Abstract
Previous genetic and biochemical studies performed with several members of the Alphaherpesvirus subfamily have shown that the UL31 and UL34 proteins are essential components of the molecular machinery that mediates the primary egress of newly assembled capsids across the nuclear membrane. Further, there is substantial evidence that BFLF2 and BFRF1, the respective positional homologs of UL31 and UL34 in the Epstein-Barr virus (EBV) genome, are also their functional homologs, i.e., that the UL31/UL34 pathway is common to distant herpesviruses. However, the low degree of protein sequence identity between UL31 and BFLF2 would argue against such a hypothesis. To further clarify this issue, we have constructed a recombinant EBV strain devoid of BFLF2 (ΔBFLF2) and show that BFLF2 is crucial for efficient virus production but not for lytic DNA replication or B-cell transformation. This defective phenotype could be efficiently restored by trans complementation with a BFLF2 expression plasmid. Detailed analysis of replicating cells by electron microscopy revealed that, as expected, ΔBFLF2 viruses not only failed to egress from the nucleus but also showed defective DNA packaging. Nonfunctional primary egress did not, however, impair the production and extracellular release of enveloped but empty viral particles that comprised L particles containing tegument-like structures and a few virus-like particles carrying empty capsids. The ΔBFLF2 and ΔUL31 phenotypes therefore only partly overlap, from which we infer that BFLF2 and UL31 have substantially diverged during evolution to fulfil related but distinct functions.
The Epstein-Barr virus (EBV), a member of the gammaherpesvirus family, establishes lifelong clinical latency (or passive latency) in its hosts. In the absence of an effective cytotoxic T-cell response, EBV-infected B cells undergo permanent proliferation upon the synthesis of the EBV nuclear antigen (EBNA) and LMP latent protein family members (also called active latency) (28). The detection of latent gene products in EBV-associated human cancers established a link between neoplastic transformation and active latency (28). Consequently, latent genes have been and continue to be studied in great detail. In contrast, knowledge of the molecular mechanisms that underlie EBV lytic replication and maturation is not as deep as it is for alphaherpesviruses such as herpes simplex virus type 1 (HSV-1) or pseudorabies (PrV). It is, however, unlikely that information gathered from the study of alphaherpesviruses can immediately be translated to EBV, even if the two virus types show a substantial degree of homology. This might be relevant for therapeutic strategies that aim at targeting viral genes and proteins involved in virus replication and infection with, e.g., inhibitory peptides or small interfering RNA.
Electron microscopy studies have led to the identification of several substructures within mature herpesvirus virions. The icosahedral viral capsid surrounds the inner nucleoprotein core, which contains the electron-dense packaged linear DNA (11, 30). Upon maturation, these viral components become enveloped in a cellular bilayered membrane into which virus glycoproteins are inserted (12, 23). The structure between the capsid and envelope, the tegument, even if more difficult to individualize morphologically, has been shown to be well organized and to host a variety of proteins with key functions during virus maturation and infection of target cells (30). Virus production results from a coordinated process requiring sequential waves of protein synthesis (immediate-early, early, and late genes) that allow newly replicated DNA to be packaged into the preassembled capsid and its internal nucleocore (28). These immature viral particles are then submitted to several rounds of envelopment-deenvelopment that allow sequential crossings of several cellular membranes until the virus reaches the extracellular milieu (23). Previous work with HSV-1 and PrV mutants has shown that crossing of the nuclear membrane, or primary egress, requires interaction between two conserved proteins, membrane protein UL34 and nuclear phosphoprotein UL31 (2, 8, 17, 27, 31, 32). These two proteins are sufficient to induce the formation of primary envelopes derived from the inner nuclear membrane in which nucleocapsids are wrapped, and genetic mutants lacking one of these proteins are sequestered in the nucleus (16). A similar scenario has been suggested for EBV, based on the identified interaction between the UL34 positional homolog BFRF1 and the UL31 positional homolog BFLF2 and the observation that UL34 or BFRF1 mutant viruses (ΔBFRF1) remain trapped in the nucleus (1, 5, 9, 18). However, the weak homology at the protein level between UL31 and BFLF2 and their different timings of expression, with BFLF2 being expressed early and all other UL31 herpesvirus homologs being expressed late during lytic replication (9), suggest a functional divergence between the two proteins. To clarify these issues, we have generated a mutant virus devoid of the gene for BFLF2 and report here its phenotypic traits, including lytic replication, infection, and B-cell immortalization in vitro.
MATERIALS AND METHODS
Eukaryotic and prokaryotic cells.
Raji is an EBV-positive Burkitt's lymphoma cell line. B95.8 is an EBV-immortalized marmoset monkey lymphoblastoid cell line (LCL) that spontaneously replicates. Recombinant EBV-wt B95.8 genomes stably transfected into 293 cells (2089; 293/EBV-wt) have been previously described (4). Peripheral blood mononuclear cells were purified from fresh buffy coat by density gradient centrifugation, and CD19-positive B cells were isolated with M-450 CD19 (Pan B) Dynabeads (Dynal), followed by detachment of the B cells with Detachabead (Dynal). LCLs were established by EBV infection of primary B cells (see below). All cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum (Biochrom). DH10B is a recA-negative Escherichia coli K-12 strain that tolerates the replication of large plasmids.
Plasmids and BFLF2 targeting vector.
p2089 is a plasmid comprising the EBV wild-type genome and the F-factor origin of replication onto which the chloramphenicol (CAM) and hygromycin resistance genes, as well as the gene encoding the green fluorescent protein (GFP), were cloned (4). The gene for kanamycin (KAN) flanked by FLP recombinase binding sites was amplified from plasmid pCP15 by a PCR-based method (26) with primers whose 5′ parts are specific to the flanking regions of BFLF2 (B95.8 coordinates 55955 to 55994 and 56975 to 56936) and whose 3′ parts are specific for KAN. With this PCR product as a targeting vector, the first 314 out of the BFLF2 318 amino acids were deleted in E. coli by homologous recombination (see below). A BFLF2 expression plasmid was constructed by cloning a 953-bp PCR fragment amplified from B95.8 genomic DNA that exactly encompasses the BFLF2 open reading frame into pRK5, as confirmed by sequencing. A BFLF1 expression plasmid was constructed by cloning a 1,574-bp PCR fragment encompassing the BFLF1 open reading frame into pRK5.
