Abstract
Human T-lymphotropic virus type 1 (HTLV-1) is the etiologic agent of adult T-cell leukemia (ATL). In Japan, the number of HTLV-1 carriers is estimated to be 1.2 million and more than 700 cases of ATL have been diagnosed every year. Considering the poor prognosis and lack of curative therapy of ATL, it seems mandatory to establish an effective strategy for the treatment of ATL. In this study, we attempted to identify the cell surface molecules that will become suitable targets of antibodies for anti-ATL therapy. The expression levels of approximately 40,000 host genes of three human T-cell lines carrying HTLV-1 genomes were analyzed by oligonucleotide microarray and compared with the expression levels of the genes in an HTLV-1-negative T-cell line. The HTLV-1-carrying T-cell lines used for experiments had totally different expression patterns of viral genome. Among the genes evaluated, the expression levels of 108 genes were found to be enhanced more than 10-fold in all of the T-cell lines examined and 11 of the 108 genes were considered to generate the proteins expressed on the cell surface. In particular, the CD70 gene was upregulated more than 1,000-fold and the enhanced expression of the CD70 molecule was confirmed by laser flow cytometry for various HTLV-1-carrying T-cell lines and primary CD4+ T cells isolated from acute-type ATL patients. Such expression was not observed for primary CD4+ T cells isolated from healthy donors. Since CD70 expression is strictly restricted in normal tissues, such as highly activated T and B cells, CD70 appears to be a potential target for effective antibody therapy against ATL.
Human T-lymphotropic virus type 1 (HTLV-1) is the etiologic agent of adult T-cell leukemia (ATL) and HTLV-1-associated myelopathy/tropical spastic paraparesis (10, 29, 40). The geographic distribution of the virus has been well defined, and the areas in the world where it is highly prevalent include Japan, Africa, the Caribbean islands, and South America (31). In Japan, the number of HTLV-1 carriers is estimated to be 1.2 million and more than 700 cases of ATL are diagnosed every year (37). Since conventional anticancer chemotherapy active against other lymphoid malignancies proved to be ineffective for treating aggressive types of ATL, combination chemotherapy designed exclusively for ATL has been examined. Although such chemotherapy considerably improved the treatment response rates in ATL patients, it could not sufficiently extend the median survival time (16, 39). Therefore, it seems still mandatory to establish an effective strategy for the treatment of ATL.
Monoclonal antibodies (MAbs) have recently gained considerable importance in the area of anticancer therapy. The first agent approved for clinical use is rituximab, which is an anti-CD20 mouse/human chimeric MAb (32). Rituximab was found to be effective for a variety of B-cell lymphomas as well as non-Hodgkin's lymphoma (12). Currently, several MAbs have been approved by the U.S. Food and Drug Administration for the treatment of lymphoma, leukemia, breast cancer, and metastatic colon cancer. One of the anticancer mechanisms of these MAbs is the induction of antibody-dependent cytotoxicity (15, 20). The antibodies bind to the surface antigens of tumor cells, while their crystallizable fragments (Fc) bind to the Fc receptors of the effector cells, such as natural killer cells and monocytes, triggering cytolysis of the target cells. In addition, complement-dependent cytotoxicity and direct induction of apoptosis are also considered anticancer mechanisms of the MAbs (20, 21).
A rationale of using MAbs for anticancer therapy is their high specificities to tumor cells. A certain number of antigens overexpressed on tumor cells have been identified as the targets of MAbs. Such antigens do not need to be completely absent from normal tissues, because their relative overexpression on tumor cells has proved to be sufficient to confer a high level of specificity of MAbs to the target cells (20). Nevertheless, MAbs with higher specificities would be preferable in terms of safety in vivo. Oligonucleotide microarray is an efficient tool for studying the comprehensive gene expression levels of tumor cells in comparison with normal tissues. In fact, several molecules overexpressed in ATL cells have been identified by this technology (7, 35). In these studies, clinical samples obtained from ATL patients were analyzed for their gene expression and compared with normal T cells. The advantage of this procedure is that the gene expression profiles of ATL cells in different disease types or stages can be analyzed directly. On the other hand, the expression profiles may be affected by several conditions of patients, such as the time of sample collection, the use of anticancer agents and/or other drugs, and the presence of complications. Therefore, the microarray analysis of primary ATL cells is not always an ideal way to identify the molecules commonly overexpressed in ATL cells.
The purpose of this study is to identify the surface molecules that will become potential targets for anti-ATL MAb therapy. To this end, the expression levels of approximately 40,000 host genes of three T-cell lines carrying HTLV-1 were analyzed by oligonucleotide microarray and compared with the levels in an HTLV-1-negative T-cell line. Among the genes that could be evaluated, the expressions of 108 genes were found to be enhanced more than 10-fold in all of the T-cell lines examined and 11 of the 108 genes were considered to generate the proteins expressed on the cell surface. In particular, the CD70 gene was upregulated tremendously (more than 1,000-fold), which was confirmed by the analysis for CD70 expression on various HTLV-1-carrying T-cell lines and primary CD4+ T cells from ATL patients.
MATERIALS AND METHODS
Cells.
The HTLV-1-carrying T-cell line S1T was established from the peripheral blood mononuclear cells (PBMCs) of an ATL patient, as described previously (2). The HTLV-1-carrying T-cell lines MT-2, MT-4, and M8166; the HTLV-1-negative T-cell lines MOLT-4, CEM, and Jurkat; and the monocytic cell lines HL-60 and U937 were also used for experiments. MT-2 and MT-4 cells are derived from umbilical cord blood lymphocytes after cocultivation with leukemia cells from ATL patients (24). MT-2 cells were reported to integrate at least eight copies, including defective types, of HTLV-1 proviral DNA in the chromosomes (18). M8166 is a subclone of C8166 cells, which were also established by cocultivation of umbilical cord blood lymphocytes with ATL cells. M8166 cells integrate one copy of provirus in the chromosome (34). All cell lines were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin G, and 100 μg/ml streptomycin. PBMCs were donated under informed consent from patients with acute-type ATL and healthy volunteers. The cells were isolated from heparinized blood with Ficoll-Paque Plus (Pharmacia, Uppsala, Sweden) to obtain PBMCs. Diagnoses of ATL were based on clinical features, hematological characterization, the presence of serum antibodies against HTLV-1, and the insertion of proviral DNA into leukemia cells.
Characterization of HTLV-1-carrying T-cell lines.
The production of viral antigens from S1T, MT-2, and M8166 cells into culture supernatants was determined by enzyme-linked immunosorbent assay (ELISA). Briefly, the cells (1 × 105 cells/ml) were incubated for 3 days at 37°C. After incubation, the culture supernatants were collected and examined for their p19 antigen levels with a sandwich enzyme-linked immunosorbent assay kit (Cellular Products, Buffalo, NY). The cells were also examined for their expression of HTLV-1 env and tax genes by reverse transcription-PCR (RT-PCR). For RT-PCR, the cells were harvested after a 3-day incubation and washed three times with ice-cold phosphate-buffered saline. Total RNA was extracted from the cells with an extraction kit (RNeasy; Qiagen, Hilden, Germany). The extracted RNA was treated with DNase I and subjected to RT-PCR. The primers used for RT-PCR were RENV1 (5′-ACGCCGGTTGAGTCGCGTTCT-3′), RENV4 (5′-CACCGAAGATGAGGGGGCAGA-3′), RPX3 (5′-ATCCCGTGGAGACTCCTCAA-3′), and RPX4 (5′-AACACGTAGACTGGGTATCC-3′). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was also amplified as an internal control by the primer pair RT-GAPDH5 (5′-CATTGACCTCAACTACATGG-3′) and RT-GAPDH3 (5′-AGTGATGGCATGGACTGTGG-3′). The samples were subjected to reverse transcription to cDNA for 30 min at 42°C and PCR amplification (95°C for 30 s, 55°C for 30 s, and 72°C for 1 min) with each primer pair. The amplified products were analyzed by the 2100 Bioanalyzer (Agilent, Santa Clara, CA).
For the detection of HTLV-1 Tax, Western blot analysis of the cells was performed as described previously (41). Briefly, the cells were incubated for 3 days and lysates were obtained by treating the cells with a low-salt extraction buffer (10 mM Tris-HCl [pH 8.0] containing 0.14 M NaCl, 3 mM MgCl2, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, and 0.5% Nonidet P-40) on ice for 20 min. The lysates were centrifuged at 12,000 × g at 4°C for 10 min. After measuring protein concentrations, the lysates (100 μg of protein) were electrophoresed on a 10% polyacrylamide gel with sodium dodecyl sulfate and transferred to a polyvinylidene difluoride membrane. The transferred proteins were reacted with the anti-p40 Tax MAb Lt-4 (38) or an anti-actin polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), followed by treatment with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Amersham Biosciences, Buckinghamshire, United Kingdom) or horseradish peroxidase-conjugated rabbit anti-goat IgG (MP Biomedicals, Solon, OH). Antibody binding was visualized with an enhanced chemiluminescence detection system (Amersham Biosciences).
