Abstract
RNA silencing is a host defense mechanism that limits the accumulation and spread of viruses in infected plants. Correspondingly, plant viruses encode suppressors of silencing. In the positive-strand RNA virus Tobacco rattle virus (TRV), the suppressor of silencing is a 16-kDa (16K) protein encoded by RNA1. The suppressor action of the 16K protein is transient and weaker than that of the P19 suppressor, encoded by tomato bushy stunt virus. Mutant TRV that does not produce its suppressor, unlike other suppressor-defective viruses, is competent to accumulate and spread systemically in the infected plant. However, this mutant virus does not exhibit the transient invasion of the meristem that is characteristic of the wild-type virus. Based on this analysis, we propose that the 16K suppressor of silencing allows TRV to transiently invade the meristem. Our data are consistent with a mechanism of long-term meristem virus exclusion that is dependent on a transient invasion of the meristem early in the infection cycle. This novel mechanism of meristem exclusion may be associated with the phenomenon of recovery in virus-infected plants in which upper leaves have little or no virus and are immune to secondary infection by the same virus.
Most plant viruses encode suppressors of RNA silencing. Mutation of viral genes that encode suppressors of RNA silencing normally disables the virus so that it accumulates at low levels in the inoculated or systemically infected leaves and induces milder symptoms than the wild-type (wt) virus. Correspondingly, if the RNA silencing machinery in the host plant is suppressed, the virus accumulates to a higher level and induces symptoms more severe than those on plants that are fully competent for RNA silencing (references 2, 30, and 33 and references therein).
Silencing suppressor proteins from different viruses do not all block the same stage in the silencing process (25, 34). Many of these proteins are RNA binding proteins, and it is possible that they interfere with the initial stages in silencing. They could prevent conversion of single-stranded RNA into double-stranded RNA (dsRNA) by an RNA-dependent RNA polymerase, or they could bind to long dsRNA and thereby prevent its cleavage into 21- or 22-nucleotide short interfering RNAs (siRNAs) by a Dicer nuclease (21, 23). RNA binding suppressors may also bind to the overhanging 3′ termini of siRNAs and prevent their transfer to an Argonaute nuclease that is the effector protein of silencing (17, 18, 20, 32, 39). Other suppressors block more-downstream stages in silencing by targeting Argonaute proteins. The cucumoviral 2b protein may prevent Argonaute nuclease activity (41), and the F box proteins of Poleroviruses mediate its degradation (4, 16).
RNA silencing suppressors may also block the silencing signal that moves between cells and through the phloem of the plant (24, 35). This signal is likely to be a dsRNA or siRNA, and in virus-infected plants it may move either with or ahead of the virus as it moves out of the initially infected cells or through the phloem (11, 40). This signal RNA would then have potential to prime the RNA silencing machinery in the recipient cell so that virus accumulation in these cells is reduced or arrested. At least two types of silencing suppressor, from a potexvirus and a cucumovirus, have properties that are consistent with a direct or indirect effect on the silencing signal. They have no effect on virus accumulation in initially infected cells, but their activity is required for either short- or long-distance movement of the virus (1, 7, 10, 36). It could be that these suppressors normally interfere with the signal that moves with or ahead of the virus so that the viral genome is not silenced when it spreads away from the site of initial infection.
To investigate aspects of viral RNA silencing, we are using tobacco rattle virus (TRV). This positive-strand-RNA virus provides a useful model for a minimal virus because plants can be systemically infected with just one of the two viral RNAs (31). In addition, because viruses in the Tobravirus group are transmitted by seed, they must be able to overcome the meristem exclusion process that restricts the spread of most viruses in infected plants (38). Previous work with potato virus X (PVX) and cucumber mosaic virus (CMV) had implicated RNA silencing in meristem exclusion (22, 26, 30), and it seemed likely that tobraviruses would have developed the ability to either evade or suppress the silencing machinery in the growing point of the infected plant.
Our analysis focused on the TRV-encoded 16-kDa putative suppressor of silencing (16K protein). This 16K protein is not needed for systemic spread of TRV (13). Our results implicate RNA silencing in meristem exclusion, in line with previous analysis with PVX and CMV (22, 26, 30). However, it seems that the mechanism of meristem exclusion is fundamentally different with PVX and TRV. With PVX, the meristematic silencing mechanism is primed by a virus-specific signal of silencing that moves from lower noninfected parts of the plant and is dependent on RDR6 (30). In contrast, with TRV and possibly with CMV (22), our evidence presented here suggests that the priming involves the transient presence of the virus in the meristematic cells and is independent of RDR6.
MATERIALS AND METHODS
Plasmids and viruses. (i) Constructs for expression of TRV genes.
The MP, 16K, and CP genes were PCR amplified and cloned into pBIN61 (5). For the MP gene, primers 5′MP (5′GGGGGGATCCATGGAAGACAAGTCATTGG) and 3′MP (5′GGGGGAATTCTTAAGACGAGTTTTTCTTATTA) were used. For the 16K gene, primers 5′16K (5′GGGGGGATCCATGACGTGTGTACTCAAGG) and 3′16K (5′GGGGGAATTCTCAAAAAGCAAACAAACGATC) were used. For the CP gene, primers 5′CP (5′GGGGGGATCCATGGGAGATATGTACGATG) and 3′CP (5′GGGGGAATTCTAGGGATTAGGACGTATC) were used. (Sequences in bold correspond to restriction sites BamHI and HindIII, used for cloning amplified fragments.)
(ii) TRV mutants.
