Abstract
X-box binding protein 1 (XBP-1), a basic leucine zipper transcription factor, plays a key role in the cellular unfolded protein response (UPR). There are two XBP-1 isoforms in cells, spliced XBP-1S and unspliced XBP-1U. XBP-1U has been shown to bind to the 21-bp Tax-responsive element of the human T-lymphotropic virus type 1 (HTLV-1) long terminal repeat (LTR) in vitro and transactivate HTLV-1 transcription. Here we identify XBP-1S as a transcription activator of HTLV-1. Compared to XBP-1U, XBP-1S demonstrates stronger activating effects on both basal and Tax-activated HTLV-1 transcription in cells. Our results show that both XBP-1S and XBP-1U interact with Tax and bind to the HTLV-1 LTR in vivo. In addition, elevated mRNA levels of the gene for XBP-1 and several UPR genes were detected in the HTLV-1-infected C10/MJ and MT2 T-cell lines, suggesting that HTLV-1 infection may trigger the UPR in host cells. We also identify Tax as a positive regulator of the expression of the gene for XBP-1. Activation of the UPR by tunicamycin showed no effect on the HTLV-1 LTR, suggesting that HTLV-1 transcription is specifically regulated by XBP-1. Collectively, our study demonstrates a novel host-virus interaction between a cellular factor XBP-1 and transcriptional regulation of HTLV-1.
Human T-lymphotropic virus type 1 (HTLV-1) is the causative agent of adult T-cell leukemia/lymphoma and the neurological disorder HTLV-1-associated myelopathy/tropical spastic paraparesis (14, 33, 34, 49). The HTLV-1 transactivator Tax activates viral transcription through three 21-bp repeats, which are known as Tax-responsive elements (TREs), located within the HTLV-1 long terminal repeat (LTR) (5, 20). Each 21-bp TRE repeat contains a cyclic AMP response element (CRE) recognized by members of the CRE binding protein/activating transcription factor (CREB/ATF) family of proteins. All CREB/ATF proteins contain a basic-region leucine zipper (bZIP) domain, which is involved in DNA binding and gene regulation (15). Tax does not bind the TRE repeats directly but interacts with CREB (or other CREB/ATF family members) to form a protein complex that associates with the DNA (1, 3, 12). The Tax-CREB complex serves as a binding site for the recruitment of cellular transcriptional coactivators, including CREB binding protein (CBP), p300, and p300/CBP-associated factor, resulting in the activation of viral transcription (16, 22, 26). CREB1 (previously known as CREB), CREB2 (also known as TREB7), ATF-1 (also known as TREB36), and ATF-2 have been identified as the cellular Tax binding proteins, suggesting that these CREB/ATF family members play a key role in the transcriptional regulation of HTLV-1 (12, 35, 45, 53).
X-box binding protein 1 (XBP-1) is a bZIP protein belonging to the CREB/ATF family of transcription factors. XBP-1 plays a major role in the cellular unfolded protein response (UPR), which is triggered by accumulation of unfolded or misfolded proteins in the endoplasmic reticulum (ER) (6). There are two isoforms of XBP-1, XBP-1U and XBP-1S. XBP-1U (also known as TREB5), which consists of 261 amino acids (aa), is translated from the unspliced mRNA of XBP-1 and is the dominant isoform under nonstress conditions. It has been reported that XBP-1U is transcriptionally inactive (46). UPR activation induces the endoribonuclease activity of inositol-requiring enzyme 1, an ER transmembrane protein, resulting in the excision of 26 bases (between nucleotides 531 and 556 of the XBP-1 mRNA) from the XBP-1 transcript. Splicing of the 26 nucleotides leads to a frameshift at aa 165 and the generation of a longer and transcriptionally active protein, XBP-1S (376 aa) (46). Both XBP-1S and XBP-1U contain basic (75 to 92 aa) and leucine zipper (93 to 133 aa) domains, which are located in the common N-terminal regions (1 to 164 aa) (48). XBP-1S activates the expression of several genes involved in protein secretion and results in increased contents of ER and Golgi complexes (27). XBP-1S plays an essential role in the up-regulation of the secretory capacity of the plasma cell to prepare it for high-level secretion of immunoglobulins (18, 19, 38). In addition, overexpression of XBP-1S has been shown to improve the productivity of pharmaceutical recombinant proteins in Chinese hamster ovary and NS0 myeloma cells by modulating the secretory pathway (25).
Recent studies show that the UPR can be induced by infection of various viruses, including Kaposi's sarcoma-associated herpesvirus (KSHV) (21), West Nile virus (WNV) (31), Japanese encephalitis virus (JEV) (40), hepatitis C virus (HCV) (41, 42), human cytomegalovirus (HCMV) (17, 43), dengue virus serotype 2 (DEN-2) (51), and severe acute respiratory syndrome coronavirus (7). Some viruses, such as JEV and DEN-2, use the ER of host cells as the primary site of glycoprotein synthesis, genomic RNA replication, and virus particle maturation and thus trigger ER stress, as well as the UPR (40, 51). In the other case, some viral proteins, such as HCMV US11, traffic to the ER of host cells and induce the UPR (43). Although viral infection results in induction of the UPR, it is notable that virus can selectively modify the outcome of the UPR signaling pathway to benefit viral replication in some cases (17).
In one of the earliest studies demonstrating the interaction between the HTLV-1 21-bp repeats and the CREB/ATF family of proteins, XBP-1U was identified as a cellular TRE binding protein in an in vitro binding experiment (50). Importantly, a higher XBP-1 mRNA level was observed in an HTLV-1-infected T-cell line while no elevated transcription of CREB1, CREB2, or ATF-1 was detected, suggesting that XBP-1 might play a more critical role in HTLV-1 transcription and replication (50). The transactivating effect of XBP-1U on HTLV-1 transcription was later reported (10, 30). However, all of the studies were carried out before the discovery of XBP-1S. Here we show that XBP-1S functions as an activator of HTLV-1 transcription through association with the HTLV-1 LTR. We also find that Tax up-regulates XBP-1 transcription and interacts with both XBP-1 isoforms, demonstrating a novel host-virus gene regulation between XBP-1 and HTLV-1 transcription.
MATERIALS AND METHODS
Cells and plasmids.
