Abstract
A highly effective attenuated equine infectious anemia virus (EIAV) vaccine (EIAVD9) capable of protecting 100% of horses from disease induced by a homologous Env challenge strain (EIAVPV) was recently tested in ponies to determine the level of protection against divergent Env challenge strains (J. K. Craigo, B. S. Zhang, S. Barnes, T. L. Tagmyer, S. J. Cook, C. J. Issel, and R. C. Montelaro, Proc. Natl. Acad. Sci. USA 104:15105-15110, 2007). An inverse correlation between challenge strain Env variation and vaccine protection from disease was observed. Given the striking differences in protective immunity, we hypothesized that analysis of the humoral and cellular immune responses to the Env protein could reveal potential determinants of vaccine protection. Neutralization activity against the homologous Env or challenge strain-specific Env in immune sera from the vaccinated ponies did not correlate with protection from disease. Cellular analysis with Env peptide pools did not reveal an association with vaccine protection from disease. However, when individual vaccine-specific Env peptides were utilized, eight cytotoxic-T-lymphocyte (CTL) peptides were found to associate closely with vaccine protection. One of these peptides also yielded the only lymphoproliferative response associated with protective immunity. The identified peptides spanned both variable and conserved regions of gp90. Amino acid divergence within the principal neutralization domain and the identified peptides profoundly affected immune recognition, as illustrated by the inability to detect cross-reactive neutralizing antibodies and the observation that certain peptide-specific CTL responses were altered. In addition to identifying potential Env determinants of EIAV vaccine efficacy and demonstrating the profound effects of defined Env variation on immune recognition, these data also illustrate the sensitivity offered by individual peptides compared to peptide pools in measuring cellular immune responses in lentiviral vaccine trials.
Equine infectious anemia virus (EIAV) is a macrophage-tropic equine lentivirus that has been used extensively as a model for human immunodeficiency virus type 1 (HIV-1) persistence and pathogenesis and for AIDS vaccine development (8, 18, 24, 25, 29, 36, 48-50, 55). Compared to other progressively degenerative lentiviral infections, EIAV is characterized by three distinct phases of infection, namely, acute, chronic, and inapparent. By 2 months post-EIAV exposure, most horses experience an acute disease episode characterized by high fever, a drop in platelets, and a high viral load. The horse then enters a chronic stage of infection characterized by recurrent disease episodes, which typically progress to the inapparent carrier stage of disease for the life span of the animal (48). During the inapparent phase, the horse has gained immunologic control of the virulent and constantly evolving lentivirus, as demonstrated by the fact that whole blood transfers from inapparent carriers to naïve horses can cause an acute episode within 2 months (31, 43, 48). Additionally, if the inapparent carrier is immunosuppressed or stressed, there can be recrudescing disease associated with a new viral quasispecies. Inapparent carriers of EIAV appear to be resistant to additional exposure to EIAV variant strains, indicating that the horse has gained a high level of prophylactic immunity (48). This type of immunity is especially desirable for lentiviral vaccines, which makes EIAV a valuable model system for studying vaccine protection.
Immune control of acute EIAV viremia has typically been associated with the appearance of cytotoxic T lymphocytes (CTL) (44) and nonneutralizing antibodies (44, 56, 62). Recently, virus-specific neutralizing antibodies have been found to develop after the resolution of the acute episode (45), and they continue to increase in titer and breadth of specificity throughout the first year of infection (5, 24, 30). We previously identified V3 and V4 of the surface envelope (Env) glycoprotein (gp90) as major neutralization epitopes, with V3 containing the principal neutralization domain of EIAV (5, 30, 36, 37). However, there has been only limited characterization of cellular immune responses to EIAV Env that are associated with protective immunity. McGuire and colleagues have thoroughly examined Gag- and Pol-specific T-helper (Th) and CTL responses (10, 22, 45, 46), but vaccines based on these broadly reactive immune determinants have failed to elicit a protective immune response against EIAV challenge (21).
Previous work with EIAV, simian immunodeficiency virus (SIV), and simian-human immunodeficiency virus (SHIV) has shown that there is a progressive maturation of Env-specific antibody responses to these various attenuated lentiviral vaccines (11-13, 25). A comparison of quantitative (titer) and qualitative (avidity and conformation) Env-specific antibodies reveals correlations between an immature immune response (low titer, low avidity, and linear epitope) and nonprotection or enhancement and between a mature immune response (high titer, high avidity, and conformational epitope) and protective vaccine immunity (11-13, 25). Based on these and other studies, we have proposed that EIAV Env is a primary determinant of vaccine efficacy and that an effective vaccine must be able to elicit broad humoral and cellular immune responses to conserved Env determinants.
Previously, we described a highly effective attenuated EIAV vaccine (EIAVD9) that protects 100% of horses from disease by homologous virulent challenge (16). In that study, we were able to identify regions of EIAV gp90 and gp45 envelope proteins that were broadly recognized by CTL and Th cells from protected horses with diverse major histocompatibility complex (MHC) haplotypes (65). It has always been assumed that Env variation poses a major obstacle to lentiviral vaccine protection; therefore, vaccine strategies have focused on the more conserved Gag proteins to elicit protective immune responses. To directly examine the role of Env variation on vaccine efficacy, 24 ponies were vaccinated with the highly effective EIAVD9 vaccine (16, 18). At 7 months postvaccination, the ponies were challenged with either a homologous Env strain (EV0) or variant Env strains with 6% (EV6) or 13% (EV13) divergence from the homologous Env. During the 120-day observation period, one EV0-, three EV6-, and five EV13-challenged ponies developed signs of equine infectious anemia (EIA), clearly revealing an inverse correlation between protection from disease and divergence from the homologous Env (17, 18). In light of the marked differences in vaccine protection observed against the variant Env challenge viruses, we hypothesized that analyses of vaccine immunity to the variant Env proteins could reveal important correlates of vaccine protection. Thus, the goal of the current study was to use this panel of experimentally immunized ponies displaying different levels of vaccine protection to evaluate potential immune correlates of protective immunity by in-depth analyses of vaccine-induced Env-specific neutralizing antibody responses, lymphoproliferative responses, and cellular cytotoxicity responses.
MATERIALS AND METHODS
Experimental vaccinations and challenge.
All equine procedures were conducted at the Gluck Equine Research Center of the University of Kentucky according to protocols approved by the University of Kentucky IACUC. Twenty-four ponies were vaccinated with the previously described attenuated EIAVD9 vaccine (16, 18, 38). Serological typing of the MHC I locus was performed as previously described (3, 4). Direct sequencing of the DRA and DQA MHC II loci was performed using previously published protocols (Table 1) (1, 20). Serum and plasma were obtained at regular intervals throughout the trial. At 7 months postvaccination, blood was drawn from each pony, and peripheral blood mononuclear cells (PBMC) were isolated and cryopreserved in autologous serum and 10% dimethyl sulfoxide (DMSO) for future use in cellular immune assays. On the following day, the vaccinated ponies (eight per group) were challenged intravenously with 103 median tissue culture infectious doses of EV0, EV6, or EV13 (eight ponies/group) (18). The challenge viruses were derived from the same reference proviral clone and differed only in the gp90 segment of the Env gene. The EV0 Env is homologous to that in the EIAVD9 vaccine strain; the EV6 and EV13 gp90 proteins diverge from EV0 by 6% and 13% in their respective amino acid sequences. The EV0, EV6, and EV13 Envs were shown to be neutralization distinct, with each being inactivated by immune sera from ponies infected with the homologous, but not heterologous, virus strain. Temperature and platelet counts were taken for 120 days postchallenge, as previously described, to monitor disease progression (18). During the observation period, one EV0-challenged pony, three EV6-challenged ponies, and five EV13-challenged ponies developed clinical signs of EIA. The remaining 15 ponies were protected from disease (Table 1).
TABLE 1.
