Abstract
In this study, we monitored the temporal breadths, frequencies, and functions of antiviral CD4 and CD8 T cells in 2 of 22 DNA/modified vaccinia virus Ankara-vaccinated macaques that lost control of a simian-human immunodeficiency virus 89.6P challenge by 196 weeks postchallenge. Our results show that both mutation and exhaustion contributed to escape. With the reappearance of viremia, responding CD8 and CD4 T cells underwent an initial increase and then loss of breadth and frequency. Antiviral gamma interferon (IFN-γ)- and interleukin 2-coproducing cells were lost before IFN-γ-producing cells and CD4 cells before CD8 cells. At euthanasia, all CD8, but no CD4, Gag epitopes detected during long-term control contained mutations.
In preclinical macaque models, recombinant human immunodeficiency virus/AIDS T-cell vaccines have primarily controlled, rather than prevented, infections (3, 6, 13, 15). For the best of these vaccines, this has resulted in chronic infections in which virus is present at the low levels associated with successful multidrug therapies in humans. In our studies with DNA/modified vaccinia virus Ankara-vaccinated and simian-human immunodeficiency virus (SHIV)-89.6P-challenged macaques, multiyear control below 300 copies of viral RNA per ml of blood has been associated with stable low breadths and low frequencies of antiviral CD8 and CD4 T lymphocytes (14).
One of the major problems in human immunodeficiency virus vaccine development has been mutational escape from CD8 T-cell responses (4, 5, 11). Mutational escape is seen in both acute and chronic infections (1, 8, 12). During chronic infections, escape can also occur due to persistent stimulation by virus, leading to exhaustion of T cells (10, 17). Exhaustion of T cells is characterized by a hierarchal loss of interleukin 2 (IL-2), then tumor necrosis factor alpha, and then gamma interferon (IFN-γ) production and ultimately apoptosis and death of the chronically stimulated cell (16).
We previously reported long-term control of a SHIV-89.6P challenge in DNA/modified vaccinia virus Ankara-vaccinated macaques (3, 14). When this trial was terminated at 190 to 200 weeks postchallenge, 90% of the macaques had retained control of their SHIV-89.6P challenge. All of these macaques had stable IFN-γ- and IL-2-coproducing CD4 and CD8 T-cell responses to epitopes mapped at week 140 postchallenge. However, 2 of the 22 animals had plasma viral RNA levels of over 1,000 copies per ml and suddenly declining levels of CD4 counts. These two animals (no. 4 and no. 5) were spared euthanasia to monitor whether they were undergoing irreversible viral escape or a transient reemergence of virus, such as had been observed earlier in the trial. Here, we report the monitoring of these two animals during what turned out to be permanent reemergences of virus and progression to opportunistic infections and AIDS.
Viral load and CD4 counts.
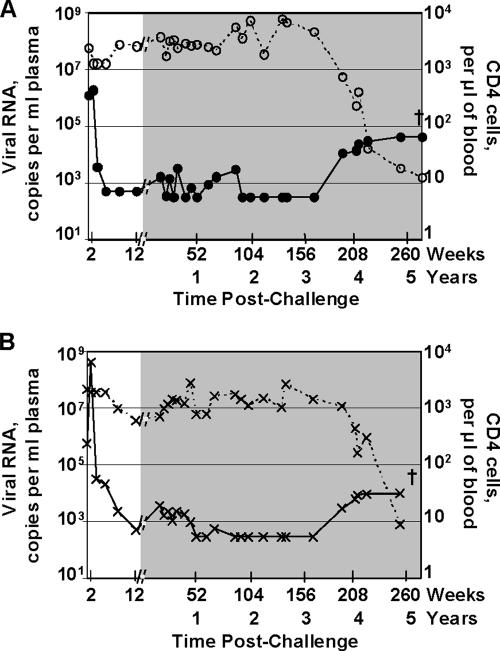
Figure 1 presents temporal postchallenge levels of viral RNA and CD4 counts for the two animals in which the infection escaped the immune response. By 12 weeks postchallenge, both animals had controlled replication of the challenge virus from peaks of 1.9 × 106 (animal no. 4) and 4.3 × 108 (animal no. 5) to ∼1,000 copies of viral RNA per ml of plasma. Between 12 and 52 weeks postchallenge, both animals had levels of viral RNA fluctuating around 1,000 copies per ml of plasma. With time, these levels further declined to <300 copies of viral RNA per ml of plasma. Between years 1 and 3 postchallenge, animals 4 and 5 ranked highest and fourth, respectively, among the 22 challenged animals for transient low levels of reemergent virus (measured by area under the viral RNA curve). Toward the end of the fourth year, both animals had reemerging virus that persisted into the fifth year and the development of AIDS.
