Abstract
Human papillomaviruses (HPVs) belonging to the Betapapillomavirus genus have recently been implicated in squamous cell carcinomas of the skin, though the mechanisms by which they initiate carcinogenesis are unclear. We show that human foreskin keratinocytes (HFKs) expressing several betapapillomavirus E6 (beta-E6) proteins display life span extension, but not to the extent seen in HFKs expressing HPV type 16 E6 (16E6). Additionally, we demonstrate that beta-E6 proteins can differentially activate telomerase. HFKs expressing 38E6 exhibit significant telomerase activity but to a lesser degree than that observed with 16E6; however, other beta-E6 proteins, including 5E6, 8E6, 20E6, and 22E6, exhibit low or background levels of telomerase activity. Utilizing glutathione S-transferase pull-down and coimmunoprecipitation experiments, the beta-E6 proteins were shown to interact with the cellular proteins E6-associated protein (E6AP) and NFX1-91, two proteins known to be important for telomerase activation by 16E6. Interestingly, the relative strength of the interaction between E6 and E6AP or NFX1-91 was proportionate to the activation of telomerase by each beta-E6 protein. To address the requirement for E6AP in telomerase activation by beta-E6 proteins, we utilized a shRNA to knock down endogenous levels of E6AP. Lysates with decreased levels of E6AP showed a reduced ability to activate telomerase, suggesting that E6AP is a necessary component. These data suggest that complex formation between E6, E6AP, and NFX1-91 is a critical step in mediating telomerase activation, which may be one contributing factor to cellular life span extension during human betapapillomavirus infection.
High-risk human papillomaviruses (HPVs) belonging to the Alphapapillomavirus genus are known to be a common cause of cervical cancer through the function of the viral oncoproteins E6 and E7 (10, 19, 26). More recently, human betapapillomaviruses (beta-HPVs; such as HPV types 5, 8, and 38) have been implicated in skin cancer progression, yet the molecular mechanisms of cancer development are still largely unknown. Beta-HPVs were first isolated from patients with the rare genetic disorder epidermodysplasia verruciformis, who suffer from life-long development of benign lesions such as warts but are also at greatly increased risk for development of squamous cell skin carcinomas (6-8). Cancers from patients with epidermodysplasia verruciformis led to the original isolation and identification of HPV type 5 (HPV5) and HPV8, whereas benign lesions from these patients contained dozens of different beta-HPV types (8). It is still unclear which, if any, beta-HPVs can be categorized as high risk and what molecular functions are associated with high-risk phenotypes.
Other evidence supporting a role for beta-HPVs in skin cancer includes previous reports that expression of HPV5 E6 (5E6) causes an inhibition of the intrinsic apoptotic cascade following UV light treatment (14, 15). These studies have shown that Bak, a BH3-containing proapoptotic factor, is degraded by 5E6, and cells expressing 5E6, as well as 18E6, display less apoptosis than control cells under the same conditions (15). In addition, transgenic mouse studies showed that expression of the HPV8 early proteins (E2, E6, and E7) caused an increased incidence of benign tumors and squamous cell carcinomas compared to control mice (33). Others have shown that the HPV38 E6 and E7 proteins expressed in transgenic mice are able to cause an increased rate of cell proliferation and squamous cell carcinomas when treated with a two-stage carcinogen protocol (9). These data suggest that the oncoproteins from some beta-HPVs are able to cause changes in cell apoptosis, an increase in cellular proliferation, and an increased incidence of tumor development in transgenic mice.
The molecular properties of E6 and E7 from the alpha-HPVs are well understood in terms of how they cause changes within the host cell that lead to altered cell growth and immortalization. In the high-risk alpha-HPVs, the major oncoproteins E6 and E7 play key roles in interfering with cell cycle checkpoint pathways. The E6 protein functions to degrade p53 and activate telomerase (21, 34), which are key steps in the immortalization of human foreskin keratinocytes (HFKs). The E7 oncoprotein from HPV16 is also crucial for cancer development through its strong interaction and inactivation of the tumor suppressor protein Rb (2, 13). In contrast, the mechanistic details of how the beta-HPVs interfere with cell cycle and apoptotic control to cause altered growth properties and cancer progression are still unknown. It has been shown that the beta-E6 proteins do not degrade p53 (3, 14), but others have suggested that inhibition of p53 may be caused by both high- and low-risk HPVs through an interaction with p300 resulting in p53 acetylation and inhibition (30, 39). Stable expression of both E6 and E7 from HPV38 is known to cause alterations in cell cycle control and induce long-lasting proliferation of primary keratinocytes in culture (4). In addition, lysates from 38E6-expressing cells have activated levels of telomerase, but not to the extent seen in lysates containing 16E6 (4). These data suggest that although HPV38 E6 does not cause p53 degradation, it may cause cell changes in conjunction with E7 that affect cell growth. The E7 proteins from HPV5 and HPV8 have been shown to have a much weaker interaction (36) with Rb and can only transform rodent cells in conjunction with activated H-ras (41). However, others have shown that HPV38 E7 can inactivate Rb, inducing an aberration in G1/S phase control (which is not observed with HPV10 or HPV20 E7 proteins) and may play a role in carcinogenesis (4), suggesting that HPV38 has a growth advantage over other beta-HPVs.
One critical step in 16E6-driven cell immortalization is the activation of telomerase (11, 19, 21). It is well understood that E6 expression during a high-risk alpha-HPV infection causes upregulation of hTERT mRNA (the catalytic subunit of telomerase) (29, 40) through its interaction with the cellular protein E6-associated protein (E6AP) (12, 16, 40). E6AP is the E3 ubiquitin ligase that in conjunction with E6 is responsible for the degradation of several cellular proteins (17, 23, 24, 27, 31), including p53 (34, 35), during an HPV infection. One protein known to be important for 16E6-mediated telomerase activity is NFX1-91 (nuclear factor binding to the X-box protein) (12). NFX1-91 is a transcriptional repressor that is predicted to bind to the X-box sequences within the hTERT promoter and cause repression of hTERT expression in normal cells (12; W. Luo, M. Xu, D. Elzi, C. Grandori, and D. Galloway, submitted for publication). Upon 16E6 expression, NFX1-91 becomes ubiquitinated and degraded, allowing expression of hTERT mRNA and activation of telomerase in 16E6-expressing cells (12, 18), which is a required step in HPV-induced immortalization.