HeLa, HEK293, 293T, Jurkat, C10/MJ, MT2, and MT4 cells were obtained from the American Type Culture Collection. HuT78 and CEM cells are available through the NIH AIDS Research and Reference Reagent Program. The human XBP-1 (accession number NM_005080) mammalian expression vector was purchased from Open Biosystems. Site-directed mutagenesis was performed to generate the XBP-1S expression vector by deleting the 26 nucleotides between positions 531 and 556 of the human XBP-1 mRNA (46). The coding regions of XBP-1(1-74), XBP-1U(134-261), and XBP-1S(134-378) (which contain XBP-1 aa 1 to 74, XBP-1U aa 134 to 261, and XBP-1S aa 134 to 378, respectively) were amplified by PCR with a wild-type XBP-1U or XBP-1S vector as the template. The amplified fragments were then subcloned into plasmid pcDNA6 (Invitrogen) to generate the XBP-1 mutant vectors. Tax expression vectors containing wild-type Tax (pcTax), the M22 mutation, and the M47 mutation were generous gifts from Warner C. Greene (37, 39). The coding region of HTLV-1 Tax was amplified by PCR with plasmid pcTax as the template and subcloned into the cytomegalovirus (CMV)-enhanced green fluorescent protein (EGFP) vector (kindly provided by Zhiwei Song, Bioprocessing Technology Institute, Singapore) to generate CMV-EGFP-Tax. The expression of EGFP-Tax was under the control of the HCMV major immediate-early (MIE) promoter. HTLV-1-LUR-GL3 firefly luciferase (HTLV-1-LTR-F-Luc) was a kind gift from Arnold Rabson (29). The vectors BiP-Luc and ATF6α(1-373) were provided by Kazutoshi Mori (47). Firefly luciferase plasmids CMV-Luc, MLV-LTR-Luc, and HIV-LTR-Luc, in which the expression of luciferase was driven by the HCMV MIE promoter, the Moloney murine leukemia virus (MLV) LTR, and the human immunodeficiency (HIV) LTR, respectively, were previously described (8, 9, 32).
To generate stable HTLV-1-LTR-F-Luc clones, we first constructed the HTLV-1-LTR-GL2 firefly luciferase vector [HTLV-1-LTR-F-Luc (GL2)], which contains the selective marker blasticidin for stable mammalian cells. The CMV MIE promoter in the CMV-F-Luc plasmid (8) was replaced with the HTLV-1 LTR to generate HTLV-1-LTR-F-Luc (GL2). HeLa cells were then transfected with the HTLV-1-LTR-F-Luc (GL2) plasmid with Fugene 6 (Roche) as described in the manufacturer's manual. Selection was performed with 12 μg/ml blasticidin (Invitrogen) to generate a stable pool of HTLV-1-LTR-F-Luc cells. Single clones displaying XBP-1S- and Tax-inducible expression of luciferase were selected.
Transient transfection and luciferase assays.
Transient transfections of HEK293, 293T, and HeLa adherent cells and stable HTLV-1-LTR-F-Luc cells were performed with Fugene 6 (Roche). To perform the luciferase-based assays, the cells were grown to 50 to 80% confluence in 96-well plates. Transfection of Jurkat cells was carried out with the Amaxa electroporation system (Amaxa). Cells were cotransfected with the expression plasmid indicated, a firefly luciferase reporter, and a Renilla luciferase plasmid, pRL-RSV (Promega). pRL-RSV was used to normalize transfection efficiency. Firefly and Renilla luciferase activities were measured at 48 h posttransfection with the Dual-Glo assay system (Promega), and the activities were determined with an Infinite 200 multiplate reader (Tecan).
Chromatin immunoprecipitation (ChIP).
The ChIP assay was performed as described previously (28). Stable HTLV-1-LTR-F-Luc cells were transfected with the XBP-1U or XBP-1S expression vector. The chromatins (which were sonicated to DNA sizes of 300 to 1,000 bp) of the transfected stable HTLV-1-LTR-F-Luc cells were isolated at 48 h posttransfection and used for ChIP analyses with normal rabbit immunoglobulin G (IgG; Upstate Biotechnology), anti-CREB1 antibody (Upstate Biotechnology), or anti-XBP-1 antibody (Santa Cruz Biotechnology). Immunoprecipitated DNA was analyzed by PCR with primers specific for the HTLV-1 LTR (5′-AAGGTCAGGGCCCAGACTAAG-3′ and 5′-GAGGTGAGGGGTTGTCGTCAA-3′) and the luciferase coding region (5′-GTTACAACACCCCAACATCTT-3′ and 5′-ATTTGGACTTTCCGCCCTTCT-3′).
Coimmunoprecipitation (co-IP) and Western blotting.
HEK293 cells were cotransfected with XBP-1S/Tax or XBP-1U/Tax expression plasmids. Lysates prepared from the transfected cells were used for co-IP. Co-IP was performed with an IP kit (according to the manufacturer's [Roche] manual) and the anti-XBP-1 and -Tax antibodies (52). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting were done according to the standard protocols.
Immunofluorescence and fluorescence microscopy.
HeLa cells were seeded onto circular coverslips in six-well plates 1 day before transfection and transfected with the expression vector indicated (i.e., EGFP-Tax, XBP-1S, or XBP-1U). The cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline at 48 h posttransfection. The samples were then incubated with anti-XBP-1 antibody. After washing with phosphate-buffered saline-0.1% Tween 20, the cells were incubated with the anti-rabbit Alexa Fluor 594 antibody (Invitrogen) for 1 h. The samples were finally mounted with 4′,6′-diamidino-2-phenylindole (DAPI) mixed ProLong Gold Antifade reagent (Invitrogen). Fluorescence microscopy was carried out with a Zeiss Axio Imager.Z1 microscope.
Quantitative reverse transcription (RT)-PCR.
RNAs of the T-cell lines and the transfected cells were isolated with TRIZOL reagent (Invitrogen). The Improm II RT system (Promega) and SYBR Green PCR Master Mix (Applied Biosystems) were used to carry out RT and real-time PCR, respectively. Amplification and detection of specific mRNAs were performed with an ABI Prism 7000 thermal cycler (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase was used as an endogenous control. To normalize the data for RNA loading, the threshold cycle value of glyceraldehyde-3-phosphate dehydrogenase was subtracted from that of each gene in the respective samples.
RESULTS
XBP-1S activates basal and Tax-dependent HTLV-1 transcription.
It was reported more than 11 years ago that XBP-1U bound to the 21-bp repeats in the HTLV-1 LTR and transactivated the viral transcription (10, 30, 50). Since the discovery of XBP-1S in 2001, most studies have demonstrated that XBP-1S is a transcription activator while XBP-1U is inactive (46). However, to date, the effect of XBP-1S on HTLV-1 transcription is still unknown.