FIG. 1.
Loss of long-term control. Temporal postchallenge viral RNA copies (solid lines) and CD4 counts (dashed lines) in animals 4 (A) and 5 (B), which showed increases in viral loads at 196 weeks postchallenge. The shaded area at the right in each panel highlights data from 12 weeks to 5 years postchallenge. The background for detection of the plasma viral RNA was 500 until 12 weeks postchallenge and was then 300. The crosses indicate euthanasia.
Both animals 4 and 5 had a dip in CD4 counts at 2 weeks postchallenge during the peak of viremia (Fig. 1). CD4 counts had recovered to prechallenge levels for animal 4 by 12 weeks postchallenge but required a year for recovery for animal 5. Over the next 3 years, animal 4 maintained prechallenge levels of CD4 counts, whereas animal 5 had transient periods of lower counts. The increase in viral loads at the end of the fourth year corresponded to a fall in CD4 counts that continued to an essentially total loss at the time of euthanasia for both animals.
Broadening of epitopes.
Responding T cells in the vaccinated animals were mapped for their CD8 and CD4 Gag and Env epitopes by enzyme-linked immunospot assay and intracellular cytokine staining (14). During the long period of viral control, IFN-γ-producing antiviral T cells were present at very low breadths and frequencies (week 140 data) (14). Of the 22 animals in the study, animal 5 had the highest number of recognized epitopes: three CD8 (one Gag and two Env) and four CD4 (three Gag and one Env). Animal 4 recognized two Gag CD8 and one Env CD4 epitopes. Following the reemergence of virus at week 196, both animals underwent marked increases in the breadths of their responses (Table 1). Animal 4 gained nine new CD8 (four Gag and five Env) and seven new CD4 (six Gag and one Env) epitopes, and animal 5 gained six new CD8 (four Gag and two Env) and five new CD4 (three Gag and two Env) epitopes. Over the next year, both animals also lost epitopes. By the time of euthanasia at week 257, animal 5 had lost its nine CD8 and nine CD4 responses. At week 264, 10 weeks before euthanasia, animal 4 had lost its eight CD4 responses and retained responses to only 5 of its 11 mapped CD8 epitopes.
TABLE 1.
Broadening of epitopes following loss of controla
Hierarchal Loss of T-cell function and exhaustion.
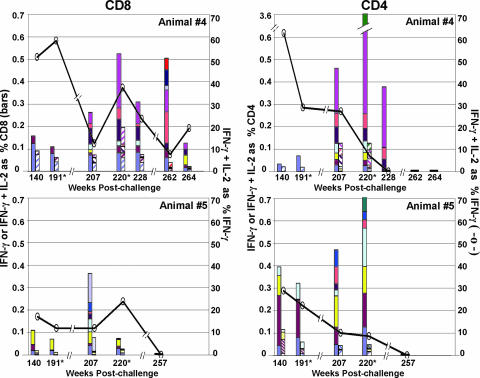
During the long period of viral control, all vaccinated animals, including animals 4 and 5, had low frequencies of IFN-γ responses with good proportions of IL-2-coproducing cells (up to 50%) (Fig. 2) (14). Among the 22 vaccinated animals, no. 5 had the lowest percentage of IFN-γ- and IL-2-coproducing T cells (18% for CD8 and 29% for CD4). With the reappearance of viremia and broadening of the T-cell epitopes, the frequencies of the IFN-γ responses increased in both animals, with the CD4 responses being higher than the CD8. The increased IFN-γ responses were not accompanied by much increase in cells coproducing IFN-γ and IL-2. In animal 4, responses to the original CD8 epitopes maintained their capacities to coproduce IFN-γ and IL-2 over time, whereas most of the newer epitopes were not fully functional. This was observed even at week 207, the first time that epitopes were mapped after escape. In animal 5, the already low levels of cells coproducing IFN-γ and IL-2 to the original epitopes decreased, and most of the new responses did not coproduce IL-2. Animal 5 had lost all functional CD8 and CD4 T cells by the time of euthanasia at week 257 postchallenge. By week 262 postchallenge, animal 4 had lost its CD4 function and had largely lost CD8 T cells capable of coproducing IFN-γ and IL-2. During escape, the levels of virus in the blood never reached the height achieved during acute viremia. This could have reflected the antiviral activities of residual immune responses, including anti-Env antibody; the loss of CD4 T cells as targets for replication; or the escape mutations affecting viral fitness, as well as other phenomena.
