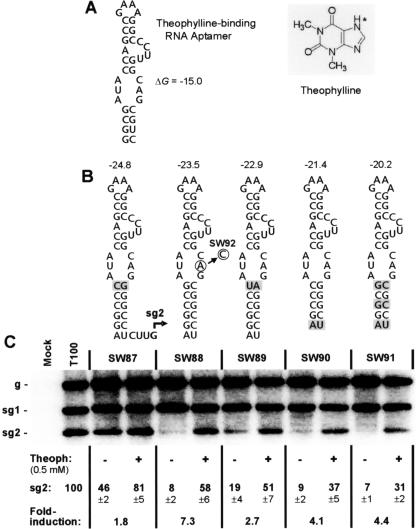
Based on this prior success, and the ability of stable SL-type structures to support sg mRNA transcription (Fig. 2), we reasoned that a similar approach could be used to modulate the function of the higher-order component of the attenuation signal and, in turn, control sg mRNA transcription. Accordingly, a theophylline-binding RNA aptamer (Fig. 5A), containing different lower stems, was introduced as the higher-order RNA component of the sg mRNA2 transcriptional attenuator (Fig. 5B). sg mRNA2 accumulation levels were then monitored in transfections of TBSV genomes containing the different aptamers, either in the presence or absence of theophylline (Fig. 5C). Levels of transcriptional induction varied for the different aptamer variants, with the best being observed for SW88 (Fig. 5C). This viral genome exhibited an ∼7-fold relative increase in sg mRNA2 to a level ∼58% that of wt in the presence of 0.5 mM ligand at an incubation temperature of 22°C (Fig. 5C). Further testing revealed that relative transcriptional induction from SW88 could be enhanced maximally to ∼10-fold by using 1.0 mM theophylline at an incubation temperature of 25°C (Fig. 6). The induction observed was also shown to be theophylline dependent, as treatment with caffeine (a structural analogue of theophylline [Fig. 5A]) did not induce transcription (Fig. 6A and B). Similarly, the induction was found to be aptamer dependent, as SW92, containing a SW88-type aptamer with an inactivating point mutation in its binding pocket (24) (Fig. 5B), was not responsive to induction by theophylline (Fig. 6A and B).
FIG. 5.
Small molecule induction of sg mRNA2 transcription. (A) Structure of a theophylline-binding RNA aptamer (6) is presented on the left and the structure of theophylline is shown to the right. The asterisk denotes the hydrogen in theophylline that is a methyl group in caffeine. (B) Modified theophylline aptamers introduced into TBSV genomes and tested for sg mRNA2 transcriptional activity. Substitutions in the lower stem, relative to SW88, are shaded. (C) Northern blot analysis of TBSV RNA accumulation. Aptamer-containing genomes were transfected into plant cell protoplasts and incubated at 22°C for 22 h in the absence (-) or presence (+) of 0.5 mM theophylline. Relative sg mRNA2 accumulation levels are presented below.
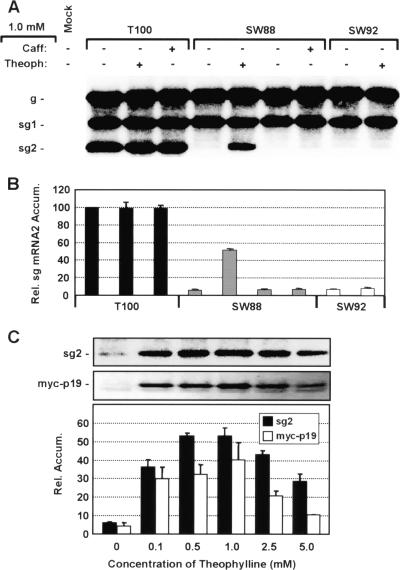
FIG. 6.