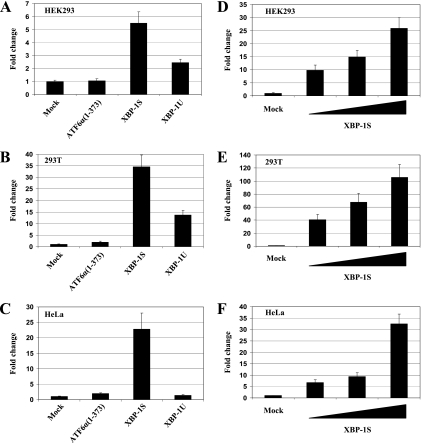
We first performed cell-based reporter assays to investigate the effect of XBP-1S on the HTLV-1 LTR. To determine the influence of XBP-1S specifically, we generated an XBP-1S expression vector by deleting the 26 nucleotides located between positions 531 and 556 of the human XBP-1 mRNA. The 26-bp deletion results in a shift in the open reading frame of the XBP-1 mRNA, generating only the XBP-1S isoform (46). HEK293, 293T, and HeLa cells were transiently transfected with HTLV-1-LTR-F-Luc and the indicated XBP-1 expression vector (i.e., XBP-1S or XBP-1U). Human ATF6α(1-373), a cleaved form of ATF6α (containing aa 1 to 373 of ATF6α) and a known transcription activator for ER chaperon genes and the XBP-1 gene, was used as a control (47). Both XBP-1S and ATF6α(1-373) have been shown to activate the transcription of an ER chaperon, BiP (47). Five- to 35-fold increases in HTLV-1-LTR-dependent transcription were detected when XBP-1S was overexpressed in HEK293, 293T, and HeLa cells (Fig. 1A to C). Overexpression of XBP-1U resulted in a significantly lower level of activation of HTLV-1 transcription in HEK293 and 293T cells and had no effect in HeLa cells (Fig. 1A to C). In all of the cell types, ATF6α(1-373) did not cause any detectable changes in HTLV-1 LTR-driven expression (Fig. 1A to C). To further confirm the activating effect of XBP-1S on the HTLV-1 LTR, a similar set of cell-based assays were carried out by titrating the amounts of XBP-1S expression plasmids. A positive correlation between the relative change in HTLV-1 LTR activation and the dosage of XBP-1S was observed in HEK293, 293T, and HeLa cells (Fig. 1D to F). Notably, a greater-than-100-fold increase in HTLV-1 LTR transactivation was detected in 293T cells (Fig. 1E).
FIG. 1.
Effects of XBP-1 on HTLV-1 LTR-dependent transcription in HEK293, 293T, and HeLa cells. HEK293 (A), 293T (B), and HeLa (C) cells were transiently transfected with HTLV-1-LTR-F-Luc and an expression vector [i.e., ATF6α(1-373), XBP-1S, or XBP-1U]. The empty plasmid (i.e., Mock) was used as a negative control. In a similar set of experiments, HEK293 (D), 293T (E), and HeLa (F) cells were transiently transfected with HTLV-1-LTR-F-Luc with the titration of XBP-1S plasmids (at twofold increments). Relative changes were determined by comparison with the reading of the negative control.
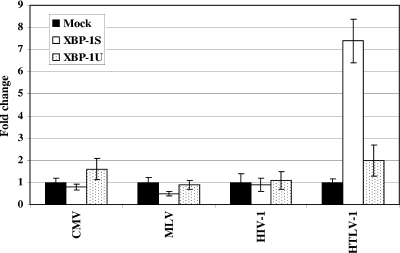
We next examined the effects of XBP-1 on other viral promoters, including the HCMV MIE promoter, the MLV LTR, and the HIV LTR. While XBP-1S and XBP-1U stimulated 7.4- and 2.0-fold increases in HTLV-1 LTR-dependent expression, respectively, little or no effect on other viral promoters was detected (Fig. 2). These results demonstrated specific activation of the HTLV-1 LTR by XBP-1.
FIG. 2.
XBP-1S up-regulates the activity of the HTLV-1 LTR specifically. HEK293 cells were cotransfected with the indicated expression vector (i.e., Mock, XBP-1S, or XBP-1U) and a luciferase reporter driven by a specific viral promoter, including CMV-Luc, MLV-LTR-Luc, HIV-LTR-Luc, and HTLV-1-LTR-Luc. The empty plasmid (i.e., Mock) was used as a negative control.
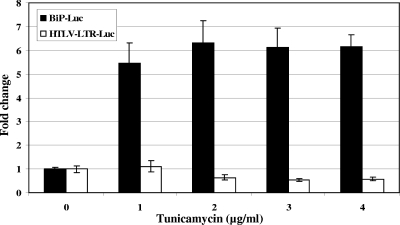
Our results showed that XBP-1S and ATF6α(1-373) exhibited different effects on the HTLV-1 LTR (Fig. 1). Since both XBP-1S and ATF6α(1-373) are known to be key regulators in the UPR, we thus further investigated the effect of UPR activation on HTLV-1 transcription. Tunicamycin was used to elicit the UPR. As the UPR is known to stimulate BiP transcription (23, 24, 46), BiP-Luc, a reporter plasmid in which the expression of firefly luciferase is driven by the BiP promoter, was used as a control (46). Cells were transiently transfected with BiP-Luc or HTLV-1-LTR-Luc in the presence of tunicamycin. Treatment with tunicamycin significantly stimulated BiP-dependent transcription, as expected (Fig. 3). In contrast, no activation on HTLV-1-mediated expression was detected (Fig. 3). Similar results were obtained when several stable HTLV-1-LTR-F-Luc cell lines were incubated with tunicamycin (data not shown).
FIG. 3.
The UPR-inducing reagent tunicamycin has no effect on HTLV-1 LTR-dependent transcription. HEK293 cells were transiently transfected with BiP-F-Luc or HTLV-1-LTR-F-Luc, followed by treatment with tunicamycin (1 to 4 μg/ml). Luciferase activity was measured at 16 h posttransfection.