Recombinant EBV plasmid and 293 cell clones.
To generate a BFLF2-negative mutant, temperature-sensitive plasmid pKD46, which carries the lambda red recombinase under the control of an arabinose-inducible promoter and the ampicillin (AMP) resistance cassette, was transiently introduced into E. coli DH10B cells previously transformed with p2089 and plated at 30°C on agar plates containing l(+)-arabinose (0.1%), AMP, and CAM to allow selection of DH10B cells containing both p2089 and pKD46 (26). These bacterial cells were then rendered electrocompetent and transformed with 2 μg of the BFLF2 targeting vector. Transformed cells were plated on CAM and KAN agar plates at 42°C to select for successfully recombined genomes and induce the elimination of pKD46. Plasmid DNA from antibiotic-resistant clones was prepared and analyzed with appropriate restriction enzymes to identify clones with the correct recombination pattern. The KAN resistance gene was excised with the FLP recombinase cloned into temperature-sensitive plasmid pCP20 (3), which also carries the AMP resistance gene. The bacterial clones that resulted from selection on CAM/AMP plates were further grown on CAM plates at 42°C to induce the loss of plasmid pCP20. Resistant clones were then submitted to restriction analysis to confirm the expected restriction pattern. Recombinant DNA from p2089 was digested in parallel and provided an appropriate control. 293 cells were then transfected with the properly recombined viral DNA with Lipofectamine (Invitrogen) as previously described (14). Selection of stable 293 cell clones carrying the EBV recombinant plasmid was performed with hygromycin (100 μg/ml). The GFP-positive cell clone that delivered the highest viral titers is referred to as 293/ΔBFLF2 in this work. To confirm the integrity of the viral genome in the 293/ΔBFLF2 cells, circular DNA molecules were extracted from 293/ΔBFLF2 cells and retransformed in E. coli DH10B by a denaturation-renaturation method as previously described (10).
qPCR-mediated DNA amplification.
The number of virus genome equivalents present in supernatants from lytically induced cells or the number of viruses bound to purified B cells was assessed by quantitative real-time PCR (qPCR) with a BALF5-specific probe and primers as described previously (6).
Virus induction and infection of target cells.
293/EBV-wt and 293/ΔBFLF2 cells were transfected in six-well cluster plates with expression plasmids containing the genes for BZLF1 and the BALF4 (0.5 μg each/well) with lipid micelles (Metafectene; Biontex) to induce lytic replication. In some experiments, 0.5 μg of the BFLF2 expression plasmid was added to the transfection mix (293/ΔBFLF2 complemented with BFLF2 are designated 293/ΔBFLF2-C). Three days after transfection, virus supernatants were harvested and filtered through a 0.45-μm filter. Functional viral titers were determined via GFP expression of infected Raji cells. To this aim, 104 Raji cells were infected in 96-well cluster plates with a serial dilution of defined virus stocks and scored for GFP expression 3 days after infection by UV microscopy. The number of green Raji units per milliliter was calculated as a measurement of the concentration of infectious particles in virus stocks. Primary B cells were infected at a multiplicity of infection (MOI) of 10 genome equivalents/cell and seeded into U-bottom 96-well plates at a density of 103 cells/well. In each well, γ-irradiated human embryonic lung fibroblasts served as a feeder layer.
Electron microscopy.
Cells were washed three times in phosphate-buffered saline (PBS), and cell pellets were fixed with 2.5% glutaraldehyde in the same buffer for 20 min at 4°C. Virus supernatant (5 ml) was ultracentrifuged for 2 h at 30,000 × g, and the pellet was fixed with glutaraldehyde. Samples were postfixed in 2% osmium tetroxide in cacodylate buffer for 2 h at 4°C, stained with 0.5% uranyl acetate for 16 h at 4°C, washed twice in distilled water, dehydrated in ethanol, and embedded in Epon 812. Thin sections were examined with a Zeiss electron microscope.
Gardella gel electrophoresis and Southern blot analysis.
Preparation of genomic DNA, Gardella gel electrophoresis, and Southern blot analysis were performed as previously described (14). Gardella gel electrophoresis, followed by Southern blotting, was used to detect viral DNA either in replicating cells or in cell-free virus supernatants. Following electrophoresis, the gel was blotted onto a Hybond XL membrane (Amersham Biosciences) and hybridized with a radioactively labeled plasmid encompassing EBV-specific sequences.
RNA isolation and Northern blot analysis.
Total RNA was isolated from lytically induced cells with an RNA isolation kit (Nucleospin; Machery & Nagel). Five micrograms of RNA was separated on a 1% formaldehyde gel, blotted onto a Hybond XL membrane (Amersham Biosciences), and hybridized with a 32P-radiolabeled probe encompassing the BFLF1 open reading frame.
EBV binding assay.
To assess EBV binding to target cells, EBV-wt-, ΔBFLF2-, or ΔBFLF2-C-containing supernatant was mixed either at an MOI of 10 virus genome equivalents/cell or at an equal volume (1 ml) with purified B cells. After a 3-h incubation on ice, cells were washed three times in PBS, after which they were analyzed by several methods, including immunostaining (in which case they were dried on glass slides), Gardella gel analysis, electron microscopy, or qPCR after DNA extraction as described previously (6).
Immunostaining.
Cells were washed three times in PBS and fixed on glass slides with acetone-methanol (1:1, vol/vol) (for BFRF1 staining) or acetone (for gp350 staining). Slides were incubated with rabbit polyclonal anti-BFRF1 (1:500) or mouse anti-gp350/220 antibody (ATCC 72A1) for 30 min, washed three times with PBS, and then incubated with a secondary Texas Red-conjugated goat anti-rabbit immunoglobulin G (Jackson) or Cy3-coupled anti-mouse antibody (Dianova) for 30 min. After several washes, BFRF1-stained cells were incubated with 1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) and coverslips were mounted face down in Mowiol (Calbiochem) whereas gp350-stained cells were embedded in 90% glycerol. gp350 immunostaining was viewed with a digital fluorescence microscope (Leica), and BFRF1 staining was viewed with a confocal laser scanning microscope (Zeiss).
Western blot analysis.
