Abstract
Research over the last few years has demonstrated the increasing role of microRNAs (miRNAs) as major regulators of gene expression in diverse cellular processes and diseases. Several viruses, particularly herpesviruses, also use the miRNA pathway of gene regulation by encoding their own miRNAs. Marek's disease (MD) is a widespread lymphomatous neoplastic disease of poultry caused by the highly contagious Marek's disease virus type 1 (MDV-1). Recent studies using virus-infected chicken embryo fibroblasts have identified at least eight miRNAs that map to the RL/RS region of the MDV genome. Since MDV is a lymphotropic virus that induces T-cell lymphomas, analysis of the miRNA profile in T-cell lymphoma would be more relevant for examining their role in oncogenesis. We determined the viral and host miRNAs expressed in MSB-1, a lymphoblastoid cell line established from an MDV-induced lymphoma of the spleen. In this paper, we report the identification of 13 MDV-1-encoded miRNAs (12 by direct cloning and 1 by Northern blotting) from MSB-1 cells. These miRNAs, five of which are novel MDV-1 miRNAs, map to the Meq and latency-associated transcript regions of the MDV genome. Furthermore, we show that miRNAs encoded by MDV-1 and the coinfected MDV-2 accounted for >60% of the 5,099 sequences of the MSB-1 “miRNAome.” Several chicken miRNAs, some of which are known to be associated with cancer, were also cloned from MSB-1 cells. High levels of expression of MDV-1-encoded miRNAs and potentially oncogenic host miRNAs suggest that miRNAs may have major roles in MDV pathogenesis and neoplastic transformation.
Marek's disease (MD) is a naturally occurring rapid-onset aggressive T-cell lymphoma of poultry. Named after the Hungarian veterinarian Jośzef Marek, who first reported the disease in 1907 (41), the disease is induced by Marek's disease virus type 1 (MDV-1), a highly contagious alphaherpesvirus belonging to the genus Mardivirus of the family Herpesviridae (31). Apart from being a major disease affecting poultry health and welfare, MD is considered to be an excellent biomedical model for virus-induced lymphoma (7, 14). Among the 100-plus genes predicted for the MDV genome (40, 47, 48), the gene for the basic leucine zipper protein Meq is considered to be the major oncogene (39, 44). Some of the functions of Meq associated with oncogenic properties, such as its interaction with CtBP, parallel those of other viral oncogenic sequences, such as adenovirus E1A and Epstein-Barr virus (EBV) nuclear antigens EBNA3A and -3C (6), highlighting the convergent evolution of oncogenic pathways in these viruses. Recent studies have also identified the role of other genes, such as the pp38 (23), viral interleukin-8 (vIL-8) (49), ICP4 (15, 38), R-LORF4 (33), UL36 (32), and MDV-encoded telomerase RNA (22, 63) genes, in pathogenesis.
Increasing evidence demonstrates that in addition to the direct role of protein-encoding genes, noncoding RNAs have profound effects in mediating neoplastic transformation (13). Among these, the 22-nucleotide microRNAs (miRNAs) have emerged as a major regulatory tier of gene expression, with the potential of targeting up to 30% of genes in humans (17, 27, 37). Given their small size with the capability for regulating multiple genes, several viruses have adopted the miRNA machinery to manipulate the cellular and viral pathways of gene regulation by encoding their own miRNAs (19, 24, 26, 42). Among the different families of viruses, herpesviruses have exploited the miRNA-mediated gene regulation pathway most successfully, since 124 of the 127 virus-encoded miRNAs in miRBase release 10.1 (http://microrna.sanger.ac.uk) are encoded by herpesviruses. It has been suggested that the miRNA-mediated regulatory mechanisms are very suited for the herpesvirus life cycle, which is characterized by nuclear replication and latent periods with minimal antigen expression (19).
Specific miRNA signatures in different types of tumors have been identified using high-throughput microarray analysis of miRNA expression (60, 64, 67). Considering the aggressive nature and rapid onset of tumors induced by MDV-1, analysis of the miRNA profile of MDV-transformed tumor cells could provide further insights into MD oncogenesis. Previous studies using small RNAs from MDV-infected chicken embryo fibroblasts (CEF) have identified several miRNAs, including eight MDV-encoded miRNAs that mapped to the Meq and the latency-associated transcript (LAT) region of the genome (8, 9). Although identification of MDV and host miRNAs in lytically infected CEF is valuable, understanding the expression profiles of miRNAs in the lymphocyte target cells of MD lymphomas would be crucial to delineate their role in neoplastic transformation. Primary MD lymphomas are often heterogeneous mixtures of neoplastic T cells and nontransformed cells of other lineages (50), so analysis of the whole tumor may not provide the miRNA profile of the transformed target cell. However, the ability to establish homogeneous clonal populations of lymphoblastoid cell lines from primary tumors has helped to gain insights into the gene expression profiles of MD tumor cells (10). We reasoned that examination of the miRNA profiles of MDV-transformed lymphoblastoid cell lines could help to analyze their roles in neoplastic transformation and in the maintenance of MDV latency in target T cells.
MSB-1 is an MDV-transformed CD4+ T-cell line derived from a spleen lymphoma induced by the BC-1 strain of MDV-1 (1, 30). The MSB-1 cell line, used in this study at passage level 13, has a CD4+ phenotype and has been shown to be coinfected with MDV-1 and MDV-2 (66). The MSB-1 cell line has both integrated and circular copies of the MDV-1 genome (56) and induces tumors when it is inoculated into susceptible chickens (21, 35). As reported for some MD tumors, these cells also showed truncated forms of p53 tumor suppressor protein (62). Northern blot analysis of the expression of MDV-induced miRNAs in MSB-1 cells showed that many of the miRNAs are expressed at much higher levels than those in infected CEF (8, 9). These results demonstrate that the MSB-1 lymphoblastoid cell line, which shares many properties of MD tumors, could be used as a model system for analyzing the molecular pathways and mechanisms of neoplastic transformation in MD tumors. We recently reported the construction of a library, using small RNAs fractionated from MSB-1 cells, to identify novel MDV-2-encoded miRNAs (66). In this paper, we describe the results of analysis of the MSB-1 “miRNAome” to examine the population of host and viral miRNAs expressed in this transformed cell line.
MATERIALS AND METHODS
Cells and viruses.
CEF prepared from 10-day-old specific-pathogen-free embryos obtained from flocks maintained at the Institute for Animal Health were used for the propagation of viruses. Low-passage-number virus stocks of RB-1B (58) grown in CEF for 72 to 96 h were used for the preparation of RNA for Northern blotting analysis. The MDV-transformed lymphoblastoid cell line MSB-1 (1) and the REV-T-transformed (16) chicken CD4+ T-cell line AVOL-1 were grown at 38.5°C in 5% CO2 in RPMI 1640 medium containing 10% fetal calf serum, 10% tryptose phosphate broth, and 1% sodium pyruvate.
Cloning and identification of miRNAs.
We have previously described the construction of a cDNA library from small RNAs prepared from MSB-1 cells (66). Concatemerized sequences of putative miRNAs from the pGEM-T Easy (Promega, Southampton, United Kingdom) library were determined using vector-specific primers. High-quality reads of small RNA sequences with both 5′ and 3′ adapters were analyzed for the characterization of miRNAs.
Northern blotting analysis.
