Abstract
The IκB kinase-related kinases, TBK1 and IKKi, were recently shown to be responsible for the C-terminal phosphorylation of IRF-3. However, the identity of the phosphoacceptor site(s) targeted by these two kinases remains unclear. Using a biological assay based on the IRF-3-mediated production of antiviral cytokines, we demonstrate here that all Ser/Thr clusters of IRF-3 are required for its optimal transactivation capacity. In vitro kinase assays using full-length His-IRF-3 as a substrate combined with mass spectrometry analysis revealed that serine 402 and serine 396 are directly targeted by TBK1. Analysis of Ser/Thr-to-Ala mutants revealed that the S396A mutation, located in cluster II, abolished IRF-3 homodimerization, CBP association, and nuclear accumulation. However, production of antiviral cytokines was still present in IRF-3 S396A-expressing cells. Interestingly, mutation of serine 339, which is involved in IRF-3 stability, also abrogated CBP association and dimerization without affecting gene transactivation as long as serine 396 remained available for phosphorylation. Complementation of IRF-3-knockout mouse embryonic fibroblasts also revealed a compensatory mechanism of serine 339 and serine 396 in the ability of IRF-3 to induce expression of the interferon-stimulated genes ISG56 and ISG54. These data lead us to reconsider the current model of IRF-3 activation. We propose that conventional biochemical assays used to measure IRF-3 activation are not sensitive enough to detect the small fraction of IRF-3 needed to elicit a biological response. Importantly, our study establishes a molecular link between the role of serine 339 in IRF-3 homodimerization, CBP association, and its destabilization.
IRF-3 is essential for the normal host response to pathogens (27; reviewed in reference 7), and its activity is regulated by phosphorylation mechanisms. Biochemical studies have clearly demonstrated that IRF-3 is a phosphoprotein that is hyperphosphorylated in response to virus infection or exposure to pathogen-activated molecular patterns (2, 5, 7, 14, 28, 29). Therefore, understanding how the transcriptional activity of IRF-3 is controlled by protein kinases has been a subject of major interest. Exposure of cells to double-stranded RNA, lipopolysaccharide, or infectious particles is known to activate several host kinases such as the IκB kinase (IKK) complex and the stress-activated protein kinases p38 and Jun N-terminal protein kinase. However, none of these kinases target IRF-3 (29). Two IKK homologs, namely, IKKe (22) (also called IKKi) (32) and Tank-binding kinase 1 (TBK1) (23), were recently shown to be activated following virus infection and to control the transcriptional activity of IRF-3 through phosphorylation of its C-terminal regulatory domain (RD) (4, 31). This domain comprises three clusters of phosphoacceptor sites, Ser 385/Ser 386 (cluster I), Ser 396/Ser 398 (cluster II), and Ser 402/Thr 404/Ser 405 (cluster III), for a total of seven potential sites: 382GGASSLENTVDLHISNSHPLSLTSDQY408. Presently, TBK1 and IKKi are the only characterized kinases responsible for the C-terminal phosphorylation of IRF-3, and different conclusions were reached about the phosphoacceptor site(s) targeted by these kinases. In vitro kinase assays using either recombinant kinases or immunoprecipitated kinases from overexpressing cells and truncated recombinant IRF-3 as a substrate (glutathione S-transferase-IRF-3 amino acids 381 to 427) suggested that cluster III is targeted by TBK1/IKKi, most likely at Ser 402 (17, 31, 38). On the other hand, crystallographic studies revealed that Ser 402 is unlikely to be an important target site because it is located in a β-strand region (36). Phosphoacceptor sites in cluster I are the most accessible amino acids, and Ser 386 could be the initial target for TBK1/IKKi (19, 36). Thus, the precise sites targeted by these kinases remain to be characterized. Despite these uncertainties, structure-function studies have clearly established a major role of the C-terminal RD region of IRF-3 in its activation (14, 16, 30, 43). Notably, mutation of the cluster I Ser 385/386 to Ala (creating IRF-3 J2A) abolishes IRF-3 activation (43). Intriguingly, mutation of Ser 385/386 to Asp (creating IRF-3 J2D) also abrogates IRF-3 activation (16). These data led to the suggestion that the Ser 385/386 cluster may serve as a recognition domain for the IKK-related kinases or alternatively that phosphomimetic substitutions do not produce a functional protein (14, 38). Mutation of the five other phosphoacceptor sites in clusters II and III to Ala (creating IRF-3 5A) reduces IRF-3 activation following virus infection (16, 34). Reciprocally, IRF-3 5D (mutation of the five Ser/Thr residues to Asp) behaves as a strong constitutively active form of IRF-3 that associates with CBP, stimulates gene transcription (14, 16), and induces apoptosis in the absence of virus infection (6, 40). Of the five phosphoacceptor sites present in clusters II and III, we further demonstrated that mutation of Ser 396 to Asp (generating IRF-3 S396D) was the minimal modification required in cluster II in order to obtain a form of IRF-3 that constitutively associates with CBP and induces gene transcription and thus is a form that mimics IRF-3 5D (28). Importantly, immunoblotting analysis with phosphospecific antibodies showed that Ser 396 and Ser 386 are targeted in vivo following virus infection (19, 28).
Based on these observations and crystallographic studies (24, 36), two models for IRF-3 activation by phosphorylation have been proposed. One model states that phosphorylation at Ser 385 or Ser 386 of IRF-3 induces its dimerization (36). The other model favors intramolecular interactions where IRF-3 exists in a latent closed conformation in the cytoplasm, the C-terminal RD interacting with the proline-rich region near the N-terminal DNA binding domain (16, 24). C-terminal phosphorylation by TBK1/IKKi is thought to abrogate these intramolecular interactions, allowing IRF-3 to homodimerize or heterodimerize with IRF-7 (8). Subsequently, the dimerized forms of IRF-3 associate with the histone acetyltransferase nuclear proteins CBP and p300, causing IRF-3, which normally shuttles in and out of the nucleus, to become predominantly nuclear (12) and induce transcription of genes encoding chemokines and type I interferon (IFN). Then, IRF-3 is targeted for degradation following its polyubiquitination by at least two intracellular signaling pathways: (i) phosphorylation at Ser 339, which induces the recruitment of the prolyl isomerase Pin1 followed by polyubiquitination (25), and (ii) phosphorylation of clusters II and III, resulting in IRF-3 recognition by a Cullin-based E3 ligase (1).
However, more recent findings suggest that the current model of IRF-3 regulation needs further refinement. For example, Collins and coworkers observed IRF-3-dependent ISG56 induction without IRF-3 hyperphosphorylation or nuclear translocation (2). It was proposed that hyperphosphorylation is related to an anti-immune defense strategy of inducing IRF-3 degradation rather than being a necessary step for the biological activation of IRF-3 (2). Other studies also reported a lack of correlation between nuclear accumulation of IRF-3 and its hyperphosphorylation, homodimerization, and CBP association state (3, 33).
Using mass spectrometry (MS) analysis coupled with comprehensive structure-activity analysis, we reevaluated the phosphoacceptor sites targeted by TBK1 and the model of IRF-3 activation. Our study reveals that Ser 396 and Ser 402 are directly targeted by TBK1 and that the current assays used to monitor the activation of IRF-3 (hyperphosphorylation on sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE], dimerization, coactivator association, and nuclear accumulation) are not directly linked to its transcriptional status but instead reflect hyperactive and unstable forms of IRF-3. Interestingly, mutation of Ser 339, which is involved in IRF-3 stability, also abrogates CBP association and dimerization without affecting gene transactivation as long as Ser 396 is available for phosphorylation. Therefore, our study establishes a molecular link between the role of Ser 339 in IRF-3 homodimerization, CBP association, and its degradation by the proteasome.
