Abstract
Over 40 different human immunodeficiency virus type 1 (HIV-1) mRNAs are produced by alternative splicing of the primary HIV-1 RNA transcripts. In addition, approximately half of the viral RNA remains unspliced and is used as genomic RNA and as mRNA for the Gag and Pol gene products. Regulation of splicing at the HIV-1 3′ splice sites (3′ss) requires suboptimal polypyrimidine tracts, and positive or negative regulation occurs through the binding of cellular factors to cis-acting splicing regulatory elements. We have previously shown that splicing at HIV-1 3′ss A1, which produces single-spliced vif mRNA and promotes the inclusion of HIV exon 2 into both completely and incompletely spliced viral mRNAs, is increased by optimizing the 5′ splice site (5′ss) downstream of exon 2 (5′ss D2). Here we show that the mutations within 5′ss D2 that are predicted to lower or increase the affinity of the 5′ss for U1 snRNP result in reduced or increased Vif expression, respectively. Splicing at 5′ss D2 was not necessary for the effect of 5′ss D2 on Vif expression. In addition, we have found that mutations of the GGGG motif proximal to the 5′ss D2 increase exon 2 inclusion and Vif expression. Finally, we report the presence of a novel exonic splicing enhancer (ESE) element within the 5′-proximal region of exon 2 that facilitates both exon inclusion and Vif expression. This ESE binds specifically to the cellular SR protein SRp75. Our results suggest that the 5′ss D2, the proximal GGGG silencer, and the ESE act competitively to determine the level of vif mRNA splicing and Vif expression. We propose that these positive and negative splicing elements act together to allow the accumulation of vif mRNA and unspliced HIV-1 mRNA, compatible with optimal virus replication.
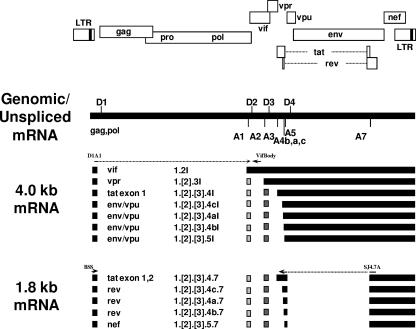
Human immunodeficiency virus type 1 (HIV-1) primary RNA transcripts are alternatively spliced, to generate over 40 different mRNAs of three different size classes, unspliced ∼9-kb mRNAs, incompletely spliced ∼4-kb mRNAs, and completely spliced ∼1.8-kb mRNAs (Fig. 1) (for a review, see reference 28). The unspliced viral RNA is used as genomic RNA and as mRNA for the Gag and Pol gene products (for a review, see reference 7). The incompletely spliced size class includes mRNAs for Vif, Vpr, single-exon Tat, and Env/Vpu, and the completely spliced size class includes mRNAs for two-exon Tat, Rev, and Nef. Four different 5′ splice sites (5′ss) and eight different 3′ splice sites (3′ss) are used to produce the different alternatively spliced HIV-1 mRNAs, which are present in different amounts in the infected cells (21). The locations and sequences of most of these splice sites are highly conserved in all clades of group M HIV-1 and in HIV-1 strains in groups N and O. The extent to which these 5′ss and 3′ss are used is dependent on the relative strengths of the splice sites and on the presence of cis splicing elements within the viral genome. The cis elements within the HIV-1 genome include both exonic splicing silencers (ESS) and intronic splicing silencers (ISS) and exonic splicing enhancers (ESE).
FIG. 1.
Viral RNA species produced within HIV-1-infected cells. HIV-1 genes are shown relative to the long terminal repeats (LTR). The viral genomic or ∼9-kb unspliced mRNA shows the location of 5′ss and 3′ss within the pNL4-3 infectious plasmid. The incompletely and completely spliced HIV-1 viral mRNAs (∼4-kb and 1.8-kb size classes) are shown as black boxes. Spliced mRNAs are denoted by the translated open reading frames and by the exon content. The incompletely spliced mRNAs, denoted by an I, are differentiated from completely spliced mRNAs by the inclusion of the intron between 5′ss D4 and 3′ss A7. Either one or both of the noncoding exons 2 and 3 are included in a fraction of all ∼4-kb and ∼1.8-kb mRNA species, with the exception of vif mRNA. The locations of the primer pairs used for qRT-PCR analysis of vif mRNA (D1A1 and VifBody) and RT-PCR analysis of 1.8-kb mRNA (BSS and SJ4.7A) are indicated.
We have previously shown that the splicing of tat mRNA, which is present in relatively low abundance, is regulated by two different ESS within the first tat coding exon that represses splicing at 3′ss A3 and in an ESS in the second tat/rev coding exon that represses splicing at 3′ss A7 (4, 11, 26, 27). In addition, an ISS upstream of 3′ss A7 regulates tat/rev mRNA splicing (30). The first tat coding exon and the second tat/rev coding exon also contain ESE that selectively increase splicing at 3′ss A3 and A7, respectively (4, 26, 27, 35). The splicing of vpr mRNA, which is also present in low abundance in infected cells, is regulated by an ESS that represses splicing at 3′ss A2 (6, 18). In addition, splicing at 3′ss A2 is facilitated by the presence of a downstream 5′ss (5′ss D3), a result predicted by the exon definition hypothesis, which proposes that the definition of exons is an early step in splicing and precedes the definition of introns (5, 10, 22). The use of the 5′ss D3 results in the inclusion of a small 74-nucleotide (nt) noncoding exon (exon 3) between 3′ss A2 and 5′ss D3 into a fraction of the HIV-1 mRNAs. Mutations that optimize 5′ss D3 result in increased splicing at 3′ss A2 and increased exon 3 inclusion (6).
vif mRNA is also present in low abundance in HIV-1-infected cells. Regulation of vif mRNA splicing may be important to maintain Vif expression at the levels necessary to inhibit the accumulation and packaging of APOBEC3G and APOBEC3F deoxycytidine deaminases (14, 20, 25, 34) without inhibiting virus protein processing and virus replication, which has been shown to occur at higher Vif levels (2). We have previously shown that splicing at 3′ss A1, the splice site used to generate vif mRNA, is limited by the presence of a suboptimal downstream 5′ss, since the mutation of 5′ss D2 to a consensus 5′ss results in an HIV-1 phenotype with excessive splicing, a dramatic increase in the expression of Vif, and the inclusion of the 50-nt noncoding exon 2 between 3′ss A1 and 5′ss D2 into a majority of the spliced viral mRNAs (17). In contrast, in the wild-type infected cells, exon 2 is included into only a fraction of viral mRNAs (21). It has also recently been shown that exon 2 contains two potential ESE elements (ESEM1 and ESEM2) with the sequence UGGAAAG; mutations of these two elements resulted in a selective decrease in exon 2 inclusion. Surprisingly, in the context of an infectious provirus, these ESE mutations were reported to not significantly affect the levels of vif mRNA and Vif expression, suggesting that ESEM1 and ESEM2 affect exon 2 definition and exon 2 inclusion but not the creation of vif mRNA (13).