Total RNA was extracted from cultured cells by using TRIzol reagent (Invitrogen) according to standard methods described by the manufacturer. RNAs were also isolated from samples of MD lymphomas as well as from livers, brains, hearts, kidneys, ovaries, lungs, thymuses, and spleens of uninfected adult chickens, using TRIzol reagent. Samples of 20 μg total RNA were resolved in a 15% polyacrylamide-urea gel and blotted onto a GeneScreen Plus membrane (Perkin-Elmer). DNA oligonucleotides with the exact complementary sequence to selected miRNAs were end labeled with [γ-32P]ATP by use of T4 polynucleotide kinase (New England Biolabs, Hertfordshire, United Kingdom) to generate high-specific-activity probes. Hybridization, washing, and autoradiography were carried out as previously described (36, 53).
RESULTS
Identification of miRNAs expressed in MSB-1 cells.
The MDV-transformed lymphoblastoid T-cell line MSB-1 has been used extensively in different laboratories for various studies, particularly for the analysis of MDV latency programs (43). As a tumor cell line latently infected with MDV-1, we chose MSB-1 to analyze the miRNA profile of MD tumor cells. Sequence analysis of ∼1,200 pGEM-T Easy clones of cDNA concatemers of small RNA sequences from the MSB-1 library identified a total of 5,099 high-quality reads. The sequences were scored as miRNAs on the basis that their flanking sequences could be predicted into a stem-loop structure with low free energyBlast (2). Homology searches of these sequences against the miRBase (25) and GenBank (5) databases were used to determine the identities of the different host- and virus-encoded small RNAs. Of the total reads, 1,641 (32.2%) matched known Gallus gallus miRNAs in the miRBase. The most abundant host miRNAs in the MSB-1 library were gga-miR-21, gga-miR-142-3p, gga-miR-142-5p, gga-let-7i, gga-miR-29b, gga-miR-16 gga-miR-17-5p, gga-miR-19a, gga-miR-19b, gga-miR-106, and gga-miR-146b. Sequence analysis of the clones from the MSB-1 library also identified six novel chicken miRNAs, which appeared to be homologs of hsa-miR-363, hsa-miR-454-3p, hsa-miR-425-5p, bta-miR-191, hsa-miR-22, and dre-miR-739. The number of reads of each host-encoded miRNA in the library and their chromosomal locations are shown in Table 1. In addition to these miRNAs and the virus-encoded miRNAs (see below), 128 (2.5%) were noncoding RNA fragments, 76 (1.5%) were mRNA fragments, and 174 (3.4%) showed no matches to any known RNAs.
TABLE 1.
Sequences, chromosomal locations, and cloning frequencies of chicken miRNAs cloned from an MSB-1 library
| Namea | Sequence | No. of hits in library | Chromosomal location | Start position | End position |
|---|---|---|---|---|---|
| gga-mir-7 | TGGAAGACTAGTGATTTTGTTG | 3 | Z_random | 12717978 | 12718086 |
| gga-mir-15a | TAGCAGCACATAATGGTTTGT | 28 | 1 | 161540787 | 161540869 |
| gga-mir-15b | TAGCAGCACATCATGGTTTGCA | 9 | 9 | 21649291 | 21649381 |
| 1 | 161540645 | 161540728 | |||
| gga-mir-16 | TAGCAGCACGTAAATATTGGTG | 82 | 9 | 21649116 | 21649209 |
| gga-mir-17-5p | CAAAGTGCTTACAGTGCAGGTAA | 55 | 1 | 140631124 | 140631208 |
| gga-mir-18a | TAAGGTGCATCTAGTGCAGATA | 10 | 1 | 140630969 | 140631061 |
| gga-mir-18b | TAAGGTGCATCTAGTGCAGTTA | 10 | 4 | 3781954 | 3782037 |
| gga-mir-19a | TGTGCAAATCTATGCAAAACTGA | 71 | 1 | 140630835 | 140630915 |
| gga-mir-19b | TGTGCAAATCCATGCAAAACTGA | 71 | 1 | 140630526 | 140630612 |
| gga-mir-20a | TAAAGTGCTTATAGTGCAGGTAG | 18 | 1 | 140630649 | 140630746 |
| gga-mir-20b | CAAAGTGCTCATAGTGCAGGTAG | 16 | 4 | 3781773 | 3781857 |
| gga-mir-21 | TAGCTTATCAGACTGATGTTGA | 249 | 19 | 6933581 | 6933677 |
| gga-mir-23b | ATCACATTGCCAGGGATTAC | 2 | Z_random | 14203199 | 14203284 |
| gga-mir-24 | TGGCTCAGTTCAGCAGGAACAG | 6 | Z_random | 14203968 | 14204035 |
| gga-mir-26a | TTCAAGTAATCCAGGATAGGC | 14 | 2 | 4034213 | 4034289 |
| gga-mir-27b | TTCACAGTGGCTAAGTTCTGC | 10 | Z_random | 14203435 | 14203531 |
| 26 | 1280249 | 1280328 | |||
| gga-mir-29b | TAGCACCATTTGAAATCAGTGTT | 98 | 1 | 204569 | 204649 |
| gga-mir-30b | TGTAAACATCCTACACTCAGCT | 1 | 2 | 141145952 | 141146038 |
| 3 | 80699454 | 80699525 | |||
| gga-mir-30c | TGTAAACATCCTACACTCTCAGCT | 1 | 23 | 4431115 | 4431203 |
| gga-mir-30d | TGTAAACATCCCCGACTGGAAGC | 16 | 2 | 141142201 | 141142264 |
| gga-mir-30a-5p | TGTAAACATCCTCGACTGGAAGCT | 13 | 3 | 80674840 | 80674911 |
| gga-mir-33 | GTGCATTGTAGTTGCATTG | 6 | 1 | 46203134 | 46203202 |
| gga-mir-34a | TGGCAGTGTCTTAGCTGGTTGTT | 7 | 21 | 3118925 | 3119033 |
| gga-miR-92 | TATTGCACTTGTCCCGGCCTGT | 9 | 1 | 140630413 | 140630490 |
| gga-mir-101 | TACAGTACTGTGATAACTGAAG | 9 | Z | 11651019 | 11651097 |
| gga-mir-106 | AAAAGTGCTTACAGTGCAGGTA | 55 | 4 | 3782085 | 3782165 |
| gga-mir-130a | CAGTGCAATATTAAAAGGGCA | 4 | 15 | 393029 | 393111 |
| gga-mir-140 | AGTGGTTTTACCCTATGGTAG | 5 | 11_random | 250924 | 251018 |
| gga-mir-142-3p | TGTAGTGTTTCCTACTTTATGGA | 224 | Un | 130069949 | 130070036 |
| gga-mir-142-5p | CCCATAAAGTAGAAAGCACTAC | 243 | Un | 130069949 | 130070036 |
| gga-mir-146b | TGAGAACTGAATTCCATAGGCG | 44 | 6 | 22212586 | 22212690 |
| gga-mir-181a | AACATTCAACGCTGTCGGTGAGTT | 5 | 8 | 1957561 | 1957664 |
| 17 | 945791 | 945881 | |||
| 8 | 1957750 | 1957838 | |||
| gga-mir-181b | AACATTCATTGCTGTCGGTGGGTTT | 6 | 17 | 944157 | 944241 |
| gga-mir-221 | AGCTACATTGTCTGCTGGGTTTC | 11 | 1 | 104369325 | 104369423 |
| gga-mir-222 | AGCTACATCTGGCTACTGGGT | 7 | 1 | 104368821 | 104368918 |
| gga-mir-301 | CAGTGCAATAATATTGTCAAAGCATT | 3 | 15 | 391803 | 391895 |
| gga-mir-456 | CAGGCTGGTTAGATGGTTGTCCT | 4 | 3 | 27735429 | 27735540 |
| gga-let-7i | TGAGGTAGTAGTTTGTGCTGT | 148 | 1 | 29988296 | 29988379 |
| 12 | 6149732 | 6149821 | |||
| gga-let-7a | TGAGGTAGTAGGTTGTATAGTT | 12 | 24 | 3263054 | 3263125 |
| 1 | 67795667 | 67795742 | |||
| gga-mir-363* | AATTGCACGGTATCCATCTGTA | 30 | |||
| gga-mir-454* | TAGTGCAATATTGCTTATAGGGT | 5 | |||
| gga-mir-425* | AATGACACGATCACTCCCGTTGA | 6 | |||
| gga-mir-191* | CAACGGAATCCCAAAAGCAGCTG | 8 | |||
| gga-mir-22* | AAGCTGCCAGTTGAAGAACTGT | 6 | |||
| gga-mir-739* | AAGGCCGAAGTGGAGAAGGGTTCCA | 1 |
*, novel miRNAs identified in chickens and assigned names on the basis of homology to miRNAs in other species.