MATERIALS AND METHODS
Reagents, antibodies, and plasmids.
Commercial antibodies were obtained from the following suppliers: anti-IRF-3 antibody and anti-pSer386 were from IBL, Japan, and anti-Flag epitope (M2) and anti-β-actin (clone AC-74) were from Sigma (Oakville, Ontario, Canada). The anti-CBP (A22) and the neutralizing anti-IFN-β were purchased from Santa Cruz (Santa Cruz, CA) and Fitzgerald (Concord, MA), respectively. The anti-mISG56 and anti-mISG54 antibodies were a kind gift of Ganes Sen (Lerner Research Institute, Cleveland, OH). A rabbit antiserum raised against proteins of Sendai virus (SeV) as well as the pFlag-IRF3wt and the pFlag-IRF3 point mutant plasmids was generously provided by John Hiscott (McGill University, Montreal, Quebec, Canada). All pFlag-IRF3 plasmids containing S339A were generated by site-directed mutagenesis (QuikChange II site-directed mutagenesis kit; Stratagene, La Jolla, CA). Plasmids encoding ΔN IRF-3, Flag-IKKi, Flag-TBK1, and the dominant-negative version Flag-IKKi K38A or Flag-TBK1 K38A were kind gifts of Rongtuan Lin (McGill University, Montreal, Quebec, Canada). The IFN-β reporter plasmid pGL3-IFN-β-LUC was described previously (13-16). Cycloheximide and leptomycin B were purchased from Calbiochem and used at final concentrations of 100 μg/ml and 100 ng/ml, respectively.
Cell culture, transfection, and infections.
HeLa, 293T, and Vero cells and IRF-3-knockout mouse embryo fibroblasts (MEFs) (kindly provided by Tom Maniatis, Harvard University, Cambridge, MA) were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum. IRF-3+/+ and IRF-3−/− MEF lines were immortalized using the 3T3 protocol (39). DNA transfections were performed with Lipofectamine 2000 (Invitrogen, Burlington, Ontario, Canada) according to the manufacturer's protocol. SeV was obtained from Specific Pathogen-Free Avian Supply (North Franklin, CT). Vesicular stomatitis virus (VSV) (Indiana strain; a kind gift of John Hiscott) was propagated in Vero cells and quantified by standard plaque assay.
Immunoblot analysis, immunoprecipitation, and native PAGE.
Preparation of whole-cell extracts (WCE), immunoprecipitation, native PAGE, and immunoblot analysis were performed as described previously (1, 5, 29). Briefly, WCE were prepared in Triton X-100 lysis buffer (50 mM Tris, pH 7.4; 150 mM NaCl; 30 mM NaF; 5 mM EDTA; 10% glycerol; 1 mM Na3VO4; 40 mM β-glycerophosphate; 0.1 mM phenylmethylsulfonyl fluoride; 5 μg/ml of leupeptin, pepstatin, and aprotinin; 1% Triton X-100). For coimmunoprecipitation studies, WCE (400 to 500 μg) were incubated with 1.5 μg of anti-CBP antibody A-22 cross-linked to 30 μl of protein A-Sepharose beads for 4 h at 4°C (Amersham, GE Health care, United Kingdom). The beads were washed five times in lysis buffer and then resuspended in denaturing SDS loading buffer. IRF-3 proteins associated with CBP were analyzed by immunoblotting as previously described (29). For native PAGE analysis, 7.5% acrylamide gels (without SDS) were prerun with 25 mM Tris and 192 mM glycine, pH 8.4, with and without 1% deoxycholate in the cathode and anode chamber, respectively, for 30 min, at 40 mA. WCE (10 to 15 μg) diluted in native sample buffer (62.5 mM Tris-HCl, pH 6.8, 15% glycerol, and bromophenol blue) were applied to the gel and subjected to electrophoresis for 60 min at 25 mA. Immunoblotting was performed using an anti-Flag monoclonal antibody.
Production of recombinant His-wild-type IRF-3 (His-wtIRF-3).
IRF-3 cDNA was subcloned in pET-15b vector (Novagen, Madison, WI), transformed in Escherichia coli BL21(DE3)pLysS bacteria. Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 3 h at an optical density of 0.6 at 600 nm. His-tagged IRF-3 protein was then purified by nickel-N-acrylotris(hydroxymethyl)aminomethane (Ni-NTA) affinity chromatography using the His Bind purification kit (Novagen) according to the manufacturer's protocol.
Immunofluorescence.
HeLa cells were transfected with the different IRF-3 constructs for 24 h and then infected with SeV. At 8 h postinfection, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min followed by permeabilization with 0.2% Triton X-100 for 10 min. Cells were washed with PBS, pH 7.2, and blocked with 5% dry milk in PBS. Anti-Flag antibody (M2; Sigma) was used at 1:1,000 in PBS-1% bovine serum albumin. Secondary fluorophore-conjugated antiserum (Alexa Fluor 488) was obtained from Molecular Probes (Eugene, OR) and used at 1:250 in PBS-1% bovine serum albumin. In the case of treatment with leptomycin B, the inhibitor was added only 3 h postinfection. The presence of the nucleus was revealed by a conventional 4′,6′-diamidino-2-phenylindole (DAPI) staining.
In vitro kinase assay.
A 1.5-μg amount of bacterially produced His-wtIRF-3 was resuspended in a kinase buffer containing 0.5 μg of recombinant His-TBK1 (Upstate Biotechnology, Lake Placid, NY) or GST-IKKi (Cell Signaling), 20 mM HEPES, pH 7.4, 20 mM MgCl2, 2 mM dithiothreitol, and 20 μM ATP. The reaction mixtures were incubated at 30°C for 30 min and then stored at −80°C for subsequent MS analysis. In vitro kinase assays were also accomplished in the presence of [γ-32P]ATP to verify the phosphotransferase activities of both recombinant TBK1 and IKKi.
MS analysis.
Phosphorylated His-wtIRF-3 was digested with aspartic acid endoprotease (AspN) at 37°C for 4 h followed by trypsin at 37°C for 4 h. Digests were reconstituted in 30 μl of 0.2% formic acid in water-acetonitril (95/5) and analyzed by liquid chromatography coupled to MS (LC-MS) on a NanoAcquity UPLC system interfaced with a Q-TOF Premier spectrometer (Waters, Milford, MA). The LC system consists of an in-house 300-μm × 5-mm C18 precolumn and a 150-μm × 10-cm in-house C18 analytical column, both packed with a Jupiter 3-μm, 300-Å stationary phase (Phenomenex, Torrance, CA). Peptide elution was performed at 600 nl/min. External calibration of the instrument was made using a Glu-Fib B (Sigma) solution of 83 fmol/μl. Data-dependent acquisition of MS-MS spectra was obtained for up to three precursor ions per survey spectrum using argon as a target gas, with collision energies ranging from 20 to 45 eV (laboratory frame of reference). Fragment ions formed in the radio frequency-only quadrupole were recorded by a time-of-flight mass analyzer.
Biological assay.
HeLa cells were first transfected with 0.5 μg of the different pFlag-IRF3 constructs in six-well plates. At 24 h posttransfection, cells were washed twice and infected with 200 hemagglutinin units (HAU)/ml of SeV in serum-free medium in order to stimulate IFN production. Two hours postinfection, cells were extensively washed and covered by 2 ml of fresh complete medium. Twenty hours later, supernatants were cleared by centrifugation at 3,000 rpm and cells were harvested in order to be analyzed by Western blotting. The supernatants of HeLa cells overexpressing the different C-terminal IRF-3 mutants (2 ml) were then split, and 500 μl was used for pretreatment (16 h) of a monolayer of confluent Vero cells seeded in a 12-well plate before further challenge with VSV at different multiplicities of infection (MOIs). The effects were evaluated through VSV-induced cell lysis using a standard plaque assay procedure, where VSV infections were performed in serum-free medium for 1 h followed by replacement with DMEM containing 1% methylcellulose. After 3 days, monolayers were fixed with 4% paraformaldehyde and stained with crystal violet. The infection step with SeV was omitted for the phosphomimetic IRF-3 mutants.