In this study, we investigated splicing elements affecting both exon 2 definition and HIV-1 Vif expression by creating mutations that strengthen or weaken 5′ss D2, mutations within exon 2, and mutations downstream of 5′ss D2. Based on our results, we propose that 5′ss D2, a GGGG splicing silencer proximal to 5′ss D2, and an ESE act competitively by interacting with host factors to determine the level of vif mRNA splicing and Vif expression. In addition, we propose that these elements act together to allow the accumulation of unspliced mRNA levels compatible with optimal virus replication.
MATERIALS AND METHODS
Plasmids.
The infectious HIV-1 molecular clone pNL4-3 was obtained from the NIH AIDS Research and Reference Program (1). pNLD2up has been described previously (17). The mutant derivatives of pNL4-3 shown in Fig. 2 were generated using a QuikChange site-directed mutagenesis kit (Stratagene) as described previously (18). The β-galactosidase expression plasmid pCMV110 was obtained from Tom Hope, Northwestern University School of Medicine.
FIG. 2.
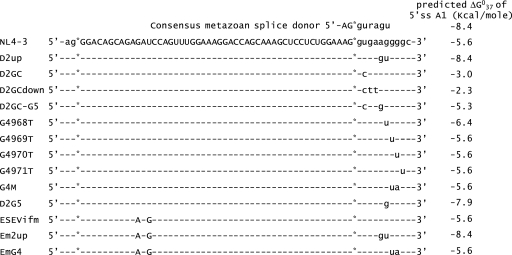
Mutations of pNL4-3 used in this study. The sequence of exon 2 is shown in uppercase letters, and the surrounding intron sequences are shown in lowercase letters. The location of the changes in sequence for each of the mutants is shown. The predicted ΔG037 in kcal/mol for the eight base pairs of U1 snRNA binding to wild-type and mutated 5′ss D2 was calculated by the method described by Serra and Turner (24).
Cell transfection.
293T cells growing in Dulbecco's modified Eagle's medium with 10% fetal bovine serum were transfected with pNL4-3 or with mutants of pNL4-3, using calcium phosphate precipitation as previously described (18). To monitor transfection efficiencies, cells were cotransfected with a lacZ expression plasmid, CMV110. The cells and culture medium were harvested at 24 to 48 h posttransfection. Aliquots of the cell extracts were used to determine β-galactosidase-specific enzyme activity (23). The remainder of the cells were disrupted by using Tri-Reagent (Molecular Research Center, Inc.), and total cellular RNA and protein were isolated according to the protocol supplied by the manufacturer.
Analysis of viral RNA species.
Reverse transcription (RT)-PCR was used to detect HIV-1 mRNA species in the ∼1.8-kb and ∼4-kb mRNA size classes, and Northern blotting analysis of the total RNA was performed as previously described (18). Blots were hybridized to a 32P-labeled probe generated from the 422-nt XhoI/BamHI restriction fragment in the 3′ untranslated region of pNL4-3, which is present in all HIV-1 mRNAs.
Quantitative real-time RT-PCR analysis of vif mRNA.
cDNA was synthesized from total cellular RNA, using Moloney murine leukemia virus reverse transcriptase (RT) and random hexamers, as previously described (6). Real-time PCR analysis to quantify vif and β-actin mRNAs was performed with an Applied Biosystems model 7300 real-time PCR system using an absolute RNA quantification method. PCR amplification was followed by detection with SYBR Green. Primers used for specific amplification of vif mRNA were D1-A1 (forward), GGCGACTGGGACAGC, and Vif body (reverse), CACACAATCATCACCTGCC. Primers used for the amplification of β-actin mRNA were β-actin-5′ (forward), GCTCCTCCTGAGCGCAAG, and β-actin-3′ (reverse), CATCTGCTGGAAGGTGGACA. Ratios of the vif mRNA concentration to the β-actin mRNA concentration were then determined. Data for the pNL4-3 mutants were then normalized to wild-type values after corrections were made for the relative transfection efficiencies based on the β-galactosidase assays of transfected cell extracts.
Immunoblotting.
Cellular proteins (50 μg) were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane via the semidry transfer method, and immunoblotted. β-Tubulin was detected with monoclonal antibody (MAb) E7 (Developmental Studies Hybridoma Bank, University of Iowa) diluted 1:1,000. Vif was detected using either MAb 319 diluted 1:50 or polyclonal Ab 2221 diluted 1:1,000. Both Vif antibodies were obtained from the NIH AIDS Research and Reference Reagent Program. Blots were developed with an ECLPlus Western blotting detection system (Amersham) using peroxidase-conjugated sheep anti-mouse immunoglobulin G (MAb E7 and MAb 319) or donkey anti-rabbit immunoglobulin G (Ab 2221).
RT assays and virus production.
Cell-free supernatants were assayed for RT activity by [α-32P]dTTP incorporation as previously described (33). Incorporation of radioactivity was quantitated using an Instant Imager (Packard) program. RT data were normalized to that of the wild type after correction was made for transfection efficiency based on β-galactosidase assays of extracts from transfected cells.
In vitro RNA splicing.
The pdsxΔE and pdsxASLV plasmids were a gift from M. Caputi, Florida Atlantic University, and have been described previously (29, 31). Plasmids for in vitro splicing templates were constructed by inserting annealed oligonucleotides into the ClaI site of dsxΔE (sequence GGACAGCAGAGATCCAGT for dsxWT and GGACAGCAGTCTAGGTGG for dsxA1M). Plasmids were linearized with HindIII for dsxΔE derivatives or with BamHI for dsxASLV and used as templates for the in vitro transcription of 32P-labeled RNA substrates, using T7 polymerase. In vitro splicing assays of purified substrates were carried out essentially as described previously (3).
UV cross-linking and immunoprecipitation.
The wild-type NL4-3 sequence (GGA CAG CAG AGA TCC AGT) or the mutant sequence corresponding to ESEVifm (GGA CAG CAG AAA GCC) were cloned into the pBluescript SK+ plasmid at the KpnI site. In vitro transcription templates (65 nt) were created by PCR amplification of the sequence between the T7 RNA promoter and the sequence downstream of the insertion. 32P-labeled RNA substrates (48 nt) were then synthesized from the PCR product. For UV cross-linking assays, 1 × 105 cpm of RNA substrates were incubated in the presence of the HeLa nuclear extracts (HNE), at the indicated volumes at 30°C for 30 min and cross-linked to bound protein by UV irradiation for 15 min. Unbound substrate was digested with 25 μg RNase A (Fermentas), and the 32P-labeled RNA-protein complexes were resolved on 10% polyacrylamide gels. For immunoprecipitation reactions, 1 × 106 cpm substrate was incubated with 20 μl HNE and cross-linked, and unbound RNA was digested with 100 μg RNase A. After samples were digested, they were incubated at 4°C for 2 h with either 2 μg of pan-SR MAb 16H3 (Zymed) or 1 μg of MAb α-SRp75 (Ptglabs). Complexes were then bound to protein A-conjugated agarose beads, and unbound proteins were removed by washing the beads four times with 20 mM Tris-HCl-0.5 M NaCl-1% NP-40 and once with 20 mM Tris-HCl-0.15 M NaCl-1% NP-40. The bound fractions were solubilized in 5 mM Tris-7 mM SDS-10 mM dithiothreitol-5% glycerol (pH 6.8), heated at 100°C for 3 min, and resolved on a 10% polyacrylamide gel. The immunoprecipitation products were then visualized by autoradiography.