For the identification of the virus-encoded miRNAs, BLAST searches were carried out against the full-length sequences of MDV-1 (Md5 strain; GenBank accession number AF243438) and MDV-2 (HPRS-24 strain; GenBank accession number AB049735). We have previously shown that 518 (10.2%) sequences from the MSB-1 library are encoded by MDV-2 (66). However, the majority of the 2,562 (50.2%) sequences from this library showed perfect sequence identity to the genome sequence of the Md5 strain. These miRNA sequences, ranging in length from 18 to 26 nucleotides, belonged to 12 distinct MDV-1-encoded miRNAs. These included the eight miRNAs (mdv-miR-M1 to -M8) identified previously from MDV-infected CEF cultures (8) and four novel MDV-1-encoded miRNAs (mdv-miR-M9 to -M12). Additionally, we identified another novel miRNA, mdv-miR-M13, using Northern blotting analysis of RNAs extracted from MSB-1 cells (see below). The genomic location and cloning frequency of each of the MDV-1-encoded miRNAs are shown in Table 2.
TABLE 2.
Sequences and genomic positions of MDV-1 miRNAsa
| Name | Sequence (5′ to 3′) | Length (nt) | No. of hits | Nucleotide positionb |
|---|---|---|---|---|
| MDV1-miR-M1-5p | UGCUUGUUCACUGUGCGGCA(UUAU) | 20-24 | 339 | 136873-136896 |
| MDV1-miR-M1-3p | (A)UGCUGCGCAUGAAAGAGCGA(A) | 21-23 | 4 | 136913-136934 |
| MDV1-miR-M2-5p | (G)UUGUAUUCUGCCCGGUAGUCC(GUUU) | 22-26 | 16 | 134231-134256 |
| MDV1-miR-M2-3p | (A)CGGACUGCCGCAGAAUAGC(UUU) | 19-22 | 11 | 134270-134292 |
| MDV1-miR-M3-5p | (CAUG)AAAAUGUGAAACCUCUC(CCGCU) | 20-25 | 390 | 134079-134104 |
| MDV1-miR-M4-5p | (UUUAA)UGCUGUAUCGGAACC(CUUCGUU) | 19-26 | 341 | 134367-134393 |
| MDV1-miR-M4-3p | (CGA)AUGGUUCUGACAGCAUGAC(CU) | 20-22 | 50 | 134403-134426 |
| MDV1-miR-M5-5p | AACCGUAUGCGAUCACAUUGAC | 22 | 0 | 133606-133628 |
| MDV1-miR-M5-3p | (U)GUGUAUCGUGGUCGUCUACU(GUU) | 21-24 | 176 | 133647-133670 |
| MDV1-miR-M6-5p | (UCU)GUUGUUCCGUAGUGUUC(UC) | 18-22 | 278 | 142335-142356 |
| MDV1-miR-M6-3p | (GAG)AUCCCUGCGAAAUGACAGU(U) | 19-23 | 14 | 142370-142392 |
| MDV1-miR-M7-5p | (UGU)UAUCUCGGGGAGAUCCC(GAU) | 19-23 | 800 | 142508-142530 |
| MDV1-miR-M8-5p | UAUUGUUCUGUGGUUGGUUUC(GA) | 21-23 | 18 | 142216-142238 |
| MDV1-miR-M8-3p | (GU)GACCUCUACGGAACAAUAG(U) | 20-22 | 36 | 142258-142279 |
| MDV1-miR-M9-5p | UUUUCUCCUUCCCCCCGGAGUU(CA) | 22-24 | 45 | 133374-133397 |
| MDV1-miR-M9-3p | AAACUCCGAGGGCAGGAAAAAG | 22 | 1 | 133414-133435 |
| MDV1-miR-M10-3p | (UCG)AAAUCUCUACGAGAUAACA(GU) | 20-23 | 6 | 142667-142690 |
| MDV1-miR-M11-5p | UUUUCCUUACCGUGUAGCUUAGA | 23 | 2 | 136053-136075 |
| MDV1-miR-M11-3p | UGAGUUACAUGGUCAGGGGAUU | 22 | 0 | 136092-136113 |
| MDV1-miR-M12-3p | (U)UGCAUAAUACGGAGGGUUCU(G) | 21-22 | 35 | 133925-133946 |
| MDV1-miR-13-3p | GCAUGGAAACGUCCUGGGAAA | 21 | 0 | 142313-142333 |
Sequence variation surrounding the recovered BC-1 miRNAs is indicated by parentheses surrounding the variable nucleotides. miRNAs derived from a single primary miRNA stem-loop precursor are indicated by a “-5p” (5′ arm) or “-3p” (3′ arm) suffix.
Based on the Md5 sequence (GenBank accession no. AF243438).
MDV-1 miRNAs fold into distinct hairpin structures.