Reporter gene assays.
Subconfluent cells in 35-mm-diameter tissue culture dishes were transfected with 100 ng of pRLTK reporter (Renilla luciferase for internal control) and 250 ng of pGL3-IFN-β-LUC with the use of Lipofectamine 2000 (Invitrogen) as detailed by the manufacturer. Cells were harvested 24 h posttransfection, lysed in passive lysis buffer (Promega, Madison, WI), and assayed for dual-luciferase activity with use of 10 μl of lysate according to the manufacturer's instructions. All firefly luciferase values were normalized to Renilla luciferase to control for transfection efficiency.
RESULTS
Role of IRF-3 phosphoacceptor sites in the establishment of an antiviral state.
IRF-3 contains seven potential phosphoacceptor sites located in the C-terminal RD (Fig. 1A). However, the precise site(s) involved in IRF-3 phosphorylation following a viral infection remain(s) to be identified. Moreover, the contribution of each phosphoacceptor site in the mechanism of IRF-3 activation is unclear (14, 17, 19, 24, 28, 36, 38, 43). We thus reevaluated the physiological relevance of the different Ser/Thr clusters by studying the effect of mutations on antiviral cytokine production. For this purpose, we took advantage of a biological assay where supernatants of HeLa cells overexpressing the different C-terminal IRF-3 mutants were collected and used to treat Vero cells before infection with VSV. The effects were evaluated through VSV-induced cell lysis using a standard plaque assay procedure. In our system, transfection of wtIRF-3 allowed enough IFN-β production in the supernatant of HeLa cells to protect Vero cells against a VSV challenge. Notably, VSV titers revealed that this protection was effective only following an infection of wtIRF-3-transfected HeLa cells with SeV (Fig. 1B, compare lane 4 to lanes 1 to 3). Consequently, involvement of endogenous IRF-3 is ruled out since SeV infection of mock-transfected HeLa cells did not result in any sign of protection (Fig. 1B, lanes 1 and 2). Importantly, the use of an anti-IFN-β antibody in the collected supernatant completely prevented the antiviral effect of overexpressed wtIRF-3 (Fig. 1D). It has been shown that the replacement of Ser residues from 396 to 405 with the phosphomimetic Asp generates a strong, constitutive transactivator named IRF-3 5D (16). As expected, transfection of this mutant conferred resistance to a further VSV challenge without any need for viral infection (Fig. 1B, compare lane 5 to lanes 4 and 6).
FIG. 1.
Analyzing IRF-3 antiviral activity by a biological assay. (A) Schematic representation of the potential phosphoacceptor sites of IRF-3. C-terminal mutations in the different IRF-3 constructs used in this study are shown. (B) HeLa cells in six-well plates were transfected with 0.5 μg of the indicated plasmid. At 24 h posttransfection, cells were left untreated or infected with SeV (200 HAU/ml; 200 HAU total) for 2 h. Then, cells were extensively washed and covered with 2 ml of fresh medium. At 20 h postinfection, supernatants containing antiviral cytokines were collected, cleared of cellular contaminants, and transferred on Vero cells plated in a 12-well plate. After 16 h of incubation, Vero cells were washed with PBS and challenged with different MOIs of VSV (0, 0.001, 0.01, and 0.1) in serum-free DMEM. After 1 hour, infection medium was removed and replaced with DMEM containing 1% methylcellulose and fetal bovine serum. The effects were evaluated through VSV-induced cell lysis using a standard plaque assay procedure. VSV titers (PFU/ml) are also shown and reflect the ability of each IRF-3 construct to confer viral resistance. (C) Immunoblot analysis of Flag-IRF-3 expression levels and viral proteins in HeLa cells 20 h postinfection. (D) wtIRF-3 was tested in the biological assay with the exception that before the incubation on Vero cells for 16 h, clarified supernatants from HeLa cells infected with SeV were incubated with no antibodies, normal rabbit serum, or neutralizing anti-IFN-β antibodies for 1 h at 4°C. (E) Different C-terminal mutants of IRF-3 were tested in the biological assay for their capacity to generate antiviral IFNs. The transactivating ability of the different IRF-3 mutants is compared to that offered by the transfection of a nonrelevant protein (GFP). The ΔN-IRF-3 construct is used as an internal control and represents the behavior of a dominant-negative IRF-3 in this assay. The expression of the different transgenes was verified by immunoblot analysis as described for panel C (data not shown). (F) Different C-terminal phosphomimetic mutants of IRF-3 were tested in the biological assay for their capacity to generate antiviral IFN as described for panel B in the absence of SeV infection. The transactivating ability of the phosphomimetic mutants is compared to that offered by the mock or IRF-3 WT-transfected HeLa cells. The expression of the different transgenes was verified by immunoblot analysis as described for panel C (data not shown). Data are representative of three independent experiments.
When different C-terminal mutants of IRF-3 were compared in this biological assay, we were able to document an important role for the three different clusters in the transactivating potential of the transcription factor. Indeed, while HeLa cells overexpressing wtIRF-3 conferred resistance to VSV infection up to an MOI of 0.1, cells overexpressing the IRF-3 J2A mutants were not able to raise a strong protection against VSV infection (Fig. 1E, compare lane 2 to lane 7). In fact, we observed a 2-log reduction of protection compared to the wild-type protein, a result similar to that with HeLa cells overexpressing a nonrelevant protein (green fluorescent protein [GFP]) or the dominant-negative version of IRF-3 that lacks the DNA binding domain (ΔN-IRF3) (Fig. 1E, compare lane 2 to lanes 7, 1, and 9). This incapacity of the IRF-3 J2A mutant to confer resistance to VSV demonstrates that these two serines are essential for IRF-3 activation. We also observed an intermediate protection (1-log reduction) with the IRF-3 2A, 3A, and 5A mutants, suggesting that the Ser/Thr clusters II and III are necessary phosphorylation sites for the achievement of a full activation of IRF-3 (Fig. 1E, compare lane 2 to lanes 4 to 6). As expected, introduction of the S385/386A mutation into IRF-3 5A (herein named IRF-3 7A) completely abolished the intermediate protection of IRF-3 5A, restoring the phenotype observed with the IRF-3 J2A mutant (Fig. 1E, compare lane 6 to lanes 7 and 8). We previously proposed that Ser 396 was the minimal phosphoacceptor site required for IRF-3 activation (28). We therefore tested if this residue was involved in the antiviral state observed in our bioassay. Surprisingly, overexpression of IRF-3 S396A conferred the same protection as did the wild-type protein (Fig. 1E, lanes 2 and 3). On the other hand, the use of different phosphomimetic mutants of IRF-3 in this biological assay clearly demonstrated that IRF-3 S396D had the strongest antiviral activity of all phosphomimetic mutants tested (Fig. 1F, lane 8). Using reporter gene assays, IRF-3 5D was previously classified as a strong constitutive gene transactivator (16). Indeed, IRF-3 5D had antiviral activities in our biological assay comparable to those of IRF-3 3D and IRF-3 S398D mutants but less than those of IRF-3 S396D and IRF-3 2D (Fig. 1F, compare lanes 5, 6, and 9 to lanes 3 and 8). Finally, our results also demonstrated that the integrity of Ser 385 and Ser 386 is primary for IRF-3 activity, as IRF-3 J2D did not behave like a phosphomimetic mutant of IRF-3 and replacement of Ser 385/386 with Asp in the IRF-3 5D backbone (generating IRF-3 7D) totally abrogated the antiviral activity of IRF-3 5D (Fig. 1F, lanes 4 and 7). Altogether, our data reconcile published data (16, 28, 41) by showing that all clusters are essential for full IRF-3 transactivation activity but at different levels of importance, raising the idea of a well-ordered cluster phosphorylation kinetic. Importantly, the results obtained using our bioassay further highlight the importance of the integrity of Ser 385/386 for IRF-3 activity, while questioning the role of Ser 396 in the activation of IRF-3.
