Abstract
Epstein-Barr virus (EBV) was the first human DNA virus to be associated with cancer. Its oncogenic potential was further demonstrated by its ability to transform primary B lymphocytes in vitro. EBV nuclear antigen 3C (EBNA3C) is one of a small subset of latent antigens critical for the transformation of human primary B lymphocytes. Although EBNA3C has been shown to modulate several cellular functions, additional targets involved in cellular transformation remain to be explored. EBNA3C can recruit key components of the SCFSkp2 ubiquitin ligase complex. In this report, we show that EBNA3C residues 130 to 190, previously shown to bind to the SCFSkp2 complex, also can strongly associate with the c-Myc oncoprotein. Additionally, the interaction of EBNA3C with c-Myc was mapped to the region of c-Myc that includes the highly conserved Skp2 binding domain. Skp2 has been shown to regulate c-Myc stability and also has been shown to function as a coactivator of transcription for c-Myc target genes. We now show that the EBV latent oncoprotein EBNA3C can stabilize c-Myc and that the recruitment of both c-Myc and its cofactor Skp2 to c-Myc-dependent promoters can enhance c-Myc-dependent transcription. This same region of EBNA3C also recruits and modulates the activity of retinoblastoma and p27, both major regulators of the mammalian cell cycle. The inclusion of c-Myc in the group of cellular targets modulated by this domain further accentuates the importance of these critical residues of EBNA3C in bypassing the cell cycle checkpoints.
Epstein-Barr virus (EBV) was the first DNA tumor virus associated with human cancers (6, 7). It is also the most ubiquitous of the eight human herpesviruses, by some estimates infecting as much as 90 to 95% of the adult population (38). All herpesviruses exhibit a remarkably high degree of host specificity (42). They have, over the millennia, coevolved with their hosts to ensure mutual coexistence (37). The life cycle of a herpesvirus has two distinct phases (39). The lytic phase results in the production of progeny virions, which expands the pool of infected cells within the same host and aids in the spread of the virus to uninfected hosts (39). After an initial lytic burst, most herpesviruses revert to the latent phase of their life cycle, in which only a small subset of viral genes is expressed (39). EBV belongs to the gamma-1 herpesvirus genus (11, 37) and can transform human primary B lymphocytes in vitro (38). This ability is dependent on the expression of a set of latent genes that includes six nuclear antigens, EBNA1, EBNA2, EBNA3A, EBNA3B, EBNA3C, and EBNA-LP, and three latent membrane proteins, LMP1, LMP2A, and LMP2B. These latent proteins are constitutively expressed in EBV-transformed lymphoblastoid cell lines (LCLs) in vitro (38).
The expression of all latent genes, known as latency type III, leads to a robust T-cell response in healthy individuals (11, 37). In the face of a T-cell response, the virus usually reverts to a lower-profile latency program, in which an even smaller subset of viral antigens is expressed (11, 37). Therefore, in immunocompetent individuals, EBV infection typically is asymptomatic (37). In situations in which the host is unable to mount an EBV-specific T-cell response, the virus is able to maintain the expression of a larger pool of viral genes, leading to the transformation and uncontrolled proliferation of infected B lymphocytes (37). EBV therefore is associated with various disease states in immunocompromised individuals. Accordingly, EBV infection is linked to endemic Burkitt lymphoma, nasopharyngeal carcinoma, and B-cell lymphomas associated with posttransplant lymphoproliferative disease (37).
Of the nine aforementioned proteins that are constitutively expressed in EBV-transformed LCLs, only four, EBNA2, EBNA3A, EBNA3C, and LMP-1, are critical for B-cell transformation in vitro (4, 13, 30, 43). Initially, this ability of EBV to transform primary B cells was attributed to transcriptional regulation by latent antigens like EBNA2, EBNA-LP, and LMP1 (2, 16, 33, 41). However, more recent studies have begun to suggest a role for EBNA3C in directly binding and regulating critical cell cycle proteins.
Initially, EBNA3C was shown to regulate retinoblastoma (pRb)-modulated pathways and to drive cells through the G1/S restriction point (32). More recently, it was demonstrated that EBNA3C can target the SCFSkp2 complex, thereby regulating the activity and stability of cyclin A/cdk2 and pRb complexes (21-24). We further explored the possible regulation of c-Myc, a critical cell cycle modulator and a known substrate of the SCFSkp2 complex (14, 15, 18, 19). It has been shown recently that c-Myc and Skp2 can cooperate in c-Myc-regulated transcription (12, 17, 45, 46). In this report, we show that the same domain of EBNA3C, amino acids 130 to 190, which binds to Skp2, also can strongly associate with c-Myc. The interaction of EBNA3C with c-Myc was mapped to the highly conserved Skp2 binding region within the amino terminus of c-Myc. The oncoprotein c-Myc has shown a broad relationship with the Skp2 ubiquitin ligase (12, 17, 45, 46). For example, the monoubiquitylation of c-Myc by Skp2 at a hitherto unidentified site can activate the transcription of c-Myc target genes (44). Polyubiquitylation, however, targets the molecule for proteasomal degradation (44). Using pulse-chase experiments, we show that EBNA3C can stabilize c-Myc. This domain of EBNA3C also recruits and modulates the activity of pRb and p27, both major regulators of the mammalian cell cycle (21-24). The inclusion of c-Myc in the group of proteins modulated by this region further demonstrates the importance of this region of EBNA3C in regulating the cell cycle in EBV infections.
MATERIALS AND METHODS
Plasmids, antibodies, and cell lines.
The pA3F vector was created by the insertion of three Flag tags followed by a stop codon between the NotI and XbaI sites of the pCDNA3.1+ vector. This vector then was used to create all Flag-tagged proteins used in the study, each with three carboxy-terminal Flag tags in frame. The multiple cloning site of the pGEX 2T vector was modified to include a NotI site downstream of the EcoRI site, and the frame was adjusted to ensure compatibility with the pA3F vector. pCDNA-c-Myc was kindly provided by Wafik el Diery (University of Pennsylvania) (29). c-Myc and its various truncations were PCR amplified and cloned between the EcoRI and NotI sites of pA3F and pGEX 2T vectors. pCDNA-EBNA3C, pGEX 2TK-EBNA3C truncations (21), and pGL3B-hTERT (20) have been described previously. pA3F-EBNA3C, pA3F-EBNA3C 1-365, pA3F-EBNA3C 366-620, and pA3F-EBNA3C 621-992 were cloned by restriction digestion of EBNA3C and its fragments from the pA3M vector, followed by ligation into pA3F vector between the EcoRI and NotI sites. Mnt and Max open reading frames were PCR amplified from pDCNA-Mnt and pSG65-Max, respectively (kindly provided by Bernard Luscher), and were cloned into pA3F between the EcoRI and NotI sites of pA3F (35).