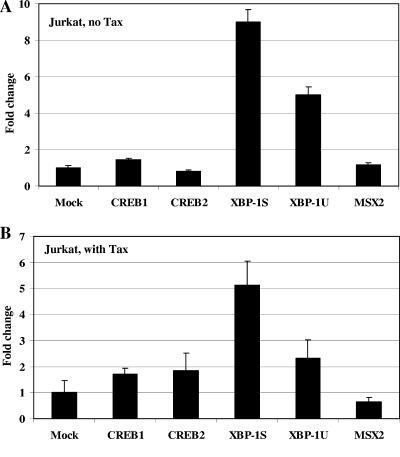
The influence of XBP-1S and XBP-1U on HTLV-1 LTR-mediated transcription was examined in the Jurkat T-cell line. Cells were separately cotransfected with HTLV-1-LTR-Luc and the expression vector indicated (i.e., CREB1, CREB2, XBP-1S, XBP-1U, or MSX2) in the presence or absence of the HTLV-1 transactivator Tax (Fig. 4). Besides the empty-plasmid control, three other controls, CREB1, CREB2, and MSX2, were included in the experiments. All three cellular proteins have been shown to associate with HTLV-1 Tax. CREB1 and CREB2 are positive regulators of HTLV-1 Tax-activated transcription, while MSX2 represses Tax transactivation (35, 44, 45). In the absence of Tax, XBP-1S and XBP-1U stimulated ninefold and fivefold increases in HTLV-1 transcription in Jurkat cells, respectively (Fig. 4A). Little or no effect on the HTLV-1 LTR was detected in the CREB1-, CREB2-, and MSX2-transfected cells (Fig. 4A). The effects of two XBP-1 isoforms on HTLV-1 Tax transactivation were investigated next. The positive regulators CREB1 and CREB2 induced a less-than-twofold change in Tax transactivation, while a 36% decrease in HTLV-1 Tax transactivation was caused by the repressor MSX2 (Fig. 4B). Stronger (i.e., fivefold) activation of HTLV-1 Tax-dependent transcription was detected when XBP-1S was overexpressed (Fig. 4B). Similar to the cellular Tax activators (i.e., CREB1 and CREB2), XBP-1U led to a twofold increase (Fig. 4B).
FIG. 4.
XBP-1S stimulates basal and Tax-activated HTLV-1 transcription in Jurkat cells. Jurkat cells were transiently transfected with HTLV-1-LTR-F-Luc and an expression vector (CREB1, CREB2, XBP-1S, XBP-1U, or MSX2) in the absence (A) or presence (B) of a Tax-producing plasmid. The empty vector was used as a negative control (i.e., Mock).
Both XBP-1S and XBP-1U bind to the HTLV-1 LTR in vivo.
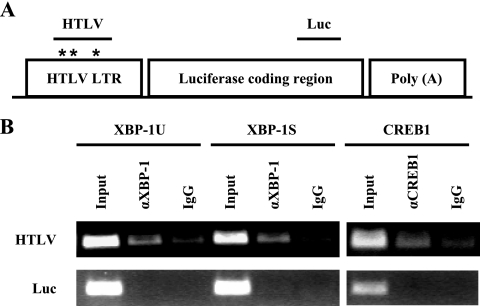
In vitro association between the 21-bp TRE repeat of HTLV-1 and XBP-1U has been reported previously (10, 50). The interaction between XBP-1S and the 21-bp repeat remains to be determined. Since both XBP-1S and XBP-1U have the common bZIP domain at their N termini, it is expected that XBP-1S may also bind to the 21-bp repeat. However, it is unknown if XBP-1S and XBP-1U bind to the 21-bp fragment or the HTLV-1 LTR in cell lines. To test the in vivo binding of XBP-1S and XBP-1U to the TRE repeat, we generated several stable HeLa-HTLV-1-F-Luc cell lines that contain an integrated HTLV-1-LTR-F-Luc DNA fragment (confirmed by Tax and XBP-1 transactivation; data not shown). ChIP analyses were conducted with cell lysates prepared from stable HTLV-1-LTR-F-Luc cells transfected with the XBP-1U, XBP-1S, and CREB1 plasmid vectors. CREB1, an HTLV-1 TRE binding protein (50), was used as a positive control. Cross-linked chromatin fragments were immunoprecipitated with anti-CREB1 antibody or anti-XBP-1 antibody, which recognizes both XBP-1 isoforms. Normal IgG was used as a negative control. XBP-1U, XBP-1S, and the positive control, CREB1, were found to associate with the LTR containing three TRE repeats (i.e., HTLV, Fig. 5), while neither of them bound to the luciferase coding region (i.e., Luc, Fig. 5), indicating an in vivo association among XBP-1S, XBP-1U, and the HTLV-1 LTR.
FIG. 5.
XBP-1S and XBP-1U interact with the HTLV-1 LTR in vivo. (A) Schematic representation of the HTLV-1-LTR-F-Luc reporter gene integrated into stable HeLa-HTLV-1-F-luc cells. The locations of the HTLV-1 LTR, the firefly luciferase coding region, the simian virus 40 poly(A) signal, and two primer sets (i.e., HTLV and Luc) for ChIP assays are indicated. The three 21-bp TRE repeats (marked by asterisks) are located within the PCR fragments amplified by the HTLV primer set. (B) Cell lysates prepared from XBP-1U-, XBP-1S-, and CREB1-expressing stable HeLa-HTLV-1-F-luc cells were analyzed by ChIP with anti-XBP-1 antibody (αXBP-1), anti-CREB1 antibody, or control rabbit IgG as described in Materials and Methods.
Both XBP-1S and XBP-1U interact with Tax.
We have shown that XBP-1 transactivated HTLV-1 Tax-dependent transcription (Fig. 4B) and bound to the HTLV-1 LTR region containing the 21-bp repeats in vivo (Fig. 5). It is known that Tax cannot bind to the HTLV-1 promoter by itself. Tax requires an additional Tax binding protein which can bind to the TRE repeat and recruit Tax to the HTLV-1 LTR to activate HTLV-1 transcription. Therefore, it is possible that XBP-1 may be a Tax interacting partner which facilitates Tax transactivation of the HTLV-1 LTR.
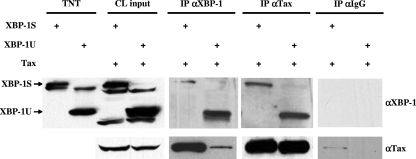
Co-IP was carried out with the lysates of cells cotransfected with Tax and one of the XBP-1 isoforms. To ensure the expression of XBP-1S and XBP-1U in the transfected cells (Fig. 6, CL input), the in vitro-synthesized XBP-1S and XBP-1U proteins were used as references (Fig. 6, TNT). IP was carried out with an anti-XBP-1 antibody which recognizes both XBP-1 isoforms, and normal IgG was used as a negative control. Tax was present in the immunoprecipitated complexes of both XBP-1S/Tax- and XBP-1U/Tax-coexpressing cells. A stronger interaction between XBP-1S and Tax was observed (Fig. 6, IP αXBP-1). Results of reciprocal IP analyses reconfirmed the interaction between XBP-1S/Tax and XBP-1U/Tax. In contrast to the previous observation, the binding affinities between Tax and the two XBP-1 isoforms were similar (Fig. 6, IP αTax). A small amount of Tax was observed in the negative control for XBP-1S/Tax-expressing cells (Fig. 6, IP αIgG), but this is insignificant compared to the amounts of Tax proteins that were found in the immunoprecipitates with anti-XBP-1 and anti-Tax antibodies (Fig. 6, IP αXBP-1 and IP αTax, XBP-1S/Tax-coexpressing cells).