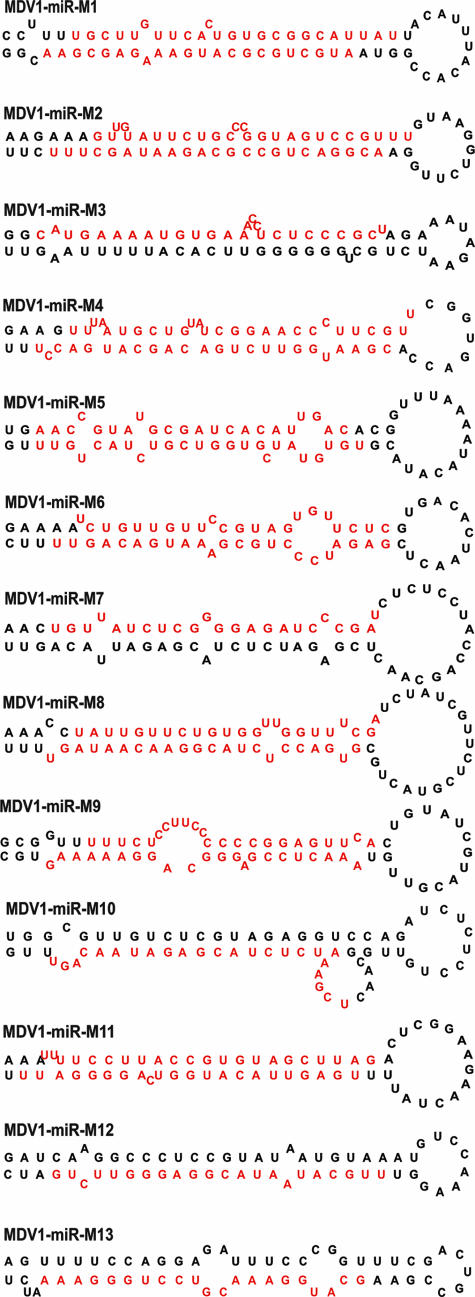
For the validation of a sequence as a miRNA, demonstration of its expression as well as its processing through the miRNA biogenesis pathway is required. One of the distinct indicators of miRNA biogenesis is the presence of adjacent complementary sequences that are able to form stable hairpins. In order to analyze the potential precursor structures of miRNAs encoded by MDV-1, the sequences of the 13 miRNAs with their adjacent 60 to 80 nucleotides were analyzed by MFOLD calculation, and the secondary structures were drawn using RNADRAW software as described previously (66). All of the MDV-1 miRNAs showed a stable hairpin with long paired stems (Fig. 1), indicating that they are bona fide miRNAs. Of the two strands of the miRNA duplex generated during biogenesis, only one of the strands, the miRNA strand, is incorporated into the RNA-induced silencing complex and guides gene regulation (3). Although the non-miRNA strand is rapidly degraded, in many instances it is also captured during cloning and may sometimes be detected with a comparable frequency to that of the miRNA strand (66). Among the MDV-1 miRNAs in MSB-1 cells, two mature forms, representing both strands of the duplex, were demonstrated by cloning or Northern blotting for 8 of the 13 candidate miRNAs, increasing the total number of miRNAs to 21. The suffixes “-5p” and “-3p” were added to designations to indicate the 5′ and 3′ arms, respectively, of the stem-loop precursor from which the miRNAs were derived (Table 2).
FIG. 1.
Secondary structures of MDV-1 pre-miRNAs predicted using the MFOLD algorithm (68). The mature miRNA strands are indicated in red.
MDV-1 miRNAs show differences in cloning frequencies.
We then examined the cloning frequency of each of the MDV-1 miRNAs as a measure of their expression levels in MSB-1 cells. The most abundantly cloned miRNAs were mdv-miR-M7-5p (800 hits), mdv-miR-M3-5p (390 hits), mdv-miR-M4-5p (341 hits), mdv-miR-M1-5p (339 hits), mdv-miR-M6-5p (278 hits), and mdv-miR-M5-3p (176 hits). Compared to this, mdv-miR-M10, -M11, and -M13 were of very low abundance, while mdv-miR-M2, -M8, -M9, and -M12 showed moderate copy numbers in the library. For most miRNAs, the non-miRNA strand of the duplex was either not cloned or had a relative frequency much lower than that of the miRNA strand. However, for some of the miRNAs, such as mdv-miR-M2, the frequencies of both strands were very similar, suggesting that both may be functional and are incorporated into the RNA-induced silencing complex.
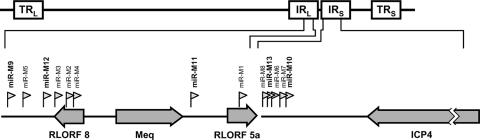
MDV-1 miRNAs are clustered in the repeat regions of the MDV-1 genome.
The nucleotide sequence positions of the different miRNAs are shown in Table 2. All 13 MDV-1-encoded miRNAs are clustered in an ∼9-kb region within the RL/RS sequences of the MDV genome (Fig. 2). The miRNAs mdv-miR-M2, -M3, -M4, -M5, -M9, and -M12 are located upstream of Meq and are antisense to the R-LORF8 transcript. The miRNAs mdv-miR-M1 and -M11 lie downstream of Meq and are embedded within the open reading frame (ORF) of the L1/LORF5a transcript (46, 59) as well as within the intron of the splice variant Meq-sp (51). MDV-1-encoded miRNAs mdv-miR-M6, -M7, -M8, -M10, and -M13 are located between the a-like sequence and the ICP4 sequence within the large intron of the LAT of MSR (15). Of this cluster, the miRNAs mdv-miR-M6 and mdv-miR-M13 were separated by only a single nucleotide. The occurrence of the miRNAs in distinct clusters in the same orientation strongly suggests that these miRNAs are likely to be processed as multicistronic pre-miRNA transcripts. Despite being processed from a single transcript, there are differences in the expression levels of the mature miRNAs, and these are thought to be due to differences in Drosha processing and/or miRNA stability.
FIG. 2.
Genomic locations of MDV-1 miRNAs. The schematic diagram shows where the MDV-1 miRNAs (small arrowheads) identified in this report map. The five novel miRNAs identified in this report are shown in bold. The TRL and IRL regions flanking the unique long region and the TRS and IRS regions flanking the unique short regions are shown. Genomic positions and orientations of MDV ORFs contained in the miRNA loci are indicated.
Analysis of miRNA expression by Northern blotting.
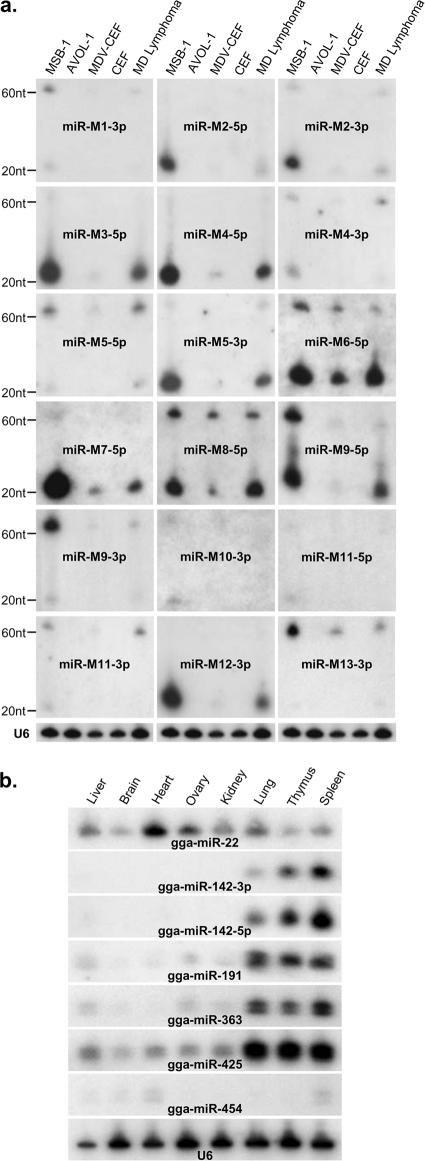
For further confirmation of the expression of miRNAs in MSB-1 cells, Northern blot hybridization with individual MDV-1 miRNA probes was carried out on RNAs extracted from MSB-1, AVOL-1 (an MDV-negative T-cell line), or uninfected or RB-1B virus-infected CEF cells and from samples of MD lymphoma. These studies confirmed that MDV-1-encoded miRNAs are expressed at high levels in MSB-1 cells and MD lymphomas and at low levels in infected CEF (Fig. 3). No signals were obtained from the RNAs extracted from AVOL-1 cells and uninfected CEF, validating the specificity of the miRNA probes. Based on the intensities of signals, the levels of expression of the majority of miRNAs were similar in both MSB-1 cells and lymphoma samples. Some of the most abundantly cloned miRNAs, such as mdv-miR-M3, -M4, -M5, -M6, and -M7, showed very strong signals by Northern blotting, validating the correlation between cloning frequency and expression level. Similarly, mdv-miR-M10, -M11, and -M13, cloned at very low frequencies from the library, gave weak signals by Northern blotting. A previous study using Northern blotting of RNAs extracted from MD lymphomas reported that mdv-miR-M6 is expressed at low levels, and mdv-miR-M7 was not detected at all (8). However, our studies using probes specific for the mdv-miR-M6-5p and mdv-miR-M7-5p strands gave strong signals for all samples, including MD lymphomas, indicating that the -5p strand of the duplex is the functional miRNA strand. The failure to detect these miRNAs in Northern blots in the previous study was likely due to the use of the non-miRNA strand as the probe. In most cases, both pre-miRNAs and mature miRNAs could be detected by Northern blotting, with the former giving much lower signals. For some of the miRNAs, such as mdv-miR-M5-5p, mdv-miR-M9-3p, and mdv-miR-M13, the signals of pre-miRNAs were higher than those of the mature miRNAs, indicating less efficient processing.