Summary of EIAVD9-vaccinated ponies
| Pony | Sex | Age (yr) | MHC I allele | MHC II allele
|
Time of acute disease (days postchallenge) | |
|---|---|---|---|---|---|---|
| DRA | DQA | |||||
| EV0-challenged ponies | ||||||
| A27 | F | 7 | A9/W11 | 101/101 | NDa | |
| C27 | F | 5 | W11/W11 | 101/JBH11 | 301/1501/1301b | |
| C28 | F | 5 | A10/A10 | 101/101 | 301/1301 | 49 |
| C34 | F | 5 | A8/A10 | 101/201 | 201/201 | |
| E31 | M | 3 | A1/A3 | 101/101 | 301/401 | |
| E33 | M | 3 | A9/A9 | 101/101 | 701/701 | |
| E34 | F | 3 | A1/A9 | 101/JBH11 | 301/1201/1301b | |
| E35 | F | 3 | A1/A1 | 201/JBH11 | 301/701 | |
| EV6-challenged ponies | ||||||
| 558 | M | 12 | A2/A9 | 101/301 | 301/1301 | 113 |
| A32 | F | 7 | A9/A9 | 101/301 | 301/1301 | |
| A33 | M | 7 | A9/A9 | 101/101 | 301/401/1301b | 4 |
| C29 | M | 5 | A3/A9 | 101/101 | 701/701 | |
| C33 | F | 5 | A1/A1 | 201/JBH11 | 1001/1201 | |
| C36 | M | 5 | A1/A1 | 101/JBH11 | 301/1201 | 47 |
| D31 | M | 4 | A2/A9 | 101/101 | 301/301 | |
| D44 | M | 4 | A2/A9 | 101/301 | 301/1501 | |
| EV13-challenged ponies | ||||||
| 886 | F | 9 | A5/A6 | 101/101 | 1301/1301 | 70 |
| B22 | M | 6 | NDa | 101/101 | 301/401 | 14 |
| B27 | M | 6 | A5/A6 | 101/101 | 1101/1101 | 36 |
| D34 | M | 4 | A5/A9 | 201/201 | 701/1001/1301b | |
| D38 | M | 4 | A2/A3 | 101/101 | 1501/1501 | 85 |
| D39 | M | 4 | A1/A2 | 101/201 | 201/201 | 17 |
| D40 | M | 4 | A2/A2 | 101/101 | 601/1501 | |
| E30 | M | 3 | A9/W11 | 101/101 | 301/301 | |
Due to insufficient PBMC, MHC typing was not performed.
Having three MHC II sequences, including the 1303 allele, for DQA is not uncommon. See reference 20.
Neutralizing antibody assay.
The development of neutralizing antibodies to EV0, EV6, and EV13 on the day of challenge (DOC) and at 4 weeks postchallenge (4 wpc) was assessed in an enzyme-linked immunosorbent assay-based infectious-center assay as previously described (24). Briefly, 105 fetal equine kidney cells were plated in 24-well plates and incubated for 18 to 24 h at 37°C. Twofold dilutions of heat-inactivated serum (56°C, 1 h) from DOC and 4 wpc were added to 100 infectious units of EV0, EV6, or EV13. After 1 hour of incubation, the serum-virus supernatant was added to the fetal equine kidney cells in triplicate. The following day, an overlay of 0.8% carboxymethyl cellulose was added to the cells to prevent the virus from spreading throughout the culture. The cultures were incubated for an additional 7 days at 37°C before being fixed with 3.7% formaldehyde and permeabilized with 1% Triton X-100. A reference anti-EIAV primary serum antibody was used at 1:200, and a secondary antibody, horseradish peroxidase-conjugated goat anti-horse immunoglobulin G (United States Biochemical Corp.) was used at 1:5,000. The labeled cells were stained with 3-amino-9-ethyl-carbazole (Sigma) in a sodium acetate buffer (pH 5.5) supplemented with H2O2 and visualized with a dissecting microscope. The 50% neutralization titer of each serum sample was determined by linear regression analysis of the log10 reciprocal dilution versus the number of apparent foci. A neutralization titer of 25 was used as the cutoff for a positive response. Assays were repeated twice to calculate the standard error.
Production of synthetic variant peptides.
EIAV Env-specific peptides (sequential 20-mers overlapping by 10 residues) were synthesized at the Biomedical Research Support Facilities Peptide Synthesis Core of the University of Pittsburgh, using an Advanced Chemtech model 396 Omega synthesizer. Variant-specific peptides were designed using EV6 clone 567IVC8 and EV13 clone 567p10 (17, 36). Previously described UK peptides served as the EV0 peptides, since EV0 and UK have identical Env amino acid sequences (65). Seventeen EV6-specific peptides and 26 EV13-specific peptides were synthesized to account for the amino acid sequence differences with EV0 (Table 2). All peptides were dissolved at 2 mg/ml in 100% DMSO and stored at −80°C. To construct variant-specific matrix pools, the variant peptides were substituted for their EV0 counterparts and added to the conserved EV0 peptides in a matrix format as previously described (65).
TABLE 2.
Variant-specific Env peptides
| EV0 peptide sequence | Variant peptide sequencea
|
|
|---|---|---|
| EV6 | EV13 | |
| MVSIAFYGGIPGGISTPITQ | ||
| PGGISTPITQQSEKSKCEEN | ||
| QSEKSKCEENTMFQPYCYNN | QSEKSKCEENTIFQPYCYNN | |
| TMFQPYCYNNDSKNSMAESK | TIFQPYCYNNDSKNSMAESK | |
| DSKNSMAESKEARDQEMNLK | ||
| EARDQEMNLKEESKEEKRRN | ||
| EESKEEKRRNDWWKIGMFLL | ||
| DWWKIGMFLLCLAGTTGGIL | ||
| CLAGTTGGILWWYEGLPQQH | ||
| WWYEGLPQQHYIGLVAIGGR | ||
| YIGLVAIGGRLNGSGQSNAI | ||
| LNGSGQSNAIECWGSFPGCR | ||
| ECWGSFPGCRPFQNYFSYET | ||
| PFQNYFSYETNRSMHMDNNT | PFQNYFSYETNRSMHIDNNT | PFQNYFSYETNRNIHIDNNT |
| NRSMHMDNNTATLLEAYHRE | NRSMHIDNNTATLLEAYHRE | NRNIHIDNNTATLLEAYHRE |
| ATLLEAYHREITFIYKSSCT | ||
| ITFIYKSSCTDSDHCQEYQC | ||
| DSDHCQEYQCKKVNLNSSDS | DSDHCQEYQCNKVNLNLNSF | DSDHCQEYQCKKVNFTVSKA |
| KKVNLNSSDSSNPVRVEDVM | NKVNLNLNSFDSSIHVEDVK | KKVNFTVSKANGSSIPSIHVGGVEDAE |
| SNPVRVEDVMNTTEYWGFKW | DSSIHVEDVKDTTEYWGFKW | NGSSIPSHIVGGVEDAETTIEYWGFKW |
| NTTEYWGFKWLECNQTENFK | DTTEYWGFKWLECNQTENFK | TTIEYWGFKWLECNQTENLK |
| LECNQTENFKTILVPENEMV | LECNQTENLKTILVPENEMV | |
| TILVPENEMVNINDTDTWIP | TILVPENEMVNINDNDTWIA | TILVPENEMVKIKNGTWTP |
| NINDTDTWIPKGCNETWARV | NINDNDTWIAKGCNETWARV | KIKNGTWTPKGCNETWARV |
| KGCNETWARVKRCPIDILYG | ||
| KRCPIDILYGIHPIRLCVQP | ||
| IHPIRLCVQPPFFLVQEKGI | IHPIRLCVQPPFFLVQEKEV | IHPIRLCVQPPFFLVQENRG |
| PFFLVQEKGIANTSRIGNCG | PFFLVQEKEVANTSRIGNCG | PFFLVQENRGDNIARIGNCG |
| ANTSRIGNCGPTIFLGVLED | DNIARIGNCGPTIFLGVLED | |
| PTIFLGVLEDNKGVVRGNYT | PTIFLGVLEDNKGVVRENYT | PTIFLGVLEDNKGVVRGSPT |
| NKGVVRGNYTACNVSRLKIN | NKGVVRENYTACNVSHLRIN | NKGVVRGSPTACNVRKLGIN |
| ACNVSRLKINRKDYTGIYQV | ACNVSHLRINRKDYTGIYQV | ACNVRKLGINRKDYTGIYQG |
| RKDYTGIYQVPIFYTCNFTN | RKDYTGIYQGPMFYTCNFTS | |
| PIFYTCNFTNITSCNNEPII | PIFYTCNFTNITSCNNESII | PMFYTCNFTSITSCNDESIT |
| ITSCNNEPIISVIMYETNQV | ITSCNNESIISVIMYETNQV | ITSCNDESITSVIMYETNQV |
| SVIMYETNQVQYLLCNNNNS | SVIMYETNQVQYLLCKYNTT | |
| QYLLCNNNNSNNYNCVVQSF | QYLLCNNNNSNHYNCVVQSF | QYLLCKYNTTDSNYTCVVQSF |
| NNYNCVVQSFGVIGQAHLEL | NHYNCVVQSFGVIGQAHLEL | DSNYTCVVQSFGVIGQAHLEL |
| GVIGQAHLELPRPNKRIRNQ | GVIGQAHLELPRPNKRIMNP | |
| PRPNKRIRNQSFNQYNCSIN | PRPNKRIMNPNFNQYNCSIN | |
| SFNQYNCSINNKTELETWKL | NFNQYNCSINNKTELETWKL | |
| NKTELETWKLVKTSGITPLP | ||
| VKTSGITPLPISSEANTGLI | ||
| ISSEANTGLIRHKRDFGISA | ||
Variant amino acids are underlined.