FIG. 6.
XBP-1S and XBP-1U interact with Tax. (A) Co-IP was performed with the indicated antibody (i.e., anti-XBP-1 or anti-Tax antibody) and cell lysates (CL) prepared from HEK293 cells transfected with XBP-1S/Tax or XBP-1U/Tax expression vectors. Normal rabbit IgG was used as a negative control. The expression of XBP-1S, XBP-1U, and Tax was confirmed by Western blotting (i.e., CL input). In vitro-translated XBP-1S and XBP-1U (i.e., TNT) were run on the same gel for comparison. The immunoprecipitated complexes were analyzed by Western blotting.
Nuclear colocalization of XBP-1 and Tax in cells.
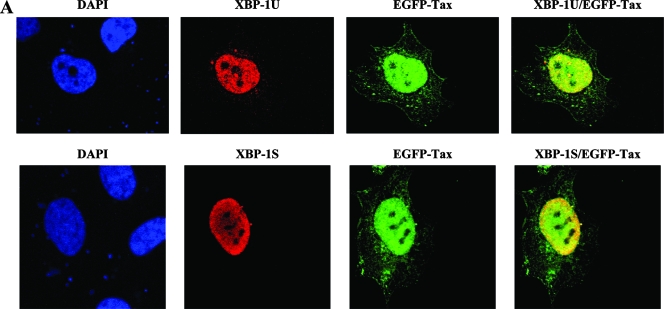
We further examined the subcellular localization of XBP-1S, XBP-1U, and Tax by immunofluorescence. Cells were cotransfected with an EGFP-Tax vector and the indicated XBP-1 plasmid (i.e., XBP-1S or XBP-1U expression vector). XBP-1U and XBP-1S were immunostained with anti-XBP-1 antibody. XBP-1U was evenly distributed within the nucleus, and a small portion of XBP-1U was detected in the cytoplasm (Fig. 7). The cytoplasmic localization of XBP-1U has already been reported (48). XBP-1S was also localized in the nucleus, with stronger labeling observed on the nuclear periphery (Fig. 7). Tax showed a predominantly nuclear localization but was also detected in the cytoplasm (Fig. 7). The cytoplasmic presence of Tax was detected in all EGFP-Tax transfected cells (data not shown). Cytoplasmic localization of Tax was expected since its localization in the ER and Golgi complex was previously reported (2). In the overlaid images, XBP-1S/Tax and XBP-1U/Tax were found to be colocalized with the nuclei (Fig. 7).
FIG. 7.
Nuclear colocalization of XBP-1 and Tax. HeLa cells were cotransfected with EGFP-Tax (green) and an XBP-1 expression plasmid. XBP-1U or XBP-1S was immunostained with anti-XBP-1 antibody (red). Colocalization of EGFP-Tax and XBP-1U (or XBP-1S) was detected as yellow color in the overlay image. Nuclei were visualized by DAPI.
Domain requirement for functional interaction between XBP-1 and Tax.
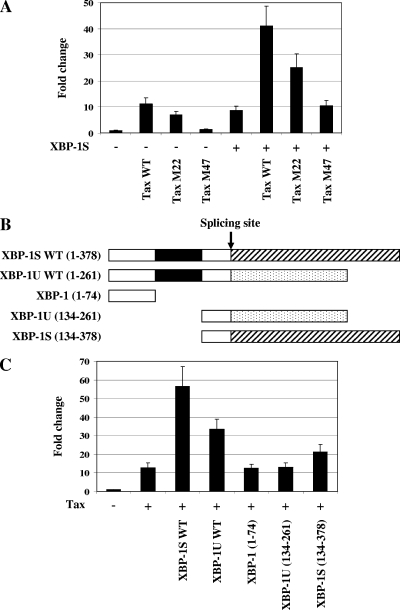
We next wished to determine the domains of HTLV-1 Tax involved in the functional association with XBP-1. To investigate the involvement of NF-κB and CREB in the activating effect of XBP-1S on the HTLV-1 LTR, the NF-κB-deficient (i.e., Tax M22) and CREB-deficient (i.e., Tax M47) Tax mutants were used in cell-based reporter assays. Jurkat cells were transiently cotransfected with an HTLV-1 LTR reporter and the Tax expression vector indicated (Fig. 8A). The transactivating abilities of Tax M22 and M47 on the HTLV-1 LTR were first examined. Consistent with a previous report (39), M22 and M47 demonstrated weaker effects on HTLV-1 LTR transcription compared to wild-type Tax (i.e., 62% ± 11% and 11% ± 2%, respectively, Fig. 8A). The influence of Tax M22 and M47 on the HTLV-1 LTR in the presence of XBP-1S was investigated next. The results showed that M22 further activated HTLV-1 LTR-dependent transcription (Fig. 8A, XBP-1S and XBP-1S/Tax M22), while no significant effect was observed when M47 was cotransfected (Fig. 8A, XBP-1S and XBP-1S/Tax M47), suggesting that the CREB domain of Tax might be important in the activating effect of XBP-1S.
FIG. 8.
Domains of HTLV-1 Tax and XBP-1 required for the functional interaction between Tax and XBP-1. (A) Jurkat cells were transiently transfected with HTLV-1-LTR-F-Luc and the indicated Tax expression vector (i.e., wild-type [WT] Tax, NF-κB-deficient mutant Tax [Tax M22], or CREB-deficient mutant Tax [Tax M47]) in the absence or presence of an XBP-1S plasmid. Cells transfected with HTLV-1-LTR-F-Luc and an empty vector (i.e., without-Tax and XBP-1S plasmids included) were used as a negative control. Relative changes were determined by comparison with the reading of the control. (B) Schematic representation of truncated XBP-1 mutant proteins. The bZIP domain (black box) and the C-terminal regions unique to XBP-1S and XBP-1U due to the UPR-induced splicing are indicated (striped and dotted boxes, respectively). (C) Jurkat cells were cotransfected with HTLV-1-LTR-F-Luc and the indicated XBP-1 expression vector with or without a wild-type Tax expression plasmid. Relative changes were determined by comparison with the reading of the negative control (i.e., without-Tax and XBP-1S plasmids transfected).