Activation of transcription is theophylline-specific and aptamer-dependent. (A) Northern blot analysis of TBSV RNA accumulation. The wt TBSV genome (T100) or genomes containing the active SW88 aptamer or inactive SW92 aptamer were tested for responsiveness to theophylline or caffeine. Aptamer-containing genomes were transfected into protoplasts and incubated at 25°C for 22 h. (B) Relative levels of accumulation of sg mRNA2. (C) Effect of increased concentration of theophylline in the incubation medium on accumulation of sg mRNA2 and myc-p19 from SW88-myc. Northern and Western blots at the top show sg mRNA2 (sg2) and myc-tagged p19 (myc-p19) accumulation levels, respectively, with corresponding average values presented graphically below. The faint band directly below myc-p19 is a cross-reacting cellular protein that was also present in control samples from uninfected cells (data not shown). Transfected protoplasts were incubated at 25°C for 22 h.
Since the function of sg mRNAs is to template the translation of their encoded protein(s), we also wanted to determine if the level of sg mRNA induced correlated with the amount of encoded protein produced. In order to detect a product from sg mRNA2, p19 was N-terminally tagged with a myc epitope. This modification was introduced into SW88, thereby creating SW88-myc, which maintained its ability to be induced by theophylline (Fig. 6C). When corresponding myc-p19 accumulation was assessed by Western blotting, the relative levels basically mirrored those for sg mRNA2 (Fig. 6C). Although intuitive, these data provide experimental evidence that the amount of sg mRNA produced in this system is roughly proportional to the amount of encoded product expressed. Accordingly, these results validate both the wt and artificial modes for controlling transcription as legitimate ways of modulating the quantity of viral protein expressed in TBSV.
Our results with the aptamer-based transcriptional control system are also significant in other respects: (i) it represents the first demonstration of small-molecule control of RdRp-mediated sg mRNA transcription, which may have applications in basic and applied research; (ii) it is the second distinct viral process shown to be controllable using RNA aptamers, thereby demonstrating the extensibility of this type of approach; (iii) the data provide definitive evidence that the higher-order RNA structure functions in the plus strand (i.e., the only strand in which the aptamer is active).
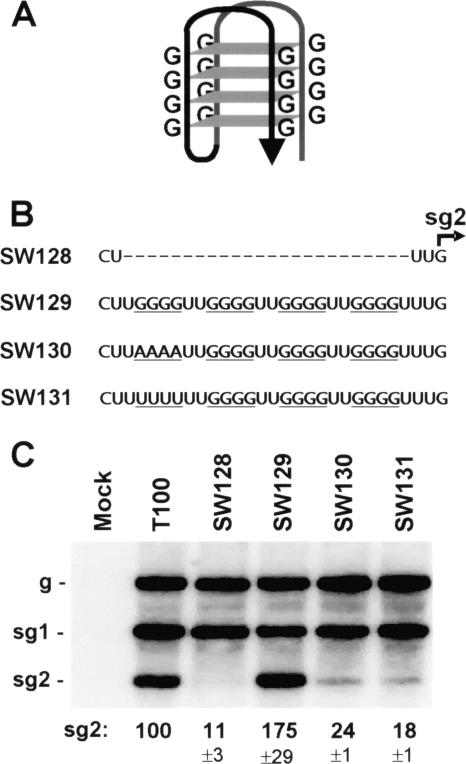
A quadruplex can serve as the higher-order RNA structural component.
To date, all of the structures shown to serve as higher-order structural components of the attenuation signal have been double helix based (Fig. 1B, 2A, and 5B). Consequently, we wanted to determine if a significantly different type of higher-order RNA structure could function in this same capacity. RNA quadruplex structures are stable, stacked, four-stranded motifs that are commonly formed by G-rich sequences (Fig. 7A) (1). These structures can exist in vivo and are able to influence gene expression from mRNAs that contain them (7, 21). The unique structure of the quadruplex prompted us to ask whether it could functionally substitute for the double helical-based structures found to operate in sg mRNA transcription. To this end, a sequence with G-quadruplex-forming potential (i.e., four tetra-G tracts, each separated by two U residues) (14) was inserted three nucleotides upstream from the sg mRNA2 initiation site (Fig. 7B). In vivo analysis of a TBSV genome containing this quadruplex sequence, SW129, indicated that it mediated very efficient sg mRNA2 accumulation to ∼175% that for wt T100 (Fig. 7C). sg mRNA2 levels were substantially lower (i.e., ∼24% or 18%, respectively) when adenylates (SW130) or uridylates (SW131) were substituted for the guanylates in the 5′-most G tract, as well as when the entire quadruplex sequence was absent (SW128; ∼11%) (Fig. 7B and C). The adenylate substitution in the 5′-most G tract is a modification known to disrupt the formation of quadruplex structure (7); therefore, the concomitantly reduced activity observed supports quadruplex formation and its participation in transcription (Fig. 7C). The higher background levels seen for the two substitution mutants (∼24% and 18%) versus that for deletion of the entire quadruplex sequence (∼11%) are likely due to the formation of weaker local/long-range RNA structures that involve the remaining quadruplex sequence. Regardless, the ∼7- to 10-fold increase in the former values compared to those in the presence of a complete quadruplex sequence is indicative that this very different type of RNA structure is functional.