RESULTS
Down mutations of 5′ss D2 decrease exon 2 inclusion and levels of vif mRNA and Vif protein expression.
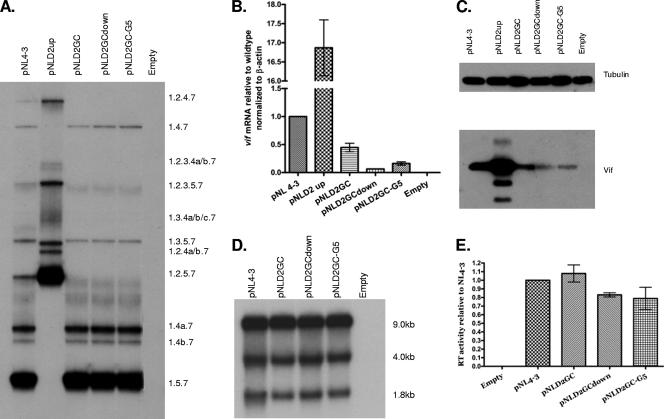
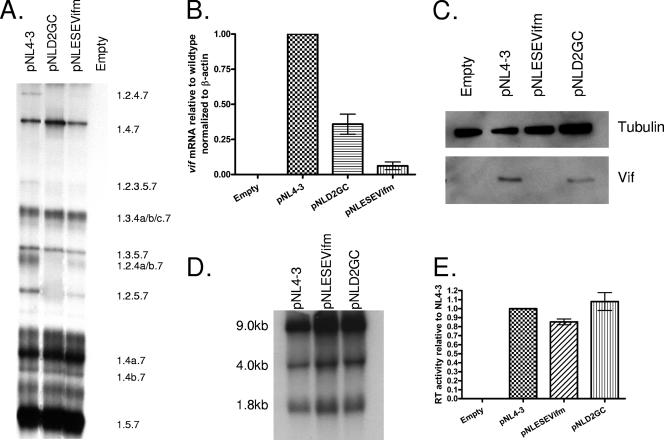
To test the role of the downstream 5′ss in Vif expression, we created 5′ss D2 mutations within the infectious HIV-1 plasmid pNL4-3, and the mutant plasmid was predicted to bind to U1 snRNP with reduced affinity compared to that of the wild type (Fig. 2). To prevent splicing at 5′ss D2, we changed the invariant GU to GC (pNLD2GC). We also changed additional bases, as shown in Fig. 2, that were predicted to either reduce or increase the affinity of U1 snRNP for 5′ss D2 compared to that of pNLD2GC (pNLD2GCdown and pNLD2GC-G5, respectively). The mutant plasmids were transfected into 293T cells, and cells and supernatants from the transfected cells were harvested at 24 to 48 h posttransfection. The extent of exon inclusion was determined by polyacrylamide gel electrophoresis of the RT-PCR products obtained from ∼1.8-kb mRNA (Fig. 3A). As controls for exon 2 inclusion, we compared the profiles of the three down mutations of 5′ss D2 to that of the wild-type pNL4-3 plasmid and to that of the previously described pNLD2up mutant plasmid. This mutant plasmid is predicted to bind to U1 snRNA with a significantly higher affinity than that of the wild type (Fig. 2). As expected from our previous results, exon 2 was spliced into most, if not all, pNLD2up ∼1.8-kb mRNAs, which include nef, rev, and tat mRNA species (17). This can be seen by the absence of RNA species 1.5.7, 1.4b, 1.4a, and 1.4.7 and by the prominence of RNA species 1.2.5.7, 1.2.4b, 1.2.4a, and 1.2.4.7. Exon 2 was also included into most of the pNLD2up plasmid ∼4.0-kb size class mRNAs (data not shown). In contrast, the three GU-to-GC mutants did not include exon 2 into the HIV-1 mRNAs. This can be seen in Fig. 3A by the absence of RNA species 1.2.5.7 and 1.2.4.7, indicating that splicing at D2 was prevented by all three of the GU-to-GC mutants. Exon 2 was also not included into mRNAs from the ∼4.0-kb mRNA size class (data not shown).
FIG. 3.
Mutations decreasing the strength of 5′ss D2 decrease splicing to 3′ss A1 and expression of Vif. (A) Total RNA samples from 293T cells transfected with the indicated plasmids were analyzed by RT-PCR using primer pair BSS and SJ4.7A, specific for completely spliced ∼1.8-kb viral mRNA. HIV-1 mRNA species are indicated on the right side. (B) Total RNA samples from transfected cells were analyzed by qRT-PCR for vif and β-actin mRNAs, and the normalized levels of vif mRNA for the indicated mutants are expressed relative to that of the wild type. (C) Proteins isolated from transfected cells of the wild-type pNL4-3 and the indicated mutants were analyzed by Western blotting using antibodies to Vif and β-tubulin. (D) Northern blotting analysis of total RNA isolated from cells transfected with the wild-type pNL4-3 or the indicated mutants. The positions of the unspliced (∼9-kb), the incompletely spliced (∼4-kb), and the completely spliced (∼1.8-kb) mRNAs are indicated. (E) RT activity of cell-free supernatants from cells transfected with the wild-type pNL4-3 or the indicated mutants.
We next determined the effect of the D2 GC mutations on vif mRNA levels by quantitative real-time RT-PCR (qRT-PCR) assays (Fig. 3B). Each of the GC mutants produced lower levels of vif mRNA than the wild type, with the highest levels seen with pNLD2GC (∼50% of the wild type) and the lowest levels seen with pNLD2GCdown (∼10% of the wild type). The pNLD2GC-G5 mutant demonstrated intermediate levels (∼25% of the wild type). In contrast, pNLD2-up, with an optimized 5′ss D2, produced approximately 17-fold more Vif mRNA than the wild type, consistent with our previous studies which showed elevated Vif expression with this mutant (17). To determine if the levels of vif mRNA were reflected by the levels of Vif protein in transfected cells, we performed Western blotting of cell extracts using Vif antibody (Fig. 3C). These results indicated that the relative levels of Vif protein were consistent with that of the qRT-PCR assays. Note that, in addition to the major Vif protein band, several lower-molecular-weight bands were detected by the anti-Vif antibody. These lower-molecular-weight bands correspond to several truncated forms of Vif, described previously, that are translated using alternative in-frame AUGs within the Vif reading frame (32). Overall, the results, as shown in Fig. 3B and C, indicated that the strength of 5′ss D2, whether splicing occurs at this site or not, is an important determinant of the level of HIV-1 Vif expression.
To test for the effects of down mutations of 5′ss D2 on total HIV-1 RNA splicing, we performed Northern blotting analysis of the total RNA isolated from transfected cells (Fig. 3D). Compared to that found with the wild type, there were no detectable differences in the levels of ∼9-kb unspliced, ∼4-kb incompletely spliced, and ∼1.8-kb incompletely spliced mRNA size classes. We also determined the effect of the down mutations of 5′ss D2 on virus production, by assaying the RT activities of the media harvested from the transfected cells (Fig. 3E). These results indicated that there were no significant differences compared to that of the wild type, indicating that virus production was not significantly affected by the lack of splicing at 5′ss D2.