To address the mechanism by which the beta-HPV E6 proteins may be causing molecular changes within cells that lead to changes in cell growth and cancer predisposition, we examined the ability of the beta-E6 proteins to prolong primary keratinocyte life span in culture and activate telomerase. Our data demonstrate that certain beta-HPV E6 proteins, especially HPV38 E6, can significantly activate telomerase through increased hTERT mRNA levels and interact with known components involved in 16E6-induced telomerase activation. This suggests that HPV38 E6 has the ability to alter cellular mechanisms similar to 16E6 that drive its ability to expand cellular life span and create a cell growth advantage above other beta-HPVs.
MATERIALS AND METHODS
Plasmid design.
The E6 gene was amplified from full-length DNA of HPV types 5, 8, 20, and 38 using PCR with the following primers: 5E6 forward, 5′AAAAAGCAGGCTTGGCAATGGCTGAGGGAG 3′, and reverse, 5′AGAAAGCTGGGTGACCTCTTTACCAATCATG 3′; 8E6 forward, 5′ AAAAAGCAGGCTTGGAAATGGACGGGCAGG 3′, and reverse, 5′ AGAAAGCT GGGTCTCTTTACCAATCATGATAC 3′; 20E6 forward, 5′ AAAAAGCAGGCTGG GACATGGCTACACCTC 3′, and reverse, 5′ AGAAAGCTGGGTATCATTATTGAAAATGCTTACAC 3′; 38E6 forward, 5′ AAAAAGCAGGCTTAATCATGGAACTACCAAAAC 3′, and reverse, 5′ AGAAAGCTGGGTCCAATCATTCTATTGCTTTGC 3′.
The E6 genes were then inserted via the Gateway recombination-based system (Invitrogen) into pLXSN for retroviral transfection and into pDEST15 to produce a recombinant glutathione S-transferase (GST)-E6 fusion protein. The hemagglutinin (HA)-tagged versions of HPV 5E6, 8E6, 16E6, and 38E6 were made by removing the C-terminal stop codon from each of the E6 genes with the following reverse primers: 5E6, 5′ AGAAAGCTGGGTACCAATCATGATAAAAATGC 3′; 8E6, 5′ AGAAAGCTGGGTACCAATCATGATACAAATGC 3′; 16E6, 5′ AGAAAGCTGGGTACAGCTGGGTTTCTCTACG 3′; 38E6, 5′ AGAAAGCTGGGTATTCTATTGCTTTGCAATGCC 3′.
PCR products were then inserted into the pLXSN-3HA Gateway vector to create an E6 protein with a C-terminal HA tag.
shRNA constructs were cloned as described in reference 12. The E6AP shRNA targeted sequences were as follows: sh1, 5′-CTAATAGAACGCTACTACCACCAGT 3′; sh2, 5′-AGAGATTGTTGAAGGCCATCACGTAT 3′. E6AP shRNAs were cloned into pBABE expression constructs for use in the G protein of vesicular stomatitis virus production protocol.
Tissue culture.
Primary HFKs and 293T cells were cultured as described elsewhere (12). HFKs were derived from neonatal foreskins and grown in EpiLife medium supplemented with calcium chloride, human keratinocytes growth supplement (Cascade Biologics), and penicillin-streptomycin. 293T cells were grown in Dulbecco's modified Eagle's medium (GIBCO-BRL) containing 10% fetal calf serum and penicillin-streptomycin.
Stable cell lines were produced using transient vesicular stomatitis virus G-pseudotyped virus as previously described (1). E6 proteins or shRNA sequences were cloned into LXSN or pBabe vectors as described above. They were cotransfected with vesicular stomatitis virus G-pseudotyped helper plasmids into 293T cells using Fugene 6 (Roche), and retrovirus was collected at 12, 24, 36, and 48 h posttransfection. Transient virus produced was concentrated by centrifugation and used to infect HFK monolayers (50 to 60% confluent) in the presence of Polybrene (8 mg/liter). Four hours postinfection, the Epilife medium was replaced. The cells were expanded when confluent and were placed under neomycin-G418 selection (50 mg/liter) or puromycin selection (0.5 mg/liter). Cells were kept under drug selection for 7 days and continually split when they reached 80 to 90% confluence. For each life span experiment, cells derived from the same human foreskin were infected with each of the beta-E6 proteins. The cells were grown and selected under identical conditions and were split 1:3 when the plate confluence was between 70 and 90%.
TRAP assays and real-time PCR.
Telomerase activity was detected using the TRAPeze kit (Chemicon International) as described elsewhere (12). In short, HFKs expressing the E6 proteins or lacking E6 expression were harvested in 3-[(3-choladmidopropyl)-dimethylammonio]-1-propanesulfoante lysis buffer as described in the TRAPeze protocol. Cell lysates (0.1 to 2 μg) were incubated with a [32P]ATP-labeled TS primer, and extension products were amplified via PCR. Telomeric repeat amplification protocol (TRAP) products were separated on a 10% nondenaturing polyacrylamide gel and visualized and quantitated via phosophorimager analysis.
Real-time PCR was used as a quantitative PCR method to detect hTERT mRNA levels. HFK cells were lysed with 1 ml of TRIzol reagent (Invitrogen, Carlsbad, CA) and purified by chloroform extraction and isopropanol precipitation. One microgram of total RNA was used to synthesize cDNA with random hexamer primers and a SuperScript II reverse transcriptase system (Invitrogen). Real-time PCR was performed on hTERT and 36b4 with sequence-specific primers previously described (11) using Sybr green on an ABI Prism 9700 and analyzed with SDS 2.2.2 software. Data analysis was performed using the comparative CT method to determine relative expression levels using 36b4 as an internal control. PCRs were done in triplicate, and error bars represent 95% confidence intervals of the ratio hTERT/36B4 as calculated by using the root mean square.