All TRV16K mutants were built into the construct pBINTRA6, the TRV RNA1 clone (27), in several steps. First, a chimeric PCR fragment carrying the mutation was generated. Second, this mutation was cloned in an intermediate vector, pBSTR3′C, which contains the 3′ (encoding the C-terminal) half of the TRV RNA1 genome from position 2698 to the 3′ end. Third, the construct was digested in pBSTR3′C and the fragment carrying the mutation was cloned into pBINTRA6. The use of pBSTR3′C was necessary because of the lack of unique sites near the 16K gene in pBINTRA6. For TRV:CH, primers 5′MP (described above) and 16KCHV2 (5′ATAAAATAAAATCATGTTTCAACACGTTTACGACA) were used to generate a 1,112-bp fragment, and primers 16KCHV1 (5′CGTGTTGAAACATGATTTTATTTTATATTGTTATCTG) and TRV2 (5′GGGGGGATCCGGGCGTAATAACGCTTACG) (the BamHI cleaving sequence is shown in bold) were used to generate a 255-bp fragment. Both fragments overlapped 26 bp and were combined and amplified in a chimeric PCR, using primers TRV2 and 5′MP, to generate a recombinant 1.34-kb fragment. The chimeric PCR product was digested with AvrII and StuI and cloned into the same sites of pBINTRA6. This deletion removed the whole basic domain of 16K from amino acid 81 to the end. For TRV:B, a fragment of 782 bp was amplified using primers 5′MP and 16KBV1 (5′TCAAGGTGACTCATATTGACAATAAATTTCTTTATG), and a fragment of 477 bp was amplified using primers 16KBV2 (5′TATTGTCAATATGAGTCACCTTGAAAAGTGTCG) and TRV2. Both fragments overlapped 24 bp and were recombined as described above to generate a 1.23-kb fragment that was digested with AvrII and StuI and cloned into the same sites of pBINTRA6. This deletion removed the whole CH domain from amino acid 2 to 68 of the 16K gene. For TRV:stop, a 1,298-bp fragment was amplified using primers TR4870 (5′ACTCACTGATTGCGTTTCCTAG) and StopR (5′GACTTCATTCACTCAACCCTTGAG), and a 669-bp fragment was amplified using primers StopF (5′CTCAAGGGTTGAGTGAATGAAGTC) and TRV2. (The mutation introduced to create the stop codon is shown in bold). Both fragments were recombined as described above to generate a 1.96-kb fragment that incorporated a point mutation at nucleotide 24 of the 16K gene, which introduced a stop codon. Chimeric PCR was digested with MluI and BamHI (introduced with the primer TRV2 [see above]) and cloned into the same sites of pBINTRA6.
(iii) PVX constructs.
PVX:GFP was described in Baulcombe et al. (3). PVX:16K was constructed by replacing GUS from pSLDB2100, which expresses PVX:GUS under a double 35S promoter (6), through chimeric PCR, given the lack of unique sites flanking the GUS gene. A 2,057-bp fragment was amplified from PVX:GUS by using primers PVX4100F (5′AAGCCAGGTCAAACCATAG) and PVX16K-R (5′ACACGTCATATTTAAATCGATGCTAGCTGGTGC). A 681-bp fragment containing either 16K or 16Kstop was amplified from pBINTRA or pSLDB2105 (TRVRNA1stop), respectively, using the primers PVX16K-F (5′AGCATCGATTTAAATATGACGTGTGTACTCAAGG) and 3′16KSal (5′GAGAGAGTCGACTCAAAAAGCAAACAAACGATC). (The SalI target sequence introduced with the primer is shown in bold). Both fragments were combined in a PCR, using primers PVX4100F and 3′16KSal, to produce a 2.7-kb PCR fragment including the 16K sequence flanked by PVX sequences, which was digested with SalI and AvrII and cloned into the same sites of pSLDB2100.
Virus inoculation. (i) Agrobacterium-mediated transient expression of viral genomes.
The binary Ti plasmid vector constructs were transformed into Agrobacterium tumefaciens strain C58C1, carrying the virulence helper plasmid pCH32 (14). The transformants were inoculated into 5 ml L broth medium supplemented with 50 μg ml/1 kanamycin and 5 μg ml/1 tetracycline and grown at 28°C overnight. Cells were centrifuged and resuspended to optical density 1 in solution containing 10 mM MgCl2, 10 mM MES (morpholineethanesulfonic acid), pH 5.6, and 150 μM acetosyringone. The cells were left at room temperature for 2 h before infiltration into Nicotiana benthamiana leaves.
(ii) Virus infections.
N. benthamiana plants at the four-leaf stage were routinely Agrobacterium infiltrated in third and fourth whole leaves. We refer to this method as agroinfiltration.
PVX:GFP/TRV coinfections.
N. benthamiana plants were first agroinfiltrated with PVX:GFP, and after 14 days, plants were reinfected with either wt TRV or TRV:stop by agroinfiltration of the two newest leaves. The presence of PVX:GFP in flowers was scored 10 days after TRV infection.
Greenhouse conditions.
All work involving virus-infected material was carried out in containment glasshouses under MAFF license PHL 24B/3654. N. benthamiana plants were germinated on a 1:1 mixture of compost and peat and then grown individually in pots at 25°C during the day and 20°C during the night. Supplementary winter lighting from halogen quartz iodide lamps provided a 16-h day length.
Nucleic acid extraction and gel blot analysis.