MS analysis of phosphorylated IRF-3 by TBK1.
The different abilities of the diverse IRF-3 mutants to yield an antiviral state prompted us to readdress the role of the IKK-related kinases in the phosphorylation and activation of IRF-3. TBK1 and IKKi were recently shown to be responsible for the C-terminal phosphorylation of IRF-3 after a viral infection (4, 31), but the exact phosphoacceptor sites on IRF-3 still remain to be identified. Therefore, we performed in vitro kinase assays using bacterially produced full-length His-tagged wtIRF-3 in the presence of recombinant TBK1. Phosphorylated wtIRF-3 was then analyzed by tandem MS (MS/MS) in order to identify the targeted phosphoacceptor sites. Figure 2A shows the MS/MS spectra of precursors m/z 800.82+ and m/z 840.82+. Both precursors correspond to IRF-3 peptide DLHISNSHPLSLTS bearing, respectively, one and two phosphate groups at residue 402 and residues 396 and 402. For both spectra, the site of phosphorylation was assigned to the Ser 402 residue, as fragment ion m/z 389.3 harbored the characteristic loss of H3PO4 moiety through a β-elimination giving rise to a dehydroalanine residue shifted by 69 Da from its adjacent y-type fragment ion at m/z 320.2. For m/z 840.82+, a supplementary phosphorylation site at Ser 396 was observed, an assignment supported by the characteristic dehydroalanine residue for the b-type fragment ion at m/z 479.3. To substantiate these results, the phosphorylation of recombinant His-IRF-3 was also analyzed using a phosphospecific antibody against Ser 396 (28). A significant phosphoserine 396 signal was detected when IRF-3 was incubated with TBK1 (Fig. 2B). Phosphorylation of Ser 402 was also observed when IKKi was used in the kinase assay (data not shown). Combined with the results obtained from the bioassay (Fig. 1), our data support the idea that clusters II and III are the first sites targeted by the IKK-related kinases, which, once phosphorylated, are likely to prime the targeting of Ser 385 and Ser 386 located in cluster I. These results suggest that sequential phosphorylation of the three clusters is necessary to elicit complete unfolding and full activation of IRF-3.
FIG. 2.
TBK1 directly phosphorylates cluster II and III of IRF-3 in vitro. (A) In vitro kinase assay using cold ATP, recombinant TBK1, and full-length His-IRF-3 as substrate with analysis by LC-MS/MS for characterization of potential phosphoacceptor sites on IRF-3. MS/MS spectra of precursors at m/z 800.82+ and 840.82+ confirming the protein identification and the sites of phosphorylation. (B) Western blot analysis of the in vitro kinase assay using an affinity-purified pS396 antibody.
The use of IRF-3 S396A reveals a lack of correlation between IRF-3 transcriptional activity and other conventional assays.
Our bioassay demonstrated that the IRF-3 S396A mutant was transcriptionally active (Fig. 1). However, our previous work showed that this mutation abolishes virus-induced IRF-3-CBP association (28), suggesting that this assay of IRF-3 activation may not reflect transcriptional potential. Thus, we have decided to test the activation of all IRF-3 mutants using conventional biochemical assays, i.e., hyperphosphorylation (retarded mobility in SDS-PAGE), dimerization, association with CBP, and nuclear accumulation. We first verified the capacity of the different IRF-3 mutants to associate with the CBP coactivator, as this step is essential for the DNA binding activity and the nuclear accumulation of IRF-3 (12, 35). Mutations in clusters I (J2A) and II (S396A and 2A) completely abolished CBP association in SeV-infected HeLa cells, whereas mutation of cluster III (IRF-3 3A) had no effect (Fig. 3A), as previously observed (14, 28). The same observation was made in HeLa cells overexpressing TBK1 (data not shown). We next verified the dimerization state of the different IRF-3 mutants using a native PAGE assay (9). As for CBP association, our data demonstrate that the dimerization of IRF-3 was restricted to wtIRF-3 and IRF-3 3A mutants. Notably, mutants lacking Ser 385/Ser 386 (J2A) or Ser 396 (2A, 5A, 7A, and S396A) phosphorylation did not show any sign of inducible dimerization following virus infection (Fig. 3B). Similar results were observed in cells overexpressing TBK1 or IKKi (data not shown). Importantly, titration experiments demonstrated that overexpression of IRF-3 S396A to six times the level of wtIRF-3 did not rescue the observed phenotype (data not shown). Interestingly, using the phosphomimetic mutants, we observed that IRF-3 2D and S396D, the two most powerful phosphomimetic transactivators (Fig. 1E), are not in a complete state of dimerization compared to IRF-3 5D (Fig. 3C).
FIG. 3.
Conventional assays of activation do not correlate with IRF-3 S396A activity. (A) Coimmunoprecipitation experiments of Flag-IRF-3 with CBP following SeV infection in HeLa cells (8 h, 200 HAU/ml; 200 HAU total). (B) SDS-PAGE and native PAGE electrophoresis analysis of the different Flag-IRF-3 constructs following SeV infection (8 h, 200 HAU/ml; 200 HAU total). Blots were probed with anti-Flag antibody to correlate the cellular expression of IRF-3 mutants (SDS-PAGE) with their dimerization state (native). (C) HeLa cells were transfected with the different IRF-3 phosphomimetic mutants for 40 h. SDS-PAGE and native PAGE were then performed as described for panel B. (D) Cellular localization of the different Flag-IRF-3 mutants following SeV infection. Immunofluorescence analyses were performed using an anti-Flag antibody. Cellular localization of the different Flag-IRF-3 mutants following SeV infection (8 h, 200 HAU/ml; 200 HAU total) in the presence of leptomycin B (LMB; 100 ng/ml) is also shown. (E) Quantification of the nuclear accumulation observed in panel D. Results are expressed as means ± standard errors of the nuclear accumulation percentage of IRF-3. Data represent three independent experiments where 100 cells were randomly counted by epifluorescence microscopy. Only cells having more than 50% of the total IRF-3 localized in the nucleus were considered for the nuclear accumulation phenotype. (F) Hyperphosphorylation profile of Flag-wtIRF3, J2A, and 3A. Flag-IRF-3 constructs were cotransfected with Flag-IKKi (kinase dead [KD] or wild type) in HEK 293T cells. At 40 h posttranfection, cells were lysed and the hyperphosphorylated form of IRF-3 (III) was resolved by SDS gel electrophoresis on a 7.5% acrylamide gel. The membrane was probed with anti-Flag antibody. Native PAGE was also performed to correlate the dimerization states of the different IRF-3 mutants with their hyperphosphorylation profiles.