Western blot analysis and coimmunoprecipitation assays.
Western blots were probed with the antibodies anti-HA (for Western blot assays; Roche), anti-HA (for coimmunoprecipitations; Covance), nucleolin (C23 [MS-3]; Santa Cruz Biotechnology), E6AP (Ab-Nova), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; AbCam). Coimmunoprecipitation assays were conducted as previously published (12).
GST protein purification and GST pull-down assays.
BL21-AI Escherichia coli cells were freshly transformed with the GST-E6 constructs and selected overnight at 37°C on LB plates containing 50 μg/ml ampicillin. Isolated colonies were used to inoculate 20 ml LB broth containing 200 μg/ml carbenicillin and grown overnight with shaking at 37°C. Ten ml of the overnight culture was added to 1 liter of fresh LB-carbenicillin and incubated at 37°C with shaking for 2.5 h. Cultures were then transferred to room temperature and incubated for 30 min with shaking, after which the optical density at 600 nm of all cultures was between 0.4 and 0.6. l-Arabinose was added to each culture at a final concentration of 0.2% to induce protein expression, followed by growth for 4 h at room temperature with shaking. Bacterial cells were then harvested by centrifugation at 6,000 rpm for 15 min at 4°C, and the resulting pellets were stored at −20°C. Bacterial pellets were resuspended in phosphate-buffered saline, 50 mM EDTA, and protease inhibitor tablets (PBS-P; Roche) and lysed via two passages through a microfluidizer, followed by a 30-min incubation with 0.1%Triton X-100 at 4°C with end-over-end rotation. Bacterial lysates were centrifuged at 14,000 rpm in a JA17 rotor for 15 min at 4°C, and the pellets were discarded. The resulting supernatants were added to preequilibrated glutathione-Sepharose bead slurries and incubated at 4°C for 1 h with end-over-end rotation. The bead slurries were then washed four times with PBS-P, followed by two consecutive elutions of the bound GST proteins with 20 mM glutathione-50 mM Tris-Cl for 1 h at 4°C with end-over-end rotation. The elutions for each GST-E6 protein were combined and dialyzed using Zeba Desalt spin columns (Pierce) in a protein buffer (5 mM Tris, 100 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 5% glycerol, 0.1% NP-40, and protease inhibitor tablets). The concentration and purity of each GST-E6 protein was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and subsequent staining with GelCode Blue reagent (Pierce).
GST pull-down assays were performed with 2 μg of fusion protein incubated with in vitro-translated radiolabeled proteins in binding buffer (1% NP-40 alternative, 2 mM dithiothreitol, Complete mini protease inhibitor tabs, PBS) to a final volume of 200 μl and incubated with agitation for 1 h at 4°C. E6AP and NFX1-91 proteins were in vitro translated using the T7 TNT Quick-coupled kit (Promega) in wheat germ extracts. Glutathione-Sepharose beads in binding buffer (30 μl) were added to each reaction mixture and incubated at 4°C for another 2 h. The beads were washed three times in binding buffer, and bound proteins were eluted by boiling in Laemmli sample buffer (20 μl). The entire elution was separated on a SDS-polyacrylamide gel, and radiolabeled proteins were visualized by phosphorimager analysis.
RESULTS
Expression of beta-E6 proteins causes life span extension and telomerase activation in primary keratinocytes.
To examine what effect the beta-E6 proteins have on cell growth in vitro, we stably expressed several beta-E6 proteins in primary human foreskin keratinocytes and passaged these cells until they stopped dividing. When compared to control cells that contained an LXSN vector, the keratinocytes expressing E6 proteins had varied degrees of life span extension (Fig. 1A). The beta-E6 proteins tested with the highest degrees of life span extension (8E6 and 38E6) grew for an average of 21 and 24 population doublings, respectively, which is significantly (P < 0.4 and P < 0.001) longer than the LXSN control (17 population doublings). Expression of other beta-E6 proteins, such as 5 and 20, had a very modest effect on life span extension (19 and 18 population doublings), and these were not shown to have statistical significance compared to LXSN control cells. However, even beta-E6-expressing cells that could prolong cellular life span (8 and 38) did not grow as long as cells expressing 16E6, which grew for an average of 28 population doublings (P < 0.0001). Error bars were calculated based on the standard deviations from three independent experiments. To determine if the differences in population doublings among the E6 cell lines were statistically significant at 66 days, a one-way analysis of variance with Bonferroni post hoc analysis was used. Data shown in Fig. 1B suggest that the life spans of primary keratinocytes expressing beta-E6 proteins are significantly different from control cells but are also different from each other.
FIG. 1.
Expression of E6 proteins extends the life span of primary keratinocytes in culture. A. Primary keratinocytes expressing beta-E6 proteins were grown in culture to compare their growth properties to control cells containing an LXSN empty vector. Cells were passaged at 80% confluence and recorded for population doublings under normal cell growth conditions. Error bars were calculated using the standard deviation of three independent experiments. B. P values for each E6 cell line compared to each other and to an LXSN control are shown. The P values were calculated using a one-way analysis of variance with Bonferroni post hoc analysis. NS, not significant.