RNA was extracted using Tri reagent (Sigma) according to the manufacturer's instructions, fractionated in a 1% (wt/vol) agarose formaldehyde gel, transferred to a nylon membrane (Hybond N), and cross-linked with UV illumination. Filters were prehybridized, hybridized, and washed as described previously (15). TRV RNA 1 was detected using a randomly primed, 32P-labeled 1.33-kb fragment expanding from position 4847 to 6181 of TRVRNA1. For siRNA analysis, RNA was separated in 8% polyacrylamide gel and detected using an in vitro-transcribed, 32P-labeled RNA probe of a cloned fragment of green fluorescent protein (GFP) as described previously (30).
In situ hybridization.
Samples were embedded in wax, sectioned, and in situ hybridized as indicated in reference 9. TRVRNA1 was detected using an in vitro-transcribed RNA fragment labeled with digoxigenin (Roche Diagnostics GmbH) and detected with antibody anti-digoxigenin conjugated to alkaline phosphatase (Roche Diagnostics GmbH). The substrate was either NBT/BCIP (Roche Diagnostics GmbH) or Fast Red (Sigma-Aldrich). The probe expands from position 5573 to 6182 of the TRVRNA1 genome. PVX was detected using a probe expanding from position 4160 to 4690 of the PVX genome.
Imaging.
PVX:GFP-infected flowers were cut by the longitudinal axis by using a razor blade. Reproductive organs were visualized using a Leica MZ-FLIII dissecting microscope with a GFP filter and recorded using a Leica DC200 digital camera. In situ hybridizations were visualized with a Nikon microphot-SA microscope and recorded using a Nikon Coolpix 990 digital camera.
RESULTS
The TRV 16K protein is a weak suppressor of RNA silencing.
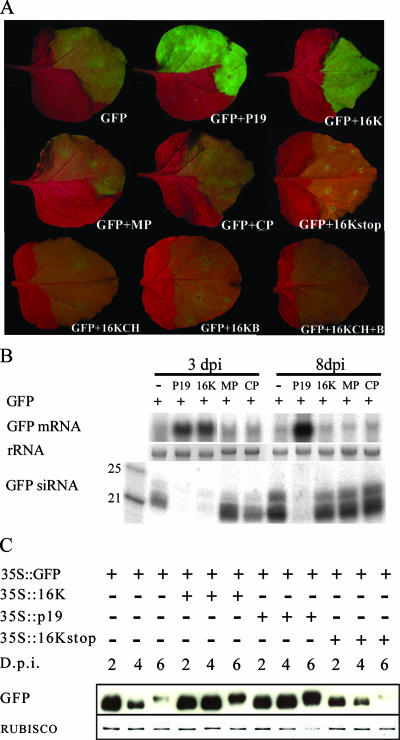
The TRV 16K protein has the expected properties of a suppressor of silencing in that it enhances virulence when expressed in a heterologous virus vector and it suppresses RNA interference in Drosophila cells (19, 29). To confirm the suppressor function of this protein, we used a transient expression assay with N. benthamiana in which the 16K protein was expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter, together with the jellyfish GFP (37). In the absence of a suppressor of silencing, the GFP fluorescence was weak due to the accumulation of GFP siRNAs that mediate silencing of the GFP mRNA (Fig. 1A and B). However, when the 16K protein was coexpressed with GFP, after 3 days of transient expression, the fluorescence was stronger, the GFP mRNA was more abundant, and the GFP siRNAs were at the limit of detection. These data are consistent with the proposed silencing suppressor activity of the 16K protein (Fig. 1A and B). However, the silencing of GFP mRNA and accumulation of GFP siRNAs were restored after 8 days of transient expression (Fig. 1B).
FIG. 1.
16K is a weak suppressor of RNA silencing. (A) Enhancement of GFP fluorescence in the presence of different TRV genes and 16K mutants. MP, TRV movement protein; CP, TRV coat protein; 16KCH, Cys-rich domain of 16K; 16KB, Basic domain of 16K; P19, TBSV p19 protein. Photographs were taken at 3 dpi. (B) Northern blot showing GFP mRNA and siRNAs in the presence of different TRV genes. Samples were taken at 3 and 8 dpi. 16K suppresses siRNA production at 3 dpi, but at 8 dpi the presence of siRNAs indicates the absence of 16K suppressor activity. (C) Western blot showing accumulation of GFP protein in the presence of 16K, TBSV p19, and the mutant 16Kstop at 2, 4, and 6 days postinfiltration. 16K produces enhancement of GFP production during 4 to 5 days after infiltration, whereas the strong suppressor TBSV p19 is still active at 6 dpi and much later (37). GFP accumulation in the presence of the 16Kstop mutant does not show additional accumulation compared to GFP expression alone. 35S, CaMV 35S promoter; 35T, CaMV 35S terminator.
A mutant 16K gene with an in-frame stop codon at the eighth nucleotide of the open reading frame did not enhance GFP expression in this transient assay, indicating that the suppression is caused by the encoded protein rather than the RNA (Fig. 1A, B, and C). Transient expression of the cysteine- and histidine-rich N-terminal domain of the 16K protein or of the basic C-terminal domain or both at the same time also did not enhance GFP expression (Fig. 1A). It is likely, therefore, that the suppressor activity of the 16K protein requires both domains to be present in the same molecule.
A second, well-characterized suppressor, p19 from tomato bushy stunt virus (TBSV), caused higher-level expression of GFP after 3 days and suppressed GFP silencing at the mRNA and siRNA levels for at least 8 days of transient expression (Fig. 1A and B), as described previously (37). We conclude therefore that the 16K protein is a weaker suppressor of RNA silencing than p19. This transient effect occurs most likely because the weak suppressor activity allows some buildup of siRNAs in the early stages of silencing. These primary siRNAs would seed secondary siRNA production so that eventually the suppressor effect is overwhelmed. This conclusion is reinforced by Western blot analysis showing that accumulation of GFP protein was higher with the 16K protein than with 16Kstop but more transient than with p19 (Fig. 1C).