FIG. 2.
Temporal heights of IFN-γ and IFN-γ plus IL-2 responses to Gag and Env epitopes. The filled bars represent the IFN-γ responses, and the hatched bars represent the IFN-γ plus IL-2 responses to individual epitopes plotted as percentages of total CD8 or CD4 T cells. The solid lines represent the average percentages of IFN-γ-producing cells that coproduced IL-2. CD8 (left) and CD4 (right) responses were mapped at 140, 207, 228, 262, and 264 weeks postchallenge for animal 4 (top) and at weeks 140, 207, and 257 postchallenge for animal 5 (bottom). At weeks 191 and 220 (*), the animals were retested by intracellular-cytokine staining for previously mapped epitopes (14). Epitope-containing sequences, designated as in the study by Sadagopal et al. (14), are shown in different colors. Animal 4 CD8: periwinkle, Gag 10/11; plum, Gag 107/108; yellow, Gag 2; light turquoise, Gag 25/26; dark purple, Gag 35/36; coral, Gag 47; ocean blue, Env 138; ice blue, Env 18; dark blue, Env 32; red, Env 43. Animal 4 CD4: periwinkle, Env 43; plum, Env 161; yellow, Gag 52; light turquoise, Gag 20; dark purple, Gag 97; coral, Gag 46; pink, Gag 25/26; sea green, Gag 47/48. Animal 5 CD8: periwinkle, Gag 35/36; plum, Env 63; yellow, Env 138; light turquoise, Gag 39/40; dark purple, Gag 59; coral, Gag 64; ocean blue, Gag 101; ice blue, Env 73. Animal 5 CD4: periwinkle, Gag 21; plum, Gag 39/40; yellow, Gag 62; light turquoise, Env 10/11; dark purple, Env 2; coral, Gag 66; ocean blue, Gag 15; sea green, Gag 101.
Mutational escape.
The role of mutational escape in the loss of viral control was investigated by sequencing the gag region of virus collected from plasma at euthanasia. Viral RNA was extracted from the plasma with the Magna Pure robotic workstation (Roche Applied Sciences, Indianapolis, IN). cDNA was synthesized with Gag-specific primers, and the gag region, amplified by two rounds of PCR, was directly sequenced (Macrogen, Rockville, MD). This bulk sequencing method detects the sequence of the predominant virus in the plasma. The viruses from animals 4 and 5 had 17 and 31 mutations, respectively, from the original SHIV-89.6P virus used in the challenge (Table 2) . Eight out of 13 coding mutations in animal 4 and 4 out of 16 in animal 5 were in Gag epitopes that had been identified in our mapping studies. Animal 4 had two mutations in each of its two original Gag CD8 epitopes, whereas animal 5 had one mutation in its single Gag CD8 epitope (Table 3). The presence of two mutations in the original CD8 epitopes in animal 4 may reflect one of the mutations compensating for the cost of viral fitness of the other (7, 9). Mutations also occurred in some of the new CD8 Gag epitopes. Two out of four new Gag CD8 epitopes in animal 4 and one out of four in animal 5 were mutated. Whereas no mutations were seen in the original CD4 epitopes, two out of six new CD4 epitopes in animal 4 and two out of three in animal 5 were mutated.
TABLE 2.
Summary of mutations in gag region of plasma virus in animals 4 and 5
| Mutation or epitope | Results for indicated animal:
|
|
|---|---|---|
| 4 | 5 | |
| Mutations | ||
| Coding | 13a | 16a |
| Noncoding | 4a | 15a |
| Total | 17a | 31a |
| Mutated epitopes | ||
| Original CD8 | 2/2b | 1/1b |
| Original CD4 | 0/0b | 0/3b |
| New CD8 | 2/4b | 1/4b |
| New CD4 | 2/6b | 2/3b |
Number of mutations.