XBP-1U and XBP-1S have the 164 aa at their N termini in common. The bZIP domain is located between aa 75 and 133 (Fig. 8B, black box). Due to the UPR-induced splicing, XBP-1U and XBP-1S contain unique C-terminal regions (XBP-1U, 165 to 261 aa; XBP-1S, 165 to 378 aa) (46) (Fig. 8B). It has been shown that the bZIP domain of CREB/ATF proteins is required for the Tax-CREB interaction (13, 45). It is possible that, as a member of the CREB/ATF protein family, XBP-1 may also bind to Tax through its bZIP domain. To determine the involvement of bZIP motifs of XBP-1 in the Tax-XBP-1 interaction, we generated three bZIP deletion mutant proteins, XBP-1(1-74), XBP-1U(134-261), and XBP-1S(134-378) (Fig. 8B).
HTLV-1 LTR reporter assays were carried out with Jurkat cells. An about 13-fold increase in HTLV-1 LTR-dependent transcription was detected in the Tax-transfected cells (Fig. 8C). Greater activation of HTLV-1 Tax transactivation was detected in the XBP-1S WT/Tax- and XBP-1U WT/Tax-expressing cells (Fig. 8C). However, XBP-1(1-74) and XBP-1U(134-261) did not demonstrate any effects on Tax transactivation. A 70% increase in Tax-dependent activation was observed in the cells coexpressing XBP-1S(134-378). It was noted that the effect of XBP-1S(134-378) on Tax-mediated transcription was significantly weaker than that of wild-type XBP-1S (Fig. 8C). The results suggest that the bZIP domain of XBP-1 proteins plays an important role in XBP-1-mediated Tax transactivation.
Tax and HTLV-1 infections up-regulate XBP-1 transcription.
It has been reported that the expression of XBP-1 (previously known as TREB5) mRNA is significantly higher in an HTLV-1-infected T-cell line (i.e., HuT102) than in an uninfected (i.e., Jurkat) cell line (50). In contrast, lower or unchanged levels of CREB (i.e., CREB1), TREB7 (i.e., CREB2), and TREB36 (i.e., ATF-1) mRNAs were detected in the HTLV-1-infected cell line (50). To further investigate the correlation between the expression of XBP-1 mRNA and HTLV-1 infection, we performed quantitative RT-PCR with the RNAs isolated from the HTLV-1-free T-cell lines (including HuT78 and CEM) and the HTLV-1-infected T-cell lines (including C10/MJ, MT2, and MT4). Similar levels of XBP-1 transcription were detected in HuT78 and CEM cells (Table 1). However, 3.5- and 2.8-fold increases in XBP-1 mRNA were detected in HTLV-1-infected C10/MJ and MT2 cells, respectively. No significant change was observed in MT4 cells (Table 1). The expression of several UPR-inducible genes, including those for BiP, the CRE binding transcription factor (C/EBP) homologous protein (CHOP), and the ER degradation-enhancing α-mannosidase-like protein (EDEM), in the T-cell lines was also determined. Higher levels of the selected UPR genes were also observed in C10/MJ and MT2 cells (Table 1). Collectively, three out of four HTLV-1-infected cell lines examined (i.e., HuT102, C10/MJ, MT2, and MT4 cells) exhibit elevated levels of XBP-1 mRNA (Table 1) (50). Furthermore, our data also suggest that HTLV-1 infection may induce the UPR in the host cells.
TABLE 1.
Expression of UPR genes in different T-cell lines analyzed by quantitative RT-PCR
| Cell line | Fold changea in mRNA of UPR gene for:
|
|||
|---|---|---|---|---|
| XBP-1 | BiP | CHOP | EDEM | |
| HTLV-1-free cells | ||||
| HuT78b | 1.0 ± 0.1c | 1.0 ± 0.1 | 1.0 ± 0.1 | 1.0 ± 0.2 |
| CEM | 1.3 ± 0.2 | 1.5 ± 0.1 | 1.9 ± 0.3 | 1.5 ± 0.2 |
| HTLV-1-infected cells | ||||
| C10/MJ | 3.5 ± 0.7 | 3.0 ± 0.4 | 3.7 ± 0.5 | 7.0 ± 1.8 |
| MT2 | 2.8 ± 0.4 | 2.9 ± 0.2 | 2.5 ± 0.2 | 2.7 ± 1.0 |
| MT4 | 0.9 ± 0.2 | 0.3 ± 0.2 | 1.6 ± 0.1 | 2.6 ± 0.5 |
The change in a particular gene was calculated by using the equation Fold change (relative to HuT78) = 2−ΔΔCT, where ΔΔCT = ΔCT(sample) − ΔCT(HuT78).
The mRNA levels of the indicated UPR genes in HuT78 cells were used as the control for comparison.
Data represent the average of triplicates with standard deviations.
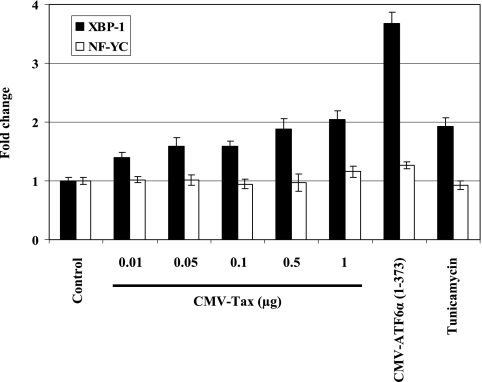
Tax has been shown to transactivate several cellular genes, such as c-myc and that for human proliferating cell nuclear antigen (11, 36). Quantitative RT-PCR was carried out to determine the effect of Tax on the expression of the XBP-1 and NF-YC mRNAs. ATF6α(1-373) and tunicamycin were used as positive controls. As shown in Fig. 9, transcription of XBP-1 was induced by ATF6α(1-373) and tunicamycin while the level of NF-YC mRNA remained unchanged. Overexpression of Tax caused a twofold increase in XBP-1 mRNA (Fig. 9). The effect of Tax on XBP-1 splicing was also investigated by RT-PCR. No significant induction of XBP-1 splicing was detected in the Tax-expressing cells (data not shown). These results suggest that XBP-1 might be a cellular target gene of Tax and its transcription could be transactivated by Tax.
FIG. 9.
Tax stimulates XBP-1 transcription. HEK293 cells were transiently transfected with the indicated amounts (0.01, 0.05, 0.1, 0.5, and 1 μg) of a Tax-producing plasmid. The empty vector was used as a negative control. Tunicamycin treatment (4 μg/ml) and an ATF6α(1-373) expression vector (1 μg) were used as positive controls. RNAs isolated from the cells were analyzed by quantitative RT-PCR to examine the expression of the genes for XBP-1 and NF-YC.