Oligonucleotide microarray.
S1T, M8166, MT-2, and MOLT-4 cells (1 × 105 cells/ml) were incubated for 3 days at 37°C. After incubation, total RNA was extracted from the cells with RNeasy (Qiagen). The quality of the total RNA was examined by the 2100 Bioanalyzer (Agilent), according to the manufacturer's protocol. The microarray processing of the samples was carried out with necessary reagent kits provided by Agilent, according to the manufacturer's one-color microarray-based gene expression analysis protocol (version 5.5). Briefly, 500 ng of the total RNA was reverse transcribed to cDNA with Moloney murine leukemia virus reverse transcriptase and T7 promoter primer. The cDNA was transcribed and amplified with T7 RNA polymerase to produce the cRNA labeled with cyanine 3. The cyanine 3-labeled cRNA was purified with RNeasy (Qiagen) and examined for its concentration and labeling quality by a spectrophotometer. The cRNA was fragmented and hybridized to Agilent whole human genome oligonucleotide microarray (4 × 44K slide format). After hybridization, the microarray was washed thoroughly and scanned with a microarray scanner (Agilent). The microarray scan data were processed with Future Extraction software (version 9.5.1; Agilent), according to its manual. Cell culture and microarray experiments were conducted simultaneously for all of the T-cell lines and repeated three times.
Data analysis.
The expression level of each gene was analyzed by GeneSpring GX software (version 7.3.1; Agilent). Briefly, after importing the processed data into the software, they were normalized based on the default normalizing settings for one-color experiments (GeneSpring 7.3 user's guide; Agilent). The normalized data were filtered on the basis of parameters in certain specific columns of the original data files to remove the control and other inappropriate spots. The genes of which expression levels were more than 10-fold in all of the three HTLV-1-carrying T-cell lines (S1T, M8166, and MT-2) compared with the levels of the control T-cell line (MOLT-4) were selected and evaluated for their statistical significance by t test (P < 0.05) with multiple testing correction.
Flow cytometric analysis.
The MAbs used for experiments were phycoerythrin (PE)-conjugated anti-human CD70 mouse MAbs (BD Biosciences, San Jose, CA [for staining cell lines] and BD Pharmingen, San Diego, CA [for staining PBMCs]), PE-conjugated anti-human CD124 mouse MAb (BD Biosciences), PE-conjugated anti-human interleukin-21 receptor (IL-21R) mouse MAb (R&D Systems, Minneapolis, MN), PE-conjugated anti-human CD151 mouse MAb (BD Biosciences), peridinin chlorophyll protein (PerCP)-conjugated anti-human CD3 mouse MAb (BD Biosciences), PerCP-conjugated anti-human CD4 mouse MAb (BD Biosciences), fluorescent isothiocyanate (FITC)-conjugated anti-human CD25 mouse MAb (Beckman Coulter, Fullerton, CA), FITC-conjugated anti-human CD8 mouse MAb (Beckman Coulter), PerCP-Cy5.5-conjugated anti-human CD19 mouse MAb (BD Biosciences), FITC-conjugated anti-human CD14 mouse MAb (BD Pharmingen), and their isotype-matched control MAbs. The test cell lines and PBMCs were washed with phosphate-buffered saline containing 1% bovine serum albumin and incubated with appropriate MAbs for 30 min at 4°C. After washing, the stained cells were analyzed by FACScan (Becton Dickinson, San Jose, CA).
Anti-cell proliferation assay.
S1T and MOLT-4 cells were incubated (1 × 104 cells/well) in a flat-bottomed microtiter plate with an anti-human CD70 mouse MAb (BD Biosciences) or its isotype-matched control MAb at a concentration of 1 μg/ml. After incubation at 37°C, the number of viable cells was determined every day by trypan blue exclusion. For primary ATL cells, PBMCs were obtained from three different ATL patients and the cells (1 × 105 cells/well) were cultured in a microtiter plate with an anti-human CD70 mouse MAb (BD Pharmingen) or its isotype-matched control MAb at various concentrations. After a 24-h incubation, 25 μl of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (1 mg/ml) was added and further incubated at 37°C for 4 h. After incubation, 20% sodium dodecyl sulfate solution was added to each well. The plate was incubated overnight at room temperature in a dark place, and specific absorbance was read at 570 nm by a microplate reader.
RESULTS
Viral gene and antigen expression in HTLV-1-carrying T-cell lines.
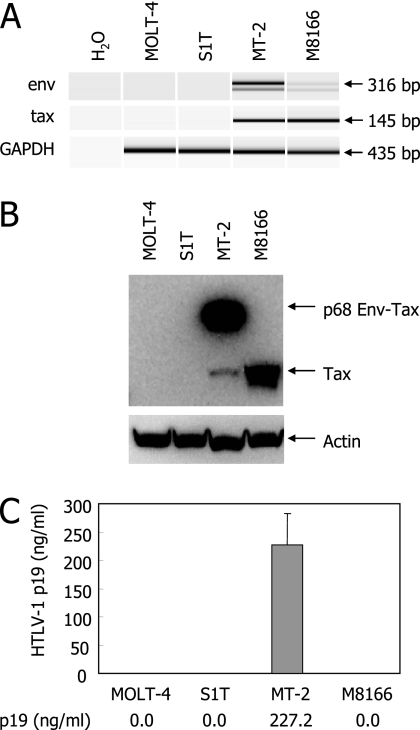
To identify the molecules selectively expressed in ATL cells, comprehensive gene expression in HTLV-1-carrying T-cell lines was examined by oligonucleotide microarray and compared with the gene expression in an HTLV-1-negative T-cell line. To this end, cell lines with various viral gene expression and replication patterns should be selected. Among the HTLV-1-carrying T-cell lines available in our laboratory, three T-cell lines, S1T, MT-2, and M8166, were selected and examined for their HTLV-1 gene expression, protein synthesis, and release of viral antigens in culture supernatants. The HTLV-1-negative T-cell leukemia line MOLT-4 was selected as a control. As shown in Fig. 1A, a strong signal of tax mRNA could be found in MT-2 and M8166 cells, whereas a significant signal of env mRNA was detected in only MT-2 cells. Like MOLT-4 cells, neither env nor tax mRNA was identified in S1T cells. These observations were confirmed by Western blot analysis of these cell lines, where sufficient amounts of p68 Env-Tax fusion protein and Tax were observed in MT-2 and M8166 cells, respectively (Fig. 1B). However, neither Tax nor Env could be detected in S1T cells. Furthermore, MT-2 cells produced and released a large amount of HTLV-1 p19 antigen, which is regarded as a component of viral particles, into culture supernatants, yet p19 production was not observed in M8166 and S1T cells (Fig. 1C). These results suggest that the three HTLV-1-carrying cell lines, of which viral gene expression patterns differ completely, are suitable tools for searching the host cellular genes and proteins commonly overexpressed in ATL cells obtained from patients.
FIG. 1.
Different patterns of viral gene expression in HTLV-1-carrying T-cell lines. (A) Detection of HTLV-1 env and tax gene expression in MOLT-4 (negative control), S1T, MT-2, and M1866 cells. Total RNA was extracted from the cells and subjected to RT-PCR with primer pairs described in Materials and Methods. GAPDH mRNA was also amplified as an internal control. The amplified products were analyzed by an Agilent Bioanalyzer. (B) Western blot analysis of the cells for detection of HTLV-1 Tax. The cell lysates were electrophoresed and transferred to a membrane, as described in Materials and Methods. The transferred proteins were reacted with an anti-p40 Tax MAb or an anti-actin polyclonal antibody, followed by treatment with the second antibody. Antibody binding was visualized with an enhanced chemiluminescence detection system. (C) The production of viral particles and antigens from the cells. The cells were incubated for 3 days. After incubation, culture supernatants were collected and examined for their p19 antigen levels by ELISA. The error bar indicates standard deviation.
Gene expression profiles in HTLV-1-carrying T-cell lines.