Nuclear accumulation is another assay frequently used to monitor the activation of IRF-3. Consistent with CBP association and the dimerization state observed by native PAGE assay, wtIRF-3 and the IRF-3 3A mutant were the only proteins able to accumulate in the nuclear compartment following virus infection (Fig. 3D and E). Accordingly, mutation of Ser 396 to Ala abrogated this accumulation, correlating with its failure to associate with CBP coactivator. Using another model, IRF-3 was reported to accumulate rapidly and transiently in the nucleus (33), but immunofluorescence analysis of cells infected for 4 h demonstrated no nuclear accumulation of IRF-3 S396A (data not shown). Importantly, addition of leptomycin B, a selective inhibitor of the CRM-1-dependent export pathway, rescued nuclear accumulation of the mutants containing mutation of Ser 396 to Ala and also the J2A mutant (Fig. 3D). This confirms that the nuclear import pathway and IRF-3 C-terminal phosphorylation are two independent mechanisms and that nuclear accumulation of IRF-3 observed in epifluorescence is dependent upon CBP binding, as previously proposed (12). Thus, the IRF-3 3A mutant behaves essentially like wtIRF-3 in CBP association, dimerization, and nuclear accumulation assays. However, IRF-3 3A is comparable to IRF-3 2A and 5A (mutants that failed to associate with CBP, dimerize, or accumulate in the nucleus) in its ability to build a moderate antiviral state (Fig. 1D). Another commonly used assay to follow IRF-3 activation is the appearance of hyperphosphorylated forms in SDS-PAGE after viral infection. Easily observable on endogenous IRF-3, these hyperphosphorylated forms of IRF-3 are harder to detect with the ectopic protein (14). However, when IKKi is coexpressed with IRF-3 in 293T cells, hyperphosphorylation becomes apparent. As shown in Fig. 3F, a hyperphosphorylated form of IRF-3, denoted as form III, was totally lost in the IRF-3 3A mutant under conditions where dimerization is still observed. The IRF-3 J2A mutant, which represents a transcriptionally inactive IRF-3 protein, is, however, hyperphosphorylated without any sign of dimerization. Altogether, these data demonstrate that the three Ser/Thr clusters are all important for full activation of IRF-3 but that the conventional biochemical assays mentioned above are not sensitive enough to monitor minimal IRF-3 activation and are likely to reveal principally the only forms of IRF-3 that are hyperactive.
Phosphorylation of Ser 339 regulates IRF-3 dimerization, coactivator association, and degradation.
The hyperactive mutant IRF-3 5D strongly associates with the CBP coactivator (16) and is mostly expressed as a dimeric form (Fig. 3C). Moreover, we recently demonstrated that this phosphomimetic mutant is highly unstable in transfected cells (1). Interestingly, the prolyl isomerase Pin1 was recently shown to bind and promote IRF-3 degradation following virus infection or poly(I:C) stimulation (25). This interaction is dependent upon phosphorylation of Ser 339 by a putative “proline-directed” kinase. Since our data demonstrate that the CBP-associated and the dimerized form of IRF-3 likely represents an unstable hyperactive version of the transcription factor (Fig. 3), we hypothesized that Ser 339 phosphorylation may be involved in the capacity of IRF-3 to homodimerize and associate with CBP. First, we verified whether Ser 339 was involved in the instability of overexpressed IRF-3 5D by using cycloheximide chase experiments. As predicted (1), IRF-3 5D was unstable compared to wtIRF-3 (Fig. 4A). Importantly, this instability was significantly rescued by mutation of Ser 339 to Ala in IRF-3 5D (IRF-3 5D S339A) (Fig. 4B). The role of Ser 339 in IRF-3 stability was also evaluated upon virus infection. The monomeric form of IRF-3 S339A was highly stable over the entire studied kinetic (Fig. 4C, right side). Importantly, the stabilization of IRF-3 S339A correlated with its incapacity to homodimerize or associate with CBP following viral infection, a profile also shared by the S396A mutant (Fig. 4C and D). Introduction of this specific mutation in IRF-3 3A, the only mutant that is still able to homodimerize upon infection (Fig. 3), also abolished its capacity to dimerize (Fig. 4E). Similar observations were made when the S339A mutation was introduced in the phosphomimetic mutants 2D, 3D, 5D, and S396D (Fig. 4F). Thus, the residue Ser 339 is also involved in the capacity of IRF-3 to homodimerize and associate with the CBP coactivator following its activation. Moreover, Ser 339 serves as a molecular link between the hyperactive dimeric and CBP-associated forms of IRF-3 and its degradation.
FIG. 4.
Mutation of IRF-3 at Ser 339 affects its dimerization and CBP association state. (A and B) HeLa cells were transfected with Flag-wtIRF3, 5D, or 5D S339A for 24 h and then treated with cycloheximide (CHX) for the indicated time. IRF-3 expression was analyzed by immunoblot analysis using anti-Flag antibody. (C) Flag-wtIRF3 and S339A were transfected in HeLa cells and then infected with SeV (8 h or 24 h, 200 HAU/ml; 200 HAU total) 24 h posttransfection. Dimerization state was analyzed by native PAGE. (D) Flag-wtIRF3, S339A, and S396A were transfected in HeLa cells for 24 h and then left untreated or infected with SeV (8 h). Coimmunoprecipitation with CBP was then monitored. (E and F) The S339A mutation was introduced in the different IRF-3 C-terminal alanine mutants (E) or aspartic mutants (F). HeLa cells were transfected and then infected with SeV for 8 h (E) or left untreated (F). Cellular expression and dimerization state of the IRF-3 mutants were then analyzed by SDS-PAGE and native PAGE, respectively, followed by Western blot analysis.
Role of Ser 339 in the transactivation activity of IRF-3.
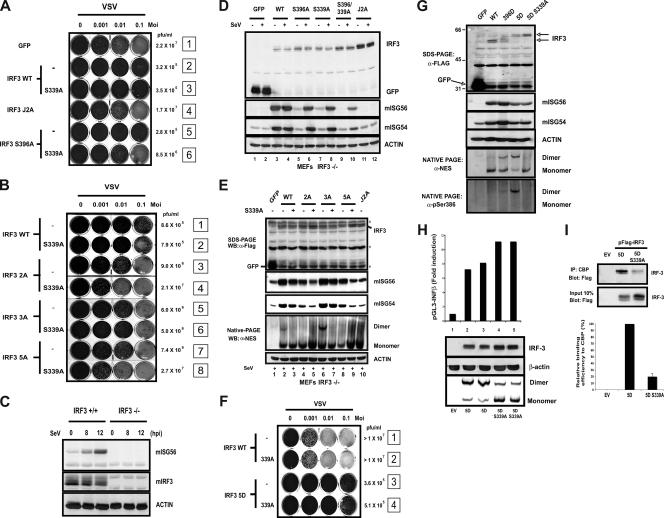
Since Ser 339 contributes to the ability of IRF-3 to homodimerize and associate with CBP, it could potentially regulate its transcriptional activity. However, our data suggest that these assays are not directly linked to the transcriptional status of IRF-3 (Fig. 3). Notably, the ability of IRF-3 S339A to establish an antiviral state in our biological assay was as efficient as those of wtIRF-3 and IRF-3 S396A (Fig. 5A, compare lanes 2, 3, and 5). On the other hand, we observed that a combination of the S339A mutation with S396A results in a transcriptionally inactive IRF-3 molecule behaving like the IRF-3 J2A mutant (Fig. 5A, compare lanes 4 and 6). The same result was obtained for every IRF-3 mutant where Ser 396 was mutated to Ala. Notably, IRF-3 2A and 5A are no longer able to confer an intermediate resistance to VSV infection while IRF-3 3A is not affected by the presence of the mutation. (Fig. 5B). In MEFs, ISG56 induction is totally dependent on the presence of IRF-3 (Fig. 5C). Therefore, we performed complementation experiments in IRF-3−/− MEFs in order to evaluate the ability of the different IRF-3 mutants to promote murine ISG56 expression. In the absence of viral infection, overexpression of wtIRF-3 was previously shown to activate the antiviral state in 2FTGH cells and rat embryonic fibroblasts (10). Indeed, we observed the induction of ISG56 and also ISG54 in IRF-3−/− MEFs complemented with wtIRF-3 in the absence of viral infection (Fig. 5D, lane 3). In line with the results from the biological assay, mISG56 induction is compromised only when Ser 396 and Ser 339 are both mutated to Ala (Fig. 5D, compare lane 4 to lane 10). As verified by the loss of the dimers and the phospho-386 signal (data not shown), introduction of the S339A mutation in IRF-3 3A totally abolished its dimerization state but did not blunt the induction of mISG56 as well as mISG54 (Fig. 5E, lanes 6 and 7), whereas mutation of Ser 339 to Ala in mutants 2A and 5A dramatically reduced the induction of mISG56 and mISG54 following virus infection (Fig. 5E, compare lanes 5 and 9 to lanes 3 and 7). When introduced in IRF-3 5D, S339A did not modify the induction of mISG56 and mISG54. However, it totally prevented its phosphorylation on Ser 386 and therefore its dimerization (Fig. 5F and G). This observation was further substantiated using a reporter gene assay where IRF-3 5D S339A was as potent as IRF-3 5D in transactivating the IFN-β promoter under conditions where both dimerization and CBP association were severely impaired (Fig. 5H and I). Thus, coactivator association and dimerization assays do not precisely reflect the transcriptional potential of IRF-3.