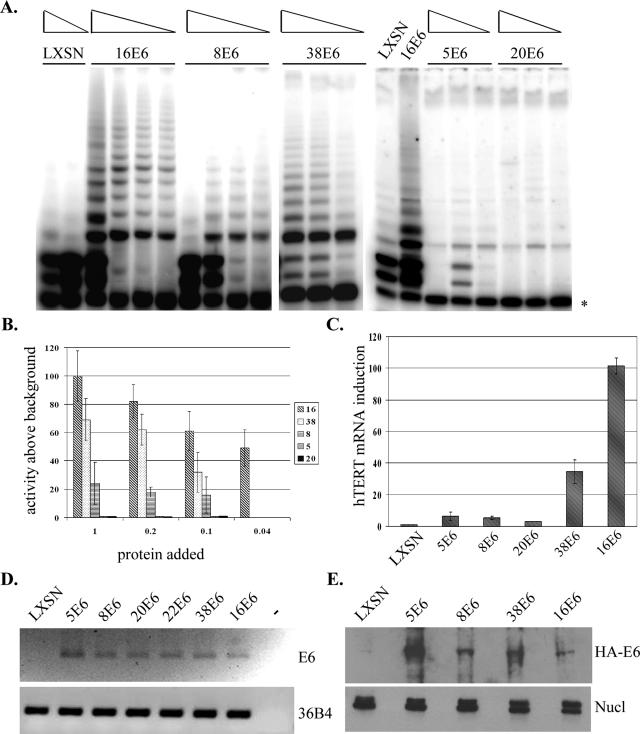
Next, we wanted to explore the mechanisms utilized by the beta-E6 proteins to extend the life span of keratinocytes. It has been well demonstrated that 16E6 activates telomerase through E6AP-mediated degradation of NFX1-91 (12, 18), and this is essential for HFK immortalization (16, 17, 20). To examine telomerase activation by the beta-E6 proteins, lysates from HFKs expressing several of the beta-E6 proteins were harvested and analyzed in TRAP assays. In these assays, a higher level of TRAP activity is indicated by the accumulation of higher-molecular-weight extension products. Figure 2A shows telomerase activity of different cell lysates containing either 16E6, beta-E6 proteins (38, 8, 5, 20), or a negative control lysate containing LXSN. Keratinocytes containing an LXSN control vector were used as a negative control to determine background levels of telomerase activity, as primary keratinocytes do not have significant levels of active telomerase. Lysates from cells expressing 16E6 showed high levels of TRAP activity, as expected (21). Interestingly, some of the beta-E6-expressing cells also demonstrated TRAP activity above background levels (Fig. 2A). Of the beta-E6 lysates tested, cells expressing 38E6 had the highest levels of telomerase activity (50 to 70% of that seen with 16E6). Lysates from cells expressing 8E6 displayed modest levels of telomerase activity, approximately 20% of that observed with 16E6 lysates (Fig. 2B). However, other beta-E6 proteins, including 5E6 and 20E6, exhibited very low or background levels of telomerase activity (Fig. 2A and B). TRAP activity among the beta-E6-containing lysates was quantitated in relation to the telomerase activation observed with 16E6-containing lysates. Quantitation of TRAP activity was performed on reaction mixtures with decreasing amounts of protein lysate and showed that TRAP activity is proportionate to the amount of protein used in each assay (Fig. 2B).
FIG. 2.
Primary keratinocytes expressing beta-E6 proteins displayed activated telomerase and induction of hTERT mRNA. A. Lysates from stable HFK cell lines expressing 16E6 or beta-E6 proteins or cells containing an empty LXSN expression vector (negative control) were analyzed in TRAP assays. The autoradiogram shows extension products from the [α-32P]ATP-labeled TS primer following incubation with various cell lysates. The band denoted by an asterisk denotes a control PCR product to show that all samples amplified to equal levels. Decreasing amounts of protein lysate (from 1 to 0.04 μg) were added to TRAP reaction mixtures. B. TRAP activity was quantitated from three independent experiments using Quantity One software. TRAP activity from 16E6 lysates was set to 100 to determine relative levels of telomerase activity in other E6-containing lysates. C. qPCR of hTERT mRNA in E6-expressing HFKs. hTERT levels in 16E6-expressing cells were set to 100 to compare the relative levels of hTERT expression in other E6 cell lines. D. Real-time PCR of E6 proteins stably expressed in HFKs. 36b4 was amplified as a loading control. The lane with a minus sign denotes a water-only control (no cDNA added). E. Western blot analysis of HA-tagged E6 proteins expressed in HFKs. Nucleolin was used as a loading control.
It is known that 16E6 transcriptionally activates hTERT mRNA (12, 21), and so we examined the induction of hTERT mRNA using quantitative real-time PCR (qPCR) in HFKs expressing beta-E6 proteins. To compare the different E6-expressing lysates for hTERT mRNA amplification, the hTERT expression level in 16E6 lysates was set to 100%. As expected, cells expressing 38E6 showed an increased level of hTERT mRNA above background, but not as high as the hTERT mRNA induction observed in 16E6 cells (approximately 30%) (Fig. 2C). In addition, cells expressing 8E6 also showed a slight increase in hTERT mRNA levels (13% of what was observed in 16E6 cells), as well as cells expressing 5E6. Keratinocytes expressing 20E6 did not show any significant increase in hTERT mRNA compared to the LXSN control cells. Thus, hTERT mRNA levels were also increased by expression of a subset of beta-E6 proteins, although not as dramatically as by 16E6 expression. These data confirm that 38E6-expressing cells activate telomerase, and the results indicate that this activation is through induction of hTERT mRNA levels. Additionally, these data suggest that both 5E6 and 8E6 exhibit a weak ability to activate hTERT transcription and that qPCR analysis is perhaps a more sensitive technique to detect low levels of hTERT induction.
To confirm that the observed differences in telomerase activity are not due to differences in E6 expression, we analyzed levels of E6 RNA expression in HFKs using reverse transcriptase PCR amplification. RNA was harvested from cell lines expressing the beta-E6 proteins, 16E6, or LXSN cells as a negative control. cDNA was synthesized and E6 was amplified using sequence-specific primers showing that all HFKs expressing E6 contained E6 RNA that was not observed in the LXSN cell lines, and the levels of RNA expression among the beta-E6 cells were similar (Fig. 2D). To confirm that equal starting material was used, the 36b4 gene transcript was amplified as a control (Fig. 2D). In addition, we examined protein expression levels of HA-tagged forms of several E6 proteins. Primary HFKs that stably expressed HA-tagged E6 proteins were examined by Western blot analysis with a monoclonal antibody to the HA epitope (Fig. 2E). Each of the cell lines tested displayed E6 expression that was not observed in LXSN control HFKs. Although levels of HA-E6 protein varied between the different cell lines, the differences in protein expression did not correlate with differences seen in TRAP activity.
Protein interactions between E6 and cellular proteins correlate with telomerase activation.