Test cells were collected by centrifugation, washed, resuspended in PBS, and lysed by sonication. The virus pellet obtained after ultracentrifugation of 5 ml of virus supernatant for 2 h at 30,000 × g or 20 μg of cellular proteins was denatured in Laemmli buffer for 5 min at 95°C, separated on a 10% sodium dodecyl sulfate-polyacrylamide gel, and electroblotted onto a nitrocellulose membrane (Schleicher & Schuell). After preincubation in 5% milk powder in 0.1% Tween in PBS, blots were incubated either with a monoclonal serum against BFLF2 (clone C1, 1:50), BFRF1 (1:1,000), BNRF1 (1:10,000), or actin (1:10,000; Sigma) for 1 h at room temperature. After several washings in 0.1% Tween in PBS, blots were incubated for 1 h with a horseradish peroxidase-conjugated secondary antibody (Pierce). After three further washings, the bound antibody was visualized with an ECL detection reagent (Roche).
RESULTS
Construction of a ΔBFLF2 mutant virus.
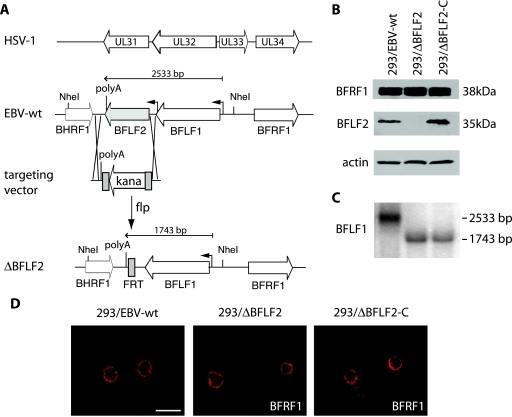
The overall organization of the HSV-1 and EBV genome fragments that carry the genes for UL31 and BFLF2 is shown in Fig. 1. This schematic representation shows similar arrangements in the two viral genomes, even though the HSV-1 locus contains an additional gene (that for UL33). ClustalW alignment of UL31 from two alphaherpesviruses, HSV and PrV, and BFLF2 illustrates the high degree of homology between the two alphaherpesvirus family members (55% identity) which does not extend to BFLF2 (15% identity). The BFLF2 open reading frame was deleted from the B95.8 complete genome cloned onto an F-factor replicon (p2089) by insertional mutagenesis via homologous recombination (Fig. 1A). The targeting vector consisted of the KAN resistance gene flanked by oligonucleotide sequences specific for the region flanking the gene for BFLF2. Clones were subjected to double selection with CAM and KAN, and properly recombined clones were transformed with FLP recombinase to delete the KAN resistance cassette. The bacterial clones that survived this selection were analyzed with several restriction enzymes to confirm successful disruption of the target gene (data not shown). Plasmid DNA from the ΔBFLF2 mutant was further amplified and stably transfected into 293 cells. Six hygromycin-resistant, GFP-positive clones carrying the recombinant EBV genome were cotransfected with BZLF1 and an expression plasmid for BFLF2. BZLF1 initiates the lytic cycle that culminates in virus production in EBV-permissive cells. A qPCR analysis of these complemented supernatants with EBV-specific primers showed virus titers ranging between 107 and 5 × 107 genome equivalents/ml in different experiments. These titers are perfectly in the range of those observed with permissive 293/EBV-wt (between 107 and 108 viral DNA equivalents/ml [data not shown]). All of the clones were therefore permissive to lytic DNA replication (data not shown), but we selected the cell clone that delivered the highest viral titers upon complementation (here referred to as 293/ΔBFLF2) for further analysis. In the next step, the viral episomes contained in 293/ΔBFLF2 cells were transferred back into E. coli DH10B cells. Restriction analysis confirmed the overall structural integrity of the mutant viral DNA in 293/ΔBFLF2 cells (data not shown). Data obtained from the DNA restriction analysis could be confirmed by Western blotting of induced 293/ΔBFLF2 cells with an antibody specific for this protein in the absence or presence of BFLF2 (Fig. 1B). As expected, no BFLF2 protein could be detected in 293/ΔBFLF2 but complementation in trans (abbreviated as 293/ΔBFLF2-C) restored normal BFLF2 expression levels. The genes for BFLF1 and BFLF2 have the same poly(A) site that was conserved in the deletion mutant. We nevertheless wished to ensure that the BFLF1 expression level had not been affected by the BFLF2 deletion. We therefore performed a Northern blot hybridization with a BFLF1-specific probe. The results of this experiment are presented in Fig. 1C and show that the BFLF1 expression level was, indeed, not significantly altered by the introduction of the BFLF2 deletion. Altogether, these results showed that 293/ΔBFLF2 cells provide a suitable genetic system to assess the function of the BFLF2 protein at the different steps of virus production.
FIG. 1.
Construction of a ΔBFLF2 mutant virus. (A) Schematic drawing of the HSV-1 and EBV loci that carry positional homologs UL31/UL34 and BFLF2/BFRF1. The EBV genome is shown before and after disruption of the BFLF2 open reading frame. One FLP recombinase target (FRT) site is left between the intact BHRF1 and BFLF1 genes after excision of the KAN resistance cassette with FLP recombinase. (B) Whole protein extracts from lytically induced 293/EBV-wt, 293/ΔBFLF2, and 293/ΔBFLF2-C cells were used to assess BFRF1 (upper panel), BFLF2 (middle panel), and actin (lower panel) protein synthesis in a Western blot analysis. The BFRF1 expression level is unchanged in the 293/ΔBFLF2 mutant. (C) Northern blot analysis of lytically induced cells. Total RNAs extracted from induced 293/ΔBFLF2, 293/ΔBFLF2-C, and 293/wt-EBV cells were separated in an agarose gel and hybridized with a BFLF1-specific probe. The BFLF1 primary transcript encoded by ΔBFLF2 is smaller than its wild-type counterpart as a result of the deletion, but expression levels are similar in all three samples. (D) Immunofluorescence analysis with a confocal laser scanning microscope of BFRF1 localization in induced 293/EBV-wt, 293/ΔBFLF2, and 293/ΔBFLF2-C cells. BFRF1 localization at the perinuclear region is not altered in 293/ΔBFLF2 cells. Scale bar, 20 μm.