FIG. 5.
Mutual compensation of Ser 339 and Ser 396 in the transactivation process of IRF-3. (A and B) The different C-terminal IRF-3 mutants harboring the S339A mutation were tested in the biological assay for their capacity to produce antiviral IFNs as described for Fig. 1E. The transactivating ability of the different IRF-3 mutants is compared to that offered by the transfection of a nonrelevant protein (GFP). VSV titers (PFU/ml) are also shown. (C) mISG56 induction profile in IRF-3−/− MEFs following viral infection. (D and E) Complementation experiment in IRF-3−/− MEFs with the different C-terminal mutants of IRF-3 in order to determine their ability to induce mISG56 and mISG54. Indicated IRF-3 constructs were transfected in IRF-3−/− MEFs. At 24 h posttransfection, cells were infected with SeV as indicated. Cellular expression of indicated proteins was analyzed by SDS-PAGE as well as native PAGE followed by Western blot analysis. Asterisks denote nonspecific signal. (F) IRF-3 5D and 5D S339A were tested in the biological assay as described for Fig. 1F. The transactivating ability of the phosphomimetic mutants is compared to that offered by HeLa cells transfected with wtIRF-3. (G) Complementation experiment in IRF-3−/− MEFs with the different IRF-3 phosphomimetic mutants. Uninfected cells were treated as described for panel D. (H) HeLa cells were transiently cotransfected with luciferase reporter constructs containing the IFN-β promoter and IRF-3 5D or 5D S339A as indicated below the bar graphs. At 36 h posttransfection, the luciferase activity was measured as described in Materials and Methods. The relative luciferase activity was measured as activation over the transfection of pFlag-CMV2 alone (EV). Each value represents the mean ± standard error of triplicate determinations. The data are representative of at least four different experiments with similar results. Cellular extracts were also analyzed by SDS-PAGE and native PAGE in order to correlate the dimerization state of the mutants and their transactivating abilities. (I) pFlag-CMV2 (EV) and pFlag-IRF3 5D and 5D S339A were transfected in HeLa cells. At 40 h posttransfection, cells were lysed and IRF-3 coimmunoprecipitation with CBP was monitored as described for Fig. 3. Quantification of coimmunoprecipitated IRF-3 was done with a gel documentation device (Typhoon scanner, GE Health care). Percent standard errors are given based on the averages of three independent experiments.
DISCUSSION
Phosphorylation mechanisms are often the rate-limiting step involved in controlling enzymatic activities and protein-protein interactions, as well as other posttranslational modifications such as acetylation and ubiquitination. Thus, several transcription factors are kept inactive in resting cells and rapidly become activated following their phosphorylation on specific subsets of phosphoacceptor sites. In resting cells, IRF-3 is already expressed as a phosphoprotein with phosphorylation largely occurring in the N-terminal region (29; J. F. Clement and M. J. Servant, unpublished results). Upon virus infection, at least nine phosphoacceptor sites (eight located in its C terminus) have been suggested to participate in the regulation of its transcriptional activity (11, 14, 19, 25, 28, 43), a complex situation that could partly explain the difficulties and discrepancies encountered in the literature about the role of the different phosphoacceptor sites of IRF-3. Our MS analysis of TBK1-phosphorylated full-length IRF-3 clearly documents the phosphorylation of at least two of these sites: residues 396 and 402, located in clusters II and III, respectively. For IKKi, only Ser 402 phosphorylation was observed (data not shown). This is the first study directly implicating the phosphorylation of Ser 396 by TBK1. Using an in vitro phosphorylation assay, tenOever et al. concluded that Ser 402 was phosphorylated in IRF-3 (37). We suspect that they did not identify Ser 396 as a targeted residue because of the use of a C-terminal fragment of IRF-3 as a substrate (residues 381 to 427), which does not represent the native protein. On the other hand, a recent study demonstrated that residue 404 or 405 is directly targeted by recombinant TBK1 (21). However, residual phosphorylation was detected in the latter C-terminal construct of IRF-3 when these two residues were mutated to alanine. Phosphorylation of these residues by TBK1 is intriguing since the crystal structure of latent IRF-3 has revealed that they are buried in a hydrophobic environment, a position not favorable for a nucleophilic attack of the gamma phosphate of the ATP. Thus, one model that could reconcile these discrepancies is an orderly phosphorylation mechanism where IRF-3 is first phosphorylated on residues 396 and 402 by TBK1 followed by phosphorylation of Ser 404 or Ser 405, which would then prime the phosphorylation of Ser 385/Ser 386 (Fig. 6B) (21). This model can also be appreciated in vivo following overexpression of IKKi and IRF-3 J2A where the generation of hyperphosphorylated forms of IRF-3 J2A was observed without any sign of dimerization (Fig. 3F, lanes 3 and 4). On the other hand, cluster III is likely responsible for the generation of the hyperphosphorylated forms of IRF-3 (Fig. 3F, lanes 5 and 6). Thus, clusters II and III can obviously be phosphorylated first, a covalent modification that would favor the phosphorylation of cluster I at Ser 386, which is essential for IRF-3 dimerization and activation.
FIG. 6.
Analysis of the Ser/Thr-to-Ala mutants of IRF-3 revealed the implication of Ser 339 in the two-step activation process of IRF-3. (A) Summary of the effect of the alanine mutants of IRF-3 and their properties in biochemical assays and in the biological activity. ND, not determined; N/A, not applicable. (B) Proposed model. Following a viral infection, the IKK-related kinases TBK1 and IKKi are activated and phosphorylate Ser 402 (cluster III) and/or Ser 396 (cluster II). This phosphorylation event allows further phosphorylation of cluster III at less accessible sites (residues 404 and 405). Modification of clusters II and III then allows cluster I to become available for phosphorylation either by TBK1 and IKKi (21) or by an atypical phosphoinositide 3-kinase pathway (20, 26). This incremental mechanism, by which phosphorylation might progressively pry off the inhibitory helix in the regulatory domain (shown in yellow), allows IRF-3 to become competent for the induction of IFN-β. However, a putative proline-directed kinase activated by the viral infection also phosphorylates IRF-3 on Ser 339. When both Ser 339 and Ser 396 are phosphorylated, IRF-3 becomes hyperactivated. Thus, homodimerization, CBP association, and nuclear accumulation are favored and become easily measurable by conventional biochemical assays. Bound to Pin1, this hyperactive form of IRF-3 will then be degraded by a Cullin-based E3 ligase.