HEK-293 cells are human embryonic kidney cells transformed with sheared adenovirus 5 DNA (8). HEK-293T cells are HEK-293 cells that stably express the simian virus 40 large T antigen. BJAB and DG-75 are Burkitt lymphoma cell lines that are negative for both Kaposi's sarcoma-associated herpesvirus and EBV (3). LCL-1 and LCL-2 are in vitro-transformed EBV-positive LCLs (28). Rat-1 cells were kindly provided by John Sedivy (Brown University). HEK-293 and HEK-293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% bovine growth serum, 50 U/ml penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine. Rat-1 fibroblasts also were maintained in DMEM with the same concentration of supplements, except that the serum concentration was 10%. All B-cell lines were grown in RPMI 1640 medium supplemented with 7% bovine growth serum, 50 U/ml penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine.
Transfection.
HEK-293 and HEK-293T cells were transfected by electroporation with a Bio-Rad Gene Pulser II electroporator. Briefly, 10 × 106 to 12 × 106 cells harvested in exponential phase were collected and washed in phosphate-buffered saline (PBS) and resuspended in 400 μl of the appropriate medium without serum containing DNA for transfection (21). Resuspended cells were transferred to a 0.4-cm gap cuvette, and electroporation was done at 975 μF and 210 V or 220 V for 293T or DG-75 cells, respectively. Transfected cells were transferred to a 100-mm petri dish containing 10 ml of complete medium and were incubated at 37°C. Cells were harvested 24 to 36 h posttransfection for analysis.
Purification of GST fusion proteins.
Escherichia coli BL21 cells were transformed with the plasmid constructs for each glutathione S-transferase (GST) fusion protein. Single colonies were picked and grown overnight in 2 ml of Luria broth. One milliliter of the overnight culture was used to inoculate a 500-ml culture. The larger culture was incubated until the optical density was approximately 0.6, at which point it was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h. The bacteria were pelleted, washed once with STE buffer (100 mM NaCl, 10 mM Tris, and 1 mM EDTA, pH 7.5), resuspended in 3 ml NETN buffer (0.5% NP-40, 100 mM NaCl, 20 mM Tris, 1 mM EDTA, pH 8.0) supplemented with protease inhibitors, and incubated on ice for 15 min. A volume of 150 μl of 1 M dithiothreitol and 1.8 ml of a 10% solution of Sarkosyl in STE buffer was added, and the suspension was sonicated (for 1 min on ice) to solubilize the proteins. The lysate was centrifuged (12,000 × g, 10 min, 4°C) to separate the unsolubilized fraction. The clear supernatant was transferred to a fresh tube, to which 3 ml of a 10% solution of Triton X-100 in STE buffer and 200 μl of glutathione Sepharose beads were added. The tube was rotated overnight at 4°C, after which the purified protein bound to glutathione was collected by centrifugation (2 min, 600 × g, 4°C) and washed five times with NETN buffer supplemented with protease inhibitors. Purified proteins were stored at 4°C.
Immunoprecipitation and Western blotting.
Transfected cells were harvested, washed with ice-cold PBS, and lysed in 1.5 ml ice-cold radioimmunoprecipitation assay (RIPA) buffer (1% NP-40, 10 mM Tris [pH 7.5], 2 mM EDTA, 150 mM NaCl) supplemented with protease inhibitors. Cell debris was removed by centrifugation (21,000 × g, 10 min, 4°C), and the supernatant was transferred to a fresh tube. Lysates then were precleared by being rotated end over end with normal mouse serum and 20 μl of a 1:1 mixture of protein A-protein G-conjugated Sepharose beads (45 min, 4°C). Beads were spun out, and supernatant was transferred to a fresh tube. Approximately 7.5% of the lysate was saved as an input control. The protein of interest was captured by rotating the remaining lysate with 1 μg of appropriate antibody overnight at 4°C. Immune complexes were captured with 30 μl of a 1:1 mixture of protein A and protein G Sepharose beads, pelleted, and washed three times with ice-cold RIPA buffer.
For Western blotting, input lysates and immunoprecipitation complexes were boiled in Laemmli buffer (25), fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to a 0.45-μm nitrocellulose membrane. The membranes then were probed with appropriate antibodies, incubated with the appropriate secondary antibodies tagged with far-red dyes, and then viewed on an Odyssey imager (LiCor Inc., Lincoln, NE).
Immunofluorescence.
U2OS cells were plated on 22- by 22-mm coverslips. Cells were transfected with pCDNA-c-Myc and pCDNA-EBNA3C using Lipofectamine 2000. Cells were fixed using 4% buffered formalin (10 min at room temperature [RT]) and washed three times with PBS. BJAB cells stably expressing EBNA3C were dried onto slides and fixed using a 1:1 mixture of acetone and methanol. Fixed cells were blocked and permeabilized with 0.2% fish skin gelatin in 1× PBS containing 0.1% Triton X-100 (30 min at RT). c-Myc was detected using Myc-reactive 9E10 mouse ascites fluid (1:1,000 dilution), and EBNA3C was detected using EBNA3C-reactive rabbit serum (1:50 dilution). Primary antibodies were diluted in blocking buffer and incubated with the cells for 1 h at RT. Cells were washed three times (for 5 min each) with blocking buffer and exposed to secondary antibodies. Goat anti-rabbit antibody conjugated to Alexa Fluor 488 and goat anti-mouse antibody conjugated to Alexa Fluor 594 were used to detect EBNA3C and c-Myc, respectively. Secondary antibodies were diluted in blocking buffer at 1:2,000 and incubated for 1 h at RT, followed by three washes (5 min each) with blocking buffer. The last wash contained 4′,6′-diamidino-2-phenylindole (DAPI) to counterstain the nuclei.
Pulse-chase assays.