Lymphoproliferation assays.
DOC PBMC from the 24 vaccinated ponies were tested for recognition of EIAV envelope peptides in a standard 7-day thymidine incorporation assay, as previously described (65). Briefly, DOC PBMC from 22 of the 24 vaccinated ponies were >80% viable upon thawing and maintained viability throughout the assays, as measured by trypan blue exclusion. DOC PBMC from ponies B22 and D34 were not viable and therefore were not used in further cellular assays. Pony C36 had abnormal DOC PBMC which yielded nonspecific hyperreactivity in the cellular assays and therefore was excluded from this study. PBMC were plated at 2 × 105 cells/well in complete RPMI 1640 (10% fetal equine serum, 1% penicillin-streptomycin, 1% l-glutamine, 55 μM β-mercaptoethanol) with 5 μM zidovudine (Sigma) and stimulated in triplicate wells with 2.5 μg/ml pokeweed mitogen (PWM), 20 μg/ml peptide matrix pools, or 10 μg/ml individual peptides. The initial scan for potentially reactive peptides utilized challenge strain-specific matrix pools to account for possible differences in T-cell recognition and reactivity. Zidovudine was replenished every other day in 5 μl of medium. Triplicate medium control wells were set up with DMSO to match the DMSO concentration in the peptide pool and peptide wells. The PBMC were incubated for 48 h with PWM and for 6 days with the peptide pools or peptides prior to being labeled with 0.75 μCi [3H]thymidine (Amersham Biosciences). After 16 to 18 h of incubation, the cells were harvested and quantified for [3H]thymidine incorporation by liquid scintillation counting. Stimulation indices (SI) were calculated by dividing the mean counts per minute (cpm) of stimulated cells by the mean cpm of nonstimulated cells. To identify all possible reactive proliferation peptides, the cutoff for peptide pools was set low, at an SI of >1. For individual peptides, we previously identified an SI of >2.5 as the cutoff for a positive proliferation response (65). Broadly reactive peptides were identified as peptides that caused positive proliferation in >50% of the ponies and that were presented by the majority of the MHC II molecules. Pony-specific peptide responses were assayed two times to calculate the standard error.
CTL assays.
DOC PBMC were used to assay CTL activity in a standard chromium release assay as previously described (65). Briefly, DOC PBMC from the vaccinated ponies were expanded in vitro for 7 to 10 days with either 2.5 μg/ml PWM (target cells) or 10 μg/ml gradient-purified EIAV (effector cells) in complete RPMI supplemented with 200 U recombinant human interleukin-2 (Hoffmann-La Roche Inc.). The target cells were labeled with 100 μCi 51Cr (Na51CrO) (MP) for 1 hour at 37°C in 5% CO2 and washed four times with 1× phosphate-buffered saline-1% horse serum. Triplicate wells of 30,000 target cells/well were pulsed with either 20 μg/ml of the peptide matrix pools or 10 μg/ml of individual peptides for 2 hours at 37°C in 5% CO2. EIAV-stimulated effector cells were added at a 20:1 effector/target (E:T) ratio and incubated for 12 to 16 h prior to being harvested. A 25-μl sample of cell supernatant was added to 175 μl of OptiPhase SuperMix scintillation fluid (Perkin-Elmer) and analyzed for 51Cr release with a MicroBeta reader (Perkin-Elmer). Maximum 51Cr release was determined by plating 51Cr-labeled target cells with the nonionic detergent Nonidet P-40 to lyse cells. Background spontaneous lysis was determined by plating 51Cr-labeled target cells with 0.1 ml of medium alone. Percent specific lysis (%SL) was calculated as follows: (51Cr release in peptide wells − spontaneous 51Cr release) × 100/(51Cr release by NP-40 − spontaneous 51Cr release) = %SL.
Effector cells were added to non-peptide-pulsed target cells to establish an average background SL of 2.7 ± 2.4 for the 12 ponies with detectable CTL activity at DOC. To identify all potentially reactive CTL peptides in the vaccinated ponies, a %SL cutoff of 5% (1 standard deviation above background) was utilized for the peptide pools. The threshold for distinguishing positive CTL peptides from nonreactive peptides was 10% (3 standard deviations above background). Broadly reactive peptides were defined as (i) being recognized by CTL in >50% of the responder ponies and (ii) having CTL recognition occurring in ponies representing the majority of MHC I haplotypes in the cohort. Pony-specific peptide responses were assayed two times to calculate the standard error.
Statistical analysis.
To determine the statistical relevance of peptide responses in the group of protected ponies compared to the nonprotected ponies, two-tailed Student's t test with unequal variances was performed. P values of <0.05 were considered statistically significant.
RESULTS
Neutralizing antibody response.
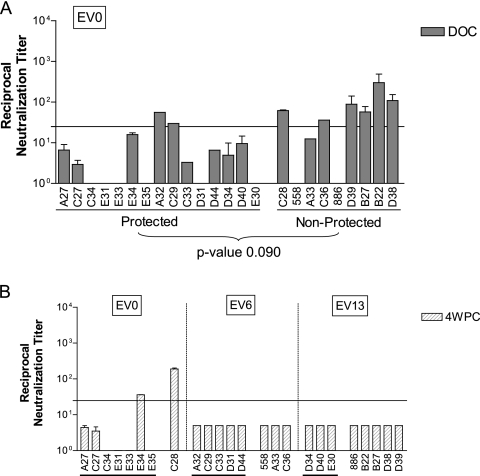
To determine if neutralizing antibodies raised against the EIAVD9 vaccine were associated with vaccine protection from disease, the level of neutralizing activity toward the vaccine envelope was measured with DOC serum from each vaccinated pony. The 24 vaccinated ponies were grouped according to those protected from disease and those that developed EIA after virulent virus challenge. The neutralizing antibody response elicited by the vaccine was quantified using DOC sera and the EV0 challenge strain, which had a homologous envelope to that of the EIAVD9 vaccine. At DOC, only 8 of the 24 vaccinated ponies had detectable neutralizing antibodies to EV0 (Fig. 1A). Only two ponies with detectable neutralizing antibodies were protected, while the other six ponies with EV0-specific neutralizing antibodies at DOC were not protected from disease after virulent challenge. While the data initially appeared to indicate that the nonprotected ponies developed better neutralizing antibody responses than did protected ponies, there was in fact no statistical difference between the neutralizing activity of the protected ponies and that of the nonprotected ponies (Student's t test P value = 0.090). Based on these assays, it appeared that EV0-specific neutralizing activity elicited by the EIAVD9 vaccine was not associated with protective vaccine immunity.
FIG. 1.
Neutralizing antibody response elicited by EIAVD9 vaccine. (A) An enzyme-linked immunosorbent assay-based infectious-center assay was used to measure EV0-specific neutralizing antibodies in DOC serum from each vaccinated pony. (B) The neutralization assay was also used to quantify the level of challenge strain-specific neutralizing antibodies in 4-wpc serum from each vaccinated pony. The ponies were grouped according to the indicated challenge strain. Underlined ponies were protected from disease.
The contribution of challenge strain-specific neutralizing antibodies to protection from disease was analyzed by measuring the level of neutralization in 4-wpc sera against the specific challenge strain for each vaccinated pony (Fig. 1B). Only two ponies, E34 and C28, had detectable challenge strain-specific neutralizing antibodies at 4 wpc; no detectable challenge strain-specific neutralizing antibodies were measured in the remaining 22 ponies. Based on the current in vitro neutralization assay, an association between vaccine protection from disease and neutralizing activity toward the respective challenge virus at 4 wpc was not evident.
Proliferation response to an EIAVD9 Env-specific peptide pool.
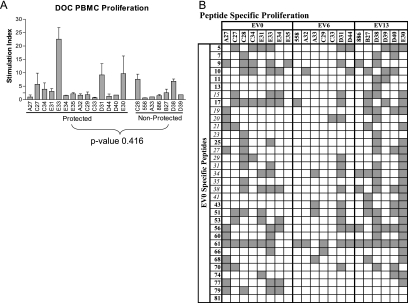
DOC PBMC from each pony were used in proliferation and CTL assays to determine if Env-specific immune responses elicited by the EIAVD9 vaccine could be associated with protection from disease. The overall Env-specific proliferative response elicited by EIAVD9 was measured in PBMC from each vaccinated pony in a [3H]thymidine incorporation assay against a comprehensive Env peptide pool. The comprehensive Env pool contained sequential EV0 Env peptides spanning the entire gp90 protein. The 21 ponies with viable DOC PBMC had variable proliferative responses to the comprehensive Env pool, as expected from an outbred population of ponies (Fig. 2A). There was no statistical difference between the proliferation responses of the protected ponies and the nonprotected ponies (Student's t test P value = 0.416), and therefore a more detailed analysis of peptide-specific proliferation was performed to determine if a relationship existed between Env-specific proliferation and protection from virulent EIAV challenge.