Up mutations of 5′ss D2 and GGGG motif mutations increase exon 2 inclusion and levels of vif mRNA and Vif protein expression.
The above-described results suggested the importance of the strength of 5′ss D2 for HIV-1 Vif expression. As shown in Fig. 2, mutations of GU to GC would be predicted to reduce the affinity of U1 snRNP, and indeed, the expression of Vif was reduced in the pNLD2GC-transfected cells compared to that of the wild type. Furthermore, Vif expression was further reduced in pNLD2GCdown-transfected cells. The affinity of U1 snRNP for 5′ss D2 in the pNLD2GC-G5 mutant was predicted to be greater than that in pNLD2GC. However, in contrast to our expectation, Vif expression in pNLD2GC-G5-transfected cells was lower than that in pNLD2GC-transfected cells. This suggested the possibility that an additional element or elements may regulate the usage of 5′ss D2. Previous studies have shown that a GGGG motif proximal to the 5′ss of the brain region-specific C1 cassette exon of the N-methyl-d- aspartic acid (NMDA)-type glutamate receptor NR1 subunit (GRIN1) transcript acts to silence inclusion of the C1 exon (9). Several GGGG motifs in the intron sequences upstream and downstream of alternative exon 4 of the HLA-DQB1 gene play a role in the differential inclusion of exon 3 into mRNA (15). In the HIV-1 genome, there is a GGGG motif which overlaps the last base of the U1 snRNP binding site and is just proximal to the 5′ss D2 sequence. In the pNLD2GC-G5 mutant, the number of G's was increased, resulting in a GGGGG sequence.
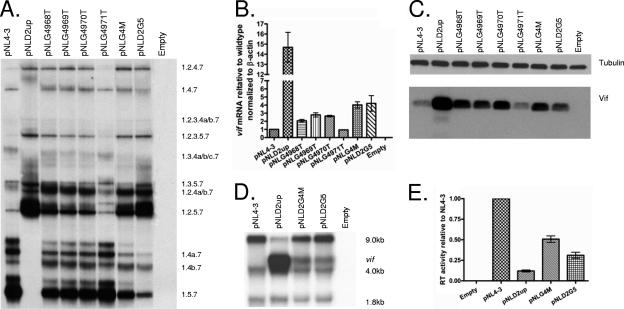
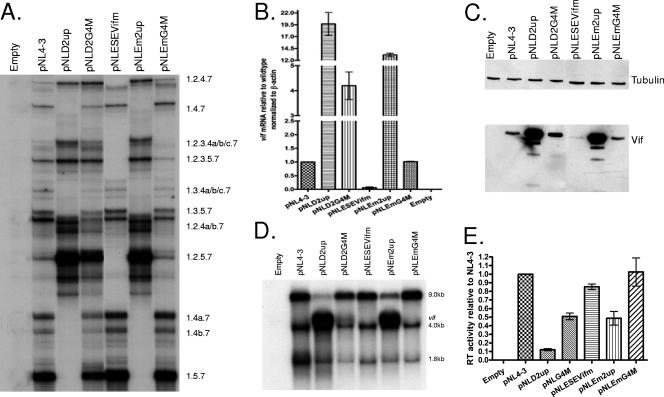
We tested for the effects of the potential 5′ss-proximal GGGG silencer motif on splicing and on Vif expression by creating the mutations shown in Fig. 2. As shown in Fig. 4A, mutations within the first three bases of the potential silencer motif (pNLG4M, pNLG4968T, pNLG4969T, and pNLG4970T) resulted in the increased inclusion of exon 2, as evidenced by increased amounts of the RNA species 1.2.5.7, 1.2.4a/b.7, and 1.2.4.7 relative to those of the wild-type pNL4-3. These mutants demonstrated significantly less exon 2 inclusion than pNLD2up, which is predicted to inactivate the putative GGGG silencer and increase the affinity of D2 for U1 snRNP. On the other hand, a G-to-T mutation of the last base of the GGGG motif (pNLG4971T) had little effect on exon 2 inclusion. The effects of the GGGG mutations were also seen with studies of vif mRNA levels (Fig. 4B) and Vif protein expression (Fig. 4C). Mutants with changes within the GGGG motif that increased exon inclusion exhibited vif mRNA levels two- to fourfold higher than the wild type but considerably lower than pNLD2up, which in these experiments expressed vif mRNA levels approximately 15-fold higher than the wild type. The pNLG4971T mutant, on the other hand, did not demonstrate an elevation of Vif expression relative to that of the wild type. Interestingly, the pNLD2-G5 mutant, in which the length of the GGGG motif was increased but in which the predicted affinity of 5′ss D2 for U1 snRNP was also increased, showed elevated inclusion of exon 2 (Fig. 4A) and increased Vif expression (Fig. 4B and C).
FIG. 4.
Mutations increasing the strength of 5′ss D2 or in a GGGG motif proximal to 5′ss D2 increase splicing at 3′ss A1 and the expression of Vif while decreasing virus production. (A) Total RNA samples from 293T cells transfected with the indicated plasmids were analyzed by RT-PCR using primers specific for completely spliced ∼1.8-kb viral mRNA. HIV-1 mRNA species are indicated on the right side. (B) Total RNA samples from transfected cells were analyzed by qRT-PCR for vif and β-actin mRNAs, and the normalized levels of vif mRNA for the indicated mutants are expressed relative to that of the wild type. (C) Proteins isolated from transfected cells of the wild-type pNL4-3 and the indicated mutants were analyzed by Western blotting using antibodies to Vif and β-tubulin. (D) Northern blotting of total RNA isolated from cells transfected with the wild-type pNL4-3 and the indicated mutants. The positions of unspliced (∼9-kb), incompletely spliced (vif, ∼4-kb), and completely spliced (∼1.8-kb) mRNAs are indicated. (E) RT activity of cell-free supernatants from cells transfected with the wild-type pNL4-3 or the indicated mutants.
We next compared the effects of the GGGG mutations and the up mutations of 5′ss D2 on total HIV-1 splicing by using Northern blotting analysis of the total RNA from transfected cells (Fig. 4D) and analysis of virus production, by RT assays of cell supernatants (Fig. 4E). We have previously reported that pNLD2up demonstrates an excessive splicing phenotype characterized by greatly reduced levels of unspliced viral RNA and reduced levels of Gag protein (17). In the experiment shown in Fig. 4D, we found that in addition to the normal band corresponding to the wild-type ∼4-kb mRNA band, a slower-migrating band was detected that corresponded to the size expected for vif mRNA. This upper band was not detectable in the wild-type RNA because of the normally low abundance of vif mRNA. The pNLG4M mutant, which inactivates the GGGG motif but does not increase the predicted affinity of D2 for U1 snRNP, and the pNLD2-G5 mutant, which is predicted to increase the affinity of D2 for U1 snRNP but not impinge on the GGGG motif, also demonstrated increased levels of ∼4-kb vif mRNAs. In addition, these two mutants produced virus particles at levels between that of the wild type and that of the pNLD2up mutant, whose virus particle production in this experiment was approximately eightfold reduced compared to that of the wild type (Fig. 4E). We concluded from these results that the level of splicing at 3′ss A1 is determined by both the affinity of U1 snRNP for 5′ss D2 and the GGGG silencer 3′ proximal to 5′ss D2. Both exon 2 inclusion and vif mRNA levels are increased by mutations within the 5′ss D2 that increase its affinity for U1 snRNP and by mutations of the GGGG silencer motif. Increased splicing at 3′ss A1 is concomitant with reduced virus production, and the extent of the reduction correlates with the extent to which the viral RNA is spliced.