HEK-293 cells were transfected with 5 μg of pA3F-c-Myc and 10 μg of either pCDNA-EBNA3C or pCDNA to balance the transfection. Twenty-four hours later, cells were harvested and similar samples were mixed and replated to avoid inconsistencies due to transfection variability. Ten percent of the cells were separated and replated for Western blot analysis. Six hours later, when the cells had attached and spread, the medium was replaced with DMEM without Met/Cys but supplemented with 35S-labeled Met/Cys mix (150 μCi/sample) for 2 h. At the end of the pulse period, the medium supplemented with radiolabel was removed, and the cells were washed twice with prewarmed PBS and incubated with regular culture medium for 30 min to allow for the depletion of the intracellular stock of the 35S-labeled Met/Cys mix. This sample then constituted that of the 0-min chase time point. Cells were harvested at regular intervals as indicated in RIPA buffer supplemented with protease inhibitor and 15 μg/ml MG-132. The immunoprecipitation was set up as described above, except that an excess (2 μg) of anti-Flag antibody was used for each sample, and the antigen was captured for 2 h. Immune complexes were resolved on a polyacrylamide gel, exposed on a phosphorimager plate, and scanned using a Storm 850 system (GE Lifesciences, Piscataway, NJ).
Reporter assays.
Rat-1 fibroblasts were cotransfected with 0.25 μg of the promoter construct and the indicated amounts of pA3F-EBNA3C using Lipofectamine 2000. The differences in the amounts of the protein construct were equalized with the corresponding vector to keep the total amount of transfected DNA constant. Twenty-four hours posttransfection, the cells were harvested, washed with PBS, and lysed in 100 μl of reporter lysis buffer (Promega, Inc., Madison, WI). A 40-μl aliquot of the lysate was transferred to a 96-well plate. Luciferase activity was measured using an LMaxII384 luminometer (Molecular Devices, Sunnyvale, CA) by injecting 25 μl of luciferase substrate into each well and integrating the luminescence for 20 s postinjection.
Half-life computation and statistical analysis.
We assumed that the decay of c-Myc followed first-order kinetics and fitted an exponential curve to the data with the y intercept set to 1. The time constant of decay and the half-life were calculated from the resulting exponential equations. For statistical significance analysis, data at corresponding time points were compared using the two-tailed Student's t test.
RESULTS
EBNA3C associates with the transcription factor c-Myc in EBV-transformed LCLs.
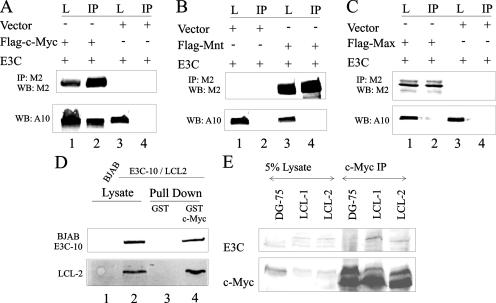
c-Myc is a transcription factor that belongs to the basic region-helix loop helix-lucine zipper (BR-HLH-LZ) family of transcription factors (44). Other members of this family are the c-Myc coactivator Max and inhibitor Mnt (27, 40). c-Myc has been shown to heterodimerize with Max to bind to its consensus sequence on target gene promoters (27, 40). Mnt also heterodimerizes with Max to prevent it from binding to c-Myc, thus acting as a negative regulator of c-Myc-mediated transcription (27, 40). We tested to see which, if any, of the members of this family of transcription regulators bind to EBNA3C. Coimmunoprecipitation experiments clearly showed that EBNA3C strongly associates with c-Myc (Fig. 1A, lane 2) but not with the other members of the BR-HLH-LZ family, Mnt and Max (Fig. 1B, lane 4, and C, lane 2, respectively). We next investigated if this interaction was of significance in a stable/endogenous setting. The interaction was tested in a GST pull-down experiment using GST-c-Myc in LCLs (LCL-2) derived from in vitro-transformed primary human B cells and in a Burkitt lymphoma cell line stably expressing EBNA3C (BJAB E3C-10). GST alone was used as a control, and the levels of interacting EBNA3C were checked by Western blot analysis (Fig. 1D, lanes 3 and 4). Also, a Burkitt lymphoma cell line, BJAB, that does not express EBNA3C was used as a negative control (Fig. 1D, lane 1). In a separate experiment, endogenous c-Myc was immunoprecipitated from two LCLs, LCL-1 and LCL-2, using anti-myc ascites fluid. An EBV-negative cell line (DG-75) was used as a control. Captured immune complexes were checked for coimmunoprecipitated EBNA3C using Western blotting (Fig. 1E, lanes 5 and 6). Results from the exogenous system as well as the stable/endogenous cell lines that express c-Myc and EBNA3C at physiological levels demonstrated the strong association of c-Myc with EBNA3C.
FIG. 1.
EBNA3C binds to overexpressed and endogenous c-Myc. (A to C) A total of 10 × 106 HEK-293T cells were cotransfected with untagged EBNA3C and Flag-tagged c-Myc (A), Mnt (B), or Max (C). In all cases, control samples were balanced with pA3F vector. Approximately 7.5% of the lysed cells were saved as the input, and the remainder were immunoprecipitated with 1 μg anti-Flag M2. (D) A total of 10 × 106 of the indicated B cells were collected and lysed in RIPA buffer. Approximately 5% of the lysates were saved as the input, and the remainder were divided into two equal parts and rotated with either GST or GST fused with c-Myc. (E) A total of 10 × 106 of the indicated B cells were collected and lysed in RIPA buffer. Approximately 5% of the lysates were saved as the input, and the remainder were immunoprecipitated with 1 μl 9E10 ascites fluid. (A to E) Samples were resolved on gels containing a suitable percentage of polyacrylamide and transferred to 0.45-μm nitrocellulose membranes. The membranes were probed with anti-EBNA3C A10 antibody followed by a secondary antibody tagged to a far-red dye and were scanned on an Odyssey imager. (A to C) The membrane was reprobed with anti-Flag M2 antibody followed by an infrared tagged secondary antibody, and it was rescanned on an Odyssey imager. L, lysate lanes; IP, immunoprecipitation lanes. All panels show representative gels from similar repeat experiments.
EBNA3C residues 130 to 190 can interact with c-Myc.