The gene expression in the HTLV-1-carrying T-cell lines S1T, MT-2, and M8166 was examined and compared with the expression in the HTLV-1-negative T-cell line MOLT-4 by Agilent whole human genome oligonucleotide microarray (4 × 44K slide format). Among all the (41,150) genes that could be analyzed, the expression levels of 3,931 genes were modulated in all of the HTLV-1-carrying T-cell lines with statistical significance (P < 0.05) (data not shown). Furthermore, among the 3,931 genes, 108 genes were upregulated more than 10-fold, respectively, in all of the HTLV-1-carrying T-cell lines relative to the control cell line MOLT-4 (Table 1). When a correlation coefficient was calculated for the relative expression levels of the 108 genes in Table 1, 0.64, 0.60, and 0.96 were obtained between S1T and MT-2 cells, S1T and M8166 cells, and MT-2 and M8166 cells, respectively (data not shown). Thus, there was a positive correlation among the highly upregulated genes of the HTLV-1-carrying T-cell lines, indicating that our strategy may be applicable for identifying the molecules commonly overexpressed in ATL cells.
TABLE 1.
Genes upregulated more than 10-fold in all of the HTLV-1-carrying T-cell lines compared with the HTLV-1-negative T-cell line MOLT-4a
| Accession no. | Symbol | Relative expression level (fold change)
|
Gene product | ||
|---|---|---|---|---|---|
| S1T | MT-2 | M8166 | |||
| NM_138410 | CKLFSF7 | 1,886.6 ± 188.7 | 293.0 ± 31.0 | 113.3 ± 19.9 | CKLF-like MARVEL transmembrane domain containing 7 isoform a |
| NM_001252 | CD70 | 1,375.3 ± 137.5 | 2,635.3 ± 263.5 | 2,594.8 ± 259.5 | Tumor necrosis factor ligand superfamily, member 7 |
| NM_005572 | FPL | 769.8 ± 88.2 | 822.0 ± 155.1 | 1,361.6 ± 136.2 | Lamin A/C isoform 2 |
| NM_004364 | CEBP | 539.9 ± 191.3 | 87.2 ± 8.7 | 56.9 ± 19.8 | CCAAT/enhancer binding protein α |
| NM_022555 | HLA-DR3B | 474.0 ± 93.8 | 14.4 ± 4.9 | 54.0 ± 45.3 | MHC II, DRβ3 precursor |
| NM_004364 | CEBP | 468.8 ± 144.7 | 72.8 ± 7.3 | 47.1 ± 17.4 | CCAAT/enhancer binding protein alpha |
| NM_002304 | NM_002304 | 336.0 ± 78.3 | 172.3 ± 36.6 | 135.3 ± 16.4 | |
| NM_002166 | GIG8 | 331.2 ± 33.1 | 265.4 ± 31.0 | 47.3 ± 24.0 | Inhibitor of DNA binding 2 |
| NM_024644 | FLJ21802 | 325.1 ± 32.5 | 108.8 ± 18.0 | 107.3 ± 35.1 | Chromosome 14 open reading frame 169 |
| NM_015392 | CAB | 275.4 ± 27.5 | 130.9 ± 13.1 | 23.4 ± 2.3 | Neural proliferation 1, differentiation and control |
| NM_014580 | GLUT8 | 221.3 ± 22.1 | 93.3 ± 13.0 | 116.4 ± 11.8 | Solute carrier family 2, (facilitated glucose transporter) member 8 |
| NM_024710 | TMEM101 | 217.8 ± 37.6 | 228.6 ± 54.0 | 245.8 ± 66.3 | Isochorismatase domain containing 2 |
| NM_015691 | BM042 | 204.4 ± 20.4 | 80.5 ± 8.0 | 34.5 ± 7.7 | WWC family member 3 |
| NM_015892 | GALNAC4S-6ST | 178.9 ± 18.2 | 40.5 ± 6.8 | 37.0 ± 3.8 | B-cell RAG-associated protein |
| NM_001017535 | NR1I1 | 140.8 ± 42.1 | 178.1 ± 81.8 | 109.3 ± 66.6 | Vitamin D (1,25-dihydroxyvitamin D3) receptor |
| NM_033518 | SN2 | 113.5 ± 26.6 | 31.5 ± 11.6 | 58.5 ± 22.2 | Amino acid transport system N2 |
| NM_181078 | NILR | 112.5 ± 11.3 | 127.1 ± 25.3 | 178.5 ± 17.8 | IL-21 receptor precursor |
| NM_012081 | ELL2 | 112.4 ± 23.2 | 52.9 ± 5.3 | 19.7 ± 2.0 | Elongation factor 2, RNA polymerase II |
| ENST00000297871 | 102.3 ± 10.2 | 82.4 ± 8.2 | 95.9 ± 9.6 | ||
| NM_004737 | MDC1D | 97.8 ± 15.4 | 15.8 ± 1.7 | 106.7 ± 26.4 | Like glycosyltransferase |
| NM_139346 | AMPH2 | 83.6 ± 8.4 | 129.0 ± 12.9 | 119.6 ± 12.6 | Bridging integrator 1 isoform 8 |
| NM_022121 | THW | 74.0 ± 7.4 | 29.3 ± 3.0 | 65.1 ± 6.6 | PERP, TP53 apoptosis effector |
| NM_014580 | GLUT8 | 71.7 ± 7.2 | 26.0 ± 3.1 | 31.0 ± 3.1 | Solute carrier family 2, (facilitated glucose transporter) member 8 |
| NM_004350 | AML2 | 67.8 ± 6.8 | 24.9 ± 2.5 | 61.0 ± 6.1 | Runt-related transcription factor 3 isoform 1 |
| NM_012465 | KIAA0932 | 65.1 ± 13.3 | 24.5 ± 7.7 | 20.2 ± 2.7 | Tolloid-like 2 |
| NM_000447 | AD4 | 64.6 ± 9.4 | 87.4 ± 19.1 | 58.6 ± 8.1 | Presenilin 2 isoform 1 |
| NM_016010 | CGI-62 | 63.1 ± 6.3 | 24.6 ± 2.5 | 23.5 ± 2.4 | Hypothetical protein LOC51101 |
| NM_000447 | AD4 | 62.2 ± 12.0 | 90.9 ± 19.7 | 57.1 ± 11.1 | Presenilin 2 isoform 1 |
| NM_000786 | LDM | 60.9 ± 8.1 | 59.2 ± 11.6 | 63.0 ± 6.3 | Cytochrome P450, family 51 |
| NM_005658 | EBI6 | 54.6 ± 35.4 | 85.3 ± 14.2 | 165.7 ± 71.1 | TNF receptor-associated factor 1 |
| NM_025195 | C8FW | 53.8 ± 19.9 | 170.5 ± 17.1 | 29.9 ± 3.7 | G-protein-coupled receptor-induced protein |
| NM_002306 | GAL3 | 52.1 ± 5.2 | 133.4 ± 14.6 | 81.8 ± 11.0 | Galectin 3 |
| AK057088 | AK057088 | 51.8 ± 5.2 | 11.4 ± 1.1 | 20.4 ± 2.0 | |
| NM_000786 | LDM | 50.9 ± 7.1 | 49.2 ± 11.5 | 52.6 ± 5.3 | Cytochrome P450, family 51 |
| NM_000958 | EP4 | 50.3 ± 10.5 | 22.7 ± 4.6 | 43.0 ± 7.0 | Prostaglandin E receptor 4, subtype EP4 |
| NM_177925 | MGC921 | 48.9 ± 4.9 | 134.3 ± 13.4 | 132.5 ± 13.3 | H2A histone family, member J isoform 1 |
| NM_003254 | EPA | 48.0 ± 15.7 | 20.1 ± 2.7 | 10.9 ± 3.2 | Tissue inhibitor of metalloproteinase 1 precursor |
| NM_018094 | GST2 | 47.4 ± 5.0 | 68.6 ± 6.9 | 68.1 ± 7.2 | Peptide chain release factor 3 |
| BC018597 | TEX264 | 43.8 ± 4.4 | 51.8 ± 5.2 | 74.4 ± 10.0 | |
| THC2440229 | 43.7 ± 9.9 | 13.5 ± 4.2 | 11.3 ± 3.1 | ||
| NM_002200 | IRF5 | 43.7 ± 5.3 | 229.9 ± 23.0 | 48.9 ± 7.3 | Interferon regulatory factor 5 isoform a |
| NM_014178 | amisyn | 38.5 ± 3.9 | 200.4 ± 32.5 | 30.6 ± 3.1 | Amisyn |
| NM_014417 | JFY1 | 38.4 ± 9.4 | 82.8 ± 14.8 | 118.3 ± 49.0 | BCL2 binding component 3 |
| NM_000199 | HSS | 37.6 ± 4.7 | 10.5 ± 1.0 | 12.8 ± 1.4 | N-Sulfoglucosamine sulfohydrolase (sulfamidase) |
| AK090416 | RXRA | 37.3 ± 8.1 | 12.3 ± 4.1 | 27.9 ± 10.2 | FLJ00318 protein |