An ESE binding selectively to SRp75 promotes exon 2 inclusion and levels of vif mRNA and Vif protein expression.
As discussed above, several ESS and ESE elements that promote or inhibit RNA splicing have previously been identified within the HIV-1 genome. Therefore, we tested for the possibility that exon 2 contained additional cis elements that contributed to the regulation of splicing at 3′ss A1. Preliminary experiments performed in the context of a gag/pol-deleted pNL4-3 plasmid indicated that point mutations and deletions within the 5′-proximal region of exon 2 resulted in a decreased inclusion of exon 2 into HIV-1 mRNAs (data not shown). Based on these data, mutations downstream of 3′ss A1 at nt 4923 and 4925 in pNL4-3 were introduced to create the pNL-ESEVifm mutant. This mutant was transfected into 293T cells, and at 24 to 48 h posttransfection, cells and cell supernatants were harvested. RT-PCR was used to detect viral RNA species in the mutant ∼1.8-kb HIV-1 mRNA size class, and this result was compared to that of the wild-type pNL4-3 and pNLD2GC (Fig. 5A). The mRNA species profile of pNL-ESEVifm was similar to that of pNLD2GC, indicating an almost complete absence of exon 2 inclusion. A decrease in exon 2 inclusion to undetectable levels was also observed with the ∼4.0-kb mRNA size class (data not shown). Analysis of total RNA from pNL-ESEVif by qRT-PCR indicated that the vif mRNA was present at a level that was approximately 3% of that of the wild type (Fig. 5B). Protein expression, as determined by Western blotting analysis of cell lysates, detected little or no Vif and was in agreement with the qRT-PCR data (Fig. 5C).
FIG. 5.
A mutation within exon 2 decreases splicing to 3′ss A1 and expression of Vif. (A) Total RNA samples from 293T cells transfected with the indicated plasmids were analyzed by RT-PCR using primers specific for completely spliced ∼1.8-kb mRNA. HIV-1 mRNA species are indicated on the right. (B) Total RNA samples from transfected cells were analyzed by qRT-PCR for vif and β-actin mRNAs, and the normalized levels of vif mRNA for the indicated mutants are expressed relative to that of the wild type. (C) Proteins isolated from transfected cells of the wild-type pNL4-3 and the indicated mutants were analyzed by Western blotting using antibodies to Vif and β-tubulin. (D) Northern blotting of total RNA isolated from cells transfected with the wild-type pNL4-3 and the indicated mutants. The positions of unspliced (∼9-kb), incompletely spliced (∼4-kb), and completely spliced (∼1.8-kb) mRNAs are indicated on the right. (E) Reverse transcriptase activity of cell-free supernatants from cells transfected with the wild-type pNL4-3 or the indicated mutants.
Analysis of pNL-ESEVifm RNA by Northern blotting indicated that the relative amounts of ∼9-kb, ∼4-kb, and ∼1.8-kb size class mRNAs were similar to those of the wild type (Fig. 5D). Furthermore, mutant virus particle production was near wild-type levels, as determined by RT assays of transfected cell supernatants (Fig. 5E). Thus, the mutations in pNL-ESEVifm, except for the reduced levels of Vif mRNA and Vif protein, did not affect virus replication.
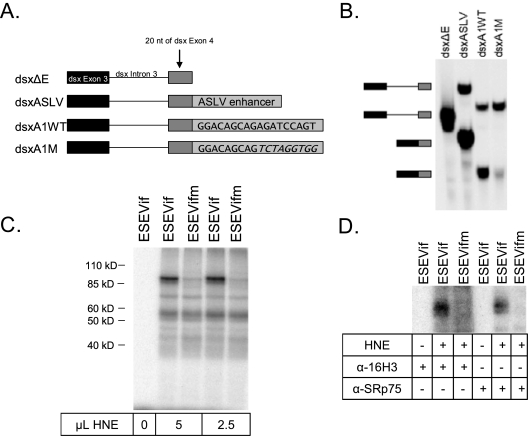
These results suggested that the 5′-proximal region of exon 2 contains an ESE. To test for the presence of an ESE in this region, we inserted the 5′-proximal 18 nt of exon 2 into a Drosophila dsx heterologous in vitro-splicing construct (Fig. 6A). In vitro splicing was performed using HNE. All host factors required for the regulation of HIV-1 splicing appear to be present in HeLa cells, since the types and levels of spliced HIV-1 mRNA species in transfected HeLa cells are similar to those of infected primary T cells (21). Splicing of RNA substrates produced from the Drosophila dsx splicing construct is dependent upon the presence of an ESE, as shown in Fig. 6B. Substrate dsxΔE, which lacks an ESE, is poorly spliced, whereas substrate dsxASLV, which contains an ESE from the avian sarcoma-leukosis virus (8), is spliced. When splicing substrates contained the wild-type but not the mutant HIV-1 sequence (dsxA1WT and dsxA1M, respectively), the spliced product of the expected size accumulated (Fig. 6). It should be noted that the sequences used in this ESE assay did not include the region of exon 2 that contained the previously described ESEM1 and ESEM2 elements (13). Our results suggest that a novel ESE downstream of HIV-1 3′ss A1 promotes the inclusion of exon 2, as well as the creation of vif mRNA.
FIG. 6.
A novel ESE within exon 2 that binds selectively to SRp75. (A) Diagram of the in vitro RNA substrates used. (B) Representative in vitro splicing assays of HNE was performed with the indicated RNA substrates. Positions of the unspliced RNA precursors and spliced products are indicated on the left. (C) UV cross-linking of the indicated substrates with different amounts of HNE was performed, and the labeled proteins were resolved by polyacrylamide gel electrophoresis. (D) UV cross-linked products were immunoprecipitated with the indicated antibodies (α-16H3 and α-SRp75).
ESE elements bind to members of the SR protein family, which are characterized by arginine-serine repeats, as well as by one or more RNA recognition motifs. To determine which SR protein binds to ESEVif, short 32P-labeled RNAs containing wild-type or mutated ESEVif were transcribed and incubated with HNE, followed by UV cross-linking of bound proteins to the RNA. A specific band with an apparent molecular mass of approximately 90 kDa was detected in the wild-type but not in the mutant RNA cross-linked product (Fig. 6C). This labeled band was immunoprecipitated with both a pan-SR antibody, 16H3, and an antibody specific for the SR protein, SRp75 (Fig. 4D). We concluded from our results that ESEVif specifically binds to SRp75 and not to other SR proteins.