Number of mutated epitopes total number of epitopes.
TABLE 3.
Putative CD8 and CD4 epitopes containing mutations and amino acid and codon changes for these epitopes in animals 4 and 5a
| Epitope | Gag peptide(s) | Epitope-containing sequence | Amino acid change
|
Codon change
|
||||
|---|---|---|---|---|---|---|---|---|
| Position | From | To | Position | From | To | |||
| Animal 4 | ||||||||
| CD8 epitopes | ||||||||
| Original | 10, 11 | LDRFGLAESLL | 46 | L | I | 671 | TTA | ATA |
| 48 | E | D | 679 | GAA | GAC | |||
| 107, 108 | DRQAGFLGLGP | 429 | D | E | 1822 | GAC | GAA | |
| 432 | A | V | 1830 | GCG | GTG | |||
| New | 25, 26 | KQIVQRHLVVE | 101 | K | R | 837 | AAA | AGA |
| 35, 36 | GGNYVHLPLSP | 145 | V | I | 968 | GTC | ATC | |
| CD4 epitopes | ||||||||
| Original | None | |||||||
| New | 25, 26 | KQIVQRHLVVE | 101 | K | R | 837 | AAA | AGA |
| 97 | RGPRKPIKCWNCGKE | 385 | R | K | 1689 | AGG | AAG | |
| Animal 5 | ||||||||
| CD8 epitopes | ||||||||
| Original | 35, 36 | GGNYVHLPLSP | 145 | V | A | 969 | GTC | GCC |
| New | 101 | HSARQCRAPRRQGCW | 402 | S | P | 1739 | TCT | CCT |
| L | 1739, 1740 | CTT | ||||||
| CD4 epitopes | ||||||||
| Original | None | |||||||
| New | 15 | CQKILSVLAPLVPTG | 59 | K | R | 711 | AAA | AGA |
| 101 | HSARQCRAPRRQGCW | 402 | S | P | 1739 | TCT | CCT | |
| L | 1739, 1740 | CTT | ||||||
Changes are in boldface.
Summary.
Here, we have shown that hierarchal loss of T-cell function, as well as mutations in CD8 epitopes, is associated with viral escape during long-term control of a SHIV-89.6P challenge in vaccinated animals. Successful long-term control was associated with low-level, low-breadth T-cell responses that were characterized by IFN-γ- and IL-2-coproducing cells (14). The escape virus had mutations in all of the original CD8 epitopes but none of the original CD4 epitopes. This suggests that the original CD8 T-cell responses exerted higher selection pressure on the virus than the original CD4 helper responses, which presumably provided support for the CD8, as well as B-cell, responses. Although much broader and higher-frequency IFN-γ-producing CD8 and CD4 responses were associated with the period of reemergent virus, these cells appeared to be less effective at controlling viremia than those that had been present during the long period of control. Also, in contrast to the T-cell response during the long period of control, these cells did not coproduce IFN-γ and IL-2. However, even though the virus was not well controlled by the new T-cell responses, the presence of mutations in some new CD8 and CD4 epitopes suggests that these cells were exerting selection on the reemergent virus.
The 2 animals with escape virus had the highest and fourth-highest levels of detectable virus among the 22 animals studied between 1 and 3 years postchallenge. This is consistent with persistently replicating virus being the source of escape mutations (4) and emphasizes the importance of the tightness of vaccine-mediated control in minimizing the frequency of escape. Other euthanized animals could also have undergone escape with time. CD8 depletion studies have clearly shown the presence of residual virus in animals in which virus could not be detected in the blood (2).
Acknowledgments
We are grateful to C. Derdeyn and P. Ahues for technical advice on viral sequencing and the NIH AIDS Research and Reference Reagent Program for the provision of peptides.
This research was supported by PHS Integrated Preclinical/Clinical AIDS Vaccine Development program projects P01 AI43045 and P01 AI 49364, Emory Center for AIDS Research P30 DA 12121, and the Yerkes National Primate Research Center base grant P51 RR00165.