BFLF2 has no obvious influence on BFRF1 synthesis and intracellular localization.
We further investigated the interplay between BFLF2 and BFRF1 by performing a Western blot assay with a BFRF1-specific antibody and induced 293/ΔBFLF2 cells. As shown in Fig. 1B, BFRF1 was properly expressed upon the induction of lytic replication, thereby demonstrating that the synthesis of this protein is independent of BFLF2. We also immunostained induced 293/ΔBFLF2 cells with the same antibody and determined BFRF1's intracellular localization by confocal microscopy. Interestingly, this assay detected most of the BFRF1-specific signals in the perinuclear region, a pattern entirely similar to the one observed in induced 293/EBV-wt cells. The picture was not modified by complementation with a BFLF2 expression plasmid in 293/ΔBFLF2, showing that perinuclear localization of BFRF1 was not obviously dependent on BFLF2 in 293 cells (Fig. 1D).
The BFLF2 protein has no influence on lytic DNA replication.
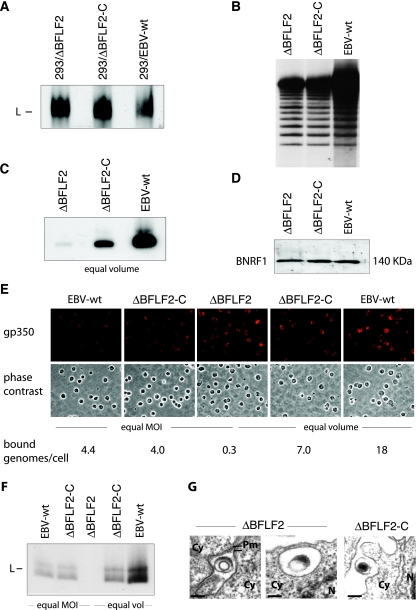
Production of mature virions is a complex process during which viral DNA is amplified, excised to form monomeric genomes, and packaged into an assembled capsid. The infectious particle is then transported from the nucleus to the cytoplasm, where further maturation steps take place (23). We first assayed viral DNA replication by submitting induced cells to a Gardella gel analysis that allows the visualization of newly replicated monomeric linear genomes (Fig. 2A). No significant differences among 293/ΔBFLF2, 293/ΔBFLF2-C, and 293/EBV-wt were noted. These findings show that the early gene for BFLF2 has no influence on DNA replication or on the production of monomeric linear genomes. Further evidence for the cleavage of newly replicated genomes was provided by the results of a Southern blot analysis with a probe specific to the EBV terminal repeats (TR). This experiment, indeed, showed the characteristic ladder produced by newly replicated genomes that contain variable numbers of TR (Fig. 2B). We then studied late gene expression by evaluating the proportion of induced gp350-expressing cells. By immunostaining with a gp350-specific antibody, we found that approximately 20% of the induced 293/ΔBFLF2 or 293/ΔBFLF2-C cells express gp350, a result which fits with our previous experience with 293/EBV-wt clones (between 10 and 25% of these express gp350 upon induction; data not shown). 293/ΔBFLF2 cells therefore appear to be as permissive to lytic replication as their wild-type counterparts.
FIG. 2.
BFLF2 is not required for DNA replication but is important for DNA packaging. (A) Gardella gel analysis of lytically induced producer cells, followed by Southern blotting with a gp350-specific DNA probe. 293/ΔBFLF2 cells produce amounts of viral DNA similar to those produced by trans-complemented (293/ΔBFLF2-C) and 293/EBV-wt cells. L, linear DNA. (B) Southern blot analysis with a TR-specific probe confirmed the presence of newly replicated monomeric linear DNA molecules in lytically induced 293/ΔBFLF2 cells. Genomic DNA was cleaved with BamHI and separated on a 0.8% agarose gel. (C) Gardella gel analysis of equal volumes of pelleted virus supernatants from the same cell lines as in panel A shows a drastic reduction in mature virus particles in the ΔBFLF2 virus stocks. The difference in signal intensities between the ΔBFLF2-C and EBV-wt virus stocks reflects the difference in MOIs (1.5 × 107 and 7 × 107 genome equivalents/ml, respectively). (D) Western blot analysis of virus supernatant showing the same amount of BNRF1 tegument protein in ΔBFLF2, ΔBFLF2-C, and EBV-wt virus particles. Protein extract from 5 ml of ultracentrifuged virus supernatant was separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and probed with an antiserum against BNRF1 tegument protein. (E) Detection of bound viruses on resting B cells. Cells were incubated with supernatants containing mutant or wild-type viruses either at an MOI of 10 or with equal volumes and immunostained with an antibody specific to gp350. B cells exposed to ΔBFLF2 are covered with a higher number of viral gp350-positive structures (red dots on the cell surface) than are B cells exposed to EBV-wt and ΔBFLF2-C virus particles at an equal MOI (left part of panel). Exposure of B cells with equal volumes of supernatants showed similar levels of staining (right part of panel). (F) Gardella gel analysis of virus bound to B cells. B cells incubated with the same virus stocks as in panel E were submitted to Southern blotting and hybridization with a gp350-specific DNA probe. No viral DNA could be detected after incubation of B cells with ΔBFLF2 supernatants. L, linear DNA. (G) ΔBFLF2 virus particles bound to B cells are devoid of DNA. B cells incubated with ΔBFLF2 or ΔBFLF2-C virus supernatant were analyzed by electron microscopy. Only VLP (left side) and L particles (right side) were identified at the surface and in intracellular vesicles of B cells, respectively, after incubation with ΔBFLF2 supernatants. In contrast, most particles bound after incubation with ΔBFLF2-C supernatant are mature and contain DNA. Cy, cytoplasm; Pm, plasma membrane; N, nucleus. Bars, 100 nm.
Supernatants from induced 293/ΔBFLF2 cells contain low virus titers.