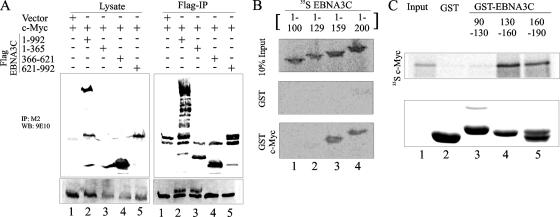
We have previously shown that EBNA3C amino acids 130 to 190 (designated EBNA3C 130-190) recruit components of the SCFSkp2 ubiquitin ligase complex. Since c-Myc is known to be modified by this same ubiquitin ligase complex, we wanted to determine if this domain of EBNA3C was important for interaction with c-Myc. As a first step, using coimmunoprecipitation experiments, we mapped the interaction with c-Myc to the amino-terminal 365 residues of EBNA3C (Fig. 2A, lane 3). To further map the interacting region within the amino terminus of EBNA3C, we in vitro translated 35S-labeled fragments of EBNA3C and tested the interaction using GST-tagged full-length c-Myc. Confirming the previous result, we found that c-Myc did not interact with EBNA3C 1-100 or EBNA3C 1-129 (Fig. 2B, lanes 1 and 2, respectively) but strongly interacted with EBNA3C 1-159 and EBNA3C 1-200 (Fig. 2B, lanes 3 and 4, respectively). All fragments of EBNA3C failed to interact with the GST control, showing that the observed interaction was specific for the c-Myc protein (Fig. 2B, lanes 1 to 4). In a reverse experiment, we used radiolabeled c-Myc in in vitro binding assays. 35S-labeled c-Myc interacted strongly with GST-EBNA3C 130-160 and GST-EBNA3C 160-190 (Fig. 2C, lanes 4 and 5, respectively) but not with the GST control or GST-EBNA3C 90-130 (Fig. 2C). Coomassie staining of a parallel gel indicated the levels of various GST proteins used in the binding assay (Fig. 2B and C). These results indicate that the domain of EBNA3C that interacts with components of the SCFSkp2 complex also interacts with c-Myc.
FIG. 2.
c-Myc binds to EBNA3C residues 130 to 190. (A) A total of 10 × 106 HEK-293T cells were cotransfected with Flag-tagged c-Myc and myc-tagged control vector (lanes 1 and 6), EBNA3C 1-992 (lanes 2 and 7), EBNA3C 1-365 (lanes 3 and 8), EBNA3C 366-620 (lanes 4 and 9), or EBNA3C 621-992 (lanes 5 and 10). Cells were harvested, and 5% of the lysed cells were saved as the input; the remainder were immunoprecipitated with 1 μg anti-Flag M2. Samples were resolved on a gel containing a suitable percentage of polyacrylamide and transferred to 0.45-μm nitrocellulose membrane. The membrane was probed with anti-myc 9E10 antibody followed by a secondary antibody tagged to a far-red dye and then was scanned on an Odyssey imager. The membrane was reprobed with anti-Flag M2 antibody followed by a secondary antibody tagged to a far-red dye and then was rescanned on an Odyssey imager. (B and C) 35S-radiolabeled E3C fragments (B) or full-length c-Myc (C) was in vitro translated using a T7 TNT translation kit. Translated protein was precleared by being rotated with GST for 30 min at 4°C. The binding reaction mixture was set up with either GST control or the indicated GST fusion proteins. Reactions were resolved on a polyacrylamide gel, exposed to a phosphorimager plate, and scanned on a Storm 850 imaging system. All panels show representative gels from similar repeat experiments.
Residues 85 to 170 of c-Myc are important for interaction with EBNA3C.
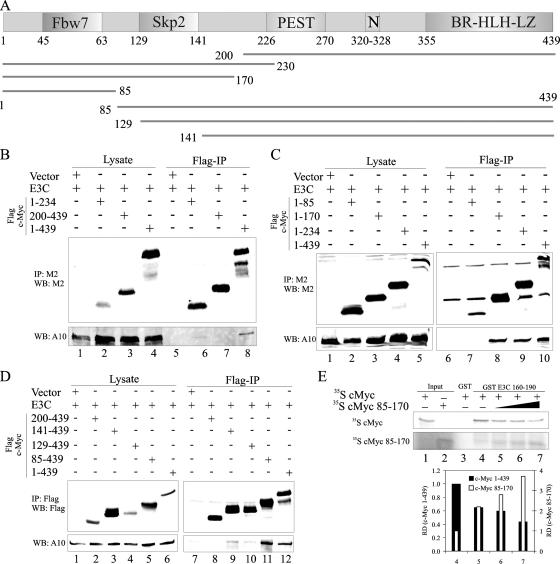
The c-Myc protein is highly labile, with a reported half-life of 20 to 30 min (36). The amino terminus of the protein has two regions that are highly conserved across species (44). The first region (residues 45 to 63) interacts with the SCFFbw7 ubiquitin ligase complex, and the second region (residues 129 to 141) interacts with the SCFSkp2 ubiquitin ligase complex (Fig. 3A) (44). The carboxy terminus of c-Myc contains the BR-HLH-LZ domain that is necessary for binding to consensus sequences within its target promoters (Fig. 3A) (44). To determine which region of c-Myc interacts with EBNA3C, we made various deletion mutants (Fig. 3A) and employed coimmunoprecipitation assays with full-length EBNA3C. We initially determined that the amino terminus of c-Myc was indeed the region that interacts with EBNA3C (Fig. 3B). The amino-terminal 230 residues of c-Myc interacted with EBNA3C, while c-Myc residues 200 to 439 failed to interact (Fig. 3B, lanes 6 and 7). To determine which of the two highly conserved regions within the amino terminus was important for this interaction, we made further deletion mutants and performed a similar experiment. c-Myc residues 1 to 85 (designated c-Myc 1-85) failed to coimmunoprecipitate EBNA3C, while c-Myc 1-170 bound strongly to EBNA3C, tentatively indicating that c-Myc amino acids 85 to 170 are important for interaction with EBNA3C (Fig. 3C). To confirm that this was indeed the case, we made some reverse deletion mutants and tested their interaction in the same assay. As expected, c-Myc residues 85 to 439 bound strongly to EBNA3C (Fig. 3D and E, lane 11), confirming that the region that interacts with EBNA3C lies within c-Myc residues 85 to 170. Interestingly, c-Myc 129-439 and c-Myc 141-439 bound weakly to EBNA3C, indicating that c-Myc residues 85 to 129 were required for high-affinity interaction with EBNA3C (Fig. 3D and E, lanes 9 and 10). EBNA3C therefore interacts with a region of c-Myc that surrounds the highly conserved Skp2 box. Having identified the region of c-Myc that interacts with EBNA3C, we made a construct that expressed just that portion of c-Myc (residues 85 to 170) to test whether it could compete with full-length c-Myc for binding to EBNA3C. In vitro-translated c-Myc 85-170 successfully competed with full-length c-Myc 1-439, in a dose-dependent manner, for binding to GST EBNA3C 130-160 (Fig. 3F and G, lanes 4 to 7). Unfortunately, we were unable to express c-Myc 85-170 in cells at appreciable levels, presumably due to the instability of the peptide in cells.