FIG. 7.
Transcription mediated by an RNA quadruplex structure. (A) Schematic example of an antiparallel four-level RNA G-quadruplex. (B) Sequences introduced into the TBSV genome corresponding to the four-level quadruplex and controls. The four G-tracts are underlined in SW129 and at corresponding positions in the controls below. (C) Northern blot analysis of TBSV RNA accumulation after a 22 h incubation at 22°C. Relative levels of accumulation of sg mRNA2 are given below.
The greater-than-wt level of sg mRNA2 accumulation seen for the quadruplex-containing SW129 (i.e., ∼175%) indicated that it is more efficient at mediating transcription than the wt tertiary structure. A similar increase in corresponding minus-strand sg mRNA2 levels (i.e., ∼184% [data not shown]) was also observed for SW129, indicating that the quadruplex functions as a very effective attenuation structure. Reverse transcription-PCR and sequence analyses of progeny SW129 viral genomes along with primer extension analysis of their associated sg mRNAs at 22 h posttransfection, respectively, showed that the quadruplex sequence was still present and that transcription had initiated from the predicted start site (data not shown); the same was also true for the aptamer-containing SW88. Accordingly, these artificial structures are stably maintained during infections and mediate sg mRNA transcription as predicted.
The functionality of the quadruplex provides important structural and mechanistic insights by arguing against both (i) the necessity for a particular type of higher-order RNA structure and (ii) a specific higher-order component-binding protein being essential for the transcriptional process. Instead, the results support the concept that “any” RNA-based structure possessing a minimum required stability, including wt long distance tertiary interactions, local hairpins, ligand-bound aptamers, and quadruplexes, can function independently as the higher-order component of the attenuation signal. Moreover, since the quadruplex is a strand-specific structure, its activity provides further corroborative evidence that the higher-order attenuation structure functions in the plus strand. Lastly, the unique structure of the quadruplex suggests that the double helix-based wt tertiary or artificial secondary structures do not play any critical role in trans to facilitate initiation of sg mRNA transcription on the minus strand template (i.e., by acting as part of the promoter or as an enhancer element).
Conclusions.
Our results have shown that both regional and overall stability within a higher-order attenuation RNA structure dictates its activity and that such structures definitely operate in the plus strand. A cutoff stability for a functional local SL attenuation structure in TBSV was determined to be approximately −16.2 kcal/mol. This value may be a useful standard for future comparisons with other viruses that use a PT mechanism (18). Moreover, recent unpublished results from our laboratory suggest that some viruses utilize localized SL structures, rather than long distance interactions, as the higher-order components of their attenuation signals. This finding adds further biological relevance to our analyses of SL structures in TBSV.
The functionality of the quadruplex structure indicates that the precise nature of the RNA attenuation structure is not critical and that its activity is primarily structure nonspecific. This flexibility suggests that the primary purpose of such structures is to act as a type of physical barrier; thereby stalling the RdRp in a position that mediates its termination. This nonspecificity also suggests that there is no fundamental requirement for a particular protein factor to specifically bind to the attenuation structure in order for it to function. This same characteristic also argues against the structure making specific functionally relevant contacts with the RdRp (e.g., causing induction of a conformational change in the RdRp that mediates its termination). Accordingly, our results point to a simple, but critical, role for the structure in mediating only the RdRp termination step in the PT mechanism by relatively nonspecific means, i.e., acting as a physical barrier.