To examine the mechanism of telomerase activation by the beta-E6 proteins, interaction studies were carried out with cellular proteins that are known to be important for telomerase activation by 16E6. Since the beta-E6 proteins had differential effects on telomerase activation, the interaction studies were focused on 5E6, 8E6, 20E6, and 38E6. We hypothesized that the differences seen in telomerase activation between alpha- and beta-E6 proteins and within different beta-E6 types may be due to differences in the relative strengths of protein-protein interactions.
In coimmunoprecipitation experiments, the interaction of stably expressed beta-E6 proteins with the endogenous cellular protein E6AP was examined in cultured keratinocytes. HFK lysates expressing HA-tagged E6 were immunoprecipitated with a monoclonal antibody to HA and then examined for the presence of endogenous E6AP (Fig. 3A). LXSN control cells did not immunoprecipitate with E6AP. The E6 proteins that activated telomerase, such as 8E6, 38E6, and 16E6, were able to interact with endogenous E6AP; however, 5E6 did not interact detectably with E6AP. The reverse interaction was tested by immunoprecipitating protein complexes with the E6AP antibody and detecting HA-E6 through Western blot analysis with the HA monoclonal antibody. Similarly, LXSN cells and 5E6 cell lysates showed no significant levels of interaction, whereas 8E6, 38E6, and 16E6 could coimmunoprecipitate with endogenous E6AP (Fig. 3B). Because we were not able to create a cell line that expresses an HA-tagged form of 20E6, this was not included in the coimmunoprecipitation experiments.
FIG. 3.
Beta-E6 proteins differentially interact with E6AP. A. HFKs expressing HA-tagged E6 proteins were immunoprecipitated with a monoclonal HA antibody (+) or mouse immunoglobulin G (-) as a negative control and immunoblotted with an E6AP antibody (αE6AP). B. Identical cell lysates were immunoprecipitated with an E6AP antibody (+) or preimmune rabbit serum as a negative control (-) and immunoblotted with an HA antibody (αHA). Input is equal to 5% of total protein lysate and is shown in the Western blot on the right hand side. C. GST pull-down assays with in vitro-translated E6AP. Input represents 20% of radiolabeled E6AP added to each reaction mixture. D. The interaction of each GST-E6 protein with E6AP was quantitated from five independent experiments. The level of interaction above the GST negative control was measured using Image J software. A one-sample two-tailed t test was used to calculate the significance of interactions. **, P < 0.0001; *, P < 0.04. E. A 0.5-μg aliquot of each GST fusion protein was analyzed with SDS-polyacrylamide gel electrophoresis and Coomassie staining. F. GST pull-down assays were repeated in an identical manner with in vitro-translated NFX1-91 protein. Input (I) represents 10% of radiolabeled NFX1-91 added to each reaction mixture, and a + denotes pull-down reactions.
To confirm these interactions in vitro, we carried out GST pull-down experiments with recombinant, purified GST-tagged E6 proteins and in vitro-translated radiolabeled E6AP. The GST proteins used in these assays are shown in a Coomassie-stained gel to demonstrate they are of equal concentration and purity (Fig. 3E); each lane represents 25% of the GST protein used in each pull-down experiment. GST and a GST-tagged form of the viral protein 16E7 were used as negative controls for nonspecific protein-protein interactions; E7 was chosen as an additional control because it is of similar size to E6 and is not known to interact with E6AP. The two negative controls, GST and GST-E7, showed a background level of interaction with E6AP; however, the beta-E6 proteins showed differential interactions with E6AP (Fig. 3C). To quantitate the differences observed in protein-protein interactions, the levels of interaction above the GST negative control from five independent pull-down experiments were determined (Fig. 3D). 5E6 did not interact with E6AP in the GST pull-down assays above background levels. In contrast, 8E6 and 20E6 interacted very weakly with E6AP. When the experiment was repeated several times, the interaction between 20E6 and E6AP was variable and not statistically above background levels. However, the interaction between 8E6 and E6AP was very weak but reproducibly above background levels (P < 0.04). 38E6 interacted significantly with E6AP (P < 0.001), and 16E6 showed a strong interaction above GST (P < 0.001) as expected (Fig. 3D). Together, these binding assays confirmed that beta-E6 proteins could differentially interact with the cellular protein E6AP, and these differences directly correlate with their ability to activate telomerase. Of the beta-E6 proteins tested, 38E6 displayed the strongest interaction with E6AP in primary cell lysates and in GST pull-down assays proportionate to its ability to activate telomerase.
Another endogenous protein previously shown to interact with the E6-E6AP complex and to be required for telomerase activation by 16E6 is the cellular protein NFX1-91 (12, 18). NFX1-91 functions as a repressor of hTERT expression, and 16E6-dependent degradation of NFX1-91 has been shown to relieve this repression and cause telomerase activation. To test if the beta-E6 proteins can interact with NFX1-91, GST pull-down assays with GST-tagged E6 proteins and in vitro-translated radiolabeled NFX1-91 were performed (Fig. 3F). As predicted, 38E6 and 16E6 interacted with NFX1-91 significantly above background levels of GST alone, correlating with their ability to efficiently activate telomerase. However, 5E6, 8E6, and 20E6 displayed no interaction with NFX1-91, suggesting that their reduced ability to interact with NFX1-91 correlates with their poor ability to activate telomerase. This is an additional piece of evidence suggesting that complex formation between E6 and the cellular proteins E6AP and NFX1-91 is a critical step in mediating telomerase activation, which may be one contributing factor to cellular life span extension during beta-HPV infection.
Beta-E6 proteins utilize E6AP to mediate telomerase activation similar to 16E6.