ESE, GGGG, and 5′ss D2 act competitively to determine splicing at 3′ss A1 and the level of Vif expression.
To determine the relative strengths of the three elements regulating splicing at 3′ss A1 and the level of Vif expression (downstream suboptimal 5′ss D2, GGGG silencer, and ESEVif), we created double mutants in the pNL4-3 backbone, with both the ESEVif mutation (Em) and either the D2up or the G4 mutation (pNLEm2up and pNLEmG4M, respectively). These two double mutants were transfected into 293T cells, and the cells were analyzed for the Vif gene expression and virus production. To determine the level of exon 2 inclusion, we used RT-PCR to amplify the ∼1.8-kb size class mRNAs (Fig. 7A). The results of this experiment indicated that the inclusion of exon 2 in the pNLEm2up double mutant was elevated and did not appear to be significantly different from than that in pNLD2up. On the other hand, the inclusion of exon 2 in the pNLEmG4M double mutant was reduced compared to that in pNLG4M and more similar to that of the wild type. This was indicated by a relative decrease in mRNA species, which include exon 2 (1.2.5.7, 1.2.4a.7, 1.2.4b.7, and 1.2.4.7), and a relative increase in mRNA species, in which exon 2 is skipped (1.5.7, 1.4a.7, 1.4b.7, and 1.4.7). The pNL-ESEVifm mutant, as shown in Fig. 5A, demonstrated little or no inclusion of exon 2 into viral mRNAs.
FIG. 7.
Negative and positive splicing elements act competitively to affect splicing to 3′ss A1, the expression of Vif, and virus production. (A) Total RNA samples from 293T cells transfected with the indicated plasmids were analyzed by RT-PCR using primers specific for completely spliced ∼1.8-kb viral mRNA. HIV-1 mRNA species are indicated on the right side. (B) Total RNA samples from transfected cells were analyzed by qRT-PCR for vif and β-actin mRNAs, and the normalized levels of vif mRNA for the indicated mutants are expressed relative to that of the wild type. (C) Proteins isolated from transfected cells of the wild-type pNL4-3 and the indicated mutants were analyzed by Western blotting using antibodies to Vif and β-tubulin. (D) Northern blotting of total RNA isolated from cells transfected with the wild-type pNL4-3 and the indicated mutants. The positions of unspliced (∼9-kb), incompletely spliced (vif, ∼4-kb), and completely spliced (∼1.8-kb) mRNAs are indicated. (E) RT activity of cell-free supernatants from cells transfected with the wild-type pNL4-3 or the indicated mutants.
We next analyzed Vif expression in cells transfected with the double mutants. We determined vif mRNA levels by qRT-PCR (Fig. 7B). This experiment indicated that pNLEm2up produced somewhat less vif mRNA (approximately 12-fold higher than that of the wild-type) than pNLD2up (19-fold higher than that of the wild type). The pNLEmG4M mutant produced wild-type levels of vif mRNA compared to that of the single-GGGG silencer pNLG4M mutant, which, in these experiments, produced vif mRNA at a level approximately fourfold higher than that of the wild type. The pNL-ESEVifm mutant, as shown in Fig. 5B, produced very little vif mRNA. As shown in Fig. 7C, the relative levels of Vif protein in extracts from transfected cells were consistent with the relative levels of mRNA seen in Fig. 7B.
We next determined the effect of the ESE double mutations on unspliced, incompletely spliced, and completely spliced viral RNA levels, by Northern blotting (Fig. 7D). As described above and elsewhere, little unspliced mRNA was present in cells transfected with the pNLD2up mutant. In addition, as shown in Fig. 4D, a prominent mRNA band was present which migrated slightly slower than the 4.0-kb mRNA band and at the size expected for single-spliced vif mRNA. Cells transfected by the pNLEm2up double mutant also exhibited a prominent vif mRNA band. However, in these cells, there was a higher level of unspliced mRNA than in pNLD2up-transfected cells. In cells transfected with the pNLG4M GGGG silencer mutant, a distinct vif mRNA band was present, as well as reduced levels of unspliced mRNA, compared to that of the wild type as shown in Fig. 4D. On the other hand, a vif mRNA band was not detected in cells transfected with the pNLEmG4M double mutant. Furthermore, these cells appeared to accumulate an increased level of unspliced mRNA comparable to that of the wild type. In contrast to the data shown in Fig. 4D, the intensity of the 1.8-kb mRNA bands of all the mutant RNA samples seen in Fig. 7D appeared to be reduced relative to that of the 4.0-kb mRNA bands. This difference was not reproduced in other repetitions of this experiment.
Finally, we compared the effect of the ESE double mutants on virus production to that of the single mutants by performing RT assays of supernatants from the transfected cells (Fig. 7E). The pNLEm2up double mutant produced significantly higher levels of particles than the pNLD2up mutant, consistent with the higher relative accumulation of 9-kb unspliced RNA in the pNLEm2up-transfected cells than in the pNLD2up-transfected cells, as shown in Fig. 7D. Similarly, the pNLEmG4M double mutant produced higher levels of particles than pNLG4M, consistent with the higher relative accumulation of unspliced mRNA observed for pNLEmG4M-transfected cells than for pNLG4M-transfected cells, as shown in Fig. 7D.
From the results with the ESEVifm/D2up and the ESEVifm/D2G4M double mutants, we concluded that the suboptimal 5′ss D2, the GGGG silencer, and ESEVif enhancer act competitively to determine the level of splicing at 3′ss A1. In this way, the levels of exon 2 inclusion and vif mRNA and Vif protein expression are regulated. In addition, the overall level of HIV-1 splicing is maintained to allow the accumulation of unspliced mRNA compatible with optimal virus production.
DISCUSSION
Based on the results of this study and previous results, we propose the model shown in Fig. 8 for the regulation of splicing at 3′ss A1. To date, three different positively acting exon 2 ESEs have been identified. Two 5′-distal ESEs (ESEM1 and ESEM2) contain the sequence UGGAAAG (13). In this report, we have shown evidence for a third ESE (ESEVif) within the 5′-proximal region of exon 2. ESEM1 and ESEM2 have been reported to facilitate exon 2 inclusion into mRNAs but not to affect the expression of vif mRNA (13). In contrast, we showed in this report that ESEVif promotes the production of vif mRNA, as well as exon 2 inclusion. We have also detected a negatively acting GGGG silencer element proximal to 5′ss D2 which represses both exon 2 inclusion and vif mRNA production. A third splicing element is the suboptimal downstream 5′ss D2 itself; the affinity of this splice site for U1 snRNP determines its influence on splicing at 3′ss A1. We have shown that the ESEVif, the GGGG silencer element, and downstream 5′ss act competitively to regulate vif mRNA levels.
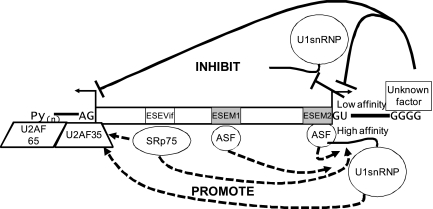
FIG. 8.