| NM_000418 | CD124 | 37.2 ± 6.2 | 43.8 ± 4.4 | 29.6 ± 9.7 | Interleukin 4 receptor α chain isoform a precursor |
| NM_015111 | N4BP3 | 36.5 ± 5.5 | 14.2 ± 1.4 | 27.5 ± 2.9 | Nedd4 binding protein 3 |
| NM_015459 | DKFZP564J0863 | 34.8 ± 6.6 | 26.2 ± 2.6 | 18.5 ± 5.4 | Hypothetical protein LOC25923 |
| NM_018664 | SNFT | 34.7 ± 3.5 | 24.6 ± 2.5 | 11.7 ± 2.5 | Jun dimerization protein p21SNFT |
| NM_013385 | CYT4 | 34.1 ± 3.4 | 16.2 ± 4.7 | 107.6 ± 34.4 | Pleckstrin homology, Sec7, and coiled-coil |
| NR_002323 | TUG1 | 33.4 ± 3.3 | 30.3 ± 3.0 | 30.1 ± 5.2 | domains 4 |
| BC004219 | MGC4604 | 33.1 ± 3.3 | 21.0 ± 2.1 | 13.0 ± 3.2 | 1-Acylglycerol-3-phosphate O-acyltransferase 3 |
| BC035647 | HLA-B | 33.0 ± 6.3 | 52.4 ± 5.2 | 36.3 ± 5.7 | |
| NM_006035 | MRCKB | 32.0 ± 3.2 | 65.7 ± 6.6 | 74.0 ± 7.4 | CDC42-binding protein kinase β |
| NM_001919 | CD79A | 31.8 ± 3.2 | 15.6 ± 1.7 | 27.2 ± 4.0 | Dodecenoyl-coenzyme A δ isomerase precursor |
| BQ189193 | ICOSLG | 31.8 ± 14.2 | 30.1 ± 3.8 | 72.7 ± 7.3 | |
| THC2401087 | 31.2 ± 3.1 | 42.5 ± 4.2 | 14.6 ± 4.0 | ||
| NM_001852 | MED | 30.7 ± 6.6 | 172.2 ± 33.3 | 52.9 ± 5.3 | α2 type IX collagen |
| NM_004357 | GP27 | 30.0 ± 4.4 | 34.0 ± 3.4 | 14.5 ± 4.5 | CD151 antigen |
| NM_006509 | I-REL | 29.9 ± 3.0 | 65.0 ± 6.5 | 43.5 ± 5.1 | Reticuloendotheliosis viral oncogene homolog B |
| AK096677 | AK096677 | 29.2 ± 3.8 | 29.7 ± 7.2 | 73.3 ± 36.4 | |
| NM_024758 | FLJ23384 | 27.7 ± 4.5 | 23.1 ± 5.6 | 13.6 ± 2.1 | Agmatine ureohydrolase (agmatinase) |
| AK021777 | FLJ00205 | 25.8 ± 2.6 | 16.8 ± 1.7 | 19.9 ± 2.0 | GalNAc transferase 10 isoform b |
| NM_001953 | TP | 25.2 ± 2.5 | 43.2 ± 4.3 | 30.8 ± 5.9 | Endothelial cell growth factor 1 (platelet-derived) |
| NM_020731 | AHH | 24.8 ± 5.3 | 10.1 ± 1.6 | 99.4 ± 42.5 | Arylhydrocarbon receptor repressor |
| NM_002413 | GST2 | 24.6 ± 5.6 | 45.4 ± 4.6 | 51.5 ± 5.2 | Microsomal glutathione S-transferase 2 |
| NM_014383 | Rog | 24.3 ± 2.4 | 41.1 ± 4.1 | 12.5 ± 3.8 | Testis zinc finger protein |
| NM_003928 | MGC117411 | 24.2 ± 2.4 | 61.8 ± 7.1 | 35.8 ± 3.6 | CAAX box 1 |
| BX362492 | BX362492 | 23.1 ± 4.0 | 26.3 ± 2.8 | 42.5 ± 11.0 | |
| NM_033375 | myr2 | 20.8 ± 2.9 | 45.4 ± 8.2 | 48.5 ± 8.3 | Myosin IC |
| NM_213589 | LPD | 20.6 ± 2.3 | 14.9 ± 3.5 | 24.8 ± 3.1 | Ras association and pleckstrin homology domains 1 isoform 2 |
| NM_004838 | HOMER-3 | 20.3 ± 2.0 | 193.6 ± 23.6 | 84.5 ± 15.8 | Homer 3, neuronal immediate early gene |
| A_23_P370707 | 18.2 ± 7.7 | 42.1 ± 4.2 | 30.1 ± 7.0 | ||
| AB033060 | AHH | 17.9 ± 3.9 | 12.2 ± 1.2 | 114.8 ± 22.4 | Arylhydrocarbon receptor repressor |
| BC024020 | VMP1 | 17.6 ± 2.6 | 39.6 ± 4.0 | 22.8 ± 6.0 | Transmembrane protein 49 |
| NM_023076 | FLJ23360 | 17.6 ± 3.1 | 21.2 ± 4.0 | 20.5 ± 2.8 | Hypothetical protein LOC65259 |
| AK097976 | DLEU2 | 17.2 ± 3.1 | 22.5 ± 5.3 | 13.6 ± 8.3 | |
| NM_003842 | DR5 | 16.7 ± 1.7 | 32.9 ± 4.0 | 37.2 ± 3.7 | TNF receptor superfamily, member 10b isoform 1 precursor |
| NM_024646 | ZYG11 | 16.5 ± 1.7 | 11.1 ± 1.1 | 15.1 ± 1.5 | Zyg-11 homolog B |
| AK092921 | HLA-B | 16.3 ± 1.6 | 25.7 ± 2.6 | 19.9 ± 2.0 | |
| NM_022152 | RECS1 | 16.2 ± 1.6 | 13.6 ± 2.9 | 14.7 ± 3.0 | Transmembrane BAX inhibitor motif containing 1 |
| AA451906 | BIN1 | 15.4 ± 1.5 | 18.2 ± 2.0 | 17.3 ± 1.7 | |
| NM_018370 | DRAM | 15.3 ± 2.5 | 80.9 ± 14.7 | 41.6 ± 10.8 | Damage-regulated autophagy modulator |
| NM_005514 | HLA B | 15.2 ± 1.7 | 32.3 ± 3.2 | 28.0 ± 4.9 | MHC I, B |
| THC2403644 | 14.9 ± 4.4 | 17.5 ± 5.9 | 15.0 ± 5.0 | ||
| NM_000152 | LYAG | 14.7 ± 1.5 | 15.8 ± 2.4 | 13.0 ± 2.1 | Acid α-glucosidase preproprotein |
| ENST00000355804 | 14.4 ± 1.4 | 23.7 ± 2.8 | 13.1 ± 1.7 | ||
| NM_001613 | ACTSA | 14.4 ± 1.6 | 35.3 ± 5.4 | 22.3 ± 2.3 | α2 actin |
| NM_018370 | DRAM | 14.3 ± 2.3 | 73.3 ± 12.4 | 33.9 ± 7.7 | Damage-regulated autophagy modulator |
| A_24_P101771 | 14.0 ± 1.5 | 21.3 ± 2.2 | 19.3 ± 3.3 | ||
| NM_017789 | SEMAI | 13.9 ± 1.7 | 20.4 ± 2.5 | 33.9 ± 13.8 | Semaphorin 4C |
| NM_002502 | LYT10 | 13.3 ± 4.3 | 15.4 ± 1.0 | 45.9 ± 2.8 | Nuclear factor of κ light polypeptide gene enhancer in B cells 2 (p49/p100) |
| NM_031419 | IKBZ | 12.6 ± 4.6 | 37.1 ± 4.9 | 134.0 ± 62.5 | Nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor, ζ isoform b |
| NM_015516 | TSK | 12.5 ± 1.2 | 12.7 ± 1.3 | 12.2 ± 1.2 | Tsukushi |
| CR594843 | HLA-B | 12.2 ± 1.2 | 11.5 ± 1.2 | 10.1 ± 1.9 | |
| NM_018950 | HLAF | 12.1 ± 1.8 | 16.7 ± 1.7 | 13.7 ± 2.0 | MHC I, F precursor |
| NM_024567 | PBHNF | 12.0 ± 1.9 | 15.3 ± 3.0 | 17.0 ± 2.8 | Homeobox containing 1 |
| NM_003764 | FHL4 | 12.0 ± 2.1 | 14.0 ± 1.4 | 11.7 ± 4.4 | Syntaxin 11 |
| THC2276547 | 11.9 ± 1.3 | 16.8 ± 4.7 | 30.9 ± 9.6 | ||
| CA431756 | CTTN | 11.3 ± 1.4 | 13.8 ± 1.5 | 12.8 ± 3.1 | |
| CR608347 | HLA-B | 11.0 ± 1.2 | 16.5 ± 1.4 | 12.7 ± 1.9 | |
| NM_005261 | KIR | 11.0 ± 4.5 | 96.2 ± 9.6 | 97.3 ± 56.7 | GTP-binding mitogen-induced T-cell protein |
| NM_130446 | FLJ00029 | 11.0 ± 1.2 | 11.6 ± 3.5 | 13.6 ± 7.1 | Kelch-like 6 |
| NM_017789 | SEMAI | 10.9 ± 1.1 | 16.4 ± 1.6 | 24.6 ± 10.4 | Semaphorin 4C |
| BC037255 | LOC389634 | 10.6 ± 1.1 | 16.8 ± 3.0 | 21.3 ± 6.4 | Hypothetical protein LOC389634 |
| AF009619 | CASH | 10.6 ± 1.7 | 16.4 ± 3.3 | 85.1 ± 38.2 | CASP8 and FADD-like apoptosis regulator |
| NM_006674 | P5-1 | 10.5 ± 1.2 | 58.0 ± 5.8 | 91.4 ± 9.3 | HLA complex P5 |
| NR_001434 | HLAHP | 10.2 ± 1.0 | 15.7 ± 1.6 | 18.5 ± 2.4 | |
The genes of which expression levels were more than 10-fold in all of the three HTLV-1-carrying T-cell lines (S1T, MT-2, and M8166) compared with the control T-cell line (MOLT-4) with statistical significance (P < 0.05) are listed and sorted by the expression level in S1T cells. All data represent means ± standard deviations for three independent microarray experiments.