FIG. 3.
(a) Northern blotting analysis for determining the expression of MDV-1 miRNAs. Twenty micrograms of total RNA from MSB-1 cells, the MDV-negative lymphoid cell line AVOL-1, MDV-1-infected CEF, uninfected CEF, or MD lymphoma tissues was separated in a 15% denaturing polyacrylamide gel, blotted, and probed with end-labeled antisense oligonucleotides to the indicated miRNAs. Size markers to indicate the positions of the pre-miRNA and the mature miRNA are shown. The cellular U6 snRNA served as the loading control, and a representative blot of this set is shown. (b) Analysis of tissue-specific expression of Gallus gallus (gga-) miRNAs identified in the MSB-1 library. Total RNAs (20 μg) extracted from different tissues of chickens were separated in polyacrylamide gels and probed with end-labeled antisense oligonucleotides specific for the individual miRNAs indicated. The cellular U6 snRNA served as the loading control.
Northern blotting was also carried out on RNAs extracted from eight different normal tissues from adult noninfected chickens to validate the expression of some of the miRNAs cloned from the MSB-1 library. Some of these miRNAs, including novel chicken miRNAs such as gga-miR-363, gga-miR-454 gga-miR-425, gga-miR-191, and gga-miR-22, could be detected by Northern blot analysis, albeit with differences in expression levels between tissues (Fig. 3b). While gga-miR-425 and gga-miR-22 showed high levels of expression in all tissues, gga-miR-454 was detected at very low levels. The expression of the miRNAs gga-miR-191, gga-miR-363, and gga-miR-425 in lymphoid organs, namely, the spleen, thymus, and the lungs (55), was at the levels observed for the lymphocyte-specific miRNA gga-miR-142 (54, 65), which gave strong signals with probes specific for either of the strands of the duplex in these tissues.
DISCUSSION
As efficient inducers of cancer, oncogenic viruses have helped to reveal several major pathways of oncogenesis. Most of these pathways involve the interactions of virus-encoded oncoproteins, such as simian virus 40 T antigen, adenovirus E1A, human papillomavirus E6/E7, and EBV EBNAs (69). In MD tumors, MDV-encoded Meq is considered to be the major oncoprotein (39), although other proteins also contribute to oncogenesis (44). The discovery of virus-encoded miRNAs in several oncogenic viruses (53) has added yet another armory to these viruses for regulating gene expression in cancer cells. Recent studies on small RNAs from infected CEF have identified eight MDV-1-encoded miRNAs that map to the Meq and LAT regions of the genome (8, 9). Furthermore, several recent studies have identified specific miRNA signatures in different tumors, providing insights into the different oncogenic pathways in these tumors (20, 64). In order to analyze the expression profiles of the host- and MDV-1-encoded miRNAs in MD tumor cells, we examined the miRNAs expressed in the MDV-transformed lymphoblastoid cell line MSB-1 by cloning and Northern blot analysis.
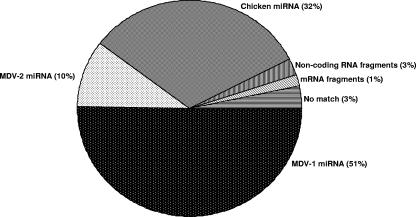
One of the conspicuous findings from the analysis of the miRNA sequences from the MSB-1 library was the very large proportion of MDV-1-encoded miRNAs (51%) in relation to the number of host miRNAs (Fig. 4). This level of expression of MDV-1 miRNAs is much higher than that identified from MDV-1-infected CEF, where only 0.6% of the nearly 172,000 reads were miRNAs encoded by MDV-1 (9). The low level of expression of MDV-1 miRNAs in CEF (also evident from the results of the Northern blotting analysis) could partly be explained by the smaller proportions of infected cells in CEF cultures in comparison to MSB-1 cell cultures, where each cell has multiple copies of the MDV genome. However, the increased expression of MDV-1 miRNAs in MSB-1 cells may also be related to the increased lymphocyte-specific expression of these miRNAs in these transformed target cells. An increased proportion of virus-encoded miRNAs over host-encoded miRNAs is not uncommon in transformed cell lines. For example, miRNAs encoded by Kaposi's sarcoma-associated herpesvirus and EBV accounted for >40% of the entire miRNA pool identified from the BC-1 cell line coinfected with these two viruses (11). Once the 518 (10%) MDV-2-encoded miRNAs that we reported previously (66) were also considered, the total proportion of virus-encoded miRNAs in the MSB-1 library was 61%, compared to the 32.2% expression of host-encoded miRNAs. The reasons for the fivefold difference in the levels of miRNAs encoded by the two viruses are not known but may be connected with the differences in relative copy numbers of the two viruses. The precise copy numbers or replication rates of the two viruses in MSB-1 cells are not known. However, on CEF cocultivated with MSB-1 cells, MDV-2 produced 10-fold more plaques than did MDV-1, suggesting that MDV-2 is better adapted for faster replication on CEF (66).
FIG. 4.
Pie chart showing proportions of MDV-1, MDV-2, and chicken miRNAs and other molecules cloned from the MSB-1 library.