We then infected the B-cell line Raji, which is susceptible to EBV wild-type infection, with supernatants from induced 293/ΔBFLF2 cells. Infection efficiency (or functional titers) was assessed by counting the GFP-positive cells in serial dilutions of the supernatants. Comparison with supernatants from 293/ΔBFLF2-C and 293/EBV-wt cells used as positive controls showed a reduction of more than 2 orders of magnitude in ΔBFLF2 functional titers (Table 1). qPCR analysis identified 20- and 35-fold reductions in the genome equivalent concentration in induced cell lines carrying the ΔBFLF2 viruses, compared to 293/ΔBFLF2-C or 293/EBV-wt cells, respectively (Table 1). Similar results were obtained after Gardella gel electrophoresis of pelleted supernatants, followed by Southern blotting with an EBV-specific probe. ΔBFLF2 virus stocks hardly contained any linear genomes, in contrast to their complemented counterparts or the wild-type virus (Fig. 2C). These results evidenced a reduction in the concentration of virions containing properly packaged DNA in supernatants from induced 293/ΔBFLF2 cells but did not exclude the possibility that they contained virions devoid of viral DNA. A first clue that this could actually be the case was provided by a Western blot analysis of pelleted ΔBFLF2, ΔBFLF2-C, and EBV-wt viruses with an antibody specific to the BNRF1 tegument protein (Fig. 2D). This assay displayed signals with the same intensity in all of the pellets analyzed, suggesting that all three types of supernatants contained similar amounts of viral proteins. We gained further support for this hypothesis by evaluating viral binding to resting B cells with ΔBFLF2, ΔBFLF2-C, and EBV-wt virus stocks. We performed these experiments by using either identical volumes of viral supernatants, knowing that the virus titers, as assessed by qPCR, were different, or an MOI of 10 genome equivalents/cell. We first immunostained these B cells with an antibody specific to the major envelope glycoprotein gp350/220. All three virus stocks, indeed, contained viral structures with the ability to bind to B cells, as evidenced by the observation of bright signals at the surface of the target cells in all of the samples tested (Fig. 2E). This type of assay does not allow precise quantitation of the number of particles bound but nevertheless showed that identical volumes of supernatant roughly gave rise to a similar number of signals at the cell surface. Exposure of cells to the same number of genome equivalents yielded a stronger signal with ΔBFLF2 viruses than with ΔBFLF2-C or EBV-wt, suggesting that ΔBFLF2 contained more virions than ΔBFLF2-C and EBV-wt at the same MOI. This would be expected in the light of our previous findings that demonstrated that 293/ΔBFLF2 supernatants contained similar amounts of viral proteins but greatly reduced amounts of properly packaged viral genomes compared to those from 293/ΔBFLF2-C and 293/EBV-wt. To test this hypothesis, we repeated the B-cell binding assay under the same conditions as described above and submitted the cells to Gardella gel analysis and a qPCR assay. Gardella gel analysis performed with equal amounts of the three types of supernatants roughly reflected the genome equivalent titers, with the strongest signal obtained with 293/EBV-wt supernatants (input MOI of 140), a weaker signal obtained with 293/ΔBFLF2-C (input MOI of 24), and a very weak signal obtained after binding with those from 293/ΔBFLF2 supernatants (input MOI of 10). The binding assay performed with equal MOIs identified equivalent amounts of viral nucleic acids bound to B cells after incubation with ΔBFLF2-C and EBV-wt virus stocks but nearly no signal from B cells incubated with ΔBFLF2 viruses (Fig. 2F). Similarly, qPCR analysis of viral particles bound to B cells detected around 4 particles/cell for the ΔBFLF2-C and EBV-wt viruses but only 0.3 ΔBFLF2 particle/cell after exposure to supernatants with an MOI of 10 genome equivalents/cell (Table 1). Thus, there is a marked discrepancy between the input MOI and the recovery of bound nucleic acids after a binding assay with 293/ΔBFLF2 supernatants but not after incubation with 293/ΔBFLF2-C and 293/EBV-wt supernatants. qPCR analysis after binding with equal volumes of supernatants again roughly paralleled the input MOI, with the strongest signals obtained with 293/EBV-wt, followed by 293/ΔBFLF2-C and finally 293/ΔBFLF2 (18, 7, and 0.3 genome equivalent bound per cell, respectively). The low recovery of bound nucleic acids at high input MOIs reflects the saturation of B-cell binding sites. Finally, we directly examined B cells after exposure to ΔBFLF2 supernatants by electron microscopy and could identify rare bound virus-like particles (VLP) containing a capsid at the surface of B cells, all of which appeared to be devoid of DNA, as evidenced by the lack of an electron-dense inner core (Fig. 2G). However, viral structures containing tegument-like material were readily identified either at the surface of the target cells or in already internalized cellular vesicles. As expected, complete virions bound to B cells could be readily seen at the surface of B cells exposed to supernatants from induced 293/ΔBFLF2-C cells (Fig. 2G). Negative stains are an integral part of the panoply used for the study of virus morphology, but this technique is inefficient with viruses such as EBV that produce low titers. We therefore embedded pelleted virions from induced 293/ΔBFLF2 and 293/ΔBFLF2-C supernatants and looked at sections from this material by electron microscopy. The results of this analysis are summarized in Table 2 and illustrated in Fig. 3A. They show that mature viruses can be readily identified in 293/ΔBFLF2-C supernatant. In contrast, only rare fully formed viruses were visible in pelleted supernatants from induced 293/ΔBFLF2 cells. Instead, they contained a large number of the already mentioned round structures limited by a clear membrane and containing tegument structures. These are reminiscent of L particles previously identified in supernatants from cells infected with herpesviruses (13, 22, 29). In addition, we detected rare VLP. The distribution of L particles, VLP, and mature particles in ΔBFLF2 pellets was about 88%, 9%, and 3%, respectively (Table 2).
TABLE 1.
Analysis of ΔBFLF2 mutant infection phenotype
| Virus | qPCR analysis (no. of genome equivalents/ml supernatant)a | Raji cell infection (no. of green Raji units/ml supernatant)b | No. of bound genome equivalents/B cellc,f | B-cell infection (% EBNA2-positive cells)d,f | B-cell transformation (% outgrowth)e,f |
|---|---|---|---|---|---|
| EBV-wt | 7 × 107 | 2 × 106 | 4.4 | 44 | 100 |
| ΔBFLF2 | 2 × 106 | 5 × 103 | 0.3 | 3 | 3 |
| ΔBFLF2-C | 4 × 107 | 1.5 × 106 | 4 | 36 | 100 |
Mean values obtained from three different virus stocks are shown.
GFP-positive cells were counted 3 days after infection. Mean values from three different infection experiments are shown.