FIG. 3.
EBNA3C interacts with c-Myc 85-170. (A) Schematic of c-Myc with identified domains shown. Potentially important lysine residues are shown in red, and serine/threonine residues are shown in black. (B to D) A total of 10 × 106 HEK-293T cells were cotransfected with untagged EBNA3C and Flag-tagged c-Myc or its truncation mutants. In all cases, control samples were balanced with pA3F vector. A total of 7.5% of the lysed cells were saved as the input, and the remainder were immunoprecipitated with 1 μg anti-Flag M2. Samples were resolved on gels containing a suitable percentage of polyacrylamide and transferred to 0.45-μm nitrocellulose membranes. The membranes were probed with anti-EBNA3C A10 antibody followed by a secondary antibody tagged to a far-red dye and then scanned on an Odyssey imager. The membranes were reprobed with anti-Flag M2 antibody followed by a secondary antibody tagged to a far-red dye and then rescanned on an Odyssey imager. (E) 35S-radiolabeled c-Myc 1-439 was mixed with increasing amounts of 35S-radiolabeled c-Myc 85-170. The total amount of reticulocyte lysate was kept constant in each reaction by adding the appropriate amount of cold reticulocyte lysate. The equivalent of 10% of each translation product used in lane 5 was used as the input (lanes 1 and 2, respectively). Binding reaction mixtures were set up with either GST (as a control) (lane 3) or GST-EBNA3C 130-190 (lanes 4 to 7). Bound radiolabeled proteins were resolved on a polyacrylamide gel, exposed to a phosphorimager plate, and scanned on a Storm 850 imaging system. All panels show representative gels from similar repeat experiments.
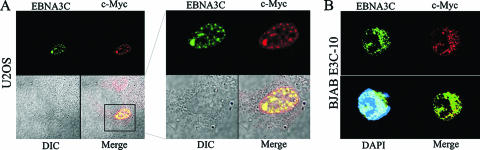
c-Myc and EBNA3C can localize to the same cellular compartments in vivo.
To visualize the interaction of c-Myc and EBNA3C in an in vivo setting, we performed colocalization experiments. c-Myc and EBNA3C were exogenously expressed in a human osteosarcoma cell line, U2OS. c-Myc colocalized with EBNA3C at a number of spots, indicating that these two proteins are in the same cellular compartments (Fig. 4A). To visualize the interaction of these proteins closer to physiological levels, we used a Burkitt lymphoma cell line stably expressing EBNA3C. At these levels of expression in B cells, EBNA3C and c-Myc displayed a distinct punctate pattern with various points that colocalized as yellow (Fig. 4B).
FIG. 4.
EBNA3C and c-Myc colocalize in vivo. Untagged c-Myc and untagged EBNA3C were either (A) exogenously expressed in human breast epithelial cell line U2OS or (B) endogenously expressed in a BJAB cell line stably expressing EBNA3C (BJAB E3C10). EBNA3C was detected using EBNA3C-reactive rabbit serum followed by anti-rabbit Alexa Fluor 488, and c-Myc was detected using mouse anti-Myc 9E10 ascites fluid followed by anti-mouse Alexa Fluor 594. The nuclei were counterstained using DAPI. The images were sequentially captured using an Olympus confocal microscope. All panels are representative pictures from similar repeat experiments.
Association with EBNA3C leads to stabilization of c-Myc.
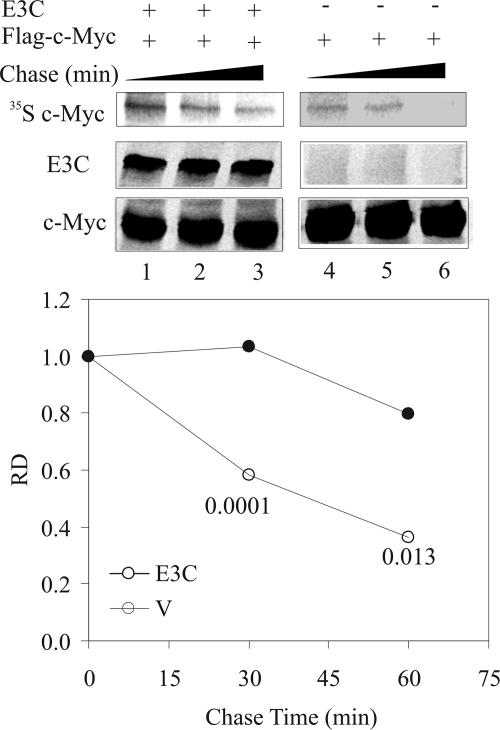
EBNA3C associates with a well-known degron (44) in c-Myc (Fig. 3A). It therefore is conceivable that this association has some effect on c-Myc stability. This study was, however, complicated by the fact that most cell lines carry mutations in c-Myc that render it more stable than its wild-type counterpart. To prevent the masking of the effect of EBNA3C coexpression on the stability of c-Myc, we used exogenously expressed wild-type c-Myc (29) in pulse-chase assays to address this question. HEK-293 cells were used for pulse-chase assays, because HEK-293T cells express the simian virus 40 large T antigen that has been shown to bind to Fbw7 and regulate c-Myc stability (48). Either Flag-tagged c-Myc alone or Flag-tagged c-Myc along with untagged EBNA3C was expressed in HEK-293 cells. Cellular proteins were radiolabeled for 2 h with 35S-labeled Met/Cys (pulse), washed, and incubated with regular culture medium for the indicated times (chase). c-Myc was immunoprecipitated with excess anti-Flag antibody from various samples, and the quantity of nondegraded radiolabeled protein remaining was ascertained to determine the half-life. In agreement with previous reports, the half-life of wild-type c-Myc in the absence of EBNA3C was ∼40 min (Fig. 5, top). In the presence of EBNA3C, however, the half-life of c-Myc was considerably longer than 60 min (∼240 min). Figure 5 shows an average quantification of multiple experiments showing similar trends.