Previous studies of MDV-1-infected CEF identified eight miRNAs that mapped to the Meq and LAT regions of the MDV genome (8, 9). We also cloned all eight of these miRNAs from the MSB-1 library. However, we also identified 5 new MDV-1 miRNAs, taking the total number of MDV-1-encoded miRNAs to 13. As in the case of the eight previously identified miRNAs, the five new MDV-1 miRNAs mapped to the Meq and LAT regions of the genome (Fig. 2). In the MDV genome, while most of the genes are transcriptionally silent in latently infected and tumor cells, the repeat (RL/RS) regions are generally active (34, 45, 61). Thus, it is not surprising that all of the miRNAs that are expressed at high levels in latently infected/tumor cells are located in a transcriptionally active region of the genome. The genomic locations of the eight previously reported MDV-1-encoded miRNAs have been described (8, 9). Two of the new miRNAs, mdv-miR-M9 and mdv-miR-M12, are also located upstream of the Meq promoter region, like the previously identified miR-M2 to miR-M5 miRNAs, suggesting that these six miRNAs are part of the same transcriptional unit in the same transcriptional orientation as Meq. The high levels of expression of all six of these miRNAs, demonstrated by strong signals in Northern blotting of RNAs from MSB-1 and tumor cells (Fig. 3), suggest that these miRNAs may have major roles in regulating the expression of viral and host genes in latently infected/transformed T cells. The transcription unit of these miRNAs is also antisense to another potential transcript, RLORF8, demonstrated in both CEF and lymphoblastoid cells (52), and hence has the potential to regulate the expression of RLORF8. A recent study demonstrated that EBV-encoded miR-BART2 can downregulate the viral DNA polymerase BALF5 via a similar mechanism (4). However, unlike miR-M2 and miR-M4, which are embedded in the RLORF8 ORF, the newly identified miRNAs miR-M9 and miR-M12 are located downstream of the ORF (Fig. 2).
One of the previously identified miRNAs, miR-M1, mapped downstream of Meq embedded in the ORF of the L1/RLORF5a transcript, although it is not clear whether it affects the expression of this transcript (33, 46, 59). We have identified a new miRNA, mdv-miR-M11, located just downstream of the Meq ORF (Fig. 2). The importance of this novel miRNA is not known, but it is expressed at only very low levels, as indicated by a low cloning frequency in the library and weak signals in Northern blot analysis.
In addition to the three miRNAs, miR-M6 to -M8, that mapped to the LAT region, our studies have revealed miR-M10 and miR-M13, two novel miRNAs encoded from this region. Because these miRNAs are located very close to miR-M6 to -M8 and are in the same transcriptional orientation, these two miRNAs are highly likely to be part of the same cluster. However, compared to miR-M6 to -M8, which are expressed at very high levels (Table 2 and Fig. 3), the levels of expression of miR-M10 and miR-M13, shown by Northern blotting and cloning frequencies, are very low. Although the reasons for their low expression levels are not known, the efficiency in processing of the mature miRNAs could be a factor, especially because of their close proximity within the cluster. For example, miR-M13 is located between the highly expressed miR-M6 and miR-M8 miRNAs, with the mature miRNA sequence of miR-M6 being separated from that of miR-M13 by only a single nucleotide (Table 2). Similarly, the newly identified miR-M10 (only 6 hits in the library) is located just adjacent to the most highly expressed miRNA, miR-M7, which had 800 hits in the library.
Although the expression levels of MDV-1-encoded miRNAs in MSB-1 cells were generally similar to those in tumor tissues, there were clear differences in expression level between infected CEF and transformed MSB-1 cells, with the latter generally expressing higher levels of all miRNAs. However, it was also interesting to see clear differences between miRNAs in the specificity of the strand expressed in infected CEF and MSB-1 cells. The most striking example of strand-specific expression was noted for miR-M7, where the mature miRNA strand, miR-M7-5p, had 800 hits, accounting for 16% of the entire MSB-1 library. Northern blot analysis also revealed very strong expression of this miRNA in both MSB-1 cells and tumor tissues. Although weak signals for miR-M7-5p were detected in the infected CEF, this strand was not identified even once among the nearly 172,000 high-quality reads of small RNA sequences from infected CEF (9), suggesting that this miRNA strand is processed only at very low levels in lytically infected CEF. This cluster of miRNAs maps antisense to the ICP4 gene and to the large intron in the 5′ end of the putative LAT, expressed at high levels in transformed cells/lymphomas as well as in the late stages of lytic infection of CEF (57). Although the reasons for the differences in processing of the two miRNA strands during miRNA biogenesis in this region between CEF and lymphocytes are not clear, it would be interesting to see whether any of the miRNAs play a role in switching between lytic replication and latency. Intriguingly, mdv-miR-M7 also showed evidence of RNA editing. However, in the absence of knowledge on the targets of any of the MDV-1-encoded miRNAs, the significance of this remains unknown.
Analysis of the miRNA repertoire from MSB-1 cells also identified several host miRNAs, some of which were cloned at high frequencies indicating high levels of expression (Table 1). Some of the more abundant host miRNAs, such as those within the miR-17-92 cluster, have been shown to be amplified in several types of cancer (28, 29). Since these miRNAs have been shown to accelerate the formation of lymphoid malignancies in mouse models (29), the increased expression of these miRNAs in MSB-1 cells could be significant. Similarly, other highly expressed miRNAs, such as miR-21 (249 hits), let-7i (148 hits), the two strands of miR-142 (224 and 243 hits), miR-15a (28 hits), and miR-16 (82 hits), have also been associated with various malignancies, including chronic lymphocytic leukemia (12, 18), suggesting that these miRNAs may contribute toward MDV oncogenicity. Currently, we are examining the roles of different host miRNAs in the induction of lymphomas by MDV. Our studies on MSB-1 cells also revealed six novel chicken miRNAs. The expression of five of these novel miRNAs could be detected by Northern blotting of different chicken tissues, although the expression of miR-454 in all tissues was very weak (Fig. 3b). The expression of both strands of miR-142 appeared to be restricted to the lymphocyte-enriched lungs, thymus, and spleen (Fig. 3b), suggesting that it is likely to be a lymphoid cell-specific miRNA.
We have carried out a study to examine the miRNAome of a herpesvirus-induced lymphoma in chickens by determining the miRNAs expressed in a lymphoblastoid cell line derived from a lymphoma. This study is the first of its kind with an avian lymphoma and has demonstrated that the analysis of the miRNA repertoire would enable investigation of some of the potential pathways used by viruses in neoplastic transformation. A major challenge in the next stage would be the identification of potential targets for some of the miRNAs overexpressed in these cells to identify networks of molecular events regulated by the altered miRNAome in these cells. The present study has not identified miRNAs that are downregulated in transformed cells, whose profiles are also very important for understanding the global events involved in transformation. Currently, we are using microarray analysis of global viral/host miRNA expression in MD tumor cells in relation to that in normal lymphocytes to determine the entire repertoire of upregulated and downregulated miRNAs to identify the extent to which MDV exploits the cellular miRNA pathways to induce neoplastic transformation.
Acknowledgments
We thank Mihaela Zavolan, Division of Bioinformatics, Biozentrum, University of Basel, Switzerland, for assistance with bioinformatic prediction of MDV-1 miRNAs and Mick Gill for assistance with digital imaging and graphics.
This work was funded by BBSRC, United Kingdom.
Footnotes
Published ahead of print on 6 February 2008.