The 16K protein is required for meristem invasion by TRV.
To investigate the influence of RNA silencing on TRV, we generated three 16K mutant forms of RNA1. These were TRV:CH, in which the basic domain coding sequence was deleted; TRV:B, in which we deleted the Cys-rich-domain-coding sequence; and TRV:stop, with a nonsense eighth codon. These mutant RNAs were able to replicate in the infiltrated leaves of N. benthamiana and to infect systemically, either as RNA1 alone (Fig. 2A) or in the presence of RNA2 (data not shown). The accumulations of wt TRV and TRV:stop RNA1 were similar in inoculated leaves at 3 days postinoculation (dpi) and at 13 and 15 dpi in systemically infected leaves but higher than that of wt RNA1 at 10 dpi in systemically infected leaves. Additionally, TRV:stop caused mild necrosis in the stem and veins of systemic leaves (Fig. 2B, panel c) that was more severe than the mild mosaic symptoms, with shortening of internodal distances induced by wt RNA1 (Fig. 2B, panels a and b). Thus, contrary to an earlier report (19) and unlike for suppressors of silencing in many other viruses, we do not find that the 16K suppressor is necessary for or enhances systemic infection. We cannot explain the discrepancy with one earlier report (19). However, our findings are fully consistent with another independent report, showing that 16K mutants of TRV RNA1 were fully competent to spread systemically (13).
FIG. 2.
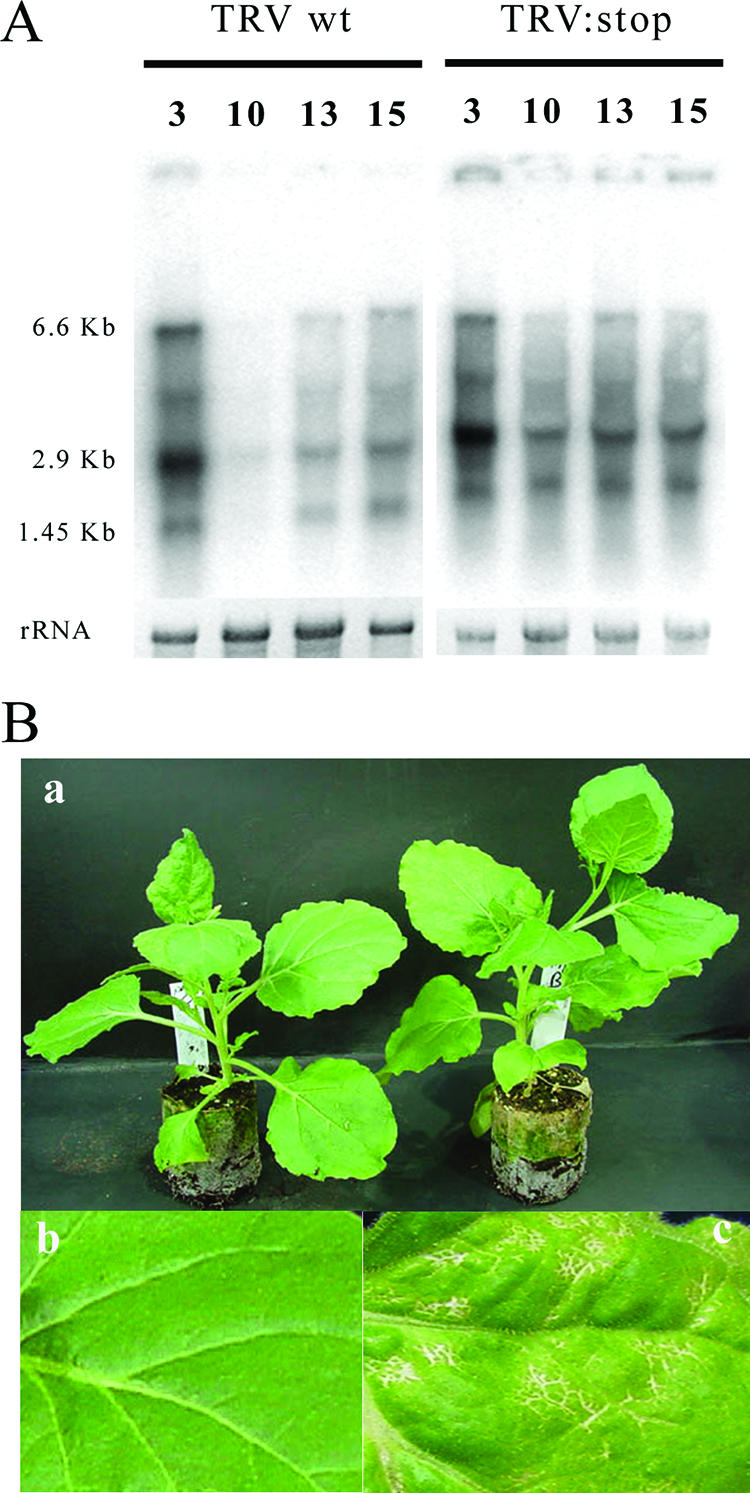
TRV 16K mutants accumulate in systemically infected leaves. (A) Northern blot showing TRV and TRV:stop RNA1 accumulation in infiltrated and systemic leaves. Both viruses accumulate by 3dpi in inoculated leaves. In systemically infected leaves, the initial accumulation (10 dpi) of wt TRV RNA1 is slower than that with TRV:stop RNA1. 6.6 Kb, genomic viral RNA; 2.9 KB, subgenomic RNA for MP; 1.45 KB, subgenomic RNA for 16K. (B) Symptoms induced by TRV RNA1. (a) N. benthamiana plants showing typical wt TRV (left) and TRV:stop (right) symptoms. (b) Detail of a wt-TRV-infected leaf. (c) Detail of a TRV:stop-infected N. benthamiana leaf showing necrosis in veins.