Previous studies showed that 16E6 activates telomerase through E6AP-mediated degradation of the cellular protein NFX1-91, and this is essential for cell immortalization (12, 22). To address the requirement for E6AP in telomerase activation by the beta-E6 proteins, we utilized shRNAs to knockdown endogenous levels of E6AP. Western blot analysis with an antibody specific to E6AP was used to determine levels of protein knockdown following shRNA treatment. LXSN cells showed endogenous levels of E6AP (Fig. 4A, lane 1), which are similar to cells containing an empty pBABE vector (shRNA-). Cells that stably express either 8E6, 38E6, or 16E6 were transduced with constructs expressing either shRNA-, shRNA 1, or shRNA 2, which have previously been shown to decrease E6AP expression levels (12). Our lab has previously shown that shRNA 1 is effective in decreasing E6AP mRNA levels by approximately 70%, whereas shRNA 2 can reduce the levels of E6AP and telomerase activity in 16E6 cells, but to a lesser extent (12). Our studies confirmed this and found that in cells expressing 16E6, shRNA 1 was more efficient at decreasing E6AP levels than shRNA 2 compared to control cells. In cells expressing 8E6 and 38E6, both shRNA 1 and shRNA 2 decreased levels of E6AP compared to the shRNA- (Fig. 4A).
FIG. 4.
Knockdown of E6AP reduces telomerase activity in 16E6-, 38E6-, and 8E6-expressing HFKs. A. shRNAs were stably expressed in E6-expressing HFKs. Knockdown was confirmed by Western blot analysis with E6AP antibody; GAPDH was used as a loading control. LXSN cells containing no shRNA were used as a positive control for E6AP expression. The shRNA- cells, containing an empty pBABE vector, show endogenous E6AP levels. B. 16E6, 8E6, and 38E6 cells expressing shRNA-, shRNA 1, or shRNA 2 were examined for TRAP activity and compared to LXSN cells as a negative control (lane 1). TRAP-postive lysates were used as a positive control in lane 11. The amplified extension products were quantitated using Quantity One software following phosphorimager scanning shown at the bottom of the gel. 16E6 with -shRNA, 8E6 with -shRNA, and 38E6 with -shRNA were set to 100 to compare the decrease in TRAP activity following E6AP shRNA treatment.
To examine the effect of reducing E6AP levels on telomerase activation in beta-E6 versus 16E6 cells, TRAP assays were performed. 16E6 lysates containing the shRNA- showed high levels of telomerase activity in TRAP assays, as expected (Fig. 4B, lane 2), and shRNA 1, which significantly reduced E6AP levels, concomitantly decreased the levels of telomerase activity (Fig. 4B, lane 3). In contrast, lysates from cells in which shRNA 2 did not affect E6AP levels still displayed high levels of telomerase activity, confirming that a decrease in E6AP is responsible for the downstream effect on telomerase activity. Using lysates from cells expressing 8E6 and 38E6, those that had the shRNA- against E6AP displayed moderate levels of telomerase activity above the LXSN background levels. In both shRNA 1 and 2 there was a decrease in E6AP levels (Fig. 4A) and a decrease in TRAP activity (Fig. 4B, lanes 6 and 7 and lanes 9 and 10) compared to background levels, suggesting that telomerase activity through 8E6 and 38E6 utilizes the same E6AP-dependent mechanism as 16E6. This experiment showed that lysates with decreased levels of E6AP were not able to sufficiently activate telomerase through 16E6 or beta-E6 proteins, suggesting that E6AP is a necessary component.
DISCUSSION
Telomerase activation by HPV16 E6 is thought to be one critical step in cellular immortalization during infection. Previous work led to the hypothesis that HPV16 E6 and E7 expression causes telomerase activation and, in concert with defects in the p53 and Rb pathways, allows cells to bypass senescence, leading to cellular immortalization (20, 32, 38). These cells are then at an increased risk for the accumulation of genetic mutations that can cause cancer progression. Published work has shown that E6 and E7 from HPV38 can cause keratinocyte immortalization when expressed in culture, but E6 proteins from other beta-HPVs (such as HPV20) do not cause immortalization (4). This raises the possibility that unique oncogenic phenotypes caused by HPV38 E6 and E7 may provide a growth advantage to infected cells and cause HPV38 to pose a greater risk regarding cancer development.
Expression of both E6 and E7 from HPV16 is required to establish immortalization of primary keratinocytes in cell culture; however, E6 expression alone will prolong the life span of cultured cells (19, 25). To directly test the role of beta-E6 proteins in cell growth, we examined the effect of beta-E6 protein expression on keratinocyte life span in culture and found that the E6 protein from several beta-HPVs allows cultured keratinocytes to grow for considerably more population doublings than control cells. This raises the question, how does beta-E6 protein expression prolong the life span of these cells?
Our data suggest that primary keratinocytes expressing beta-E6 proteins that have a prolonged life span in culture, such as HPV38, also contain high levels of telomerase activity. In addition, the amount of telomerase activity correlates with the protein-protein interactions observed between E6, E6AP, and NFX1-91. Previous work revealed that for HPV16, the E6-E6AP complex is responsible for ubiquitinating and degrading a telomerase repressor, NFX1-91 (12, 18, 22), allowing increased synthesis of the hTERT mRNA transcript. Protein interaction studies showed that beta-E6 proteins, especially 38E6, can form a direct interaction with both E6AP and NFX1-91, similar to what is observed with 16E6, and that the stability of the interactions is above what is observed with the other beta-E6 proteins. This provided initial evidence that beta-E6 proteins, particularly 38E6, utilize an E6AP- and NFX1-91-dependent mechanism for telomerase activation. By decreasing expression of E6AP (using shRNAs), we have shown that beta-E6 proteins require E6AP to activate telomerase. These results argue that the beta-E6 proteins are capable of activating telomerase with differing efficiencies, and they utilize similar protein factors that are required by 16E6. It does not eliminate the possibility that additional factors may be involved in 16E6- or beta-E6-mediated telomerase activation.
The differences between the beta-E6 proteins in their ability to extend the life span of keratinocytes, activate telomerase, and interact with cellular proteins are summarized in Table 1. In general, the ability of E6 to activate telomerase correlates with how well it extends the life span of primary keratinocytes and their ability to interact with E6AP and NFX1-91. HPV8 E6 moderately prolongs the life span of cultured cells and activates telomerase, as observed through TRAP assays. It is able to interact with E6AP in primary keratinocytes, as shown by coimmunoprecipitation assays, but displays weak or no interaction with E6AP and NFX1-91 in GST pull-down assays. These data suggest that 8E6 cannot efficiently interact in in vitro assays, possibly due to misfolding or low specific activity levels of our recombinant protein. Alternatively, it is possible that a cellular interaction with E6AP is required for telomerase activation and a direct interaction between E6 and NFX1-91 boosts the extent of telomerase activation, leading to activities observed with 38E6 and 16E6.