Finally, we have reported a novel strategy to control the activity of an attenuation structure using a small-molecule ligand and aptamer technology. This ability allows for modulation of the timing and levels sg mRNA transcription which, in turn, provides a unique means to control viral protein accumulation in vivo. Accordingly, this system should prove to be useful for assessing quantitative and temporal effects related to viral protein expression in TBSV-infected cells. Moreover, the approach may be applicable to other viral systems that utilize a PT mechanism or SL-type structures to control sg mRNA transcription.
Acknowledgments
We thank lab members for reviewing the manuscript and Baodong Wu for technical assistance.
This work was supported by NSERC, a Steacie Fellowship, and a Canada Research Chair.
Footnotes
Published ahead of print on 6 February 2008.
REFERENCES
- 1.Burge, S., G. N. Parkinson, P. Hazel, A. K. Todd, and S. Neidle. 2006. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 345402-5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi, I. R., M. Ostrovsky, G. Zhang, and K. A. White. 2001. Regulatory activity of distal and core RNA elements in Tombusvirus subgenomic mRNA2 transcription. J. Biol. Chem. 27641761-41768. [DOI] [PubMed] [Google Scholar]
- 3.Choi, I. R., and K. A. White. 2002. An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J. Biol. Chem. 2773760-3766. [DOI] [PubMed] [Google Scholar]
- 4.Fabian, M. R., H. Na, D. Ray, and K. A. White. 2003. 3′-terminal RNA secondary structures are important for accumulation of tomato bushy stunt virus DI RNAs. Virology 313567-580. [DOI] [PubMed] [Google Scholar]
- 5.Hearne, P. Q., D. A. Knorr, B. I. Hillman, and T. J. Morris. 1990. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 177141-151. [DOI] [PubMed] [Google Scholar]
- 6.Jenison, R. D., S. C. Gill, A. Pardi, and B. Polisky. 1994. High-resolution molecular discrimination by RNA. Science 2631425-1429. [DOI] [PubMed] [Google Scholar]
- 7.Kumari, S., A. Bugaut, J. L. Huppert, and S. Balasubramanian. 2007. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 3218-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin, H. X., and K. A. White. 2004. A complex network of RNA-RNA interactions controls subgenomic mRNA transcription in a tombusvirus. EMBO J. 233365-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, H. X., W. Xu, and K. A. White. 2007. A multicomponent RNA-based control system regulates subgenomic mRNA transcription in a tombusvirus. J. Virol. 812429-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masquida, B., and E. Westhof. 2000. On the wobble G-U and related pairs. RNA 69-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 12.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 2731-8. [DOI] [PubMed] [Google Scholar]
- 13.Na, H., and K. A. White. 2006. Structure and prevalence of replication silencer-3′ terminus RNA interactions in Tombusviridae. Virology 345305-316. [DOI] [PubMed] [Google Scholar]
- 14.Rachwal, P. A., T. Brown, and K. R. Fox. 2007. Effect of G-tract length on the topology and stability of intramolecular DNA quadruplexes. Biochemistry 463036-3044. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281829-832. [DOI] [PubMed] [Google Scholar]
- 17.Wang, S., and K. A. White. 2007. Riboswitching on RNA virus replication. Proc. Natl. Acad. Sci. USA 10410406-10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White, K. A. 2002. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304147-154. [DOI] [PubMed] [Google Scholar]
- 19.White, K. A., and P. D. Nagy. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acids Res. Mol. Biol. 78187-226. [DOI] [PubMed] [Google Scholar]
- 20.White, K. A., and T. J. Morris. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 6814-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wieland, M., and J. S. Hartig. 2007. RNA quadruplex-based modulation of gene expression. Chem. Biol. 14757-763. [DOI] [PubMed] [Google Scholar]
- 22.Wu, B., and K. A. White. 2007. Uncoupling tombusvirus genome replication from transcription via the polymerase: functional and evolutionary insights. EMBO J. 265120-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, G., V. Slowinski, and K. A. White. 1999. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA 5550-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann, G. R., C. L. Wick, T. P. Shields, R. D. Jenison, and A. Pardi. 2000. Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA 6659-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. RNA biochemistry and biotechnology. Kluwer, Dordrecht, The Netherlands.