FIG. 2.
Env-specific proliferation responses of vaccinated ponies. (A) Proliferation against a comprehensive Env peptide pool spanning the entire EV0 gp90 sequence was tested against DOC PBMC from vaccinated ponies in a thymidine incorporation assay. An SI of 2.5 was the cutoff for a positive response. (B) Thymidine incorporation was used to measure the proliferation of DOC PBMC from the 21 EIAVD9-vaccinated ponies in response to peptides indicated by the matrix pools. Variant peptides are shown in italics, and conserved peptides are shown in bold. Shaded blocks identify ponies with positive proliferation (SI > 2.5) in response to specific peptides.
Env peptide-specific proliferation.
Challenge strain-specific peptide matrix pools were tested against the PBMC from the 21 vaccinated ponies in a thymidine incorporation assay to execute an inclusive scan of all Env peptides. Specific peptides were chosen from the matrix pools to be tested individually if they were reactive (SI > 1.0) in 11 or more of the 21 ponies tested. Following these criteria, 33 EV0-specific peptides, along with 8 EV6-specific peptides and 11 EV13-specific peptides, were indicated to be tested individually (data not shown).
To identify peptide-specific proliferation that was potentially associated with protective immunity, the 33 EV0-specific peptides indicated by the peptide matrix were tested against DOC PBMC from the 21 vaccinated ponies in a thymidine incorporation assay. Peptides 17 and 61 caused a positive proliferation response (SI > 2.5) in 11 or more of the 21 ponies (Fig. 2B) and were presented by the majority of the represented MHC II alleles. Therefore, these vaccine Env-specific peptides were considered to be broadly reactive in this group of ponies.
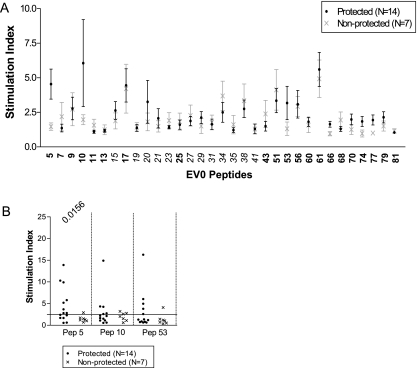
Peptide-specific proliferation responses potentially associated with vaccine protection were identified by grouping the ponies according to protection or progression to disease after virulent EIAV challenge. For each peptide, the SI of the 14 protected ponies were averaged and the SI of the 7 nonprotected ponies were averaged (Fig. 3A). A clear separation between the two groups of ponies was observed for peptides 5, 10, and 53. A more thorough analysis of these peptides was achieved by plotting the SI values of the individual ponies within the two groups (Fig. 3B). The only statistically significant difference between the individual peptide responses of the protected and nonprotected ponies was for peptide 5 (Student's t test P value = 0.0156).
FIG. 3.
Peptide-specific proliferation responses in protected and nonprotected ponies. (A) SI for each peptide from the thymidine incorporation assay were averaged for the 14 protected ponies and the 7 nonprotected ponies to highlight differences in peptide responses. Variant peptides are shown in italics, and conserved peptides are shown in bold. (B) Specific reactivity for each pony to the peptides of interest in the thymidine incorporation assay. The P value from Student's t test is listed for each statistically significant peptide.
Peptide-specific CTL reactivity.
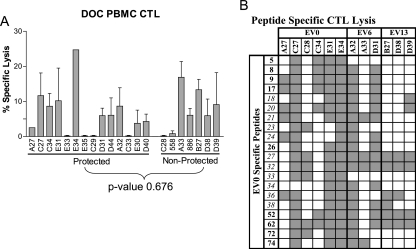
The Env-specific CTL response was examined to determine if the EIAVD9 vaccine elicited peptide-specific cytotoxic responses associated with vaccine protection from disease after virulent challenge. In an initial evaluation, the CTL responses to a comprehensive Env pool of peptides were tested in DOC PBMC from the 21 vaccinated ponies (Fig. 4A). Cytolytic activity was detected in 12 of the 21 vaccinated ponies at DOC, presumably reflecting natural fluctuations in virus-specific CTL activity during persistent EIAV infection (24). Of these 12 ponies, 7 were protected from disease and 5 developed EIA after virulent EIAV challenge. Differences in cytolytic activity between the protected and nonprotected ponies in response to the Env peptide pool were not statistically significant (Student's t test P value = 0.676).
FIG. 4.
Env-specific CTL responses of vaccinated ponies. (A) CTL lysis of DOC PBMC from vaccinated ponies pulsed with a comprehensive peptide pool spanning the entire EV0 gp90 sequence was measured in a chromium release assay. A %SL of 5 was the cutoff for a positive response. (B) Chromium release assays were used to measure CTL lysis of DOC PBMC from the 12 vaccinated ponies in response to peptides indicated by the matrix pools. Variant peptides are shown in italics, and conserved peptides are shown in bold. Shaded blocks represent positive cytolytic peptide responses (%SL > 10%) by the ponies. The E:T ratio was 20:1.
A more comprehensive study to identify individual peptide responses potentially associated with vaccine protection from disease was performed next. DOC PBMC from the 21 vaccinated ponies were tested in a chromium release assay against challenge strain-specific peptide matrix pools to identify potentially reactive CTL peptides (data not shown). Of the 21 ponies tested, 12 vaccinated ponies had detectable CTL reactivity against the peptide matrix pools. Results from the matrix pool analysis identified 20 EV0-, 9 EV6-, and 11 EV13-specific peptides. Peptide-specific CTL responses elicited by the vaccine were determined by testing the EV0 peptides against the DOC PBMC of the 12 vaccinated ponies in a chromium release assay. The nine ponies without CTL responses to the peptide pools were excluded from individual peptide analysis. Of the 20 EV0 peptides, peptides 8, 9, 17, 18, 20, 21, 27, 32, 36, 38, 52, and 62 were reactive (>10% SL) in >50% of the ponies tested (Fig. 4B). Additionally, these peptides, with the exception of peptides 18 and 21, were recognized by CTL in ponies representing the majority of the MHC I haplotypes in this cohort and were therefore considered to be broadly reactive. Peptides 8, 9, 17, 20, 27, 32, 36, 38, 52, and 62 most likely contained multiple epitopes capable of being recognized by the MHC I molecules within the represented haplotypes. Peptides 18 and 21 appeared to contain only epitopes presented by molecules within the A1, A9, and W11 haplotypes and therefore were not broadly reactive.
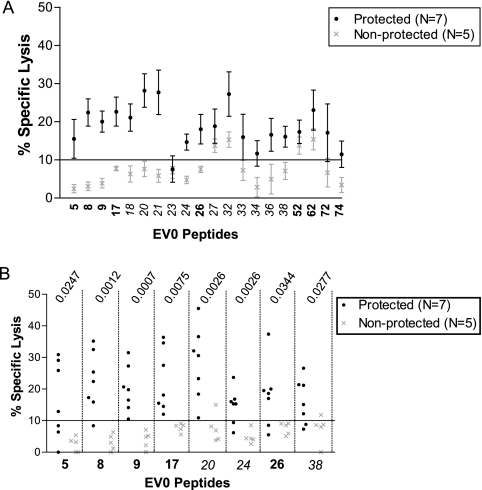
CTL responses associated with protective immunity.
Peptide-specific CTL responses potentially associated with protective vaccine immunity were identified by grouping the vaccinated ponies according to disease development. For this comparison, the %SL of the seven protected ponies and five nonprotected ponies were averaged for each reactive peptide (Fig. 5A). There was an apparent demarcation between the two groups of ponies for peptides 5, 8, 9, 17, 18, 20, 21, 24, 26, 36, and 38. Closer analysis revealed statistically significant differences between the protected and nonprotected ponies for peptides 5, 8, 9, 17, 20, 24, 26, and 38 (t test P values ranged from 0.0007 to 0.0344) (Fig. 5B). There was no statistical significance between the %SL of the protected and nonprotected ponies for peptide 36. Because peptides 18 and 21 likely contained fewer epitopes in the 20-mer sequence than the other peptides, reactivities to peptides 18 and 21 segregated with the corresponding haplotypes and not with vaccine protection. From this study, it appeared that peptides 5, 8, 9, 17, 20, 24, 26, and 38 were associated with protective vaccine immunity.
FIG. 5.
Peptide-specific CTL responses in protected and nonprotected ponies. (A) The %SL for each peptide in the chromium release assay were averaged for the seven protected ponies and the five nonprotected ponies to highlight the different peptide responses. Variant peptides are shown in italics. (B) Specific reactivity for each pony to the peptides of interest in the chromium release assay. The P value from Student's t test is listed for each statistically significant peptide. The E:T ratio was 20:1.