Proposed model for vif mRNA splicing regulation. The results presented in this study and in a previous study (13) indicate that splicing at 3′ss A1 and 5′ss D2 is regulated at an early step of splicing by positively acting ESEs that promote splicing (dashed lines) and a negatively acting GGGG silencer (solid line) that inhibits splicing. In addition, the wild-type 5′ss D2 is suboptimal; mutagenesis that lowers the affinity of 5′ss D2 for U1 snRNP inhibits splicing at 3′ss A1 (solid line), whereas mutagenesis that increases the affinity of 5′ss D2 for U1 snRNP promotes splicing at 3′ss A1 (dashed line). ESEM1 and ESEM2 have been previously reported to be binding sites for the SR protein SF2/ASF (13). ESEVif binds selectively to the SRp75 protein. The silencing activity of the GGGG motif is thought to be mediated by a cellular factor or factors that have not yet been identified. We cannot conclude yet whether the major effects of the GGGG motif and ESEVif are on 5′ss recognition, 3′ss recognition, or recognition of both splice sites. The overall outcome of these competing positive and negative factors binding downstream of 3′ss A1 may be to determine the binding affinity of U2AF65/U2AF35 to the polypyrimidine tract (Py)n and 3′ss A1, which is one of the earliest steps in splicing.
In the model shown in Fig. 8, we propose that the regulation of splicing at 3′ss A1 involves the interaction of multiple cellular factors binding to the cis splicing elements within the viral genome. It has been reported that ESEM1 and ESEM2 bind to the SR protein ASF/SF2 (13). We have shown herein that ESEVif specifically binds to the SR protein SRp75, and this suggests that this protein is responsible for the activation of splicing at 3′ss A1. It has been previously shown by Han et al. (9) that the hnRNP H and F proteins bind to GGGG motifs proximal to 5′ss of the C1 exon of the GRIN1 transcript but that, in this context, these proteins appear to act as antagonists of splicing silencing rather than as silencers themselves. This result suggested that the 5′ss proximal GGGG motif may mediate silencing by binding to another cellular protein or proteins. It was speculated by Han et al. that hnRNP A1 may mediate silencing by cooperative binding to GGGG motifs and UAGG ESS elements within the C1 exon of the GRIN1 transcript (9). This does not appear to be the case with the HIV-1 5′ss proximal GGGG silencer, since there are no UAGG sequences within HIV-1 exon 2, and to date, we have not identified any other sequences within HIV-1 noncoding exon 2 that would allow such cooperative interactions with hnRNP A1.
We showed that G-to-T mutations of the second and third G residue of the GGGG motif (G4969T and G4970T) resulted in increased exon 2 inclusion and a three- to fourfold increase in vif mRNA levels relative to that of the wild type. A G-to-T mutation of the first G residue resulted in a somewhat lower increase of vif mRNA level, whereas a G-to-T mutation of the fourth G residue of the GGGG motif had little or no effect on exon 2 inclusion or vif mRNA level. It should be noted that mutations of the first residue of the GGGG motif are also predicted to change the affinity of 5′ss D2 for U1 snRNP. Therefore, the contribution of each of the two elements to the splicing changes produced by these mutations cannot be discerned. It has previously been shown that several GGGG motifs in the intron sequences upstream and downstream of the alternative exon 4 of the HLA-DQB1 gene play a role in the differential inclusion of exon 4 into mRNA. Mutations of the upstream GGGG motifs result in decreased exon 4 inclusion, suggesting that they act as intronic splicing enhancers. Alternatively, these mutations may change the structural conformation necessary for splicing. In contrast, mutations of the downstream 5′ss-proximal GGGG motif result in increased splicing, suggesting that in this case, the motif acts as a splicing silencer. It was found in this study that point mutations within the second and third G residues had the most effect on splicing enhancement, but similar mutations in the 5′ss-proximal GGGG silencer were not tested (15). In the case of the brain region-specific C1 cassette exon of the GRIN1 transcript, mutation of the central two G residues of the 5′-proximal GGGG motif was shown to inactivate the silencer activity, but the effect of mutating only the first or fourth G residues of the GGGG motif was not tested (9).
Our studies indicate that, in general, the levels of vif mRNA in the absence of changes in ESEVif and the GGGG silencer vary according to the predicted binding affinity of U1 snRNP for 5′ss D2. An exception to this general rule is the pNLD2GC-G5 mutant, which would be expected to express levels of vif mRNA higher than those of pNLD2GC, based on its predicted U1 snRNP binding. The anomalous behavior of pNLD2GC-G5 may be explained by the increased length of the GGGG motif, resulting in increased silencer activity relative to that of U1 snRNP binding. Interestingly, this same G5 mutant in the context of the wild-type GU sequence (pNLD2-G5) demonstrated an elevated splicing phenotype, as expected from the predicted increased affinity of the mutated 5′ss D2 to U1 snRNP. This suggests that in the context of pNLD2-G5, U1 snRNP binding is dominant over the extended GGGG motif created by the G5 nt change. Further characterization of the GGGG silencer and its hypothetical binding protein(s) is required to fully understand this phenomenon.
It is possible that the 5′ss mutations could affect HIV-1 mRNA stability in addition to their effects on RNA splicing. Indeed, it has been shown previously that in the context of a subgenomic HIV-1 env expression construct containing only 5′ss D5 and 3′ss A7, mutations inactivating 5′ss D5 result in destabilization of the unspliced env RNA (12). In the context of the complete HIV-1 genome, mutations that inactivated a normally cryptic 5′ss within the gag/pol gene resulted in a dramatic reduction in the steady-state level of unspliced RNA (16). In our experiments, we have not seen a reduction in steady-state levels of unspliced RNA as a result of inactivating 5′ss D2. We have also not detected significant differences between the wild type and the 5′ss D2 mutants in terms of the rate of decay of vif mRNA after the addition of the transcription inhibitor actinomycin D (Z. Feng and C. M. Stoltzfus, unpublished data). Thus, our results suggest that the primary effects of the mutations we have studied are on HIV-1 mRNA splicing and not on RNA stability.
Both the removal of repression of splicing at 3′ss A1 by inactivation of the GGGG silencer element and the increase in the strength of 5′ss D2 resulted in decreased virus production. This presumably is due to a skewing of the balance of normal HIV-1 splicing, resulting in a relative decrease in the level of unspliced mRNA and a consequent decrease in the level of Gag and Gag-Pol proteins as we have previously shown for the D2up mutant (17) and the D2A3 mutant, which has a G-to-A change at position +3 of D2 (19). The negative effects of the GGGG mutations on virus production can be partially or completely abrogated by second-site mutations within ESEVif. This is further evidence that the splicing elements act competitively to allow optimal levels of virus replication.
The results presented here and elsewhere have indicated that the presence or absence of noncoding exon 2 or 3 within the 5′ leaders of viral mRNAs does not significantly affect virus production or mRNA stability or the expression of viral mRNAs (17, 18). Thus, the inclusion of exon 2 and exon 3 into some of the HIV-1 mRNAs during virus replication may be a by-product and reflect the necessity of a functional downstream 5′ss D2 and D3 for the expression of optimum levels of Vif and Vpr, respectively. In the case of Vif, optimal levels of expression in infected cells may be necessary both to prevent the accumulation and packaging of APOBEC3G and APOBEC3F deoxycytidine deaminases (14, 20, 25, 34) and to avoid the accumulation of excess Vif, which has been shown to inhibit virus protein processing and virus replication (2).