From the 108 genes, 11 genes of which products were considered to be expressed on the cell surface were listed in Fig. 2. These include the genes of tumor necrosis factor (TNF) ligand superfamily member 7 (CD70), major histocompatibility complex class II (MHC II) DRβ3 (HLA-DR3B), glucose transporter member 8 (GLUT8), IL-21R (NILR), prostaglandin E receptor 4 subtype EP4 (EP4), IL-4 receptor α chain isoform a (CD124), dodecenoyl-coenzyme A δ isomerase (CD79A), CD151 antigen (GP27), TNF superfamily member 10b isoform 1 (DR5), semaphorin 4C (SEMAI), and MHC I F (HLAF). Above all, the expression of the CD70 gene was enhanced more than 1,000-fold in all of the HTLV-1-carrying T-cell lines (Table 1 and Fig. 2). Therefore, we examined whether such high upregulation of the CD70 gene was reflected in the expression of the CD70 molecule on the surfaces of the cell lines.
FIG. 2.
Genes upregulated more than 10-fold in the HTLV-1-carrying T-cell lines S1T, MT-2, and M8166 compared with the genes in HTLV-1-negative T-cell line MOLT-4. The genes of which products are considered to be expressed on the cell surface are shown. All data represent means ± standard deviations (error bars) for three independent microarray experiments.
CD70 expression in HTLV-1-carrying T-cell lines.
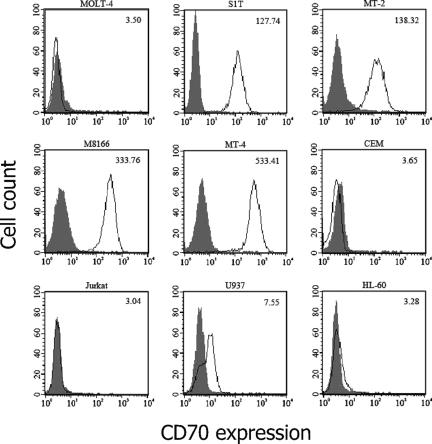
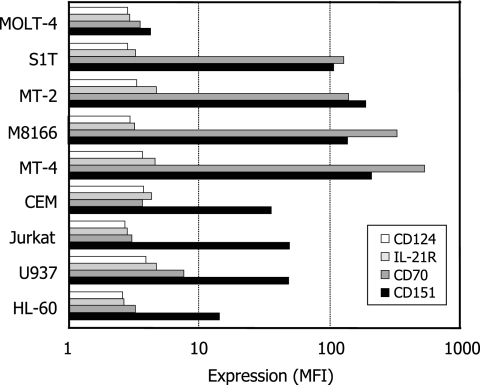
As shown in Fig. 3, MOLT-4 cells did not express CD70 on their surfaces, whereas this molecule was highly expressed on the HTLV-1-carrying cell T-lines S1T, MT-2, M8166, and MT-4. Like the case for MOLT-4 cells, CD70 expression was scarcely observed for other HTLV-1-negative T-cell lines (CEM, Jurkat, and the monocytic cell lines U937 and HL-60), suggesting that CD70 is selectively expressed in HTLV-1-carrying T-cell lines. Such selectivity was also confirmed by the analysis of these cell lines for the expression of CD124, IL-21R, and CD151 on the surface. The gene expression of not only CD70 but also CD124, IL-21R, and CD151 was highly upregulated in all of the HTLV-1-carrying T-cell lines (Table 1 and Fig. 2). However, there was no significant difference of CD124 and IL-21R expression among the nine cell lines (Fig. 4). Like CD70, CD151 was strongly expressed on the HTLV-1-carrying T-cell lines compared with MOLT-4 cells, yet this molecule was also highly expressed on CEM, Jurkat, and U937 cells, indicating that CD151 expression was not selective enough to HTLV-1-carrying T-cell lines.
FIG. 3.
CD70 expression on various cell lines. The cells were strained with an anti-human CD70 MAb (white histogram) or its isotype control MAb (gray histogram) and analyzed by laser flow cytometry. The number in each histogram indicates the mean fluorescence intensity of the cells.
FIG. 4.
Expression of CD124, IL-21R, CD70, and CD151 on various cell lines. The cells were strained with appropriate MAbs described in Materials and Methods and analyzed by laser flow cytometry. The expression level of each molecule is expressed as mean fluorescence intensity (MFI).
CD70 expression in leukemic cells from ATL patients.
To determine whether CD70 is a potential target for anti-ATL MAb therapy, the selective expression of CD70 has to be demonstrated in the primary ATL cells isolated from patients. When PBMCs were isolated from an HTLV-1-negative healthy donor and examined for their CD4 and CD70 expression by laser flow cytometry, a small number (approximately 1.8%) of CD4+ cells, which were regarded as T cells because of their being gated by forward and side scattering intensities, were CD70+ (Fig. 5A). Under the same conditions, 69.2% of the CD4+ cells isolated from an acute-type ATL patient were CD70+ (Fig. 5B). Therefore, we extended the analysis to PBMCs obtained from an additional five HTLV-1-negative healthy donors and five acute-type ATL patients. As shown in Table 2, the average numbers of CD70+ cells were 3.2 and 79.3% of the total CD4+ T cells obtained from the healthy donors and ATL patients, respectively, which was statistically significant (P < 0.00095). In contrast, there was no practical difference of CD70 expression on B cells and monocytes between healthy donors and ATL patients. Although certain difference of CD70 expression was observed for CD8+ T-cells, it was not statistically significant. Furthermore, difference of CD70 expression on CD8+ T cells, B cells, and monocytes varied from one patient to another. These results suggest that CD70 is predominantly expressed on the CD4+ T cells, presumably leukemia cells, from acute-type ATL patients.
FIG. 5.
CD70 expression on CD4+ T cells isolated from healthy donors and ATL patients. PBMCs were isolated from (A) an HTLV-1-negative healthy donor (HD-1 in Table 1) and (B) an acute-type ATL patient (ATL-1 in Table 1). The cells were examined for their CD4 and CD70 expression by laser flow cytometry after being gated by their forward and side scattering intensities. The percentage of CD70+ cells among CD4+ cells was calculated by the following formula: percentage of upper right quadrant/(percentage of upper right quadrant + percentage of lower right quadrant).
TABLE 2.