Effects of Env variation on immune reactivity.
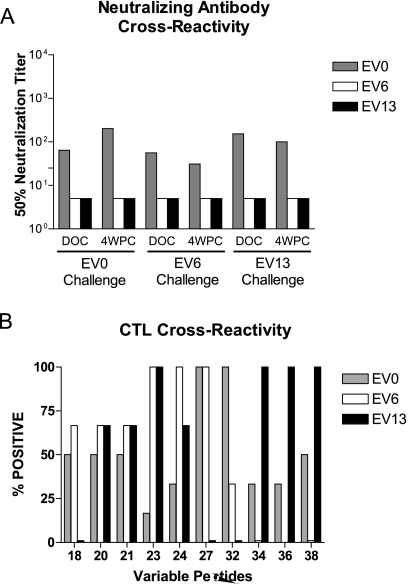
Amino acid sequence variations can alter the specificity of humoral and cellular responses to a protein antigen. The above experiments focused on the immune responses specific to the vaccine Env. To determine if these vaccine immune responses were cross-reactive with the EV6 and EV13 challenge strains, immune responses to strain-specific Env species were investigated. As demonstrated above, 8 of the 24 vaccinated ponies had EV0-specific neutralizing antibodies at DOC, and only two ponies, both of which were EV0 challenged, developed a challenge strain-specific neutralization response by 4 wpc (cf. Fig. 1A and B). To ascertain if the EIAVD9 vaccine was capable of eliciting cross-reactive neutralizing antibodies, DOC and 4-wpc sera from each vaccinated pony were tested against all three challenge strains in the cell-based neutralization assay. Data for a representative vaccinated pony from each challenge group are illustrated in Fig. 6A. Previous studies demonstrated EV0, EV6, and EV13 to be neutralization distinct, with each being inactivated by immune sera from ponies infected with the homologous, but not heterologous, virus (18). In this case, it appeared that sera from EIAVD9-vaccinated ponies are only capable of neutralizing EV0, from the homologous Env challenge strain, indicating that EIAVD9 does not induce cross-reactive neutralizing antibodies to EV6 or EV13.
FIG. 6.
Effects of Env variation on immune recognition. (A) Cross-reactivity of DOC and 4-wpc neutralizing antibodies against the three variant challenge strains. The graph shows data for a representative pony from each challenge group. (B) Variant peptides corresponding to amino acid differences between EV0, EV6, and EV13 were used in a chromium release assay against DOC PBMC from EV6- and EV13-challenged ponies. % Positive, percentage of ponies tested that had positive %SL (>10%) for the specified peptide.
For the cellular immune assays, strain-specific peptides indicated by the variant matrices were tested against DOC PBMC from the EV6- and EV13-challenged ponies to establish if cross-reactive cellular immune responses were generated by EIAVD9. In the proliferation assays, 8 EV6- and 11 EV13-specific peptides were indicated in the matrix studies. The amino acid changes in these strain-specific peptides did not alter the observed level of proliferation responses (data not shown). These data suggest that the sequence variation of these peptides does not affect PBMC proliferation, at least within the sensitivity provided by the current thymidine incorporation assay.
The CTL assays with challenge strain-specific peptide matrix pools identified 9 EV6- and 11 EV13-specific peptides. Three of the eight CTL peptides associated with vaccine protection from disease, namely, peptides 20, 24, and 38, were included in the identified variant peptides. To ascertain if these peptides were reactive against DOC PBMC from the vaccinated ponies, the variant-specific peptides identified through the matrix studies were tested against DOC PBMC from the vaccinated ponies (Fig. 6B).
While the amino acid variations for peptides 20 and 21 did not affect CTL activity, sequence variation in the remaining peptides altered the observed CTL responses (Fig. 6B). All of the ponies tested against EV0-32 had a positive CTL response, whereas PBMC from one-third of the ponies recognized EV6-32 and none of the ponies tested recognized EV13-32. For peptides 18 and 27, the EV0- and EV6-specific sequences were recognized equally by the PBMC from the vaccinated ponies, while the EV13-specific sequence did not allow CTL recognition in any of the tested ponies. Conversely, the EV0- and EV13-specific sequences for peptide 38 produced lytic activity in 50% and 100% of the ponies tested, respectively, while EV6-38 was not recognized by the PBMC from any of the vaccinated ponies tested. Fewer than 50% of the vaccinated ponies reacted to the EV0-specific sequences for peptides 23, 24, 34, and 36; however, this did not preclude recognition of EV6- or EV13-specific sequences for these peptides. More than 65% of the ponies tested had cytolytic activity in response to the EV6- and EV13-specific peptides 23 and 24. All of the ponies tested recognized the EV13-specific sequences for peptides 34 and 36 (Fig. 6B).
DISCUSSION
Using EIAV variants with divergent Env proteins to challenge EIAVD9-vaccinated ponies provided the unique opportunity to address the effects of defined Env variation on vaccine efficacy and also to identify potential determinants of protective immunity. A study by Craigo et al. (18) definitively revealed Env as a primary determinant of lentiviral vaccine efficacy, indicating a significant inverse relationship between the extent of challenge strain Env divergence from the vaccine strain Env and the observed vaccine protection. In the current study, we have taken advantage of this unique vaccine trial to characterize the humoral and cellular immune responses to Env that may be associated with vaccine protective immunity. Thus, the focus of this study was to identify potential Env determinants of protection and to address the effects of Env variation on immune recognition. Neutralizing antibody titers, Th proliferation, and CTL lysis of EIAVD9-vaccinated ponies protected from disease were compared to those of ponies that were not protected to evaluate potential determinants of vaccine immunity. For the first time, Th and CTL envelope peptides associated with protective EIAV vaccine immunity have been identified. We also illustrate how minor amino acid changes in Env can alter immune recognition and ultimately affect vaccine efficacy.
Like the case for other lentiviral model systems (HIV, SIV, and SHIV), there is considerable debate about the role of neutralizing antibodies in protective immunity. Various studies with EIAV, like those with SIV, have failed to establish a correlation between neutralizing antibody development and vaccine protection (14, 16, 23, 25, 51, 52). While a range of studies have shown that passive immunization with neutralizing antibodies can protect against HIV or SHIV challenge (40, 41, 59), there have been other studies demonstrating that passively administered immune serum cannot protect animals from SIV infection (2, 47, 51, 52, 64). When the neutralizing antibody titers of the 24 vaccinated ponies were measured against the homologous Env strain, EV0, only 8 ponies had detectable neutralization titers at DOC, but these responses could not be correlated with vaccine protection from disease. The antibodies were also specific to EV0, as they were unable to cross-neutralize EV6 or EV13. It has long been assumed that inapparent carrier ponies are protected from virulent challenge due to broadly cross-reactive neutralizing antibodies, and recent studies with other lentiviruses have reported cross-reactive neutralizing antibodies against HIV and SHIV (35, 67). Therefore, it was surprising that neutralizing antibodies generated by EIAVD9 were unable to neutralize EV6 or EV13. Because an in vitro assay was used to measure neutralization titers, the possibility that EV6 and EV13 neutralizing antibodies were present in the vaccinated ponies but undetectable in our neutralization assay could not be discounted.
With no established relationship between antibody neutralization and vaccine protection, the cellular immune response elicited by EIAVD9 was analyzed. Initially, comprehensive Env peptide pools were utilized to analyze PBMC proliferation and CTL responses in the vaccinated ponies at DOC. There was no statistical difference detected between the protected and nonprotected ponies in the response to comprehensive peptide pools in either cellular assay. Only when individual peptides were utilized did differences in cellular immune responses between the two groups of vaccinated ponies become apparent. These data suggest that using large peptide pools to track vaccine-induced cellular immunity may not have the sensitivity to differentiate between protected and nonprotected subjects and that individual peptide analysis may be required.
Effective vaccines against viruses that cause chronic infection, such as Epstein-Barr virus and cytomegalovirus, elicit polyfunctional Th and CTL responses (26). During HIV infection, there is a skewing of the Th response, such that CD4+ T cells with proliferative capabilities are destroyed, while CD4+ T cells secreting gamma interferon are abundant (58). Targeting an appropriate Th proliferation response is therefore a desirable quality for lentiviral vaccines. The thymidine incorporation assay for PBMC proliferation identified one Env-specific peptide which was associated with vaccine protection from disease. Peptide 5 was located in a more conserved region of gp90 (Fig. 7) and had an identical amino acid sequence across the three variant challenge strains. CTL analysis also determined that cytolytic activity to peptide 5 was associated with vaccine protection from disease. With the current assays, we cannot determine if peptide 5 contains both CTL and T-helper epitopes or if the observed proliferation response was due to CTL expansion. As equine-specific reagents become available to study the cytokine profile of the Th cells, it will be of interest to study the cytokine response evoked by the EIAVD9 vaccine to determine if there is an association with vaccine immunity.