It is curious that the level of splicing at 3′ss A1 is regulated by such a complex set of splicing elements, which involve three different ESEs, the GGGG silencer, and a suboptimal downstream 5′ss. The importance of these three elements is suggested by their sequence conservation in all HIV-1 clades. There are several possibilities to explain the evolution of this seemingly cumbersome system to regulate vif splicing. First, exon 2 and the flanking vif mRNA intron sequences overlap the integrase protein reading frame. Because of this overlap, the HIV-1 sequence must be compatible with both the requirements for optimum Vif levels and for integrase function. Thus, the necessity of both functions for efficient virus replication may require multiple positive and negative cis splicing elements to achieve the necessary level of splicing to produce optimal levels of vif mRNA. Second, the presence of multiple splicing elements may be important to allow regulation of the levels of Vif in different cell types or at different stages of virus life cycle. This regulation may be affected by the relative concentrations of the RNA binding proteins that bind to the cognate negative and positive RNA splicing elements.
Acknowledgments
We thank Dibyakanti Mandal for critical evaluation of the manuscript. We acknowledge the NIH AIDS Research and Reference Reagent Program for supplying HIV-1-related reagents. We thank Max Caputi, Florida Atlantic University, for the dsx splicing template plasmids and Tom Hope, Northwestern University School of Medicine, for the pCMV110 β-galactosidase expression plasmid.
HeLa cells were obtained from the National Cell Culture Center, which is sponsored by the National Center for Research Resources of the NIH. Monoclonal antibody E7 was obtained from the Development Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA.
This research was supported by PHS grant AI36073 from the National Institute of Allergy and Infectious Diseases.
C.M.E. was supported by predoctoral training grant T32AI007533 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akari, H., M. Fujita, S. Kao, M. A. Khan, M. Shehu-Xhilaga, A. Adachi, and K. Strebel. 2004. High level expression of human immunodeficiency virus type-1 Vif inhibits virus infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J. Biol. Chem. 27912355-12362. [DOI] [PubMed] [Google Scholar]
- 3.Amendt, B. A., D. Hesslein, L.-J. Chang, and C. M. Stoltzfus. 1994. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of the human immunodeficiency virus type 1. Mol. Cell Biol. 143960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amendt, B. A., Z.-H. Si, and C. M. Stoltzfus. 1995. Presence of exon splicing silencers within HIV-1 tat exon 2 and tat/rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 154606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 2702411-2414. [DOI] [PubMed] [Google Scholar]
- 6.Bilodeau, P. S., J. K. Domsic, A. Mayeda, A. R. Krainer, and C. M. Stoltzfus. 2001. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 758487-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butsch, M., and K. Boris-Lawrie. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 763089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, X.-D., R. A. Katz, A. M. Skalka, and T. Maniatis. 1991. The role of branchpoint and 3′-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 5211-220. [DOI] [PubMed] [Google Scholar]
- 9.Han, K., G. Yeo, P. An, C. B. Burge, and P. J. Grabowski. 2005. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 3e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman, B. E., and P. J. Grabowski. 1992. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 62554-2568. [DOI] [PubMed] [Google Scholar]
- 11.Jacquenet, S., A. Mereau, P. S. Bilodeau, L. Damier, C. M. Stoltzfus, and C. Branlant. 2001. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J. Biol. Chem. 27640464-40475. [DOI] [PubMed] [Google Scholar]
- 12.Kammler, S., C. Leurs, M. Freund, J. Krummheuer, K. Seidel, T. O. Tange, M. K. Lund, J. Kjems, A. Scheid, and H. Schaal. 2001. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA 7421-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kammler, S., M. Otte, I. Hauber, J. Kjems, J. Hauber, and H. Schaal. 2006. The strength of the HIV-1 3′ splice sites affects Rev. function. Retrovirology 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 7711398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kralovicova, J., and I. Vorechovsky. 2006. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J. Immunol. 1762381-2388. [DOI] [PubMed] [Google Scholar]
- 16.Lutzelberger, M., L. S. Reinert, A. T. Das, B. Berkhout, and J. Kjems. 2006. A novel splice donor site in the gag-pol gene is required for HIV-1 RNA stability. J. Biol. Chem. 28118644-18651. [DOI] [PubMed] [Google Scholar]
- 17.Madsen, J. M., and C. M. Stoltzfus. 2006. A suboptimal 5′ splice site downstream of HIV-1 splice site A1 is required for unspliced viral mRNA accumulation and efficient virus replication. Retrovirology 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madsen, J. M., and C. M. Stoltzfus. 2005. An exonic splicing silencer downstream of 3′ splice site A2 is required for efficient human immunodeficiency virus type 1 replication. J. Virol. 7910478-10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandal, D., Z. Feng, and C. M. Stoltzfus. 2008. Gag-processing defect of human immunodeficiency virus type 1 integrase E246 and G247 mutants is caused by activation of an overlapping 5′ splice site. J. Virol. 821600-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 91398-1403. [DOI] [PubMed] [Google Scholar]
- 21.Purcell, D. F. J., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication and infectivity. J. Virol. 676365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robberson, B. L., G. J. Cote, and S. M. Berget. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 1084-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Serra, M., and D. Turner. 1995. Predicting thermodynamic properties of RNA. Methods Enzymol. 259242-261. [DOI] [PubMed] [Google Scholar]
- 25.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 41397-1400. [DOI] [PubMed] [Google Scholar]
- 26.Si, Z.-H., D. Rauch, and C. M. Stoltzfus. 1998. The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol. 185404-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staffa, A., and A. Cochrane. 1995. Identification of positive and negative splicing regulatory elements within the terminal tat/rev exon of HIV-1. Mol. Cell. Biol. 154597-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoltzfus, C. M., and J. M. Madsen. 2006. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV Res. 443-55. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, K., A. Watakabe, and Y. Shimura. 1994. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol. Cell. Biol. 141347-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tange, T. O., C. K. Damgaard, S. Guth, J. Valcarcel, and J. Kjems. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 205748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian, M., and T. Maniatis. 1993. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell 74105-114. [DOI] [PubMed] [Google Scholar]
- 32.Wang, H., A. Sakurai, B. Khamsri, T. Uchiyama, H. Gu, A. Adachi, and M. Fujita. 2005. Unique characteristics of HIV-1 Vif expression. Microbes Infect. 7385-390. [DOI] [PubMed] [Google Scholar]
- 33.Willey, R. L., D. H. Smith, L. A. Lasky, T. S. Theodore, P. L. Earl, B. Moss, D. J. Capon, and M. A. Martin. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J. Virol. 62139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X.-F. Yu. 2003. Induction of APOBEC3G ubiquitation and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]
- 35.Zahler, A. M., C. K. Damgaard, J. Kjems, and M. Caputi. 2004. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 27910077-10084. [DOI] [PubMed] [Google Scholar]