The research was conducted in compliance with all relevant federal and institutional policies.
Footnotes
Published ahead of print on 30 January 2008.
REFERENCES
- 1.Allen, T. M., M. Altfeld, S. C. Geer, E. T. Kalife, C. Moore, M. O'Sullivan, K. I. Desouza, M. E. Feeney, R. L. Eldridge, E. L. Maier, D. E. Kaufmann, M. P. Lahaie, L. Reyor, G. Tanzi, M. N. Johnston, C. Brander, R. Draenert, J. K. Rockstroh, H. Jessen, E. S. Rosenberg, S. A. Mallal, and B. D. Walker. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 7913239-13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amara, R. R., C. Ibegbu, F. Villinger, D. C. Montefiori, S. Sharma, P. Nigam, Y. Xu, H. M. McClure, and H. L. Robinson. 2005. Studies using a viral challenge and CD8 T cell depletions on the roles of cellular and humoral immunity in the control of an SHIV-89.6P challenge in DNA/MVA-vaccinated macaques. Virology 343246-255. [DOI] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H.-L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 29269-74. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, J. Glowczwskie, K. J. Kunstman, M. A. Egan, F. W. Peyerl, S. Santra, M. J. Kuroda, J. E. Schmitz, K. Beaudry, G. R. Krivulka, M. A. Lifton, D. A. Gorgone, S. M. Wolinsky, and N. L. Letvin. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 777367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barouch, D. H., J. Kuntsman, M. J. Kuruda, J. E. Schmitz, S. Santra, F. W. Peyeri, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gargonne, D. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415335-339. [DOI] [PubMed] [Google Scholar]
- 6.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290486-492. [DOI] [PubMed] [Google Scholar]
- 7.Friedrich, T. C., C. A. Frye, L. J. Yant, D. H. O'Connor, N. A. Kriewaldt, M. Benson, L. Vojnov, E. J. Dodds, C. Cullen, R. Rudersdorf, A. L. Hughes, N. Wilson, and D. I. Watkins. 2004. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 782581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4630-640. [DOI] [PubMed] [Google Scholar]
- 9.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kostense, S., K. Vandenberghe, J. Joling, D. Van Baarle, N. Nanlohy, E. Manting, and F. Miedema. 2002. Persistent numbers of tetramer+ CD8+ T cells, but loss of interferon-gamma+ HIV-specific T cells during progression to AIDS. Blood 992505-2511. [DOI] [PubMed] [Google Scholar]
- 11.Mortara, L., F. Letourneur, H. Gras-Masse, A. Venet, J. G. Guillet, and I. Bourgault-Villada. 1998. Selection of virus variants and emergence of virus escape mutants after immunization with an epitope vaccine. J. Virol. 721403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connor, D., T. Friedrich, A. Hughes, T. M. Allen, and D. Watkins. 2001. Understanding cytotoxic T-lymphocyte escape during simian immunodeficiency virus infection. Immunol. Rev. 183115-126. [DOI] [PubMed] [Google Scholar]
- 13.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106539-549. [DOI] [PubMed] [Google Scholar]
- 14.Sadagopal, S., R. R. Amara, D. C. Montefiori, L. S. Wyatt, S. I. Staprans, N. L. Kozyr, H. M. McClure, B. Moss, and H. L. Robinson. 2005. Signature for long-term vaccine-mediated control of a SHIV-98.6P challenge: stable low-breadth and low-frequency T-cell response capable of coproducing gamma interferon and interleukin-2. J. Virol. 793243-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiver, J. W., T. Fu, L. Chen, D. Casimiro, M. E. Davies, R. K. Evans, Z.-Q. Zhang, A. Simon, W. L. Trigona, S. Dubey, L. Huang, V. A. Harris, R. S. Long, L. Xiaoping, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Iospi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency virus immunity. Nature 415331-335. [DOI] [PubMed] [Google Scholar]
- 16.Wherry, E. J., J. N. Blattman, K. Murali-Krishna, R. van der Most, and R. Ahmed. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 774911-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zajac, A. J., J. N. Blattman, K. Murali-Krishna, D. J. Sourdive, M. Suresh, J. D. Altman, and R. Ahmed. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 1882205-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]