FIG. 7.
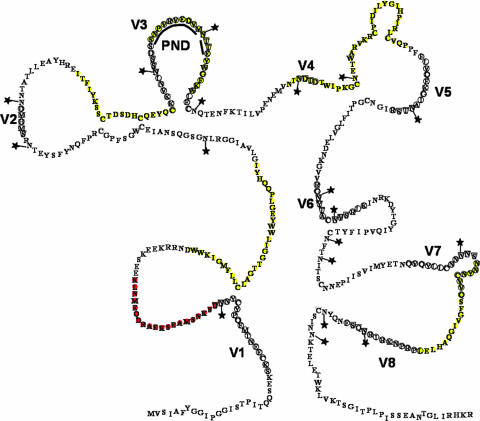
Env-specific peptide responses associated with vaccine protection. A schematic model of gp90 is shown. Variable domains are circled and labeled V1 to V8. N-linked glycosylation sites are marked with stars. PND is the principal neutralization domain. CTL peptides associated with protection are highlighted in yellow. The Th and CTL peptide 5 is highlighted in red. (Adapted from reference 5 with permission.)
The proliferation assay also distinguished broadly reactive regions of EIAV Env. A group of peptides was recently described as broadly reactive for 12 EIAVD9-vaccinated horses demonstrating protective immunity from disease (65). One of the peptides identified in the current trial, peptide 61, overlaps with a previously described broadly reactive peptide, peptide 60. Additionally, when the two EIAVD9 trials are combined, peptides 9, 17, 27, and 70 can be described as broadly reactive peptides for the two groups of ponies. It should be noted that ponies having the same DRA/DQA alleles in this study did not always have the same peptide-specific proliferation responses. This may be attributed to the third MHC II allele, DRB. There is not complete sequence information for DRB, and therefore it was not characterized in these ponies (19). As with a Th response, a polyfunctional CTL response is regarded as necessary for a protective lentiviral vaccine (28). For HIV, various studies have shown a correlation between CTL levels, long-term nonprogressors (27, 61), and control of acute (9, 34) and chronic (53) viremia. Additionally, in the SIV model of infection, depletion of CD8+ T cells results in a loss of immune control (32, 42, 63). While several peptides associated with Mamu A*01 have been correlated with SIV vaccine protection (6, 7, 33), broadly reactive CTL peptides associated with HIV or SIV vaccine protection have yet to be identified. The bulk of HIV and SIV CTL research has relied upon gamma interferon and tetramer staining, but it is becoming increasingly clear that determining the functional diversity of the CTL response is equally important to quantifying the CTL response by these techniques (54, 66). In the current study, a functional chromium release assay was utilized to test PBMC from 12 vaccinated ponies with measurable DOC CTL activity to identify potentially protective CTL peptide-specific responses elicited by the EIAVD9 vaccine. Broadly reactive CTL peptides elicited by the EIAVD9 vaccine were previously described (65). Again, without a more discriminating assay to type the ELA-A allele, firm MHC restrictions cannot be assigned to identified peptides (65, 68). However, the peptides identified in the current vaccine trial do not seem to be MHC restricted and confirm four of the broadly reactive peptides from the previous trial, i.e., peptides 9, 17, 27, and 52 (65). Additionally, when data from the two EIAVD9 trials are combined, peptides 5, 32, and 62 are also found to be broadly reactive.
In defining CTL peptide responses associated with vaccine protection from disease, our analysis revealed a statistically significant association between protection from disease and CTL responses to peptides 5, 8, 9, 17, 20, 24, 26, and 38. In an independent study, lipopeptides corresponding to the amino acid sequences of EIAV gp90 peptides 20 and 21 were used to vaccinate three horses. While the vaccine did not prevent infection, it was capable of reducing the severity of disease, indicating that the CTL responses associated with these peptides can have some protective effect (60). The cytolytic response to peptides 5, 8, 9, 17, 20, 24, 26, and 38 can be targeted in future lentiviral vaccine trials to enhance protective immune responses.
Five (peptides 5, 8, 9, 17, and 26) of the eight CTL peptides associated with vaccine protection from disease were found in more conserved regions of gp90 (Fig. 7), with each challenge strain having identical amino acid sequences for these five peptides. Additionally, there were no N-linked glycosylation sites found in these five peptides. The three other CTL peptides were variable between the three challenge strains. CTL peptide 20 spanned the V3 region of gp90, peptide 24 included the V4 region, and peptide 38 contained part of the V7 region. The variant challenge strains each had unique amino acid sequences for these peptides and were also variable in the number and placement of N-linked glycosylation sites.
While variation affected numerous challenge strain peptide-specific CTL responses, the amino acid changes present in peptide 20 did not drastically alter CTL reactivity (Fig. 6B and Table 3). This was quite surprising, as EV13-20 contained seven additional amino acids. This indicates that the CTL epitope for peptide 20 is most likely in the C terminus. There were minimal differences between the three challenge strains for peptide 24, but the amino acid changes were enough to drastically alter the cytolytic activity compared to the observed EV0 response. The most surprising observation was with peptide 38. The majority of the ponies tested responded to the EV0- and EV13-specific peptides but not to the EV6-specific sequence. There was one amino acid difference between EV0 and EV6 in this peptide. The second amino acid was changed from an asparagine (N) to a histidine (H). This indicates that N may be a critical anchor residue for peptide 38. While the EV13 sequence does not have an N at position 2, there is an N at position 3, which could serve as the anchor for that peptide. These data illustrate how a single amino acid substitution within a CTL epitope can abrogate recognition, either by decreasing the binding affinity of the peptide for the MHC I molecule or by altering the peptide-MHC complex so that the T-cell receptor cannot bind (15, 57). Without knowing the necessary anchor residues for MHC I presentation, we could not determine if the loss in CTL reactivity was due to low peptide-MHC affinity or to the inability of the T-cell receptor to recognize the variant peptide-MHC complex. Further studies are required to define the cause of CTL reactivity loss. While Env variation may have profound effects on CTL recognition, these data also suggest that CTL are capable of recognizing viral sequences not present in the original vaccine inoculum (39).
TABLE 3.
Variant peptide sequences and effects on CTL reactivity
| Peptide | Sequencea | CTL reactivityb |
|---|---|---|
| EV0-18 | DSDHCQEYQCKKVNLNSSDS | + |
| EV6-18 | ****************LNSF | + |
| EV13-18 | **************FTV*KA | − |
| EV0-20 | SNP------VR-VEDVMNTTEYWGFKW | + |
| EV6-20 | DSS------IH-****KD********* | + |
| EV13-20 | NGSSIPSIHVGG***AE**I******* | + |
| EV0-21 | NTTEYWGFKWLECNQTENFK | + |
| EV6-21 | D******************* | + |
| EV13-21 | **I***************L* | + |
| EV0-23 | TILVPENEMVNINDTDTWIP | − |
| EV6-23 | **************N****A | ++ |
| EV13-23 | **********K*KN-G**T* | ++ |
| EV0-24 | NINDTDTWIPKGCNETWARV | − |
| EV6-24 | ****N****A********** | ++ |
| EV13-24 | K*KN-G**T*********** | + |
| EV0-27 | IHPIRLCVQPPFFLVQEKGI | ++ |
| EV6-27 | ******************EV | ++ |
| EV13-27 | *****************NRG | − |
| EV0-32 | ACNVSRLKINRKDYTGIYQV | ++ |
| EV6-32 | *****H*R************ | − |
| EV13-32 | ****RK*G************ | − |
| EV0-34 | PIFYTCNFTNITSCNNEPII | − |
| EV6-34 | *****************S** | − |
| EV13-34 | *M*******S*****D*S*T | ++ |
| EV0-36 | SVIMYETNQVQYLLCNNNNS | − |
| EV13-36 | ***************KY*TT | ++ |
| EV0-38 | NNYNCVVQSFGVIGQAHLEL | + |
| EV6-38 | *H****************** | − |
| EV13-38 | DSN*T*************** | ++ |
Asterisks denote conserved amino acids, and dashes denote deleted amino acids.
Average specific lysis of >10% (+), >15% (++), or <10% (−).
In conclusion, the current study used the EIAV vaccine system to evaluate potential Env-specific immune responses associated with the development of protective vaccine-mediated immunity. While the current study focused on the critical role of Env in vaccine protection, it should be emphasized that this study does not preclude a role for Gag- or Pol-specific immune responses in vaccine protection. As discussed previously by Craigo et al. (18), it is possible and likely that other virus-specific immune responses are necessary but insufficient for vaccine protection. Further experiments are necessary to define the relative contributions of Env-, Gag-, and Pol-specific immunity to vaccine protection by attenuated EIAV and to determine the ability of exclusive Env-specific vaccine immunity to protect against EIAV infection and disease. These studies can provide further insights into the critical determinants of EIAV vaccine efficacy that may be applicable to other lentiviruses, including HIV-1.