Resting B cells were incubated for 3 h on ice with virus supernatant, and the number of bound mature particles per cell was determined by qPCR after DNA extraction.
Primary B cells were stained for EBNA2 expression at 3 days postinfection.
Outgrowth of LCL clones (total of 288 wells per supernatant) was visually scored at 4 weeks after infection.
Infection experiments were performed at an MOI of 10 qPCR genome equivalents of virus/cell, and mean values obtained from three independent experiments are shown.
TABLE 2.
Quantitative analysis of cell-free virus particles in pelleted supernatantsa
Percentage of virus particle types observed in pellets from the indicated supernatants by electron microscopy.
L particles are defined as enveloped particles containing only tegument-like structures.
VLP are defined as enveloped particles containing capsid structures devoid of DNA.
Mature particles are defined as enveloped particles with DNA-filled capsids.
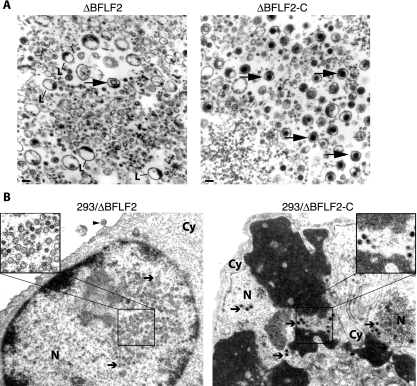
FIG. 3.
BFLF2 is required for efficient nuclear egress and DNA packaging. (A) Electron micrographs of pelleted ΔBFLF2 and ΔBFLF2-C particles. L particles dominated the picture in the ΔBFLF2 supernatant, but rare VLP could also be identified (arrow, left side). ΔBFLF2-C supernatant mainly contained mature virions (arrows, right side). Bar, 100 nm. L, L particles. (B) Electron micrograph of lytically induced 293/ΔBFLF2 cells showing empty A capsids and B capsids (arrows, left side) in the nucleus. One virus-like empty particle could be identified at the surface of these cells (arrowhead). In contrast, induced 293/ΔBFLF2-C cells contain a normal number of electron-dense C capsids (arrows, right side). Cy, cytoplasm; N, nucleus.
BFLF2 is important for both nuclear egress and DNA packaging.
The data accumulated thus far suggested that the ΔBFLF2 mutant was defective with regard to virus production. This prompted us to monitor the different steps of virus maturation at the single-cell level by electron microscopy. Two major differences between induced 293/ΔBFLF2 and 293/EBV-wt cells could be pinpointed. First, numerous A and B capsids but only a few mature C capsids were visible in the nuclei of induced 293/ΔBFLF2 cells, which contrasts with the picture seen in 293/EBV-wt or 293/ΔBFLF2-C cells, in which all three capsid types were readily identified (Fig. 3B and data not shown). A summary of the results obtained with 100 replicating cells of each induced cell line is presented in Table 3. Second, capsids were nearly exclusively detected in the nuclei of induced 293/ΔBFLF2 cells. Only a few particles containing capsids were found in the cytoplasm, and only a single enveloped yet empty particle could be identified in the extracellular milieu near the cell surface after the examination of 100 induced 293/ΔBFLF2 cells (Fig. 3B, left panel). Again, lytically induced 293/EBV-wt or 293/ΔBFLF2-C cells differed from the mutant in that they contained viruses both in the nucleus and in the cytoplasm, as well as on the surface. Most of the viral structures contained a capsid, and the majority of the virions identified outside the nucleus also carried viral DNA. We conclude from these findings that BFLF2 is crucial for efficient viral DNA packaging and for primary egress across the nuclear membrane.
TABLE 3.
Quantitative analysis of intranuclear virus capsid forms in induced cellsa
| Cell line | % (no.) of A capsids | % (no.) of B capsids | % (no.) of C capsids |
|---|---|---|---|
| 293/EBV-wt | 32 (362) | 48 (536) | 20 (220) |
| ΔBFLF2 | 12 (265) | 84 (1,740) | 4 (74) |
| ΔBFLF2-C | 21 (306) | 56 (807) | 23 (340) |
Capsid types were observed in different induced cell types by electron microscopy.
Analysis of theΔBFLF2 infection phenotype in primary B cells.
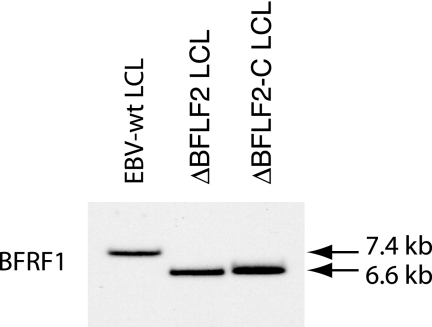
We extended our analysis by assessing the ability of ΔBFLF2 virions to infect and transform primary B cells, one of the main targets of EBV infection in vivo. The results of the B-cell infection experiments with mutant and wild-type supernatants at an MOI of 10 genome equivalents/cell are summarized in Table 1. Approximately 44% of the primary B cells exposed to EBV-wt showed nuclear expression of EBNA2 3 days after infection. The infection rate, however, dropped to 3% in B cells infected with ΔBFLF2 viruses but could be restored to nearly wild-type levels by trans complementation with BFLF2. This implies that, even at the same MOI, infection with ΔBFLF2 was less efficient than with complemented or wild-type virus. This reduced infection rate was clearly reflected in the ability of viruses to immortalize B cells. Whereas cell outgrowth was observed in all wells containing 103 B cells infected at an MOI of 10 with either wild-type or trans-complemented ΔBFLF2 mutants, only 3% of the wells infected with ΔBFLF2 contained immortalized B-cell clones. However, this shows that the small number of EBNA2-positive lymphocytes efficiently underwent transformation. Southern blot analysis with a probe specific to the gene for BFRF1 confirmed that the immortalized B-cell lines carried the mutant virus, as evidenced by a rearranged BFLF2 locus (Fig. 4). This set of experiments therefore provided definite evidence that BFLF2 is dispensable for primary B-cell infection and transformation.
FIG. 4.