CD70 expression in PBMCs isolated from healthy donors and ATL patientsa
| Donor | WBCb (cells/mm3) | Expression on indicated marker-positive cells (%)c
|
||||||
|---|---|---|---|---|---|---|---|---|
| CD3+ CD70+ | CD4+ CD70+ | CD4+ CD25+ | CD4+ CD25+ CD70+ | CD8+ CD70+ | CD19+ CD70+ | CD14+ CD70+ | ||
| HD-1 | 2.6 | 1.8 | ND | 0.3 | 0.6 | 16.5 | 0 | |
| HD-2 | 2.9 | 2.7 | 1.5 | 0.4 | 4.2 | 17.9 | 0.1 | |
| HD-3 | 2.4 | 1.9 | 3.6 | 0.4 | 0.2 | 13.6 | 0 | |
| HD-4 | 6.3 | 4.2 | 7.1 | 1.2 | 0.6 | 20.3 | 0.5 | |
| HD-5 | 9.4 | 5.8 | 7.7 | 1.3 | 3.9 | 15.8 | 0.3 | |
| HD-6 | 5.5 | 2.8 | 14.2 | 1.3 | ND | 27.5 | 0.3 | |
| ATL-1 | 414,000 | 67.5 | 69.2 | 73.6 | 56.6 | ND | 14.5 | 0 |
| ATL-2 | 28,300 | 98.0 | 98.6 | 97.5 | 98.5 | 91.3 | 18.3 | 0 |
| ATL-3 | 296,000 | 66.6 | 84.3 | 40.6 | 36.9 | 0 | 0 | 0 |
| ATL-4 | 9,100 | 99.1 | 98.7 | 58.7 | 58.8 | 84.6 | 8.7 | 0.5 |
| ATL-5 | 81.6 | 31.5 | 10.4 | 5.7 | 74.6 | 21.5 | 33.9 | |
| ATL-6 | 94.3 | 93.4 | 73.1 | 69.6 | 24.5 | 16.7 | 1.5 | |
PBMCs obtained from healthy donors (HD) and acute-type ATL patients were stained with appropriate MAbs (see Materials and Methods). After staining, the cells were analyzed by laser flow cytometry.
WBC, white blood cell count.
Mean ± standard deviation values for healthy donors were 4.9 ± 2.8 for CD3+ CD70+ cells, 3.2 ± 1.5 for CD4+ CD70+ cells, 6.8 ± 4.8 for CD4+ CD25+ cells, 0.8 ± 0.5 for CD4+ CD25+ CD70+ cells, 1.9 ± 2.0 for CD8+ CD70+ cells, 18.6 ± 4.9 for CD19+ CD70+ cells, and 0.2 ± 0.2 for CD14+ CD70+ cells. Mean ± standard deviation values for ATL patients were 84.5 ± 14.9 for CD3+ CD70+ cells (statistically significant [P < 0.01] by t test), 79.3 ± 25.9 for CD4+ CD70+ cells (statistically significant [P < 0.01] by t test), 59.0 ± 30.3 for CD4+ CD25+ cells (statistically significant [P < 0.01] by t test), 54.4 ± 31.2 for CD4+ CD25+ CD70+ cells (statistically significant [P < 0.01] by t test), 55.0 ± 40.4 for CD8+ CD70+ cells, 13.3 ± 7.8 for CD19+ CD70+ cells, and 6.0 ± 13.7 for CD14+ CD70+ cells. ND, not determined.
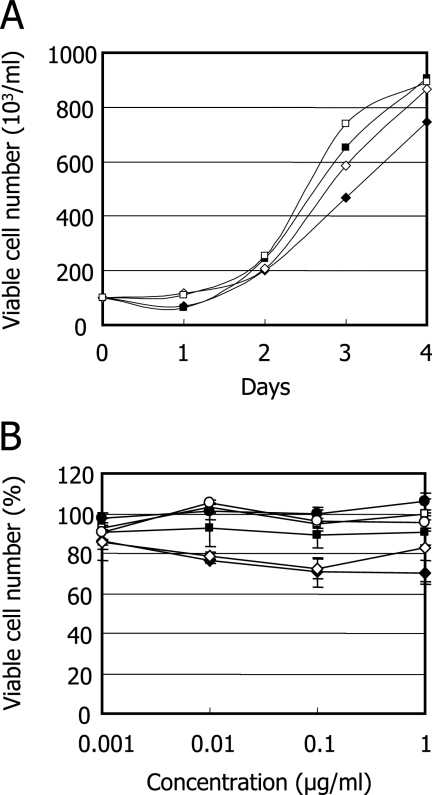
Effect of anti-human CD70 MAb on ATL cells.
When S1T and MOLT-4 cells were incubated with a commercially available anti-human CD70 MAb, the MAb did not affect the proliferation of these cell lines at a concentration of 1 μg/ml during a 4-day incubation period (Fig. 6A). The effect of an anti-human CD70 MAb for the viability of primary ATL cells was also examined. No significant reduction of cell viability was observed at concentrations of up to 1 μg/ml for all of the PBMCs obtained from three different ATL patients (Fig. 6B).
FIG. 6.
Effect of anti-human CD70 MAb on the growth and viability of ATL cells. (A) S1T (diamonds) and MOLT-4 (squares) cells were incubated with an anti-CD70 MAb (filled symbols) or its isotype-matched control MAb (open symbols) at a concentration of 1 μg/ml. After a 4-day incubation, the number of viable cells was determined by trypan blue exclusion. (B) PBMCs obtained from three different ATL patients (circles, squares, and diamonds) were incubated with an anti-CD70 MAb (closed symbols) or its isotype-matched control MAb (open symbols) at various concentrations. After a 24-h incubation, the number of viable cells was determined by the MTT method. Error bars indicate standard deviations.
DISCUSSION
Human oligonucleotide microarrays have been used to examine gene expression patterns of PBMCs infected with HTLV-1 (11), HTLV-1-transformed T-cell lines (8, 30), Jurkat cells expressing either p12I (26) or p30II (23), and the Jurkat cell line JPX-9 that can be induced to express higher levels of Tax-1 (27). In addition, activated PBMC cDNA has been used in subtraction hybridization studies with cDNA from cultured ATL cells from a patient (33). The complexity of the data from these studies and differences in chip composition preclude a full definition of genes that are affected by viral infection. However, there is a consensus on the expression of some cellular genes. Enhanced expression of cell cycle and antiapoptotic genes includes the cyclin B1, p21WAF1/CIP1, and Bcl-X(L) genes, confirming prior biological/biochemical findings (1, 5, 25, 28). In contrast, caspase-8 appears to be consistently downregulated. Among the interleukins and their receptors, the upregulation of IL-2Rα, but not IL-2, is also consistently detected. In contrast, IL-15Rα appears to be upregulated in only some HTLV-1-infected T-cell lines and PBMCs. Similarly, IL-15 is not upregulated in all cell lines and IL-15 expression does not appear to be induced by Tax-1 in Jurkat cells.
There is a criticism that limited or biased information regarding the molecules selectively expressed in ATL cells will be obtained when HTLV-1-carrying T-cell lines, instead of primary ATL cells, are used for oligonucleotide microarray analysis (35). This criticism may be appropriate from one aspect, since such HTLV-1-carrying T-cell lines generally express the viral transactivator protein Tax that considerably affects viral and cellular gene expression. In fact, our study demonstrated that MT-2 and M8166 cells strongly expressed Env-Tax fusion protein and Tax, respectively (Fig. 1B). Both cell lines were established by cocultivation of healthy human cord blood T cells with ATL cells (24). Therefore, it is not surprising that unlike primary ATL cells, these in vitro-transformed T-cell lines still retain functional Tax. This may be a reason for the high correlation coefficient (0.96) in relative expression levels of the 108 genes between MT-2 and M8166 cells (Table 1). On the other hand, S1T cells were directly established from primary ATL cells by cultivation with IL-2 (2). Consequently, S1T cells did not express env or tax gene as well as Env or Tax (Fig. 1).
In this point of view, if HTLV-1-carrying T-cell lines with totally different origins could be included for oligonucleotide microarray analysis, it would become an efficient approach to determining the molecules selectively expressed in ATL cells. In the present study, 108 genes were found to be upregulated more than 10-fold in different HTLV-1-carrying T-cell lines relative to a control T-cell line (Table 1). Among them, tremendous (more than 1,000-fold) upregulation was observed for the CD70 gene, of which product should be expressed on the cell surface (Fig. 2). In fact, the CD70 molecule was strongly and selectively expressed on various HTLV-1-carrying T-cell lines and CD4+ T-cells obtained from ATL patients but not on HTLV-1-negative T-cell lines, monocytic cell lines, or CD4+ T-cells obtained from HTLV-1-negative healthy donors (Fig. 4 and 5 and Table 2).