FIG. 5.
c-Myc stability is enhanced in the presence of EBNA3C. A total of 10 × 106 HEK-293 cells per sample were cotransfected with Flag-tagged c-Myc and either vector control or untagged EBNA3C. Approximately 30 h later, cellular proteins were radiolabeled for 2 h using Met/Cys-free medium supplemented with 150 μCi 35S-labeled Met/Cys per sample. Cells were washed with PBS and chased with regular growth medium for the indicated times before harvest. c-Myc was immunoprecipitated from harvested samples using excess (2 μg) anti-Flag M2 antibody. Immune complexes were resolved on an appropriate acrylamide gel, which then was exposed on a phosphorimager plate. The plate was scanned on a Storm 850 scanner, and the signal was quantified using ImageQuant software. Parallel cold samples were used to detect the expression levels of c-Myc and EBNA3C by Western blot analysis. All panels show representative gels from similar repeat experiments. The asterisks indicate a statistically significant difference in protein levels in the presence and absence of EBNA3C compared to the corresponding chase times. This suggests that EBNA3C can enhance the stability of c-Myc.
EBNA3C can modulate the transcription of c-Myc target genes.
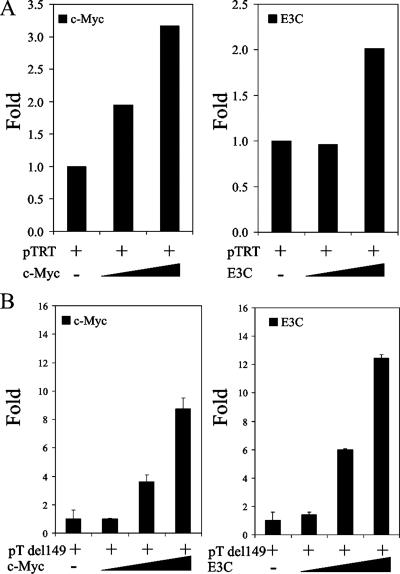
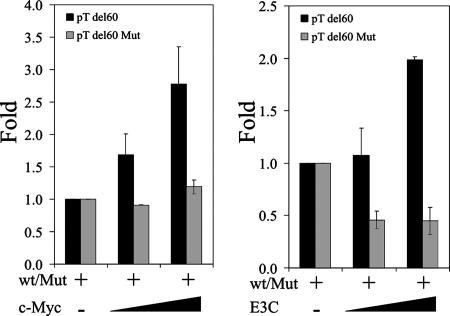
c-Myc has been shown to bind to more than 3,000 loci on the human genome (5). It has been shown to regulate major genes involved in cell cycle control (15). Skp2 has been shown to be a cofactor in c-Myc-regulated transcription from a number of target promoters, particularly the telomerase reverse transcriptase promoter (TRTp) (17). Since EBNA3C binds both c-Myc and Skp2, we tested to see if EBNA3C could regulate the promoters of some c-Myc target genes. Our results show that EBNA3C can regulate the transcription of the telomerase reverse transcriptase (TERT) promoter construct in a dose-dependent manner (Fig. 6A, right). Importantly, the regulation profile is identical to that observed with an increasing dose of c-Myc (Fig. 6A, left) (9, 34, 49). c-Myc has been shown to upregulate transcription from the TERT promoter by a number of investigators (9, 49). This upregulation has been attributed to the presence of a well-documented E box (at positions −30 to −24) within the promoter sequence (49). To narrow down the regulation of the human TERT promoter by EBNA3C to this critical E box, we used a truncation mutant of the promoter, pTdel149, in which promoter sequences upstream of −149 from the transcription initiation site were deleted. We observed similar dose-dependent transcriptional profiles for both c-Myc and EBNA3C by using this truncation mutant (Fig. 6B), confirming that the EBNA3C-modulated element within the promoter was contained within this truncation mutant. To further confirm that EBNA3C could target c-Myc genes, we constructed a reporter construct using the sequence flanking E box 1 of the human TERT promoter (49). c-Myc has been shown to bind to this region of the TERT promoter and to regulate TERT transcriptional activity (49). In parallel, we also constructed a reporter construct containing the same sequence, except that we replaced the E box in this construct with a series of adenines. The cotransfection of this truncated reporter construct, along with increasing levels EBNA3C or c-Myc, show that both EBNA3C and c-Myc are able to upregulate transcription from the human TERT promoter at similar levels (Fig. 7). Increasing levels of c-Myc did not, however, have any effect on transcription from the mutated TERT promoter, and EBNA3C repressed transcription from this construct, suggesting a loss of endogenous c-Myc activity at the mutated site that results in repression by EBNA3C in the absence of c-Myc bound to the E box.
FIG. 6.
EBNA3C can modulate transcription from c-Myc target promoters. Increasing amounts (0 to 200 ng) of either EBNA3C or c-Myc expression constructs were cotransfected with 0.25 μg of the c-Myc-responsive promoter/reporter construct pTERT (A) or pTERTdel149 (B). Differences in protein and reporter construct levels between samples were balanced by the corresponding empty vectors. Cells were harvested ∼24 h later and tested for luciferase reporter activity using a luminometer. Plots are representative of data from similar repeat experiments.
FIG. 7.
EBNA3C and c-Myc synergistically modulate transcription from c-Myc target promoters. Increasing amounts (0 to 200 ng) of either c-Myc or EBNA3C expression constructs were cotransfected with 0.25 μg of either a wild-type or a mutant c-Myc-responsive promoter/reporter construct. Differences in protein and reporter construct levels between samples were balanced by the corresponding empty vectors. Cells were harvested ∼24 h later and tested for luciferase reporter activity using a luminometer. Plots are representative of data from similar repeat experiments.
DISCUSSION
A great deal of attention has been focused on c-Myc translocation to the immunoglobulin G loci in Burkitt lymphoma and its consequent amplification. This translocation was only the second genetic aberration associated with cancer, and at the time it underscored the importance of c-Myc in tumorigenesis and in EBV biology (26). Since then, it has been reported that, in most Burkitt lymphoma cell lines, not only is c-Myc amplified but it also is stabilized due to mutations in conserved c-Myc degron 1 at T48 (10). These observations, however, do not provide any clear insights on the ability of EBV to transform B lymphocytes in vitro. In most such cases of in vitro transformation, it is reasonable to assume that c-Myc is wild type and not amplified. A few viral proteins ostensibly are able to transform these cells. It is conceivable, then, that the virus has other modes of regulating c-Myc that come to the fore in these cells carrying the wild-type c-Myc allele.