Acknowledgments
This work was funded by NIH/NIAID grant RO1 AI25850, by funds from the Lucille P. Markey Charitable Trust, and by the University of Kentucky Agricultural Experiment Station.
We thank Shannon Barnes and Timothy Sturgeon for helping with the extensive amount of blood processing.
Footnotes
Published ahead of print on 30 January 2008.
REFERENCES
- 1.Albright-Fraiser, D. G., R. Reid, V. Graber, and E. Bailey. 1996. Polymorphism of DRA among equids. Immunogenetics 43315-317. [PubMed] [Google Scholar]
- 2.Almond, N., R. Rose, R. Sangster, P. Silvera, R. Stebbings, B. D. Walker, and E. J. Stott. 1997. Mechanisms of protection induced by attenuated simian immunodeficiency virus. I. Protection cannot be transferred with immune serum. J. Gen. Virol. 781919-1922. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, E. 1980. Identification and genetics of horse lymphocyte alloantigens. Immunogenetics 11499-506. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, E. 1983. Population studies on the ELA system in American standardbred and thoroughbred mares. Anim. Blood Groups Biochem. Genet. 14201-211. [DOI] [PubMed] [Google Scholar]
- 5.Ball, J. M., K. E. Rushlow, C. J. Issel, and R. C. Montelaro. 1992. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J. Virol. 66732-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barouch, D. H., T. M. Fu, D. C. Montefiori, M. G. Lewis, J. W. Shiver, and N. L. Letvin. 2001. Vaccine-elicited immune responses prevent clinical AIDS in SHIV(89.6P)-infected rhesus monkeys. Immunol. Lett. 7957-61. [DOI] [PubMed] [Google Scholar]
- 7.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T.-M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M.-E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290486-492. [DOI] [PubMed] [Google Scholar]
- 8.Bogers, W. J. M., C. Cheng-Mayer, and R. C. Montelaro. 2000. Developments in preclinical AIDS vaccine efficacy models. AIDS 45141-5152. [PubMed] [Google Scholar]
- 9.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 686103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, C., R. H. Mealey, and T. C. McGuire. 2005. Evaluation of high functional avidity CTL to Gag epitope clusters in EIAV carrier horses. Virology 342228-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clements, J. E., R. C. Montelaro, M. C. Zink, A. M. Amedee, S. Miller, A. M. Trichel, B. Jagerski, D. Hauer, L. N. Martin, and R. P. Bohm. 1995. Cross-protective immune responses induced in rhesus macaques by immunization with attenuated macrophage-tropic simian immunodeficiency virus. J. Virol. 692737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, K. S., M. Murphey-Corb, O. Narayan, S. V. Joag, G. M. Shaw, and R. C. Montelaro. 1998. Common themes of antibody maturation to simian immunodeficiency virus, simian-human immunodeficiency virus, and human immunodeficiency virus type 1 infections. J. Virol. 727852-7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole, K. S., J. L. Rowles, B. A. Jagerski, M. Murphey-Corb, T. Unangst, J. E. Clements, J. Robinson, M. S. Wyand, R. C. Desrosiers, and R. C. Montelaro. 1997. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J. Virol. 715069-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor, R. I., D. C. Montefiori, J. M. Binley, et al. 1998. Temporal analyses of virus replication, immune responses, and efficacy in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Virol. 727501-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couillin, I., B. Culmann-Penciolelli, E. Gomard, J. Choppin, J. P. Levy, J. G. Guillet, and S. Saragosti. 1994. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J. Exp. Med. 1801129-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craigo, J. K., S. Durkin, T. J. Sturgeon, T. Tagmyer, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2007. Immune suppression of challenged vaccinates as a rigorous assessment of sterile protection by lentiviral vaccines. Vaccine 25834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craigo, J. K., T. J. Sturgeon, S. J. Cook, C. J. Issel, C. Leroux, and R. C. Montelaro. 2006. Apparent elimination of EIAV ancestral species in a long-term inapparent carrier. Virology 344340-353. [DOI] [PubMed] [Google Scholar]
- 18.Craigo, J. K., B. S. Zhang, S. Barnes, T. L. Tagmyer, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2007. Envelope variation as a primary determinant of lentiviral vaccine efficacy. Proc. Natl. Acad. Sci. USA 10415105-15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser, D. G., and E. Bailey. 1996. Demonstration of three DRB loci in a domestic horse family. Immunogenetics 44441. [DOI] [PubMed] [Google Scholar]
- 20.Fraser, D. G., and E. Bailey. 1998. Polymorphism and multiple loci for the horse DQA gene. Immunogenetics 47487-490. [DOI] [PubMed] [Google Scholar]
- 21.Fraser, D. G., S. R. Leib, B. S. Zhang, R. H. Mealey, W. C. Brown, and T. C. McGuire. 2005. Lymphocyte proliferation responses induced to broadly reactive Th peptides did not protect against equine infectious anemia virus challenge. Clin. Diagn. Lab. Immunol. 12983-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraser, D. G., J. L. Oaks, W. C. Brown, and T. C. McGuire. 2002. Identification of broadly recognized, T helper 1 lymphocyte epitopes in an equine lentivirus. Immunology 105295-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gundlach, B. R., S. Reiprich, S. Sopper, R. E. Means, U. Dittmer, K. Matz-Rensing, C. Stahl-Hennig, and K. Uberla. 1998. Env-independent protection induced by live attenuated simian immunodeficiency virus vaccines. J. Virol. 727846-7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond, S. A., S. J. Cook, D. L. Lichtenstein, C. J. Issel, and R. C. Montelaro. 1997. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J. Virol. 713840-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond, S. A., M. L. Raabe, C. J. Issel, and R. C. Montelaro. 1999. Evaluation of antibody parameters as potential correlates of protection or enhancement by experimental vaccines to equine infectious anemia virus. Virology 262416-430. [DOI] [PubMed] [Google Scholar]
- 26.Harari, A., S. Petitpierre, F. Vallelian, and G. Pantaleo. 2003. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1 infected subjects with progressive disease: changes after antiretroviral therapy. Blood 103966-972. [DOI] [PubMed] [Google Scholar]
- 27.Harrer, T., E. Harrer, S. A. Kalams, T. Elbeik, S. I. Staprans, M. B. Feinberg, Y. Cao, D. D. Ho, T. Yilma, and A. M. Caliendo. 1996. Strong cytotoxic T cell and weak neutralizing antibody responses in a subset of persons with stable nonprogressing HIV type 1 infection. AIDS Res. Hum. Retrovir. 12585. [DOI] [PubMed] [Google Scholar]
- 28.Heeney, J. L., and S. A. Plotkin. 2006. Immunological correlates of protection from HIV infection and disease. Nat. Immunol. 71281-1284. [DOI] [PubMed] [Google Scholar]
- 29.Howe, L., J. K. Craigo, C. J. Issel, and R. C. Montelaro. 2005. Specificity of serum neutralizing antibodies induced by transient immune suppression of inapparent carrier ponies infected with a neutralization-resistant equine infectious anemia virus envelope strain. J. Gen. Virol. 86139-149. [DOI] [PubMed] [Google Scholar]
- 30.Howe, L., C. Leroux, C. J. Issel, and R. C. Montelaro. 2002. Equine infectious anemia virus envelope evolution in vivo during persistent infection progressively increases resistance to in vitro serum antibody neutralization as a dominant phenotype. J. Virol. 7610588-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Issel, C. J., and L. Coggins. 1979. Equine infectious anemia virus: current knowledge. J. Am. Vet. Med. Assoc. 174727-733. [PubMed] [Google Scholar]