CD70 is the only known ligand for its receptor CD27 that belongs to the TNF receptor superfamily 7. In general, this molecule is expressed on strongly activated T and B cells (4) and some hematological malignancies, such as non-Hodgkin's lymphoma (42). In fact, when PBMCs were isolated and stimulated with phytohemagglutinin, approximately 18 and 32% of the cells became CD70+ after 7 and 12 days of cultivation, respectively (data not shown). However, there has been no definitive report describing the selective expression of CD70 in ATL cells. CD70 is also highly expressed on some solid tumors, including renal cell carcinoma (9, 17) and glioblastoma (6, 43). In contrast, CD70 expression is highly restricted in normal tissues (19). Therefore, CD70 has been considered to be an attractive target of MAbs and MAb-drug conjugates for selective anticancer therapy. It was recently shown that the administration of an engineered anti-CD70 MAb significantly prolonged the survival of severe combined immunodeficient mice bearing CD70+-disseminated human non-Hodgkin's lymphoma xenografts (22). In this study, treatment with control IgG did not prolong median survival (21 days). In contrast, median survival was increased to 72 days when the mice were treated with the anti-CD70 MAb at a dose of 4 mg/kg of body weight. Furthermore, anti-CD70 antibody-drug conjugates were effective against tumor growth in mice bearing human renal cell carcinoma xenografts (6). These results suggest that irrespective of drug conjugates, anti-CD70 MAbs deserve to be investigated for their anticancer activities against ATL in vitro and in vivo.
In addition to CD70, we have also identified 10 genes of which products should be highly expressed on the HTLV-1-carrying T-cell lines (Fig. 2). Among these, three molecules, CD124, IL-21R, and CD151, could be evaluated for their expressions on various cell lines, since MAbs for these molecules were commercially available. CD151 was indeed highly expressed on the HTLV-1-carrying T-cell lines, yet it was also expressed in other T-cell and monocytic cell lines, except MOLT-4 (Fig. 4). CD151 is a member of the tetraspanin family and is a broadly expressed molecule. It is also noted for its strong molecular associations with integrins (44). CD151 was initially identified as a marker of human acute myeloid leukemia cells, platelets, and vascular endothelial cells (3). The upregulation of the CD151 gene in HTLV-1-carrying T-cell lines has already been reported and investigated for its pathological role (13, 14). Our microarray analysis has confirmed these reports. Since CD151 is broadly expressed by a variety of cell types (36), it does not seem to be a suitable target for anticancer therapy with MAbs. Further studies are in progress to identify other molecules selectively expressed on primary ATL cells obtained from patients.
At present, there is no evidence indicating that commercially available anti-CD70 MAbs are capable of inhibiting cell proliferation or inducing apoptosis of primary ATL cells obtained from patients as well as the S1T cells (Fig. 6). It is possible that these anti-CD70 MAbs are not optimized to exert their biological functions and may be required for structural modification. However, a company in New Jersey has recently obtained permission from the U.S. Food and Drug Administration to use a fully human MAb directed against CD70 in a phase I clinical trial for treatment of clear cell renal cell carcinoma (Medarex). Considering this fact and the poor prognosis and lack of curative therapy for ATL, CD70 should be further perused as a potential target in anticancer therapy against ATL.
Acknowledgments
The anti-p40 Tax monoclonal antibody Lt-4 was kindly provided by Y. Tanaka (University of the Ryukyus, Okinawa, Japan). We thank T. Uto and M. Tokitou for their technical assistance.
This work was supported by a grant from the Frontier Science Research Center, Kagoshima University, and a grant-in-aid for Scientific Research (B) from the Japan Society for the Promotion of Science (grant no. 19390153).
Footnotes
Published ahead of print on 6 February 2008.
REFERENCES
- 1.Akagi, T., H. Ono, and K. Shimotohno. 1996. Expression of cell-cycle regulatory genes in HTLV-I infected T-cell lines: possible involvement of Tax1 in the altered expression of cyclin D2, p18(Ink4) and p21(Waf1/Cip1/Sdi1). Oncogene 121645-1652. [PubMed] [Google Scholar]
- 2.Arima, N., J. A. Molitor, M. R. Smithe, J. H. Kim, Y. Daitoku, and W. C. Greene. 1991. Human T-cell leukemia virus type I Tax induces expression of the Rel-related family of κB enhancer-binding proteins: evidence for a pretranslational component of regulation. J. Virol. 656892-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashman, L. K., G. Aylett, P. Mehrabani, L. Bendall, S. Niutta, A. C. Cambareri, S. R. Cole, and M. Berndt. 1991. The murine monoclonal antibody, 14A2.H1, identifies a novel platelet surface antigen. Br. J. Haematol. 79263-270. [DOI] [PubMed] [Google Scholar]
- 4.Borst, J., J. Hendriks, and Y. Xiao. 2005. CD27 and CD70 in T cell and B cell activation. Curr. Opin. Immunol. 17275-281. [DOI] [PubMed] [Google Scholar]
- 5.Cereseto, A., J. C. Mulloy, and G. Franchini. 1996. Insights on the pathogenicity of human T-lymphotropic/leukemia virus types I and II. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13(Suppl. 1)S69-S75. [DOI] [PubMed] [Google Scholar]
- 6.Chahlavi, A., P. Rayman, A. L. Richmond, K. Biswas, R. Zhang, M. Vogelbaum, C. Tannenbaum, G. Barnett, and J. H. Finke. 2005. Glioblastomas induce T-lymphocyte death by two distinct pathways involving gangliosides and CD70. Cancer Res. 655428-5438. [DOI] [PubMed] [Google Scholar]
- 7.Choi, Y. L., K. Tsukasaki, M. C. O'Neill, Y. Yamada, Y. Onimaru, K. Matsumoto, J. Ohashi, Y. Yamashita, S. Tsutsumi, R. Kaneda, S. Takada, H. Aburatani, S. Kamihira, T. Nakamura, M. Tomonaga, and H. Mano. 2007. A genomic analysis of adult T-cell leukemia. Oncogene 261245-1255. [DOI] [PubMed] [Google Scholar]
- 8.de La Fuente, C., L. Deng, F. Santiago, L. Arce, L. Wang, and F. Kashanchi. 2000. Gene expression array of HTLV type 1-infected T cells: up-regulation of transcription factors and cell cycle genes. AIDS Res. Hum. Retrovir. 161695-1700. [DOI] [PubMed] [Google Scholar]
- 9.Diegmann, J., K. Junker, B. Gerstmayer, A. Bosio, W. Hindermann, J. Rosenhahn, and F. von Eggeling. 2005. Identification of CD70 as a diagnostic biomarker for clear cell renal cell carcinoma by gene expression profiling, real-time RT-PCR and immunohistochemistry. Eur. J. Cancer 411794-1801. [DOI] [PubMed] [Google Scholar]
- 10.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de Thé. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet 2407-410. [DOI] [PubMed] [Google Scholar]
- 11.Harhaj, E. W., L. F. Good, G. T. Xiao, and S. C. Sun. 1999. Gene expression profiles in HTLV-I-immortalized T cells: deregulated expression of genes involved in apoptosis regulation. Oncogene 181341-1349. [DOI] [PubMed] [Google Scholar]
- 12.Harris, M. 2004. Monoclonal antibodies as therapeutic agents for cancer. Lancet Oncol. 5292-302. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa, H., Y. Utsunomiya, K. Kishimoto, K. Yanagisawa, and S. Fujita. 1996. SFA-1, a novel cellular gene induced by human T-cell leukemia virus type 1, is a member of the transmembrane 4 superfamily. J. Virol. 703258-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa, H., T. Nomura, K. Kishimoto, K. Yanagisawa, and S. Fujita. 1998. SFA-1/PETA-3 (CD151), a member of the transmembrane 4 superfamily, associates preferentially with α5β1 integrin and regulates adhesion of human T cell leukemia virus type 1-infected T cells to fibronectin. J. Immunol. 1613087-3095. [PubMed] [Google Scholar]