EBNA3C is one of the four EBV proteins that are critical for the transformation of B lymphocytes in vitro (37). Previous studies have demonstrated the ability of EBNA3C to target cell cycle regulators, resulting in the deregulation of the mammalian cell cycle (21-24, 32). EBNA3C targets the key cell cycle regulators p27 and pRb by recruiting components of the SCFSkp2 ubiquitin ligase and facilitating the degradation of these inhibitors of the mammalian cell cycle (21, 23). In this report, we investigated the role of EBNA3C in regulating another key component of the mammalian cell cycle control, c-Myc (15). Like p27 and pRb, c-Myc also is a target of the SCFSkp2 complex (31). Unlike p27 and pRb, however, the association of c-Myc with SCFSkp2 is somewhat more complex. Not only has Skp2 been shown to regulate the stability of c-Myc but it also is a cofactor in the c-Myc-regulated transcription of its target genes (12, 17, 46).
Our results show that c-Myc binds to EBNA3C. EBNA3C associates with c-Myc via the same domain, amino acids 130 to 190, that has been implicated in recruiting the SCFSkp2 complex. Residues of c-Myc that associate with EBNA3C overlap those that have been shown to bind Skp2. It is possible, then, that EBNA3C recruits both c-Myc and its transcriptional cofactor Skp2 to the same domain, thus bringing them together and facilitating c-Myc-regulated transcription. Indeed, we find that EBNA3C can modulate transcription from the promoter of a well-known c-Myc target gene, the telomerase reverse transcriptase. Whether EBNA3C can bind to the promoter via c-Myc or requires additional factors currently is under investigation in our laboratory. Recent reports on human papillomavirus E7, which also recruits the SCFSkp2 complex and similarly regulates the stability of p27 and pRb, have shown that E7 also binds to c-Myc and promotes its transcriptional activity (47).
Previous data from our laboratory show that the recruitment of SCFSkp2 by EBNA3C leads to the degradation of targets of the ubiquitin ligase complex (23, 24). Paradoxically, here we found that EBNA3C enhances the stability of c-Myc. A possible explanation for this might be that the ubiquitylation of c-Myc by SCFSkp2 is necessary for transcriptional activation by c-Myc (17, 45, 46). The coupling of ubiquitylation and transcription under normal circumstances ensures rapid degradation after the G1/S transition and acts as a safety mechanism to ensure that cells do not continuously cycle (44). Whether the lysine residues that are involved in transcriptional activation and degradation are unique or the same remains an open question. The exact mechanism of how EBNA3C disrupts this coupling of ubiquitylation/degradation and the specific lysine residues involved in each currently is under investigation in our laboratory.
This work constitutes an initial report showing the modulation of c-Myc activity by EBNA3C. We believe this report points to a possible mechanism of c-Myc deregulation in EBV-infected cells that does not involve chromosomal translocation or amplification. This work also underscores the importance of the EBNA3C region of amino acids 130 to 190 in cell cycle deregulation. Further elucidation of the mechanism by which EBNA3C is able to deregulate c-Myc would not only be important for understanding EBV pathogenesis but also would shed light on mammalian cell cycle control.
Acknowledgments
S.C.V. is supported by the Lady Tata Memorial Trust. This work was supported by grants from the National Institutes of Health: NCI CA108461 and CA072510 and NIDCR grant DE014136. K.L. is a special fellow and E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
Footnotes
Published ahead of print on 6 February 2008.
REFERENCES
- 1.Reference deleted.
- 2.Arvanitakis, L., N. Yaseen, and S. Sharma. 1995. Latent membrane protein-1 induces cyclin D2 expression, pRb hyperphosphorylation, and loss of TGF-beta 1-mediated growth inhibition in EBV-positive B cells. J. Immunol. 1551047-1056. [PubMed] [Google Scholar]
- 3.Ben-Bassat, H., N. Goldblum, S. Mitrani, T. Goldblum, J. M. Yoffey, M. M. Cohen, Z. Bentwich, B. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75). Int. J. Cancer 1927-33. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 869558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang, C. V., K. A. O'Donnell, K. I. Zeller, T. Nguyen, R. C. Osthus, and F. Li. 2006. The c-Myc target gene network. Semin. Cancer Biol. 16253-264. [DOI] [PubMed] [Google Scholar]
- 6.Epstein, M. A., B. G. Achong, and J. H. Pope. 1967. Virus in cultured lymphoblasts from a New Guinea Burkitt lymphoma. Br. Med. J. 2290-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein, M. A., Y. M. Barr, and B. G. Achong. 1965. Studies with Burkitt's lymphoma. Wistar Inst. Symp. Monogr. 469-82. [PubMed] [Google Scholar]
- 8.Graham, F. L. 1977. Biological activity of tumor virus DNA. Adv. Cancer Res. 251-51. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg, R. A., R. C. O'Hagan, H. Deng, Q. Xiao, S. R. Hann, R. R. Adams, S. Lichtsteiner, L. Chin, G. B. Morin, and R. A. DePinho. 1999. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene 181219-1226. [DOI] [PubMed] [Google Scholar]
- 10.Gregory, M. A., and S. R. Hann. 2000. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol. Cell. Biol. 202423-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hislop, A. D., G. S. Taylor, D. Sauce, and A. B. Rickinson. 2007. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu. Rev. Immunol. 25587-617. [DOI] [PubMed] [Google Scholar]
- 12.Jin, J., and J. W. Harper. 2003. A license to kill: transcriptional activation and enhanced turnover of Myc by the SCF(kp2) ubiquitin ligase. Cancer Cell 3517-518. [DOI] [PubMed] [Google Scholar]
- 13.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 909150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly, K., and U. Siebenlist. 1986. The regulation and expression of c-myc in normal and malignant cells. Annu. Rev. Immunol. 4317-338. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, K., and U. Siebenlist. 1985. The role of c-myc in the proliferation of normal and neoplastic cells. J. Clin. Immunol. 565-77. [DOI] [PubMed] [Google Scholar]