- 32.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson, P. R., B. C. Schnepp, M. J. Connell, D. Rohne, S. Robinson, G. R. Krivulka, C. I. Lord, R. Zinn, D. C. Montefiori, N. L. Letvin, and K. R. Clark. 2005. Novel adeno-associated virus vector vaccine restricts replication of simian immunodeficiency virus in macaques. J. Virol. 79955-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune response with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 684650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraft, Z., N. R. Derby, R. A. McCaffrey, R. Niec, W. M. Blay, N. L. Haigwood, E. Moysi, C. J. Saunders, T. Wrin, C. J. Petropoulos, M. J. McElrath, and L. Stamatatos. 2007. Macaques infected with a CCR5-tropic simian/human immunodeficiency virus (SHIV) develop broadly reactive anti-HIV neutralizing antibodies. J. Virol. 816402-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leroux, C., J. K. Craigo, C. J. Issel, and R. C. Montelaro. 2001. Equine infectious anemia virus genomic evolution in progressor and nonprogressor ponies. J. Virol. 754570-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leroux, C., C. J. Issel, and R. C. Montelaro. 1997. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J. Virol. 719627-9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, F., J. K. Craigo, L. Howe, J. D. Steckbeck, S. Cook, C. Issel, and R. C. Montelaro. 2003. A live attenuated equine infectious anemia virus proviral vaccine with a modified S2 gene provides protection from detectable infection by intravenous virulent virus challenge of experimentally inoculated horses. J. Virol. 777244-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malhotra, U., F. Li, J. Nolin, M. Allison, H. Zhao, J. I. Mullins, S. Self, and M. J. McElrath. 2007. Enhanced detection of human immunodeficiency virus type 1 (HIV-1) Nef-specific T cells recognizing multiple variants in early HIV-1 infection. J. Virol. 815225-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mascola, J., M. G. Lewis, G. Stiegler, D. Harris, T. VanCott, D. Hayes, M. Louder, C. Brown, C. Sapan, S. Frankel, Y. Lu, M. Robb, H. Katinger, and D. Birx. 1999. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6PD by passive transfer of neutralizing antibodies. J. Virol. 734009-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mascola, J., G. Stiegler, T. VanCott, H. Katinger, C. Carpenter, C. Hanson, H. Beary, D. Hayes, S. Frankel, D. Birx, and M. G. Lewis. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6207-210. [DOI] [PubMed] [Google Scholar]
- 42.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGuire, T. C., T. B. Crawford, and J. B. Henson. 1971. Immunofluorescent localization of equine infectious anemia virus in tissue. Am. J. Pathol. 62283-294. [PMC free article] [PubMed] [Google Scholar]
- 44.McGuire, T. C., D. B. Tumas, K. M. Byrne, M. T. Hines, S. R. Leib, A. L. Brassfield, K. I. O'Rourke, and L. E. Perryman. 1994. Major histocompatibility complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J. Virol. 681459-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mealey, R. H., A. Sharif, S. A. Ellis, M. H. Littke, S. R. Leib, and T. C. McGuire. 2005. Early detection of dominant Env-specific and subdominant Gag-specific CD8+ lymphocytes in equine infectious anemia virus-infected horses using major histocompatibility complex class I/peptide tetrameric complexes. Virology 339110-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mealey, R. H., B. Zhang, S. R. Leib, M. H. Littke, and T. C. McGuire. 2003. Epitope specificity is critical for high and moderate avidity cytotoxic T lymphocytes associated with control of viral load and clinical disease in horses with equine infectious anemia virus. Virology 313537-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Metzner, K. J., W. J. Moretto, S. M. Donahoe, X. Jin, A. Gettie, D. C. Montefiori, P. A. Marx, J. M. Binley, D. F. Nixon, and R. I. Connor. 2005. Evaluation of CD8+ T-cell and antibody responses following transient increased viremia in rhesus macaques infected with live, attenuated simian immunodeficiency virus. J. Gen. Virol. 863375-3384. [DOI] [PubMed] [Google Scholar]
- 48.Montelaro, R. C., J. M. Ball, and K. E. Rushlow. 1993. Equine retroviruses, p. 257-360. In J. A. Levy (ed.), The Retroviridae. Plenum Press, New York, NY.
- 49.Montelaro, R. C., and D. P. Bolognesi. 1995. Vaccines against retroviruses, p. 605-656. In J. A. Levy (ed.), The Retroviridae, vol. 4. Plenum Press, New York, NY. [Google Scholar]
- 50.Montelaro, R. C., K. S. Cole, and S. A. Hammond. 1998. Maturation of immune responses to lentivirus infection: implications for AIDS vaccine development. AIDS Res. Hum. Retrovir. 14S225-S229. [PubMed] [Google Scholar]
- 51.Nilsson, C., B. Makitalo, R. Thorstensson, S. Norley, D. Binninger-Schinzel, M. Crange, E. Rud, G. Biberfeld, and P. Putkonen. 1998. Live attenuated simian immunodeficiency virus (SIV)mac in macaques can induce protection against mucosal infection with SIVsm. AIDS 122261-2270. [DOI] [PubMed] [Google Scholar]
- 52.Norley, S., B. Beer, D. Binninger-Schinzel, C. Cosma, and R. Kurth. 1996. Protection from pathogenic SIVmac challenge following short-term infection with a nef-deficient attenuated virus. Virology 219195-205. [DOI] [PubMed] [Google Scholar]
- 53.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 2792103-2106. [DOI] [PubMed] [Google Scholar]
- 54.Pantaleo, G., and R. A. Koup. 2004. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat. Med. 10806-810. [DOI] [PubMed] [Google Scholar]
- 55.Payne, S. L., F. D. Fang, C. P. Liu, B. R. Dhruva, P. Rwambo, C. J. Issel, and R. C. Montelaro. 1987. Antigenic variation and lentivirus persistence: variations in envelope gene sequences during EIAV infection resemble changes reported for sequential isolates of HIV. Virology 161321-331. [DOI] [PubMed] [Google Scholar]
- 56.Perryman, L. E., K. O'Rourke, and T. C. McGuire. 1988. Immune responses are required to terminate viremia in equine infectious anemia lentivirus infection. J. Virol. 623073-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. M. Bangham, C. R. Rizza, and A. J. McMichael. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354453-459. [DOI] [PubMed] [Google Scholar]
- 58.Pitcher, C. J., C. Quittner, D. M. Peterson, M. Connors, R. A. Koup, V. C. Maino, and L. J. Picker. 1999. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 5518-525. [DOI] [PubMed] [Google Scholar]
- 59.Polacino, P., V. Stallard, D. C. Montefiori, C. R. Brown, B. A. Richardson, W. R. Morton, R. E. Benveniste, and S.-L. Hu. 1999. Protection of macaques against intrarectal infection by a combination immunization regimen with recombinant simian immunodeficiency virus SIVmne gp160 vaccines. J. Virol. 733134-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridgely, S. L., B. Zhang, and T. C. McGuire. 2003. Response of ELA-A1 horses immunized with lipopeptide containing an equine infectious anemia virus ELA-A1-restricted CTL epitope to virus challenge. Vaccine 21491-506. [DOI] [PubMed] [Google Scholar]
- 61.Rinaldo, C., X. L. Huang, Z. F. Fan, M. Ding, L. Beltz, A. Logar, D. Panicali, G. Mazzara, J. Liebmann, and M. Cottrill. 1995. High levels of anti-human immunodeficiency virus type 1 (HIV-1) memory cytotoxic T-lymphocyte activity and low viral load are associated with lack of disease in HIV-1-infected long-term nonprogressors. J. Virol. 695838-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rwambo, P. M., C. J. Issel, K. A. Hussain, and R. C. Montelaro. 1990. In vitro isolation of a neutralization escape mutant of equine infectious anemia virus (EIAV). Arch. Virol. 111275-280. [DOI] [PubMed] [Google Scholar]
- 63.Schmitz, J., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283857-860. [DOI] [PubMed] [Google Scholar]
- 64.Sharpe, S. A., A. Cope, S. Dowall, N. Berry, C. Ham, J. L. Heeney, D. Hopkins, L. Easterbrook, M. Dennis, N. Almond, and M. Cranage. 2004. Macaques infected long-term with attenuated simian immunodeficiency virus (SIVmac) remain resistant to wild-type challenge, despite declining cytotoxic T lymphocyte responses to an immunodominant epitope. J. Gen. Virol. 852591-2602. [DOI] [PubMed] [Google Scholar]
- 65.Tagmyer, T. L., J. K. Craigo, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2007. Envelope-specific T-helper and cytotoxic T-lymphocyte responses associated with protective immunity to equine infectious anemia virus. J. Gen. Virol. 881324-1336. [DOI] [PubMed] [Google Scholar]
- 66.Yang, O. O., P. T. N. Sarkis, A. Trocha, S. A. Kalams, R. P. Johnson, and B. D. Walker. 2003. Impacts of avidity and specificity on the antiviral efficiency of HIV-1-specific CTL. J. Immunol. 1713718-3724. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, P. F., F. Cham, M. Dong, A. Choudhary, P. Bouma, Z. Zhang, Y. Shao, Y.-R. Feng, L. Wang, N. Mathy, G. Voss, C. C. Broder, and G. V. Quinnan, Jr. 2007. Extensively cross-reactive anti-HIV-1 neutralizing antibodies induced by gp140 immunization. Proc. Natl. Acad. Sci. USA 10410193-10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, W., S. M. Lonning, and T. C. McGuire. 1998. Gag protein epitopes recognized by ELA-A-restricted cytotoxic T lymphocytes from horses with long-term equine infectious anemia virus infection. J. Virol. 729612-9620. [DOI] [PMC free article] [PubMed] [Google Scholar]