The phenotype of the TRV 16K mutants indicates that RNA silencing has only a slight effect on gross levels of TRV accumulation. This phenotype was reminiscent of our previous finding that overall levels of PVX were unaffected by suppression of a silencing pathway through down-regulation of a silencing-related RDR6 (30). In these experiments with PVX, although there was no effect of the silencing suppression on overall levels of virus, there was an effect of silencing suppression on the amount of virus in the meristem and growing point of the plant: plants with full silencing excluded PVX from the meristem, whereas on the RDR6 knockdown plants they did not (30). This similarity suggested to us that the 16K silencing suppressor might influence the ability of TRV to invade the meristems of infected plants. This is an attractive hypothesis because an orthologue of the 16K protein in pea early browning virus affects transmission of the virus through the seed (38), a process that would be dependent on meristem invasion.
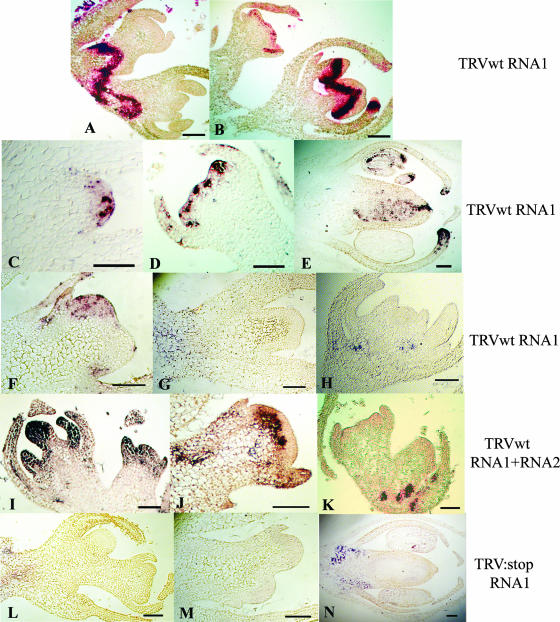
The distribution of wt or mutant TRV RNA in meristems of N. benthamiana was tested by in situ hybridization using an RNA1 probe. The findings showed that the virus moves as a band of infected tissue in the growing point that is below the meristematic zone in the period up to 6 to 7 dpi. After 7 dpi, the wt TRV RNA1 invaded the meristematic regions of the growing point in apical and lateral buds and in floral primordia. This pattern is consistent with our hypothesis that the RNA1-encoded 16K protein allows the virus to overcome meristem exclusion. Out of 35 samples examined, there were 34 in which the viral RNA could be detected in the meristem between 7 and 10 dpi, invading different layers of the meristematic tissue, including tunica and corpus. Strikingly, the TRV RNA was more concentrated in the meristem than in the surrounding regions of the growing point and, consistent with the infected L2 layer of the tunica giving rise to the reproductive organs (8), both ovaries and anthers were invaded in flowers and floral primordia (Fig. 3E).
FIG. 3.
Transient invasion of the meristem by TRV. In situ hybridizations of growing point regions in plants infected with TRV RNA1 (A to H), RNA1 plus RNA2 (I to K), or TRV:stop RNA1 (L to N). (A and B) Growing points showing a band of virus close and invading the meristem at 7 dpi. (C) Axillary meristem, 8 dpi. (D) Floral meristem, 8 dpi. (E) Floral primordium, 8 dpi. (F) Apical meristem, 10 dpi. (G) Secondary meristem, 13 dpi. (H) Floral primordium, 25 dpi. (I) Growing point, 8 dpi. (J) Meristem showing the first few cell layers free of virus, 14 dpi. (K) Floral meristem, 14 dpi, with the virus already completely out of meristematic layers. (L) Lateral meristem, 9 dpi. (M) Lateral and floral meristems, 13 dpi. (N) Floral primordium, 13 dpi. Blue and red indicate the presence of viral RNA. Bars = 100 μm.
However, at later time points, after 11 dpi, we failed to detect wt TRV in any of the 20 growing points of TRVRNA1-infected plants examined (Fig. 3G and H). Thus, the TRV meristem invasion is transient in the period up to and including 10 dpi. In parallel experiments using the TRV (RNA1 plus RNA2) inocula, we similarly observed transient invasion of the meristem, although the timing was different: TRV was present in growing points at 8 dpi (four invaded out of four) (Fig. 3I), although with a more dispersed localization than the RNA1 virus, invading all areas or the meristem and including leaf primordia. By 14 dpi, the RNA1-plus-RNA2 virus was being excluded from the meristem or was already excluded (Fig. 3J and K). Combined, these data show that TRV is allowed into the meristem by expression of 16K and that this invasion is transient. Transient meristem invasion has also been described to occur in CMV-infected tobacco plants. CMV invades the meristem with low efficiency, mainly corpus cells but sometimes tunica cells. In contrast, TRV invades all layers of all meristems tested (Fig. 3I).
In contrast, and consistent with our prediction about the role of the 16K protein, none of the 33 TRV:stop RNA1 or 20 TRV:stop mutants were found in meristems or in floral primordia of infected plants at any time postinfection (Fig. 3L to N). This mutant viral RNA moved to the base of the meristem or floral primordia, but we did not detect it in the meristems or reproductive organs (Fig. 3N). The same result was obtained in in situ hybridizations of growing points of nine TRV:CH-infected and seven TRV:B-infected growing points (data not shown). Therefore, these data are consistent with our proposal that TRV 16K is the viral factor that allows invasion of meristems by TRV RNA through its activity as a suppressor of silencing.