Several biochemical assays have been used in order to monitor IRF-3 activation. Higher-migrating forms of IRF-3 on SDS gels, which correlate with its C-terminal phosphorylation (14, 28, 29), and association with coactivators CBP and p300, as well as electrophoretic mobility shift assay analysis, were among the first proposed assays because they correlated with nuclear accumulation of IRF-3 and gene transactivation. The ability of IRF-3 to homodimerize following its phosphorylation was then introduced as another readout of activation, followed by the use of phosphospecific antibodies to monitor IRF-3 phosphorylation by immunoblot analysis (9, 19, 28). However, recent reports in the literature have challenged the validity of these biochemical assays. Notably, in response to UV-treated SeV or low MOI, Collins and coworkers failed to observe hyperphosphorylation, nuclear localization, or subsequent degradation of IRF-3 and yet readily observed induction of the IRF-3-responsive gene ISG56 (2). Another study also reported early nuclear accumulation of IRF-3 without any sign of hyperphosphorylation, homodimerization, or CBP association (33). The discrepancy between the different assays in measuring IRF-3 activation was also observed in cells infected with NS1 mutants of the influenza B virus where nuclear accumulation of IRF-3 occurred in the absence of measurable CBP association (3).
Using a comprehensive combination of different IRF-3 mutants, our study clearly demonstrates that the conventional assays are not sensitive enough to detect the small fraction of IRF-3 needed to elicit a biological response (Fig. 6A). Notably, the IRF-3 S396A and S339A mutants are active in our bioassay (Fig. 1 and 5), despite the fact that they are greatly impaired in their capacity to dimerize, associate with CBP, and accumulate into the nucleus following virus infection (Fig. 3 to 5 and data not shown). Intriguingly, a lack of sensitivity of the methods used to follow IRF-3 activation was previously shown by Mori et al., where overexpression of IRF-3 S396D in 293T cells did not result in either dimerization or phosphorylation of Ser 386 (19) despite its strong transcriptional activity (this study and reference 28). Using complementation assays in IRF-3−/− MEFs, we further demonstrate that overexpression of S396D resulted in strong induction of both ISG56 and ISG54 under conditions where the phospho-386 signal is almost absent (Fig. 5G). We therefore propose that the hyperphosphorylation, homodimerization, CBP association, and nuclear accumulation observed following virus infection are measurable only when IRF-3 is hyperactivated, a condition that leads to its degradation (1).
We and others have demonstrated that the hyperphosphorylated/dimerized forms of IRF-3 are unstable in infected cells (1, 2, 5, 11, 14, 25, 28, 29). These hyperphosphorylated and dimerized forms of IRF-3, detected by a decreased mobility in SDS-PAGE and in native PAGE assays, respectively, are artificially reproduced following overexpression of IRF-3 5D (1, 14, 28). Ser 339 is involved in IRF-3 degradation following virus infection (25), and the observed instability of IRF-3 5D is abrogated when Ser 339 is replaced by Ala (IRF-3 5D S339A), a condition that also affects the ability of IRF-3 5D to homodimerize and associate with CBP (Fig. 4 and 5). These observations suggest that IRF-3 5D is constitutively phosphorylated on Ser 339. The effect of mutating Ser 339 to Ala also abrogates virus-mediated IRF-3 homodimerization, CBP association, and degradation (Fig. 4). Thus, through phosphorylation of Ser 339, the dimerized and CBP-associated form of IRF-3 is more likely to represent a highly transcriptionally active but also unstable version of IRF-3. Therefore, our study establishes a molecular link between the role of Ser 339 in IRF-3 homodimerization, CBP association, and its destabilization. Interestingly, this working model was also suggested for the transcriptional regulator steroid receptor coactivator 3 (SRC-3), where phosphorylation allows interaction with Pin1, which in turn enhances SRC-3-CBP/p300 interaction, promoting the turnover of the activated SRC-3 oncoprotein (42). We are currently investigating the role of Pin1 in the ability of IRF-3 to interact with CBP.
Since the homodimerization and CBP association assays do not reflect the transcriptional status of IRF-3, we were not surprised to find that IRF-3 S339A and IRF-3 S396A, two mutants completely devoid of these activities, had antiviral properties similar to those of wtIRF-3 (Fig. 1 and 5). However, when introduced in IRF-3 mutants in which Ser 396 is mutated to Ala (IRF-3 S396A, IRF-3 2A, and IRF-3 5A), S339A completely suppresses the protection provided by these mutants (Fig. 5). These results suggest that Ser 339 and Ser 396 share redundant roles at the level of IRF-3 transcriptional activity. Together, they are essential for the transcriptional activity of IRF-3, as replacement of these residues with nonphosphorylable sites completely suppressed virus-induced production of IFN-β in our bioassay or greatly reduced the ability of IRF-3 to induce mISG56 expression. This conclusion is also supported by the fact that mutation of Ser 339 to Ala in mutant IRF-3 3A (IRF-3 3A S339A) did not dramatically affect the transactivation potential of the molecule (Fig. 5B, compare lanes 5 and 6; Fig. 5E, compare lanes 6 and 7). The availability of Ser 396 in this mutant likely explains why the transcriptional activity of IRF-3 is not affected.
A model for the activation of IRF-3 is presented in Fig. 6. Following virus infection, activation of the IKK-related kinases TBK1 and IKKi results in IRF-3 phosphorylation of sites accessible on cluster II (Ser 396) and cluster III (Ser 402) that will subsequently lead to the phosphorylation of Ser 385/386 for a significant activation and antiviral gene transactivation. Since phosphorylation of Ser 396 precedes the phosphorylation of Ser 339 (25), we propose that phosphorylation of Ser 396 on cluster II further enhances the antiviral response by presumably exposing Ser 339 and favoring its subsequent phosphorylation by an unknown proline-directed kinase, a modification that results in the generation of a hyperactive version of IRF-3 that is easily measurable through CBP association and homodimerization assays. Once generated, this hyperactive form of IRF-3 is recognized by Pin1, a process leading to its polyubiquitination and subsequent degradation by the proteasome (1, 25). Our results thus favor the two-step activation model of IRF-3 as previously suggested (26, 33), and we propose that phosphorylation of Ser 339 is another important covalent modification involved in that process. As demonstrated by our MS data, TBK1 preferably targets clusters II and III. We did not detect phosphorylation of cluster I by recombinant TBK1, thus favoring the idea that Ser 385/Ser 386 are likely to be the target of an atypical phosphoinositide 3-kinase-regulated kinase pathway (20, 26). On the other hand, it is also possible that Ser 385/Ser 386 become targets of TBK1 following the phosphorylation of clusters II and III (21). Despite these uncertainties and taking into consideration the important role of Ser 385/386 in the activation of IRF-3, phosphorylation of Ser 396 by TBK1 and phosphorylation of Ser 339 by a putative “proline-directed kinase” would be two essential covalent modifications for the dimerization and the formation of a hyperactive form of IRF-3.
Since we clearly observe antiviral gene expression in the almost total absence of CBP association with IRF-3 S339A or IRF-3 S396A mutants (Fig. 1E, 4D, and 5), one could argue that IRF-3-CBP association is not absolutely required for the induction of the IFN-β gene as recently proposed (18). While our study did not address the role of coactivators in the antiviral properties of IRF-3, our results suggest that current biochemical assays do not always accurately reflect the transcriptional activity of IRF-3. The inductions of IRF-3 response genes such as ISG56 and type I IFN still represent the most reliable assays.
Acknowledgments
We thank John Hiscott, Rongtuan Lin, Ganes Sen, Tom Maniatis, and Benjamin TenOever for reagents used in this study. We are also grateful to members of Sylvain Meloche's laboratory for helpful discussions.