REFERENCES
- 1.Akiyama, Y., and S. Kato. 1974. Two cell lines from lymphomas of Marek's disease. Biken J. 17105-116. [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 4.Barth, S., T. Pfuhl, A. Mamiani, C. Ehses, K. Roemer, E. Kremmer, C. Jaker, J. Hock, G. Meister, and F. A. Grasser. 2008. Epstein Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2007. GenBank. Nucleic Acids Res. 35D21-D25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, A. C., S. J. Baigent, L. P. Smith, J. P. Chattoo, L. J. Petherbridge, P. Hawes, M. J. Allday, and V. Nair. 2006. Interaction of MEQ protein and C-terminal-binding protein is critical for induction of lymphomas by Marek's disease virus. Proc. Natl. Acad. Sci. USA 1031687-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgess, S. C., J. R. Young, B. J. Baaten, L. Hunt, L. N. Ross, M. S. Parcells, P. M. Kumar, C. A. Tregaskes, L. F. Lee, and T. F. Davison. 2004. Marek's disease is a natural model for lymphomas overexpressing Hodgkin's disease antigen (CD30). Proc. Natl. Acad. Sci. USA 10113879-13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnside, J., E. Bernberg, A. Anderson, C. Lu, B. C. Meyers, P. J. Green, N. Jain, G. Isaacs, and R. W. Morgan. 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 808778-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnside, J., and R. W. Morgan. 2007. Genomics and Marek's disease virus. Cytogenet. Genome Res. 117376-387. [DOI] [PubMed] [Google Scholar]
- 10.Buza, J. J., and S. C. Burgess. 2007. Modeling the proteome of a Marek's disease transformed cell line: a natural animal model for CD30 overexpressing lymphomas. Proteomics 71316-1326. [DOI] [PubMed] [Google Scholar]
- 11.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 1025570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin, G. A., and C. M. Croce. 2007. Investigation of microRNA alterations in leukemias and lymphomas. Methods Enzymol. 427191-213. [DOI] [PubMed] [Google Scholar]
- 13.Calin, G. A., Y. Pekarsky, and C. M. Croce. 2007. The role of microRNA and other non-coding RNA in the pathogenesis of chronic lymphocytic leukemia. Best Pract. Res. Clin. Haematol. 20425-437. [DOI] [PubMed] [Google Scholar]
- 14.Calnek, B. W. 1986. Marek's disease: a model for herpesvirus oncology. CRC Crit. Rev. Microbiol. 12293-320. [DOI] [PubMed] [Google Scholar]
- 15.Cantello, J. L., A. S. Anderson, and R. W. Morgan. 1994. Identification of latency-associated transcripts that map antisense to the ICP4 homolog gene of Marek's disease virus. J. Virol. 686280-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, I. S., T. W. Mak, J. J. O'Rear, and H. M. Temin. 1981. Characterization of reticuloendotheliosis virus strain T DNA and isolation of a novel variant of reticuloendotheliosis virus strain T by molecular cloning. J. Virol. 40800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho, W. C. 2007. OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cowland, J. B., C. Hother, and K. Gronbaek. 2007. MicroRNAs and cancer. APMIS 1151090-1106. [DOI] [PubMed] [Google Scholar]
- 19.Cullen, B. R. 2006. Viruses and microRNAs. Nat. Genet. 38(Suppl.)S25-S30. [DOI] [PubMed] [Google Scholar]
- 20.Cummins, J. M., Y. He, R. J. Leary, R. Pagliarini, L. A. Diaz, Jr., T. Sjoblom, O. Barad, Z. Bentwich, A. E. Szafranska, E. Labourier, C. K. Raymond, B. S. Roberts, H. Juhl, K. W. Kinzler, B. Vogelstein, and V. E. Velculescu. 2006. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 1033687-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doi, K., A. Kojima, Y. Akiyama, and S. Kato. 1976. Pathogenicity for chicks of line cells from lymphoma of Marek's disease. Natl. Inst. Anim. Health Q. (Tokyo) 1616-24. [PubMed] [Google Scholar]
- 22.Fragnet, L., M. A. Blasco, W. Klapper, and D. Rasschaert. 2003. The RNA subunit of telomerase is encoded by Marek's disease virus. J. Virol. 775985-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gimeno, I. M., R. L. Witter, H. D. Hunt, S. M. Reddy, L. F. Lee, and R. F. Silva. 2005. The pp38 gene of Marek's disease virus (MDV) is necessary for cytolytic infection of B cells and maintenance of the transformed state but not for cytolytic infection of the feather follicle epithelium and horizontal spread of MDV. J. Virol. 794545-4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grey, F., H. Meyers, E. A. White, D. H. Spector, and J. Nelson. 2007. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 3e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths-Jones, S., H. K. Saini, S. van Dongen, and A. J. Enright. 2008. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36D154-D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta, A., J. J. Gartner, P. Sethupathy, A. G. Hatzigeorgiou, and N. W. Fraser. 2006. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature 44282-85. [DOI] [PubMed] [Google Scholar]
- 27.Hagan, J. P., and C. M. Croce. 2007. MicroRNAs in carcinogenesis. Cytogenet. Genome Res. 118252-259. [DOI] [PubMed] [Google Scholar]
- 28.Hayashita, Y., H. Osada, Y. Tatematsu, H. Yamada, K. Yanagisawa, S. Tomida, Y. Yatabe, K. Kawahara, Y. Sekido, and T. Takahashi. 2005. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 659628-9632. [DOI] [PubMed] [Google Scholar]
- 29.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirai, K., M. Yamada, Y. Arao, S. Kato, and S. Nii. 1990. Replicating Marek's disease virus (MDV) serotype 2 DNA with inserted MDV serotype 1 DNA sequences in a Marek's disease lymphoblastoid cell line MSB1-41C. Arch. Virol. 114153-165. [DOI] [PubMed] [Google Scholar]
- 31.ICTVdB Management. 25 April 2006, posting date. 00.031.1.03. Mardivirus. In C. Búchen-Osmond (ed.), ICTVdB—The universal virus database, version 4. Columbia University, New York, NY.
- 32.Jarosinski, K., L. Kattenhorn, B. Kaufer, H. Ploegh, and N. Osterrieder. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 10420025-20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarosinski, K. W., N. Osterrieder, V. K. Nair, and K. A. Schat. 2005. Attenuation of Marek's disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J. Virol. 7911647-11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, D., L. Lee, J. L. Liu, H. J. Kung, and J. K. Tillotson. 1992. Marek's disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc. Natl. Acad. Sci. USA 894042-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, L. F., K. Nazerian, and J. A. Boezi. 1975. Marek's disease virus DNA in a chicken lymphoblastoid cell line (MSB-1) and in virus-induced tumours, p. 199-204. In G. de Thé, M. A. Epstein, and H. zur Hausen (ed.), Oncogenesis and herpesviruses II. IARC, Lyon, France. [PubMed]
- 36.Lee, R. C., and V. Ambros. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294862-864. [DOI] [PubMed] [Google Scholar]
- 37.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 12015-20. [DOI] [PubMed] [Google Scholar]
- 38.Li, D.-S., J. Pastorek, V. Zelnik, G. D. Smith, and L. J. N. Ross. 1994. Identification of novel transcripts complementary to the Marek's disease virus homologue of the ICP4 gene of herpes simplex virus. J. Gen. Virol. 751713-1722. [DOI] [PubMed] [Google Scholar]
- 39.Lupiani, B., L. F. Lee, X. Cui, I. Gimeno, A. Anderson, R. F. Silva, R. L. Witter, H. J. Kung, and S. M. Reddy. 2004. Marek's disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. USA 10111815-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupiani, B., L. F. Lee, and S. M. Reddy. 2001. Protein-coding content of the sequence of Marek's disease virus serotype 1. Curr. Top. Microbiol. Immunol. 255159-190. [DOI] [PubMed] [Google Scholar]
- 41.Marek, J. 1907. Multiple Nervenentzuendung (polyneuritis) bei Huehnern. Dtsch. Tierarztl. Wochenschr. 15417-421. [Google Scholar]
- 42.McElroy, J. P., J. C. Dekkers, J. E. Fulton, N. P. O'Sullivan, M. Soller, E. Lipkin, W. Zhang, K. J. Koehler, S. J. Lamont, and H. H. Cheng. 2005. Microsatellite markers associated with resistance to Marek's disease in commercial layer chickens. Poult. Sci. 841678-1688. [DOI] [PubMed] [Google Scholar]
- 43.Morgan, R. W., Q. Xie, J. L. Cantello, A. M. Miles, E. L. Bernberg, J. Kent, and A. Anderson. 2001. Marek's disease virus latency. Curr. Top. Microbiol. Immunol. 255223-243. [PubMed] [Google Scholar]
- 44.Nair, V., and H. J. Kung. 2004. Marek's disease virus oncogenicity: molecular mechanisms, p. 32-48. In F. Davison and V. Nair (ed.), Marek's disease, an evolving problem. Elsevier Academic Press, Oxford, United Kingdom.