Southern blot analysis of LCLS obtained by infection of resting B cells with ΔBFLF2, ΔBFLF2-C, or EBV-wt supernatants. Genomic DNA was cleaved with BamHI and separated by gel electrophoresis. Hybridization with a BFRF1-specific probe detected a 6.6-kb BamHI F fragment corresponding to the deleted BFLF2-encoding gene in both 293/ΔBFLF2 and 293/ΔBFLF2-C LCLs. LCLs infected with the wild-type virus carry an intact 7.4-kb BamHI F fragment.
DISCUSSION
Primary egress through the nuclear membrane is an obligatory maturation step for all of the herpesviruses studied so far. This process requires HSV-1 protein UL31 or UL34 or its homolog from PrV or cytomegalovirus (2, 8, 17, 21, 25, 31). UL31 and UL34 form a complex at the nuclear membrane that induces the formation of primary envelopes in the absence of any other viral component (16). A similar role has been proposed for EBV proteins BFRF1 and BFLF2, but genetic evidence implicating the latter protein in primary egress is still lacking. This prompted the present study, in which we constructed a ΔBFLF2 mutant and analyzed its phenotype in an attempt to gather further information on BFLF2's function during the EBV replicative cycle and EBV active latency. Here we report our findings, from which we infer that BFLF2 assumes at least a dual role in the early phases of DNA packaging and virion maturation. Electron microscopic observation of induced 293/ΔBFLF2 cells, indeed, showed a rather unexpected nucleocapsid maturation impairment, as evidenced by the accumulation of A and B capsids with only a minority of C capsids containing viral DNA visible in the cell nuclei. It should be noted, however, that an HSV-1 mutant lacking UL31 also showed a reduced proportion of DNA-containing C capsids (2). This finding was ascribed to a reduced synthesis of monomeric viral DNA genomes, but the possibility of an additional defect in DNA packaging could not be excluded. We did not find any evidence for impaired viral DNA replication and monomeric genome production as assessed by Gardella gel and Southern blot analysis of induced 293/ΔBFLF2 cells. Aside from the abnormal capsid maturation, it also became apparent that capsids synthesized during lytic replication of 293/ΔBFLF2 cells accumulated in the nuclei of induced cells and that mature virions were only rarely visible in the cytoplasm or at the cell surface, a result largely expected from the collective literature on UL31 and UL34. The observed alterations in the primary egress of mature virions could be further confirmed by two assays showing a marked reduction in the production of infectious EBV virions. First, qPCR analysis of supernatants from induced cells showed 20- and 35-fold reductions in EBV DNA equivalents obtained through the induction of 293/ΔBFLF2 cells compared to cells carrying complemented mutant and wild-type viruses, respectively. Second, we found that ΔBFLF2 defective particles, either in cell-free supernatants or bound to B cells, contained only marginal amounts of full-length monomeric linear EBV DNA, as assessed by Gardella gel analysis. A qPCR analysis of bound viruses confirmed that ΔBFLF2 virions, indeed, contained little viral DNA. We could exclude the possibility that the ΔBFLF2 phenotype resulted from down-regulation of BFRF1 by showing the normal expression and localization of this protein in induced 293/ΔBFLF2 cells. These results therefore confirmed earlier reports that assigned an important role to BFLF2 in primary egress and suggest that BFLF2 has functions in common with UL31 (9, 18). However, they also identified an additional role for BFLF2 as a member of the group of proteins directly or indirectly required for viral DNA packaging. This phenotypic trait is not shared with the ΔBFRF1 mutant, which does not show any alteration of the packaging process (5). A role of BFLF2 during DNA packaging would fit with the previously described diffuse nuclear localization of BFLF2 prior to its interaction with BFRF1 (9, 18). One model of primary egress posited that UL31 is a primary tegument protein that interacts with the viral capsid (24). A variation on this model could be conceived for EBV in which BFLF2, as part of the tegument, could interact with proteins responsible for DNA packaging but would also mediate contact between the capsid and the inner nuclear membrane through its interactions with BFRF1. In this respect, it is interesting that whereas ΔBFRF1 capsids were mostly aligned along the nuclear membrane (5, 9), ΔBFLF2 capsids were homogeneously distributed throughout the nucleus, suggesting that migration of the capsids toward the nuclear membrane before primary egress was impaired. We consider it unlikely that impairment of intranuclear migration and primary egress was a direct consequence of improper viral DNA packaging. Indeed, induction of the lytic cycle in an EBV mutant devoid of packaging signals (ΔTR) is followed by the production of large amounts of VLP containing capsids devoid of DNA in the extracellular milieu, thereby showing that the two functions are largely independent (7). In all probability, BFLF2 therefore cannot be considered a direct functional homolog of UL31, as could have already been suspected on the basis of the low level of homology at the protein level and the different temporal expression patterns of the two proteins during replication. Proteins required for DNA packaging, which are also frequently involved in the cleavage of newly replicated DNA concatemers, are poorly characterized in gammaherpesviruses. Genetic analysis of HSV-1 mutants has identified a group of seven proteins involved in viral DNA cleavage/packaging (UL6, UL15, UL17, UL25, UL28, UL32, UL33) (30), five of which possess homologs in the EBV genome (BBRF1, BGRF1, BGLF1, BVRF1, BFLF1). Neither genetic nor biochemical information about the functions of these EBV proteins during lytic replication is available. The present findings suggest that the molecular mechanisms that control this cascade of events around viral DNA processing and packaging might differ in different herpesvirus subfamilies. It is noteworthy that the gene for BFLF1, whose tentative function involves monomeric viral genome production and packaging, is directly adjacent to that for BFLF2 on the viral genome (19). Dysfunctions in BFLF1 synthesis, e.g., as a result of the BFLF2 knockout construction, could, in principle, impair packaging, as observed in the 293/ΔBFLF2 mutant. Aside from the fact that BFLF1's function is still entirely hypothetical, we consider this hypothesis to be very unlikely to be true because the 293/ΔBFLF2 phenotype was so straightforwardly complemented by a BFLF2 expression plasmid devoid of any BFLF1 sequences. In addition, induction of the lytic cycle in 293/ΔBFLF2 cells resulted in BFLF1 expression as expected. These results, however, do not exclude the possibility that the ΔBFLF2 phenotype can be partly explained by interactions between BFLF2 and proteins involved in DNA packaging.