This work was supported by research grants from the Canadian Institutes of Health Research (CIHR) to M.J.S. (MOP-53282) and S.M. (MOP-14168) and a CIHR Research Resource Grant (PRG 80293) to P.T. M.J.S. is a recipient of a Rx&D/CIHR Health Research Foundation Career Award in Health Sciences. S.M., P.T., and N.G. are recipients of Canada Research Chairs. J.-F.C. and S.-P.G. are both recipients of a studentship from the Fonds de la Recherche en Santé du Québec (FRSQ).
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Bibeau-Poirier, A., S. P. Gravel, J. F. Clement, S. Rolland, G. Rodier, P. Coulombe, J. Hiscott, N. Grandvaux, S. Meloche, and M. J. Servant. 2006. Involvement of the IκB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J. Immunol. 1775059-5067. [DOI] [PubMed] [Google Scholar]
- 2.Collins, S. E., R. S. Noyce, and K. L. Mossman. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J. Virol. 781706-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dauber, B., J. Schneider, and T. Wolff. 2006. Double-stranded RNA binding of influenza B virus nonstructural NS1 protein inhibits protein kinase R but is not essential to antagonize production of alpha/beta interferon. J. Virol. 8011667-11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4491-496. [DOI] [PubMed] [Google Scholar]
- 5.Gravel, S. P., and M. J. Servant. 2005. Roles of an IκB kinase-related pathway in human cytomegalovirus-infected vascular smooth muscle cells: a molecular link in pathogen-induced proatherosclerotic conditions. J. Biol. Chem. 2807477-7486. [DOI] [PubMed] [Google Scholar]
- 6.Heylbroeck, C., S. Balachandran, M. J. Servant, C. DeLuca, G. N. Barber, R. Lin, and J. Hiscott. 2000. The IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J. Virol. 743781-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-κB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010237-248. [DOI] [PubMed] [Google Scholar]
- 8.Honda, K., and T. Taniguchi. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6644-658. [DOI] [PubMed] [Google Scholar]
- 9.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-κB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6375-388. [DOI] [PubMed] [Google Scholar]
- 10.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factory-3. Proc. Natl. Acad. Sci. USA 959837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karpova, A. Y., M. Trost, J. M. Murray, L. C. Cantley, and P. M. Howley. 2002. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 992818-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 204159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, R., C. Heylbroeck, P. Genin, P. M. Pitha, and J. Hiscott. 1999. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol. Cell. Biol. 19959-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 182986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin, R., Y. Mamane, and J. Hiscott. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 27534320-34327. [DOI] [PubMed] [Google Scholar]
- 16.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 192465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McWhirter, S. M., K. A. Fitzgerald, J. Rosains, D. C. Rowe, D. T. Golenbock, and T. Maniatis. 2004. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. USA 101233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mokrani, H., O. Sharaf el Dein, Z. Mansuroglu, and E. Bonefoy. 2006. Binding of YY1 to proximal region of the murine beta interferon promoter is essential to allow CBP recruitment and K8H4/K14H3 acetylation on the promoter region after virus infection. Mol. Cell. Biol. 268551-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori, M., M. Yoneyama, T. Ito, K. Takahashi, F. Inagaki, and T. Fujita. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 2799698-9702. [DOI] [PubMed] [Google Scholar]
- 20.Noyce, R. S., S. E. Collins, and K. L. Mossman. 2006. Identification of a novel pathway essential for the immediate-early, interferon-independent antiviral response to enveloped virions. J. Virol. 80226-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panne, D., S. M. McWhirter, T. Maniatis, and S. C. Harrison. 2007. Interferon response factor 3 is regulated by a dual phosphorylation-dependent switch. J. Biol. Chem. 28222816-22822. [DOI] [PubMed] [Google Scholar]
- 22.Peters, R., S. M. Liao, and T. Maniatis. 2000. IKKe is part of a novel PMA-inducible IkB kinase complex. Mol. Cell 5513-522. [DOI] [PubMed] [Google Scholar]
- 23.Pomerantz, J. L., and D. Baltimore. 1999. NF-κB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 186694-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin, B. Y., C. Liu, S. S. Lam, H. Srinath, R. Delston, J. J. Correia, R. Derynck, and K. Lin. 2003. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat. Struct. Biol. 10913-921. [DOI] [PubMed] [Google Scholar]
- 25.Saitoh, T., A. Tun-Kyi, A. Ryo, M. Yamamoto, G. Finn, T. Fujita, S. Akira, N. Yamamoto, K. P. Lu, and S. Yamaoka. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7598-605. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar, S. N., K. L. Peters, C. P. Elco, S. Sakamoto, S. Pal, and G. C. Sen. 2004. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 111060-1067. [DOI] [PubMed] [Google Scholar]
- 27.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13539-548. [DOI] [PubMed] [Google Scholar]
- 28.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 2789441-9447. [DOI] [PubMed] [Google Scholar]
- 29.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276355-363. [DOI] [PubMed] [Google Scholar]
- 30.Servant, M. J., B. Tenoever, and R. Lin. 2002. Overlapping and distinct mechanisms regulating IRF-3 and IRF-7 function. J. Interferon Cytokine Res. 2249-58. [DOI] [PubMed] [Google Scholar]
- 31.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 3001148-1151. [DOI] [PubMed] [Google Scholar]
- 32.Shimada, T., T. Kawai, K. Takeda, M. Matsumoto, J. Inoue, Y. Tatsumi, A. Kanamaru, and S. Akira. 1999. IKK-i, a novel lipopolysaccharide-inducible kinase that is related to IκB kinases. Int. Immunol. 111357-1362. [DOI] [PubMed] [Google Scholar]
- 33.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 792079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suhara, W., M. Yoneyama, T. Iwamura, S. Yoshimura, K. Tamura, H. Namiki, S. Aimoto, and T. Fujita. 2000. Analyses of virus-induced homomeric and heteromeric protein associations between IRF-3 and coactivator CBP/p300. J. Biochem. (Tokyo) 128301-307. [DOI] [PubMed] [Google Scholar]
- 35.Suhara, W., M. Yoneyama, I. Kitabayashi, and T. Fujita. 2002. Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 27722304-22313. [DOI] [PubMed] [Google Scholar]
- 36.Takahasi, K., N. N. Suzuki, M. Horiuchi, M. Mori, W. Suhara, Y. Okabe, Y. Fukuhara, H. Terasawa, S. Akira, T. Fujita, and F. Inagaki. 2003. X-ray crystal structure of IRF-3 and its functional implications. Nat. Struct. Biol. 10922-927. [DOI] [PubMed] [Google Scholar]
- 37.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 763659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.tenOever, B. R., S. Sharma, W. Zou, Q. Sun, N. Grandvaux, I. Julkunen, H. Hemmi, M. Yamamoto, S. Akira, W. C. Yeh, R. Lin, and J. Hiscott. 2004. Activation of TBK1 and IKKɛ kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J. Virol. 7810636-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todaro, G. J., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver, B. K., O. Ando, K. P. Kumar, and N. C. Reich. 2001. Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53. FASEB J. 15501-515. [DOI] [PubMed] [Google Scholar]
- 41.Yang, H., C. H. Lin, G. Ma, M. Orr, M. O. Baffi, and M. G. Wathelet. 2002. Transcriptional activity of interferon regulatory factor (IRF)-3 depends on multiple protein-protein interactions. Eur. J. Biochem. 2696142-6151. [DOI] [PubMed] [Google Scholar]
- 42.Yi, P., R. C. Wu, J. Sandquist, J. Wong, S. Y. Tsai, M. J. Tsai, A. R. Means, and B. W. O'Malley. 2005. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1). Mol. Cell. Biol. 259687-9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukada, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 171087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]