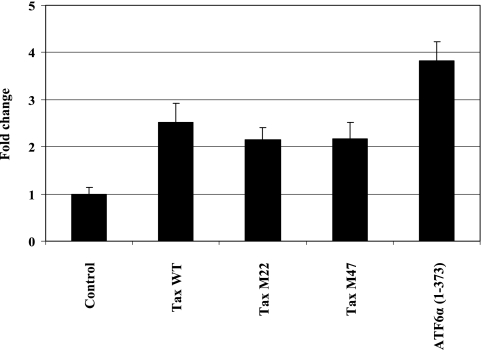
We further investigated if the induction of XBP-1 by HTLV-1 Tax was NF-κB or CREB dependent. HEK293 cells were transiently transfected with a wild-type Tax, Tax M22, Tax M47, or ATF6α(1-373) expression vector. The empty vector was used as a negative control, while the wild-type Tax and ATF6α(1-373) plasmids were used as positive controls (Fig. 10). Cell lysates prepared from the transfected cells were analyzed by quantitative RT-PCR to examine the expression of XBP-1. Similar increases in XBP-1 mRNA were detected when wild-type Tax, Tax M22, and Tax M47 were overexpressed (2.52-fold ± 0.40-fold, 2.15-fold ± 0.26-fold, and 2.17-fold ± 0.35-fold, respectively) (Fig. 10), suggesting that NF-κB and CREB might not be required for Tax-mediated XBP-1 expression.
FIG. 10.
Effects of mutant Tax proteins on XBP-1 expression. HEK293 cells were transiently transfected with 1 μg of the indicated plasmid to express wild-type Tax (Tax WT), an NF-κB-deficient mutant Tax protein (Tax M22), or a CREB-deficient mutant Tax protein (Tax M47). The empty vector was used as a negative control, while an ATF6α(1-373) expression vector (1 μg) was used as a positive control. RNAs isolated from the cells were analyzed by quantitative RT-PCR to examine the expression of XBP-1.
DISCUSSION
It has been shown that infection of KSHV, WNV, JEV, HCV, HCMV, DEN-2, and severe acute respiratory syndrome coronavirus induces the UPR to various degrees in host cells (7, 17, 21, 31, 40-43, 51). Since XBP-1 is a key regulator involved in the UPR, the correlation between viral infection and XBP-1 expression has been investigated in detail. Elevated expression of XBP-1 was detected in the cells infected with KSHV (21). Splicing of XBP-1 was induced by HCV, WNV, and HCMV infections; however, transcriptional activation of XBP-1S is inhibited or is not required for viral replication (17, 31, 41). Recently, Bhende, et al. showed that XBP-1S activates the transcription of two Epstein-Barr virus (EBV) immediate-early gene promoters (i.e., the BZLF1 and BRLF1 promoters) and induces lytic viral gene expression in EBV-positive cell lines (4). Here we demonstrate that XBP-1 stimulates HTLV-1 transcription specifically (Fig. 2). We reconfirm the transactivation of the HTLV-1 LTR by XBP-1U reported previously (10) and identify XBP-1S as a stronger transcriptional activator of HTLV-1 (Fig. 1 and 4).
The stimulating effect of XBP-1U on the HTLV-1 LTR was reported by two groups before the identification of XBP-1S (10, 30). However, almost all of the published work suggests that XBP-1U is an inactive transcription factor, including a recent study showing that XBP-1U does not enhance the activity of the EBV BZLF1 and BRLF1 promoters (4). We found that overexpression of XBP-1U resulted in 2- to 13-fold activation of HTLV-1 LTR-dependent transcription in HEK293, 293T, and Jurkat cells (Fig. 1A and B and 4A). Activation of the HTLV-1 LTR in the T-cell (i.e., Jurkat cell) line suggested a role for XBP-1 in HTLV-1 transcription and replication (Fig. 4A). However, no significant influence was detected in XBP-1U-expressing HeLa cells (Fig. 1C). It is possible that XBP-1U requires additional cellular cofactors to regulate the activity of the HTLV-1 LTR and the expression of the XBP-1U-interacting factors is cell specific. However, it is still unknown why XBP-1U can activate HTLV-1 transcription. The mechanism of XBP-1U activation requires further investigation.
XBP-1S demonstrates stronger activating effects on both basal and Tax-dependent HTLV-1 transcription than XBP-1U (Fig. 1 and 4). This is not surprising since XBP-1S has been shown to have higher transcriptional activity (46). Because of the UPR-induced splicing, the C terminus of XBP-1S, but not that of XBP-1U, contains a transactivation domain (46, 48). We also compared the activating effect of XBP-1S and XBP-1U to two known positive regulators of HTLV-1 Tax-activated transcription, CREB1 and CREB2. XBP-1S induced a fivefold increase in Tax-dependent transcription, while less than twofold activation was caused by CREB1 and CREB2 (Fig. 4B). Both CREB1 and CREB2 had little or no effect on basal HTLV-1 transcription, while XBP-1S and XBP-1U resulted in nine- and fivefold activation of HTLV-1 LTR, respectively (Fig. 4A). Previously, elevated expression of XBP-1 mRNA was detected in HTLV-1-infected T cells (i.e., HuT102 cells) (50). Compared with the transcriptional levels of the genes for CREB1 and CREB2 in uninfected T cells (i.e., Jurkat cells), those in HuT102 cells were unchanged or significantly lower (50). In our study, we also detected higher levels of XBP-1 mRNA in two HTLV-1-infected cell lines (Table 1). Collectively, XBP-1 may play a more important role than CREB1 and CREB2 in the transcriptional regulation of HTLV-1.
The interaction between the Tax and XBP-1 proteins was examined by IP experiments. Compared to XBP-1U, more Tax was precipitated with XBP-1S, suggesting a stronger interaction between XBP-1S and Tax (Fig. 6, IP αXBP-1). However, in reciprocal IP assays, the amounts of XBP-1S and XBP-1U coimmunoprecipitated by Tax were similar (Fig. 6, IP αTax). One possible explanation is that the epitope recognized by the anti-XBP-1 antibody is not required for Tax-XBP-1S interaction and remains accessible to the antibody when associated with Tax. Therefore, the anti-XBP-1 antibody may precipitate most of the Tax-XBP-1S complexes. However, the epitope recognized by the anti-Tax antibody may be involved in the protein-protein interaction between XBP-1S and Tax or become inaccessible to the antibody when complexing with XBP-1S. Therefore, the anti-Tax antibody may only pull down a small portion of the Tax-XBP-1S complexes.