Despite its multifunctional role, we could not find any evidence that implicates BFLF2 in lytic DNA replication, B-cell immortalization, or infection. The latter fits with the previously described absence of this protein in mature virions (9, 15). Detailed analysis of the viral particles present in supernatants from induced 293/ΔBFLF2 cells confirmed the rare production of mature virions by these cells. However, they swarmed with round viral structures limited by a membrane encircling tegument-like structures. These are probably related or identical to L particles initially described in HSV-1- and cytomegalovirus-infected cells, even if we do not have evidence yet for the particular floating properties that characterize them (2, 13, 29). Rare VLP containing an empty capsid could also be identified. Both wild-type viruses and these defective viral particles contained equal amounts of BNRF1 and gp350/220, two EBV proteins present in the tegument and envelope of mature EBV virions, respectively, and bound to B cells, as shown by several assays. These results also suggest that, as previously described, e.g., for HSV-1 or PrV, primary egress and production of L particles are two independent processes (2, 8, 22). The observation of rare VLP in the extracellular milieu also shows that BFLF2 is not strictly required for the nuclear egress of these defective particles that contain an immature capsid.
Assessment of viral titers by qPCR, Gardella gel analysis, and infection assays with Raji or primary B cells yielded discrepant results. Infection assays, indeed, showed a more pronounced decrease in functional infectious titers (400-fold reduction) than expected from the genome equivalent concentrations that we calculated on the basis of the qPCR data (35-fold reduction). The data gathered from the various binding assays we performed at the same MOI or with equal volumes of supernatants from 293/ΔBFLF2, 293/ΔBFLF2-C, and 293/EBV-wt cells clearly showed that all three types of supernatants contained a large amount of viral structures but also that the ratio of the input viral genomes (in the supernatant) to the viral genomes bound to B cells (binding efficiency) was much higher with 293/EBV-wt and 293/ΔBFLF2-C (40%) than with 293/ΔBFLF2 supernatants (3%) after infection at the same input MOI. Taking into account the results of the Western blotting of supernatant pellets and of their direct examination by electron microscopy, which showed similar amounts of viral proteins in all three supernatants, we consider it likely that the lower viral DNA-binding efficiency of ΔBFLF2 reflects unequal competition for binding sites at the surface of target cells between abundant defective and rare viral DNA-containing particles.
The 293/ΔBFLF2 mutant also afforded us an opportunity to further study the interplay between BFLF2 and BFRF1 in the context of a replication-competent genome. Both BFRF1 production and intracellular localization were unaltered by BFLF2's absence. These results differ from those previously reported for CV1 cells transfected with BFLF2 and BFRF1 expression plasmids in which BFRF1 alone was diffusely found in the cytoplasm and the perinuclear region but was retargeted to the nuclear rim following the introduction of BFLF2 (18). These differences might stem from the different experimental systems used, i.e., transient plasmid expression versus mutant analysis in the context of the viral genome. Alternatively, they may reflect the use of different cell lines, i.e., CV1 and 293 cells, to perform the experiments. Previous reports have, indeed, shown that the cellular background influences UL34 intracellular localization (20).
Comparisons between the UL31 and BFLF2 proteins more generally illustrate how positional homologs diverged during evolution to fulfil related but distinct functions. Further analysis of EBV maturation pathways is likely to reveal more differences between alpha- and gammaherpesviruses.
Acknowledgments
We thank H. Bannert and H. Lips for expert technical assistance.
This work was supported by institutional grants.
Footnotes
Published ahead of print on 20 February 2008.
REFERENCES
- 1.Calderwood, M. A., K. Venkatesan, L. Xing, M. R. Chase, A. Vazquez, A. M. Holthaus, A. E. Ewence, N. Li, T. Hirozane-Kishikawa, D. E. Hill, M. Vidal, E. Kieff, and E. Johannsen. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. USA 1047606-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 718307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1589-14. [DOI] [PubMed] [Google Scholar]
- 4.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 958245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H. J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 793703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feederle, R., B. Neuhierl, G. Baldwin, H. Bannert, B. Hub, J. Mautner, U. Behrends, and H. J. Delecluse. 2006. Epstein-Barr virus BNRF1 protein allows efficient transfer from the endosomal compartment to the nucleus of primary B lymphocytes. J. Virol. 809435-9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feederle, R., C. Shannon-Lowe, G. Baldwin, and H. J. Delecluse. 2005. Defective infectious particles and rare packaged genomes produced by cells carrying terminal-repeat-negative Epstein Barr virus. J. Virol. 797641-7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonnella, R., A. Farina, R. Santarelli, S. Raffa, R. Feederle, R. Bei, M. Granato, A. Modesti, L. Frati, H. J. Delecluse, M. R. Torrisi, A. Angeloni, and A. Faggioni. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 793713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffin, B. E., E. Bjorck, G. Bjursell, and T. Lindahl. 1981. Sequence complexity of circular Epstein-Bar virus DNA in transformed cells. J. Virol. 4011-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grünewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 3021396-1398. [DOI] [PubMed] [Google Scholar]
- 12.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7107-122. [DOI] [PubMed] [Google Scholar]
- 13.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130118-133. [DOI] [PubMed] [Google Scholar]
- 14.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H.-J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 7410142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 10116286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klupp, B. G., H. Granzow, W. Fuchs, G. M. Keil, S. Finke, and T. C. Mettenleiter. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. USA 1047241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 7410063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 32099-106. [DOI] [PubMed] [Google Scholar]
- 19.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 722463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, L., M. Tanaka, Y. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex virus 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 32968-76. [DOI] [PubMed] [Google Scholar]
- 21.Lötzerich, M., Z. Ruzsics, and U. H. Koszinowski. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 8073-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73269-276. [DOI] [PubMed] [Google Scholar]
- 23.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106167-180. [DOI] [PubMed] [Google Scholar]
- 24.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297854-857. [DOI] [PubMed] [Google Scholar]
- 26.Neuhierl, B., and H. J. Delecluse. 2005. Molecular genetics of DNA viruses: recombinant virus technology. Methods Mol. Biol. 292353-370. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 29.Rixon, F. J., C. Addison, and J. McLauchlan. 1992. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J. Gen. Virol. 73277-284. [DOI] [PubMed] [Google Scholar]
- 30.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses, p. 2501-2601. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 31.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye, G.-J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 9711002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]