We can envision two scenarios in which transient meristem invasion would lead to a long-term meristem exclusion mechanism. First, it could be that continuous low-level TRV RNA accumulation in the meristem is sufficient to provide a substrate for Dicer and siRNA production. This low-level accumulation would have to be below the level of detection in the in situ hybridizations (Fig. 3). Alternatively, it could be that a host-encoded RDR protein is able to replicate subgenomic fragments of the viral genome. The dsRNA forms of this subgenomic RNA would be the viral siRNA precursors that would exclude virus from the meristematic cells. At present, we have no data to justify a preference for either alternative. However, as shown in Fig. 4, TRV accumulation is unaffected by down-regulation of RDR6 and it invades the meristem as in wt plants, which implies that such a mechanism would have to involve another RDR homologue.
FIG. 4.
TRV accumulation is unaffected by downregulation of RDR6. (A) In situ hybridizations of growing points of TRV RNA1-infected N. benthamiana wt and RDR6 plants taken after 7 dpi. (B) Northern blots of TRV RNA1-infected leaves taken after 3, 10, and 17 dpi.
A predicted consequence of the “transient-accumulation” mechanism would affect the pattern of virus accumulation in the leaves of the infected plant. In the leaves derived from the transiently infected meristem, the virus would persist and continue to accumulate. Conversely, in the leaves derived from the “posttransient-accumulation meristem,” the persistent silencing mechanism would continue to exclude virus or keep its accumulation at a low level. In fact, consistent with our prediction, there is abundant TRV RNA1 in leaves sampled at 17 dpi that are derived from an invaded meristem. In contrast, the leaves taken at 25 dpi that are derived from a “posttransient-accumulation meristem” were almost free of TRV RNA1 (Fig. 5A).
FIG. 5.
Transient meristem invasion leads to recovery. (A) Northern blots from upper N. benthamiana leaves collected after 10, 17, and 25 dpi with wt TRV RNA1. (B) Schematic representation showing how the kinetics of TRV RNA1 meristem invasion influence the TRV RNA accumulation in the upper leaves of the plant. Invaded meristems at 10 dpi generate highly infected leaves that are fully developed at 17 dpi. The TRV-free meristems at 17 dpi are then the precursors of leaves that are fully developed at 25 dpi with a very small amount of viral RNA. Dark blue indicates abundant viral RNA, and light blue indicates less-abundant RNA.
The patterns of TRV accumulation in different leaves of an infected plant are illustrated diagrammatically in Fig. 5B. In N. benthamiana used for this experiment, TRV does not induce symptoms. However, if it did, the early infected leaves would be symptomatic whereas the late leaves would not. This pattern would be characteristic of the “recovery” phenomenon that has been already characterized as a manifestation of RNA silencing (28). Thus, according to this idea, “recovery” could be a side effect of transient meristem invasion by a virus.
The 16K protein acts in trans.
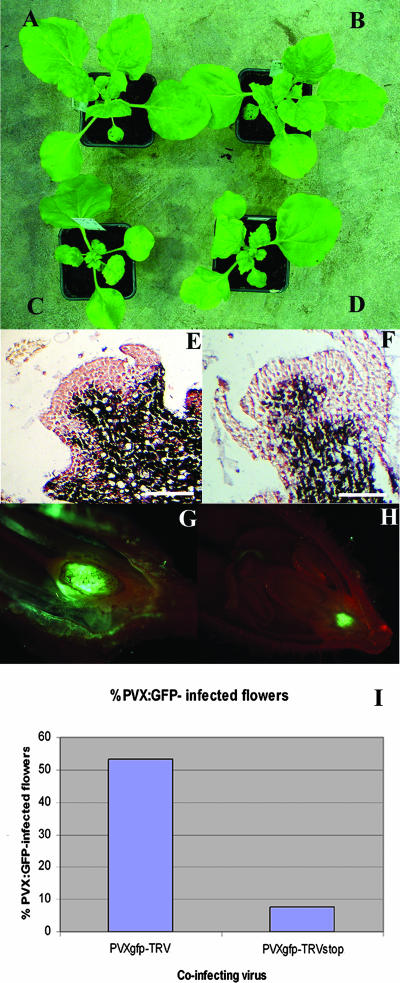
To find out whether the 16K protein could mediate meristem invasion by a heterologous virus, we inserted the 16K open reading frame into a PVX vector (3). N. benthamiana plants infected with this PVX:16K construct showed symptoms that were more severe than the normal mosaic observed with PVX and also PVX:16Kstop (Fig. 6A to D). These plants were smaller, had curled leaves, and exhibited severe mosaic, as reported previously for a similar construct (19). The enhanced symptoms indicate that the 16K protein was expressed in the infected plants, but in situ hybridization failed to detect differences in meristem invasion (Fig. 6E and F).
FIG. 6.