- 16.Kempkes, B., D. Pich, R. Zeidler, B. Sugden, and W. Hammerschmidt. 1995. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J. Virol. 69231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, S. Y., A. Herbst, K. A. Tworkowski, S. E. Salghetti, and W. P. Tansey. 2003. Skp2 regulates Myc protein stability and activity. Mol. Cell 111177-1188. [DOI] [PubMed] [Google Scholar]
- 18.Klein, G. 1986. Chromosomal translocations in B-cell derived tumors. Princess Takamatsu Symp. 17159-170. [PubMed] [Google Scholar]
- 19.Klein, G. 1986. Constitutive activation of oncogenes by chromosomal translocations in B-cell derived tumors. AIDS Res. 2(Suppl. 1)S167-S176. [PubMed] [Google Scholar]
- 20.Knight, J. S., M. A. Cotter, Jr., and E. S. Robertson. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 27622971-22978. [DOI] [PubMed] [Google Scholar]
- 21.Knight, J. S., and E. S. Robertson. 2004. Epstein-Barr virus nuclear antigen 3C regulates cyclin A/p27 complexes and enhances cyclin A-dependent kinase activity. J. Virol. 781981-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight, J. S., N. Sharma, D. E. Kalman, and E. S. Robertson. 2004. A cyclin-binding motif within the amino-terminal homology domain of EBNA3C binds cyclin A and modulates cyclin A-dependent kinase activity in Epstein-Barr virus-infected cells. J. Virol. 7812857-12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight, J. S., N. Sharma, and E. S. Robertson. 2005. Epstein-Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc. Natl. Acad. Sci. USA 10218562-18566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight, J. S., N. Sharma, and E. S. Robertson. 2005. SCFSkp2 complex targeted by Epstein-Barr virus essential nuclear antigen. Mol. Cell. Biol. 251749-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 26.Leder, P., J. Battey, G. Lenoir, C. Moulding, W. Murphy, H. Potter, T. Stewart, and R. Taub. 1983. Translocations among antibody genes in human cancer. Science 222765-771. [DOI] [PubMed] [Google Scholar]
- 27.Lüscher, B., and L. G. Larsson. 1999. The basic region/helix-loop-helix/leucine zipper domain of Myc proto-oncoproteins: function and regulation. Oncogene 182955-2966. [DOI] [PubMed] [Google Scholar]
- 28.Maruo, S., E. Johannsen, D. Illanes, A. Cooper, and E. Kieff. 2003. Epstein-Barr virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J. Virol. 7710437-10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell, K. O., M. S. Ricci, T. Miyashita, D. T. Dicker, Z. Jin, J. C. Reed, and W. S. El-Deiry. 2000. Bax is a transcriptional target and mediator of c-myc-induced apoptosis. Cancer Res. 606318-6325. [PubMed] [Google Scholar]
- 30.Moorthy, R. K., and D. A. Thorley-Lawson. 1993. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of rat-1 fibroblasts. J. Virol. 671638-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama, K. I., and K. Nakayama. 2005. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin. Cell Dev. Biol. 16323-333. [DOI] [PubMed] [Google Scholar]
- 32.Parker, G. A., T. Crook, M. Bain, E. A. Sara, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 132541-2549. [PubMed] [Google Scholar]
- 33.Peng, M., and E. Lundgren. 1992. Transient expression of the Epstein-Barr virus LMP1 gene in human primary B cells induces cellular activation and DNA synthesis. Oncogene 71775-1782. [PubMed] [Google Scholar]
- 34.Philipp, A., A. Schneider, I. Vasrik, K. Finke, Y. Xiong, D. Beach, K. Alitalo, and M. Eilers. 1994. Repression of cyclin D1: a novel function of MYC. Mol. Cell. Biol. 144032-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulverer, B., A. Sommer, G. A. McArthur, R. N. Eisenman, and B. Luscher. 2000. Analysis of Myc/Max/Mad network members in adipogenesis: inhibition of the proliferative burst and differentiation by ectopically expressed Mad1. J. Cell Physiol. 183399-410. [DOI] [PubMed] [Google Scholar]
- 36.Ramsay, G., G. I. Evan, and J. M. Bishop. 1984. The protein encoded by the human proto-oncogene c-myc. Proc. Natl. Acad. Sci. USA 817742-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rickinson, A., and E. Kieff. 2002. Epstein-Barr virus, p. 2575-2627. In D. M. Knipe and P. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA.
- 38.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2447. In B. N. Fields et al. (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven, Philadelphia, PA.
- 39.Roizman, B. 1996. Herpesviridae, p. 2221-2231. In B. N. Fields et al. (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven, Philadelphia, PA.
- 40.Rottmann, S., and B. Luscher. 2006. The Mad side of the Max network: antagonizing the function of Myc and more. Curr. Top. Microbiol. Immunol. 30263-122. [DOI] [PubMed] [Google Scholar]
- 41.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 133321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skinner, G. R., A. Ahmad, and J. A. Davies. 2001. The infrequency of transmission of herpesviruses between humans and animals; postulation of an unrecognised protective host mechanism. Comp. Immunol. Microbiol. Infect. Dis. 24255-269. [DOI] [PubMed] [Google Scholar]
- 43.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 672014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vervoorts, J., J. Luscher-Firzlaff, and B. Luscher. 2006. The ins and outs of MYC regulation by posttranslational mechanisms. J. Biol. Chem. 28134725-34729. [DOI] [PubMed] [Google Scholar]
- 45.von der Lehr, N., S. Johansson, and L. G. Larsson. 2003. Implication of the ubiquitin/proteasome system in Myc-regulated transcription. Cell Cycle 2403-407. [PubMed] [Google Scholar]
- 46.von der Lehr, N., S. Johansson, S. Wu, F. Bahram, A. Castell, C. Cetinkaya, P. Hydbring, I. Weidung, K. Nakayama, K. I. Nakayama, O. Soderberg, T. K. Kerppola, and L. G. Larsson. 2003. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell 111189-1200. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y. W., H. S. Chang, C. H. Lin, and W. C. Yu. 2007. HPV-18 E7 conjugates to c-Myc and mediates its transcriptional activity. Int. J. Biochem. Cell Biol. 39402-412. [DOI] [PubMed] [Google Scholar]
- 48.Welcker, M., and B. E. Clurman. 2005. The SV40 large T antigen contains a decoy phosphodegron that mediates its interactions with Fbw7/hCdc4. J. Biol. Chem. 2807654-7658. [DOI] [PubMed] [Google Scholar]
- 49.Wu, K. J., C. Grandori, M. Amacker, N. Simon-Vermot, A. Polack, J. Lingner, and R. Dalla-Favera. 1999. Direct activation of TERT transcription by c-MYC. Nat. Genet. 21220-224. [DOI] [PubMed] [Google Scholar]