- 45.Ohashi, K., P. H. O'Connell, and K. A. Schat. 1994. Characterization of Marek's disease virus BamHI-A-specific cDNA clones obtained from a Marek's disease lymphoblastoid cell line. Virology 199275-283. [DOI] [PubMed] [Google Scholar]
- 46.Ohashi, K., W. Zhou, P. H. O'Connell, and K. A. Schat. 1994. Characterization of a Marek's disease virus BamHI-L-specific cDNA clone obtained from a Marek's disease lymphoblastoid cell line. J. Virol. 681191-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osterrieder, K., and J. F. Vautherot. 2004. The genome content of Marek's disease-like viruses, p. 17-31. In F. Davison and V. Nair (ed.), Marek's disease, an evolving problem. Elsevier Academic Press, Oxford, United Kingdom.
- 48.Osterrieder, N., J. P. Kamil, D. Schumacher, B. K. Tischer, and S. Trapp. 2006. Marek's disease virus: from miasma to model. Nat. Rev. Microbiol. 4283-294. [DOI] [PubMed] [Google Scholar]
- 49.Parcells, M. S., S. F. Lin, R. L. Dienglewicz, V. Majerciak, D. R. Robinson, H. C. Chen, Z. Wu, G. R. Dubyak, P. Brunovskis, H. D. Hunt, L. F. Lee, and H. J. Kung. 2001. Marek's disease virus (MDV) encodes an interleukin-8 homolog (vIL-8): characterization of the vIL-8 protein and a vIL-8 deletion mutant MDV. J. Virol. 755159-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Payne, L. N., K. Howes, M. Rennie, J. M. Bumstead, and A. W. Kidd. 1981. Use of an agar culture technique for establishing lymphoid cell lines from Marek's disease lymphomas. Int. J. Cancer 28757-766. [DOI] [PubMed] [Google Scholar]
- 51.Peng, Q., and Y. Shirazi. 1996. Characterization of the protein product encoded by a splicing variant of the Marek's disease virus Eco-Q gene (Meq). Virology 22677-82. [DOI] [PubMed] [Google Scholar]
- 52.Peng, Q., and Y. Shirazi. 1996. Isolation and characterization of Marek's disease virus (MDV) cDNAs from a MDV-transformed lymphoblastoid cell line: identification of an open reading frame antisense to the MDV Eco-Q protein (Meq). Virology 221368-374. [DOI] [PubMed] [Google Scholar]
- 53.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304734-736. [DOI] [PubMed] [Google Scholar]
- 54.Ramkissoon, S. H., L. A. Mainwaring, Y. Ogasawara, K. Keyvanfar, J. P. McCoy, Jr., E. M. Sloand, S. Kajigaya, and N. S. Young. 2006. Hematopoietic-specific microRNA expression in human cells. Leukoc. Res. 30643-647. [DOI] [PubMed] [Google Scholar]
- 55.Reese, S., G. Dalamani, and B. Kaspers. 2006. The avian lung-associated immune system: a review. Vet. Res. 37311-324. [DOI] [PubMed] [Google Scholar]
- 56.Rhiza, H.-J., and B. Bauer. 1984. Persistence of viral DNA in Marek's disease virus-transformed lymphoblastoid cell lines, p. 481-488. In G. Wittman, R. M. Gaskell, and H.-J. Rhiza (ed.), Latent herpesvirus infections in veterinary medicine. Martinus Nijhoff, Boston, MA.
- 57.Ross, N. L. 1999. T-cell transformation by Marek's disease virus. Trends Microbiol. 722-29. [DOI] [PubMed] [Google Scholar]
- 58.Schat, K. A., B. W. Calnek, and J. Fabricant. 1982. Characterisation of two highly oncogenic strains of Marek's disease virus. Avian Pathol. 11593-605. [DOI] [PubMed] [Google Scholar]
- 59.Schat, K. A., B. J. Hooft van Iddekinge, H. Boerrigter, P. H. O'Connell, and G. Koch. 1998. Open reading frame L1 of Marek's disease herpesvirus is not essential for in vitro and in vivo virus replication and establishment of latency. J. Gen. Virol. 79841-849. [DOI] [PubMed] [Google Scholar]
- 60.Subramanian, S., W. O. Lui, C. H. Lee, I. Espinosa, T. O. Nielsen, M. C. Heinrich, C. L. Corless, A. Z. Fire, and M. van de Rijn. 8 October 2007. MicroRNA expression signature of human sarcomas. Oncogene. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed]
- 61.Sugaya, K., G. Bradley, M. Nonoyama, and A. Tanaka. 1990. Latent transcripts of Marek's disease virus are clustered in the short and long repeat regions. J. Virol. 645773-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takagi, M., T. Takeda, Y. Asada, C. Sugimoto, M. Onuma, and K. Ohashi. 2006. The presence of a short form of p53 in chicken lymphoblastoid cell lines during apoptosis. J. Vet. Med. Sci. 68561-566. [DOI] [PubMed] [Google Scholar]
- 63.Trapp, S., M. S. Parcells, J. P. Kamil, D. Schumacher, B. K. Tischer, P. M. Kumar, V. K. Nair, and N. Osterrieder. 2006. A virus-encoded telomerase RNA promotes malignant T cell lymphomagenesis. J. Exp. Med. 2031307-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volinia, S., G. A. Calin, C. G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 1032257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu, H., X. Wang, Z. Du, and N. Li. 2006. Identification of microRNAs from different tissues of chicken embryo and adult chicken. FEBS Lett. 5803610-3616. [DOI] [PubMed] [Google Scholar]
- 66.Yao, Y., Y. Zhao, H. Xu, L. P. Smith, C. H. Lawrie, A. Sewer, M. Zavolan, and V. Nair. 2007. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation with those encoded by MDV-1. J. Virol. 817164-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zanette, D. L., F. Rivadavia, G. A. Molfetta, F. G. Barbuzano, R. Proto-Siqueira, and W. A. Silva, Jr. 2007. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz. J. Med. Biol. Res. 401435-1440. [DOI] [PubMed] [Google Scholar]
- 68.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 313406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.zur Hausen, H. 2001. Oncogenic DNA viruses. Oncogene 207820-7823. [DOI] [PubMed] [Google Scholar]