TRV-assisted invasion of meristems by PVX:GFP. (A) PVX-infected N. benthamiana at 13 dpi. (B) PVX:16Kstop-infected N. benthamiana at 14 dpi. (C and D) PVX:16K-infected N. benthamiana at 14 dpi. (E and F) In situ hybridization of PVX:16K- and PVX:16Kstop-infected growing points of N. benthamiana showing the absence of virus in the central zone of the meristem. (G) PVX:GFP-invaded flower from a coinfection with wt TRV-PVX:GFP showing GFP fluorescent ovules. (H) Non-PVX:GFP-invaded flower from a TRVstop-PVX:GFP coinfection showing GFP accumulation at the base of the flower. (I) Percentage of PVX:GFP-invaded flowers in a plant inoculated doubly with PVX:GFP and TRV or TRV:stop. Data are averages from two independent experiments, with values of 54.28% and 52% for TRV-PVX:GFP coinfections (n = 35 and 25, respectively) and 7.5% and 8% for TRVstop-PVX:GFP coinfections (n = 40 and 24, respectively).
However, when TRV was coinoculated with PVX-GFP there was GFP fluorescence in the ovaries and ovules of 53% of the infected plants, indicating that the PVX vector had entered the growing point (Fig. 6G and I). The oldest flowers showed the most fluorescence, and normally the ovules had been invaded rather than the stamens (Fig. 6G). In contrast, in plants coinfected with TRV:stop and PVX:GFP or with PVX:GFP alone, only 8% or 4%, respectively, of flowers were green fluorescent in ovules (Fig. 6H and I). It is likely that infection of the ovules would depend on the virus's being able to invade the meristematic zone of floral primordia, and these findings are therefore consistent with the idea that the 16K protein can act in trans, promoting meristem entry of a heterologous virus. However, it is likely that the 16K protein acts with other components of TRV because the PVX:16K construct did not invade the growing point regions of infected plants.
DISCUSSION
Our analysis shows that TRV 16K allows TRV to enter meristems and to invade the reproductive organs. At present, we cannot rule out that 16K is a multifunctional protein and that its silencing suppressor activity is separate from its ability to invade meristems. It could be, for example, that 16K opens a barrier that prevents virus entry into the meristem and suppresses silencing independently. However, we do not favor this type of explanation, because results with other systems indicate that the growing point and meristem can be invaded by viral RNA if silencing is suppressed (12, 30).
In another tobravirus, pea early browning virus (38), the orthologue of the 16K protein, mediates seed transmission. This finding is certainly consistent with our proposal that the 16K protein function allows transient invasion of the meristem: the presence of the virus in the meristem would facilitate its entry into embryonic tissue of the seed. However, TRV is not seed transmissible in N. benthamiana and we propose that the transience of the meristem invasion allowed by this protein varies in different host plants or even between plants. In TRV-infected N. benthamiana, the meristem invasion would be highly transient and followed by a phase of meristem exclusion. In contrast, with pea infected with PEBV, it could be that the meristem exclusion fades so that the virus invades the primordia that give rise to reproductive organs.
To explain how “transient accumulation” of virus in the meristem leads to longer-term virus exclusion, we propose that the weakness of the16K suppressor activity is a crucial factor. If the suppressor activity were strong, the virus would accumulate to high levels in the meristematic cells and there would be severe damage to the infected plant, as is the case of N. benthamiana transgenic plants expressing ectopically a suppressor of silencing (12). Conversely, in the absence of a silencing suppressor, as in TRV:stop (Fig. 3), the unsuppressed meristematic silencing would eliminate the viral RNA and the viral RNA would not accumulate in the meristem. However, with a weak suppressor, the viral RNA would accumulate in the meristem at an intermediate level, to a lower level than with a strong suppressor, so that damage to the meristem would be limited but more abundant than in the absence of a suppressor. The weakness of the suppressor would allow silencing to eventually reduce the levels of viral RNA below the level of detection by in situ hybridization. In the “posttransient-invasion” phase, the molecular memory of the viral RNA would be provided by RDR-replicated fragments of viral RNA or subliminal replication of the viral genome, as discussed above.
Acknowledgments
This work was supported by British Biotechnology Science and Research Council grant 83/P15101.
We thank Natalya Elina for her help, Desmond Bradley for his advice with in situ hybridizations, and Juan José Lopez-Moya for his helpful critical reading.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Angell, S. M., C. Davies, and D. C. Baulcombe. 1996. Cell-to-cell movement of potato virus X is associated with a change in the size exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii. Virology 216197-201. [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe, D. 2004. RNA silencing in plants. Nature 431356-363. [DOI] [PubMed] [Google Scholar]
- 3.Baulcombe, D. C., S. Chapman, and S. Santa Cruz. 1995. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 71045-1053. [DOI] [PubMed] [Google Scholar]
- 4.Baumberger, N., C. H. Tsai, M. Lie, E. Havecker, and D. C. Baulcombe. 2007. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr. Biol. 171609-1614. [DOI] [PubMed] [Google Scholar]
- 5.Bendahmane, A., G. Farnham, P. Moffett, and D. C. Baulcombe. 2002. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32195-204. [DOI] [PubMed] [Google Scholar]
- 6.Brigneti, G., A. M. Martin-Hernandez, H. Jin, J. Chen, D. C. Baulcombe, B. Baker, and J. D. Jones. 2004. Virus-induced gene silencing in Solanum species. Plant J. 39264-272. [DOI] [PubMed] [Google Scholar]
- 7.Brigneti, G., O. Voinnet, W. X. Li, L. H. Ji, S. W. Ding, and D. C. Baulcombe. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 176739-6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Carpenter, R., and E. S. Coen. 1995. Transposon induced chimeras show that floricaula, a meristem identity gene, acts non-autonomously between cell layers. Development 12119-26. [DOI] [PubMed] [Google Scholar]