- 15.Iannello, A., and A. Ahmad. 2005. Role of antibody-dependent cell-mediated cytotoxicity in the efficacy of therapeutic anti-cancer monoclonal antibodies. Cancer Metastasis Rev. 24487-499. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa, T. 2003. Current status of therapeutic approaches to adult T-cell leukemia. Int. J. Hematol. 78304-311. [DOI] [PubMed] [Google Scholar]
- 17.Junker, K., W. Hindermann, F. von Eggeling, J. Diegmann, K. Haessler, and J. Schubert. 2005. CD70: a new tumor specific biomarker for renal cell carcinoma. J. Urol. 1732150-2153. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, N., H. Konishi, H. Sabe, K. Shigesada, T. Noma, T. Honjo, and M. Hatanaka. 1984. Genomic structure of HTLV (human T-cell leukemia virus): detection of defective genome and its amplification in MT-2 cells. EMBO J. 31339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law, C. L., K. A. Gordon, B. E. Toki, A. K. Yamane, M. A. Hering, C. G. Cerveny, J. M. Petroziello, M. C. Ryan, L. Smith, R. Simon, G. Sauter, E. Oflazoglu, S. O. Doronina, D. L. Meyer, J. A. Francisco, P. Carter, P. D. Senter, J. A. Copland, C. G. Wood, and A. F. Wahl. 2006. Lymphocyte activation antigen CD70 expressed by renal cell carcinoma is a potential therapeutic target for anti-CD70 antibody-drug conjugates. Cancer Res. 662328-2337. [DOI] [PubMed] [Google Scholar]
- 20.Lin, M. Z., M. A. Teitell, and G. J. Schiller. 2005. The evolution of antibodies into versatile tumor-targeting agents. Clin. Cancer Res. 11129-138. [PubMed] [Google Scholar]
- 21.Marcus, R., and A. Hagenbeek. 2007. The therapeutic use of rituximab in non-Hodgkin's lymphoma. Eur. J. Haematol. S. 675-14. [DOI] [PubMed] [Google Scholar]
- 22.McEarchern, J. A., E. Oflazoglu, L. Francisco, C. F. McDonagh, K. A. Gordon, I. Stone, K. Klussman, E. Turcott, N. van Rooijen, P. Carter, I. S. Grewal, A. F. Wahl, and C. L. Law. 2007. Engineered anti-CD70 antibody with multiple effector functions exhibits in vitro and in vivo antitumor activities. Blood 1091185-1192. [DOI] [PubMed] [Google Scholar]
- 23.Michael, B., A. M. Nair, H. Hiraragi, L. Shen, G. Feuer, K. Boris-Lawrie, and M. D. Lairmore. 2004. Human T lymphotropic virus type-1 p30II alters cellular gene expression to selectively enhance signaling pathways that activate T lymphocytes. Retrovirology 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyoshi, I., I. Kubonishi, S. Yoshimoto, T. Akagi, Y. Ohtsuki, Y. Shiraishi, K. Nagata, and Y. Hinuma. 1981. Type C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T cells. Nature 294770-771. [DOI] [PubMed] [Google Scholar]
- 25.Mori, N., M. Fujii, G. Cheng, S. Ikeda, Y. Yamasaki, Y. Yamada, M. Tomonaga, and N. Yamamoto. 2001. Human T-cell leukemia virus type I tax protein induces the expression of anti-apoptotic gene Bcl-xL in human T-cells through nuclear factor-κB and c-AMP responsive element binding protein pathways. Virus Genes 22279-287. [DOI] [PubMed] [Google Scholar]
- 26.Nair, A., B. Michael, H. Hiraragi, S. Fernandez, G. Feuer, K. Boris-Lawrie, and M. Lairmore. 2005. Human T lymphotropic virus type 1 accessory protein p12I modulates calcium-mediated cellular gene expression and enhances p300 expression in T lymphocytes. AIDS Res. Hum. Retrovir. 21273-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng, P. W., H. Iha, Y. Iwanaga, M. Bittner, Y. Chen, Y. Jiang, G. Gooden, J. M. Trent, P. Meltzer, K. T. Jeang, and S. L. Zeichner. 2001. Genome-wide expression changes induced by HTLV-1 Tax: evidence for MLK-3 mixed lineage kinase involvement in Tax-mediated NF-κB activation. Oncogene 204484-4496. [DOI] [PubMed] [Google Scholar]
- 28.Nicot, C., R. Mahieux, S. Takemoto, and G. Franchini. 2000. Bcl-X(L) is up-regulated by HTLV-I and HTLV-II in vitro and in ex vivo ATLL samples. Blood 96275-281. [PubMed] [Google Scholar]
- 29.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet 110310-11032. [DOI] [PubMed] [Google Scholar]
- 30.Pise-Masison, C. A., M. Radonovich, R. Mahieux, P. Chatterjee, C. Whiteford, J. Duvall, C. Guillerm, A. Gessain, and J. N. Brady. 2002. Transcription profile of cells infected with human T-cell leukemia virus type I compared with activated lymphocytes. Cancer Res. 623562-3571. [PubMed] [Google Scholar]
- 31.Proietti, F. A., A. B. Carneiro-Proietti, B. C. Catalan-Soares, and E. L. Murphy. 2005. Global epidemiology of HTLV-I infection and associated diseases. Oncogene 246058-6068. [DOI] [PubMed] [Google Scholar]
- 32.Reff, M. E., K. Carner, K. S. Chambers, P. C. Chinn, J. E. Leonard, R. Raab, R. A. Newman, N. Hanna, and D. R. Anderson. 1994. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood 83435-445. [PubMed] [Google Scholar]
- 33.Ruckes, T., D. Saul, J. Van Snick, O. Hermine, and R. Grassmann. 2001. Autocrine antiapoptotic stimulation of cultured adult T-cell leukemia cells by overexpression of the chemokine I-309. Blood 981150-1159. [DOI] [PubMed] [Google Scholar]
- 34.Salahuddin, S. Z., P. D. Markham, F. Wong-Staal, G. Franchini, V. S. Kalyanaraman, and R. C. Gallo. 1983. Restricted expression of human T-cell leulemia-lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology 12951-64. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki, H., I. Nishikata, T. Shiraga, E. Akamatsu, T. Fukami, T. Hidaka, Y. Kubuki, A. Okayama, K. Hamada, H. Okabe, Y. Murakami, H. Tsubouchi, and K. Morishita. 2005. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood 1051204-1213. [DOI] [PubMed] [Google Scholar]
- 36.Sincock, P., G. Mayrhofer, and L. K. Ashman. 1997. Localization of the transmembrane 4 superfamily (TM4SF) member PETA-3 (CD151) in normal human tissues: comparison with CD9, CD63, and α5β1 integrin. J. Histochem. Cytochem. 45515-525. [DOI] [PubMed] [Google Scholar]
- 37.Takatsuki, K. 2005. Discovery of adult T-cell leukemia. Retrovirology 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka, Y., A. Yoshida, Y. Takayama, H. Tsujimoto, A. Tsujimoto, M. Hayami, and H. Tozawa. 1990. Heterogeneity of antigen molecules recognized by anti-tax1 monoclonal antibody Lt-4 in cell lines bearing human T cell leukemia virus type I and related retroviruses. Jpn. J. Cancer Res. 81225-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, G. P., and M. Matsuoka. 2005. Natural history of adult T-cell leukemia/lymphoma and approaches to therapy. Oncogene 246047-6057. [DOI] [PubMed] [Google Scholar]
- 40.Uchiyama, T., J. Yodoi, K. Sagawa, K. Takatsuki, and H. Uchino. 1977. Adult T-cell leukemia: clinical and hematologic features of 16 cases. Blood 50481-492. [PubMed] [Google Scholar]
- 41.Wang, X., H. Miyake, M. Okamoto, M. Saito, J. Fujisawa, Y. Tanaka, S. Izumo, and M. Baba. 2002. Inhibition of the tax-dependent human T-lymphotropic virus type I replication in persistently infected cells by the fluoroquinolone derivative K-37. Mol. Pharmacol. 611359-1365. [DOI] [PubMed] [Google Scholar]
- 42.Widney, D., G. Gundapp, J. W. Said, M. van der Meijden, B. Bonavida, A. Demidem, C. Trevisan, J. Taylor, R. Detels, and O. Martinez-Maza. 1999. Aberrant expression of CD27 and soluble CD27 (sCD27) in HIV infection and in AIDS-associated lymphoma. Clin. Immunol. 93114-123. [DOI] [PubMed] [Google Scholar]
- 43.Wischhusen, J., G. Jung, I. Radovanovic, C. Beier, J. P. Steinbach, A. Rimner, H. Huang, J. B. Schulz, H. Ohgaki, A. Aguzzi, H. G. Rammensee, and M. Weller. 2002. Identification of CD70-mediated apoptosis of immune effector cells as a novel immune escape pathway of human glioblastoma. Cancer Res. 622592-2599. [PubMed] [Google Scholar]
- 44.Wright, M. D., S. M. Geary, S. Fitter, G. W. Moseley, L. M. Lau, K. C. Sheng, V. Apostolopoulos, E. G. Stanley, D. E. Jackson, and L. K. Ashman. 2004. Characterization of mice lacking the tetraspanin superfamily member CD151. Mol. Cell. Biol. 245978-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]