TABLE 1.
Abilities of beta-E6 proteins to activate telomerase and interact with required cellular proteinsa
| E6 protein | Extension of life span | TRAP activity | hTERT mRNA induction | Interacts with E6AP | Interacts with NFX1-91 |
|---|---|---|---|---|---|
| LXSN | − | − | − | − | − |
| HPV5 | + | − | +/− | − | − |
| HPV8 | ++ | + | +/− | + | − |
| HPV20 | − | − | − | − | − |
| HPV38 | +++ | ++ | ++ | ++ | ++ |
| HPV16 | ++++ | +++ | +++ | ++++ | ++ |
−, no detectable activity; the number of + symbols indicates the relative level of activity in each assay.
The experiments conducted thus far cannot determine if telomerase activity is the direct cause of the prolonged cellular life span observed. Previous work suggests that E6 expression in cell culture may not alone cause telomere extension that would lead to cellular immortalization (20). 16E6 cells, despite high levels of telomerase, still exhibit telomere shortening; however, it is thought that the cells maintain minimum telomere lengths which allow them to avoid cellular senescence, immortalize, and recover lengthened telomeres at later population doublings in culture (32, 38). There are several possibilities that may explain the connection between E6 protein expression, telomerase activity, and cellular life span extension. Firstly, the activation of telomerase has been shown to result in cellular growth advantages that could cause life span extension (for review, see references 5 and 37) and in vivo would result in the accumulation of mutations important for cancer development. Alternatively, there is the possibility that E6 protein expression in primary cells affects cell growth through a mechanism independent of telomerase activation. It is known that several of the beta-E6 proteins cause an increase in cell growth as well as a decrease in apoptosis, which could explain the role of E6 protein expression in the cellular life span (4, 14, 15).
One interesting aspect of beta-E6 protein function is the possibility that a subset of beta-E6 proteins contain properties that cause them to function as high risk in skin cancer progression. Our data suggest that telomerase activation differs between the beta-E6 proteins, and future work will focus on determining sequence and structure differences between the beta-E6 proteins that may provide insight into their functional differences. A structural model for 16E6 based on nuclear magnetic structural resonance structural data has recently been proposed (28). Using this structure, we can identify domains of amino acid sequence similarity (and proposed structural homology) between 38E6 and 16E6. Future studies will be required to determine if these domains are responsible for telomerase activation by 16E6 and 38E6. Our lab and others have previously utilized mutagenesis to define amino acids important for protein-protein interactions between HPV16 E6 and E6AP (11). However, the recent information gained from structural studies on 16E6 have suggested that several of the mutations affecting E6-E6AP binding may cause overall structural instability within E6. In the future, it will be important to identify what amino acids are crucial for the protein-protein interactions and how these residues are conserved among the beta-E6 proteins.
HPV38 has been shown to have additional unique characteristics, such as cellular immortalization and high levels of telomerase activation, which may create a cell growth advantage over other beta-HPVs. Our work provides strong evidence that telomerase activation by E6 from some of the beta-HPVs (especially HPV38) is one plausible mechanism that imparts a cellular growth advantage for immortalization and potential cancer development.
Acknowledgments
Full-length DNA from the beta-HPV types was kindly provided by E. M. de Villiers and Michel Favre. We thank members of the Galloway lab for helpful suggestions regarding experimental design and data analysis and Rachel Katzenellenbogen for critical reading of the manuscript.
This work was supported by grants from the NIH, including T32CA09229 (K.M.B.), T32DC000018 (M.P.U.), T32AI07140 (H.L.H.), and RO1 CA 064795 and PO1CA042792 (D.A.G.).
Footnotes
Published ahead of print on 6 February 2008.
REFERENCES
- 1.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12337-342. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 564620-4624. [PubMed] [Google Scholar]
- 3.Caldeira, S., R. Filotico, R. Accardi, I. Zehbe, S. Franceschi, and M. Tommasino. 2004. p53 mutations are common in human papillomavirus type 38-positive non-melanoma skin cancers. Cancer Lett. 209119-124. [DOI] [PubMed] [Google Scholar]
- 4.Caldeira, S., I. Zehbe, R. Accardi, I. Malanchi, W. Dong, M. Giarre, E. M. de Villiers, R. Filotico, P. Boukamp, and M. Tommasino. 2003. The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties. J. Virol. 772195-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung, H. K., C. Cheong, J. Song, and H. W. Lee. 2005. Extratelomeric functions of telomerase. Curr. Mol. Med. 5233-241. [DOI] [PubMed] [Google Scholar]
- 6.de Oliveira, W. R., N. C. Festa, P. L. Rady, and S. K. Tyring. 2003. Clinical aspects of epidermodysplasia verruciformis. J. Eur. Acad. Dermatol. Venereol. 17394-398. [DOI] [PubMed] [Google Scholar]
- 7.de Oliveira, W. R., Q. He, P. L. Rady, T. K. Hughes, C. F. Neto, E. A. Rivitti, and S. K. Tyring. 2004. HPV typing in Brazilian patients with epidermodysplasia verruciformis: high prevalence of EV-HPV 25. J. Cutan. Med. Surg. 8110-115. [DOI] [PubMed] [Google Scholar]