- 9.Coen, E. S., J. M. Romero, S. Doyle, R. Elliott, G. Murphy, and R. Carpenter. 1990. floricaula: a homeotic gene required for flower development in antirrhinum majus. Cell 631311-1322. [DOI] [PubMed] [Google Scholar]
- 10.Ding, S. W., W.-X. Li, and R. H. Symons. 1995. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 145762-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunoyer, P., C. Himber, and O. Voinnet. 2005. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 371356-1360. [DOI] [PubMed] [Google Scholar]
- 12.Foster, T. M., T. J. Lough, S. J. Emerson, R. H. Lee, J. L. Bowman, R. L. Forster, and W. J. Lucas. 2002. A surveillance system regulates selective entry of RNA into the shoot apex. Plant Cell 141497-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilford, P. J., V. Ziegler-Graff, and D. C. Baulcombe. 1991. Mutation and replacement of the 16-kDa protein gene in RNA-1 of tobacco rattle virus. Virology 182607-614. [DOI] [PubMed] [Google Scholar]
- 14.Hamilton, C. M., A. Frary, C. Lewis, and S. D. Tanksley. 1996. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc. Natl. Acad. Sci. USA 939975-9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, A. L., I. E. Johansen, S. J. Bean, I. Bach, and A. J. Maule. 1998. Specificity of resistance to pea seed-borne mosaic potyvirus in transgenic peas expressing the viral replicase (Nlb) gene. J. Gen. Virol. 793129-3137. [DOI] [PubMed] [Google Scholar]
- 16.Kapranov, P., J. Cheng, S. Dike, D. A. Nix, R. Duttagupta, A. T. Willingham, P. F. Stadler, J. Hertel, J. Hackermuller, I. L. Hofacker, I. Bell, E. Cheung, J. Drenkow, E. Dumais, S. Patel, G. Helt, M. Ganesh, S. Ghosh, A. Piccolboni, V. Sementchenko, H. Tammana, and T. R. Gingeras. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 3161484-1488. [DOI] [PubMed] [Google Scholar]
- 17.Lakatos, L., T. Csorba, V. Pantaleo, E. J. Chapman, J. C. Carrington, Y.-P. Liu, V. Dolja, L. F. Calvino, J. J. Lopez-Moya, and J. Burgyan. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 252768-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lakatos, L., G. Szittya, D. Silhavy, and J. Burgyan. 2004. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 23876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, H., B. Reavy, M. Swanson, and S. A. MacFarlane. 2002. Functional replacement of the tobacco rattle virus cysteine-rich protein by pathogenicity proteins from unrelated plant viruses. Virology 298232-239. [DOI] [PubMed] [Google Scholar]
- 20.Ma, J. B., K. Ye, and D. J. Patel. 2004. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mérai, Z., Z. Kerényi, A. Molnar, E. Barta, A. Válóczi, G. Bisztray, Z. Havelda, J. Burgyán, and D. Silhavy. 2005. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 797217-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mochizuki, T., and S. T. Ohki. 2004. Shoot meristem tissue of tobacco inoculated with Cucumber mosaic virus is infected with the virus and subsequently recovers from infection by RNA silencing. J. Gen. Plant Pathol. 70363-366. [Google Scholar]
- 23.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 9514687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palauqui, J. C., T. Elmayan, J. M. Pollien, and H. Vaucheret. 1997. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 164738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pantaleo, V., G. Szittya, and J. Burgyan. 2007. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 813797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu, F., X. Ye, G. Hou, S. Sato, T. E. Clemente, and T. J. Morris. 2005. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 7915209-15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratcliff, F., A. M. Martin-Hernandez, and D. C. Baulcombe. 2001. Technical advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25237-245. [DOI] [PubMed] [Google Scholar]
- 28.Ratcliff, F. G., S. A. MacFarlane, and D. C. Baulcombe. 1999. Gene silencing without DNA. rna-mediated cross-protection between viruses. Plant Cell 111207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reavy, B., S. Dawson, T. Canto, and S. A. MacFarlane. 2004. Heterologous expression of plant virus genes that suppress post-transcriptional gene silencing results in suppression of RNA interference in Drosophila cells. BMC Biotechnol. 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwach, F., F. E. Vaistij, L. Jones, and D. C. Baulcombe. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 1381842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanson, M., H. Barker, and S. A. MacFarlane. 2002. Rapid vascular movement of tobraviruses does not require coat protein: evidence from mutated and wild-type viruses. Ann. Appl. Biol. 141259-266. [Google Scholar]
- 32.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. Tanaka Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115799-811. [DOI] [PubMed] [Google Scholar]
- 33.Vaucheret, H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20759-771. [DOI] [PubMed] [Google Scholar]
- 34.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6206-220. [DOI] [PubMed] [Google Scholar]
- 35.Voinnet, O., and D. C. Baulcombe. 1997. Systemic signalling in gene silencing. Nature 389553. [DOI] [PubMed] [Google Scholar]
- 36.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103157-167. [DOI] [PubMed] [Google Scholar]
- 37.Voinnet, O., S. Rivas, P. Mestre, and D. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33949-956. [DOI] [PubMed] [Google Scholar]
- 38.Wang, D., S. A. MacFarlane, and A. J. Maule. 1997. Viral determinants of pea early browning virus seed transmission in pea. Virology 234112-117. [DOI] [PubMed] [Google Scholar]
- 39.Ye, K., L. Malinina, and D. J. Patel. 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo, B. C., F. Kragler, E. Varkonyi-Gasic, V. Haywood, S. Archer-Evans, Y. M. Lee, T. J. Lough, and W. J. Lucas. 2004. A systemic small RNA signaling system in plants. Plant Cell 161979-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, X., Y. R. Yuan, Y. Pei, S. S. Lin, T. Tuschl, D. J. Patel, and N. H. Chua. 2006. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 203255-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]