- 8.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. H. zur. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 9.Dong, W., U. Kloz, R. Accardi, S. Caldeira, W. M. Tong, Z. Q. Wang, L. Jansen, M. Durst, B. S. Sylla, L. Gissmann, and M. Tommasino. 2005. Skin hyperproliferation and susceptibility to chemical carcinogenesis in transgenic mice expressing E6 and E7 of human papillomavirus type 38. J. Virol. 7914899-14908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis, D. A., S. I. Schmid, and P. M. Howley. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 742679-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 757198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gewin, L., H. Myers, T. Kiyono, and D. A. Galloway. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 182269-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helt, A. M., and D. A. Galloway. 2001. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 756737-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson, S., C. Harwood, M. Thomas, L. Banks, and A. Storey. 2000. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 143065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, S., and A. Storey. 2000. E6 proteins from diverse cutaneous HPV types inhibit apoptosis in response to UV damage. Oncogene 19592-598. [DOI] [PubMed] [Google Scholar]
- 16.James, M. A., J. H. Lee, and A. J. Klingelhutz. 2006. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int. J. Cancer 1191878-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao, W. H., S. L. Beaudenon, A. L. Talis, J. M. Huibregtse, and P. M. Howley. 2000. Human papillomavirus type 16 E6 induces self-ubiquitination of the E6AP ubiquitin-protein ligase. J. Virol. 746408-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katzenellenbogen, R. A., E. M. Egelkrout, P. Vliet-Gregg, L. C. Gewin, P. R. Gafken, and D. A. Galloway. 2007. NFX1-123 and poly(A) binding proteins synergistically augment activation of telomerase in human papillomavirus type 16 E6-expressing cells. J. Virol. 813786-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Klingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 39684-88. [DOI] [PubMed] [Google Scholar]
- 20.Klingelhutz, A. J., S. A. Barber, P. P. Smith, K. Dyer, and J. K. McDougall. 1994. Restoration of telomeres in human papillomavirus-immortalized human anogenital epithelial cells. Mol. Cell. Biol. 14961-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 38079-82. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., H. Yuan, B. Fu, G. L. Disbrow, T. Apolinario, V. Tomaic, M. L. Kelley, C. C. Baker, J. Huibregtse, and R. Schlegel. 2005. The E6AP ubiquitin ligase is required for transactivation of the hTERT promoter by the human papillomavirus E6 oncoprotein. J. Biol. Chem. 28010807-10816. [DOI] [PubMed] [Google Scholar]
- 23.Mantovani, F., P. Massimi, and L. Banks. 2001. Proteasome-mediated regulation of the hDlg tumour suppressor protein. J. Cell Sci. 1144285-4292. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto, Y., S. Nakagawa, T. Yano, S. Takizawa, K. Nagasaka, K. Nakagawa, T. Minaguchi, O. Wada, H. Ooishi, K. Matsumoto, T. Yasugi, T. Kanda, J. M. Huibregtse, and Y. Taketani. 2006. Involvement of a cellular ubiquitin-protein ligase E6AP in the ubiquitin-mediated degradation of extensive substrates of high-risk human papillomavirus E6. J. Med. Virol. 78501-507. [DOI] [PubMed] [Google Scholar]
- 25.Munger, K., and P. M. Howley. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89213-228. [DOI] [PubMed] [Google Scholar]
- 26.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 634417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa, S., and J. M. Huibregtse. 2000. Human Scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 208244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nomine, Y., M. Masson, S. Charbonnier, K. Zanier, T. Ristriani, F. Deryckere, A. P. Sibler, D. Desplancq, R. A. Atkinson, E. Weiss, G. Orfanoudakis, B. Kieffer, and G. Trave. 2006. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol. Cell 21665-678. [DOI] [PubMed] [Google Scholar]
- 29.Oh, S. T., S. Kyo, and L. A. Laimins. 2001. Telomerase activation by human papillomavirus type 16 E6 protein: induction of human telomerase reverse transcriptase expression through Myc and GC-rich Sp1 binding sites. J. Virol. 755559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 185061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pim, D., M. Thomas, R. Javier, D. Gardiol, and L. Banks. 2000. HPV E6 targeted degradation of the discs large protein: evidence for the involvement of a novel ubiquitin ligase. Oncogene 19719-725. [DOI] [PubMed] [Google Scholar]
- 32.Plug-DeMaggio, A. W., T. Sundsvold, M. A. Wurscher, J. I. Koop, A. J. Klingelhutz, and J. K. McDougall. 2004. Telomere erosion and chromosomal instability in cells expressing the HPV oncogene 16E6. Oncogene 233561-3571. [DOI] [PubMed] [Google Scholar]
- 33.Schaper, I. D., G. P. Marcuzzi, S. J. Weissenborn, H. U. Kasper, V. Dries, N. Smyth, P. Fuchs, and H. Pfister. 2005. Development of skin tumors in mice transgenic for early genes of human papillomavirus type 8. Cancer Res. 651394-1400. [DOI] [PubMed] [Google Scholar]
- 34.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75495-505. [DOI] [PubMed] [Google Scholar]
- 35.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 631129-1136. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt, A., J. B. Harry, B. Rapp, F. O. Wettstein, and T. Iftner. 1994. Comparison of the properties of the E6 and E7 genes of low- and high-risk cutaneous papillomaviruses reveals strongly transforming and high Rb-binding activity for the E7 protein of the low-risk human papillomavirus type 1. J. Virol. 687051-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shay, J. W., and W. E. Wright. 2005. Senescence and immortalization: role of telomeres and telomerase. Carcinogenesis 26867-874. [DOI] [PubMed] [Google Scholar]
- 38.Stoppler, H., D. P. Hartmann, L. Sherman, and R. Schlegel. 1997. The human papillomavirus type 16 E6 and E7 oncoproteins dissociate cellular telomerase activity from the maintenance of telomere length. J. Biol. Chem. 27213332-13337. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, M. C., and C. M. Chiang. 2005. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol. Cell 17251-264. [DOI] [PubMed] [Google Scholar]
- 40.Veldman, T., I. Horikawa, J. C. Barrett, and R. Schlegel. 2001. Transcriptional activation of the telomerase hTERT gene by human papillomavirus type 16 E6 oncoprotein. J. Virol. 754467-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita, T., K. Segawa, Y. Fujinaga, T. Nishikawa, and K. Fujinaga. 1993. Biological and biochemical activity of E7 genes of the cutaneous human papillomavirus type 5 and 8. Oncogene 82433-2441. [PubMed] [Google Scholar]