Quantitative RT-PCR analyses of HTLV-1-infected cells revealed that HTLV-1 infection may trigger the UPR in host cells (Table 1). We examined the effects of the UPR on the HTLV-1 LTR promoter by overexpressing the UPR regulator ATF6α(1-373) or by tunicamycin treatment. Although ATF6α(1-373) and tunicamycin up-regulate XBP-1 transcription and/or induce the generation of XBP-1S, neither of them caused any significant changes in HTLV-1 transcription (Fig. 1 and 3). Besides the activation of XBP-1, ATF6α(1-373) and tunicamycin have profound effects on UPR-inducible genes and UPR signaling pathways. It is possible that other signaling pathways or gene expression induced by ATF6α(1-373) or tunicamycin may directly or indirectly repress the transactivation of the HTLV-1 LTR.
Besides its nuclear localization, Tax has been found to be present in the medium of Tax-transfected cells and to localize in the organelles associated with protein secretion, including the ER and Golgi complex (2). It is possible that Tax may trigger the UPR, resulting in up-regulation of XBP-1 expression due to its presence in the ER. Another possibility is that Tax may activate XBP-1 transcription directly. Several cellular target genes of Tax, such as c-myc and the gene for the human proliferating cell nuclear antigen, have been reported (11, 36).
The domains of Tax and XBP-1 required for functional Tax-XBP-1 interaction were investigated. Since XBP-1 belongs to the CREB/ATF protein family, it is possible that XBP-1 may use the same molecular mechanism in binding to Tax and stimulating HTLV-1 Tax transactivation as other Tax binding CREB proteins. As expected, CREB-deficient Tax protein M47 failed to activate XBP-1S-mediated HTLV-1 LTR transactivation (Fig. 8A). In addition, it has been shown that the bZIP domains of CREB proteins are essential for CREB-Tax interaction (13, 45). Our results obtained with the bZIP deletion mutant forms of XBP-1 demonstrated the involvement of the bZIP domain of XBP-1 in HTLV-1 LTR transactivation (Fig. 8B and C). However, XBP-1S(134-378) still could induce a 70% increase in Tax transactivation (Fig. 8C). A study has shown that the C terminus of XBP-1S, which is unique to the XBP-1S isoform, contains a transcriptional activation domain (46). It is possible that the C terminus of XBP-1S may induce a specific set of cellular genes which directly or indirectly regulate HTLV-1 transcription without interacting with Tax.
Although Tax M22 and M47 demonstrated different effects on XBP-1-dependent HTLV-1 Tax transactivation, the mutant and wild-type Tax proteins showed similar levels of activation of XBP-1 transcription (Fig. 10). The results exclude the requirement of the NF-κB and CREB proteins for the regulation of XBP-1 transcription. It is known that Tax cannot bind to DNA directly. Therefore, it is clear that Tax requires an unidentified DNA binding cellular factor to bind to the promoter and up-regulate the expression of XBP-1. However, we could not rule out the possibility that Tax can induce XBP-1 transcription indirectly.
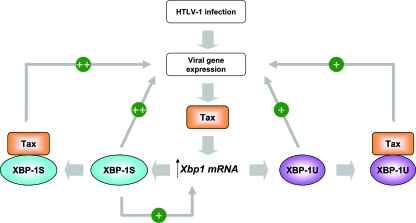
On the basis of our study, we propose a model of the host-virus interaction between XBP-1 and HTLV-1 (Fig. 11). Higher expression of XBP-1 mRNA was detected in HTLV-1-infected cells, resulting in the generation of XBP-1S and XBP-1U. We did not detect a significant increase in XBP-1 splicing in HTLV-1-infected cells by RT-PCR (data not shown). It was technically difficult to determine the expression of XBP-1S mRNA quantitatively. However, higher levels of mRNA for EDEM, which is encoded by an XBP-1S target gene (27), were detected in HTLV-1-infected cells by quantitative RT-PCR (data not shown), suggesting that HTLV-1 infection might induce the splicing of XBP-1 as well. Higher expression of XBP-1 would be expected then since the gene for XBP-1 has been identified as a target of XBP-1S (27). We identified Tax as an activator of XBP-1 transcription, suggesting that the gene for XBP-1 could be a cellular target of Tax (Fig. 11). XBP-1S and XBP-1U were found to interact with Tax and bind to the HTLV-1 LTR in vivo (Fig. 5 and 6). Both XBP-1 isoforms stimulated HTLV-1 basal, as well as Tax-mediated, transcription through the interaction with Tax (Fig. 1, 2, 4, and 6). More work is required to investigate the involvement of XBP-1 in the viral replication of HTLV-1.
FIG. 11.
Model of cell-virus interaction between a cellular factor XBP-1 and transcriptional regulation of HTLV-1.
Acknowledgments
We thank Warner C. Greene, Kazutoshi Mori, Arnold Rabson, and Zhiwei Song for providing plasmids; Peter Nissom for critical review of the manuscript; and Yong Xiao and Grant Tan for expert technical assistance.
This work was supported by the Agency for Science, Technology and Research (Singapore) and National Institutes of Health grant AI043894 to F.K.
Footnotes
Published ahead of print on 20 February 2008.
REFERENCES
- 1.Adya, N., and C. Z. Giam. 1995. Distinct regions in human T-cell lymphotropic virus type I Tax mediate interactions with activator protein CREB and basal transcription factors. J. Virol. 691834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alefantis, T., K. Mostoller, P. Jain, E. Harhaj, C. Grant, and B. Wigdahl. 2005. Secretion of the human T cell leukemia virus type I transactivator protein Tax. J. Biol. Chem. 28017353-17362. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, M. G., and W. S. Dynan. 1994. Quantitative studies of the effect of HTLV-I Tax protein on CREB protein-DNA binding. Nucleic Acids Res. 223194-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhende, P. M., S. J. Dickerson, X. Sun, W. H. Feng, and S. C. Kenney. 2007. X-box-binding protein 1 activates lytic Epstein-Barr virus gene expression in combination with protein kinase D. J. Virol. 817363-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady, J., K. T. Jeang, J. Duvall, and G. Khoury. 1987. Identification of p40x-responsive regulatory sequences within the human T-cell leukemia virus type I long terminal repeat. J. Virol. 612175-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewer, J. W., and L. M. Hendershot. 2005. Building an antibody factory: a job for the unfolded protein response. Nat. Immunol. 623-29. [DOI] [PubMed] [Google Scholar]
- 7.Chan, C. P., K. L. Siu, K. T. Chin, K. Y. Yuen, B. Zheng, and D. Y. Jin. 2006. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 809279-9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao, S. H., J. N. Harada, F. Hyndman, X. Gao, C. G. Nelson, S. K. Chanda, and J. S. Caldwell. 2004. PDX1, a cellular homeoprotein, binds to and regulates the activity of human cytomegalovirus immediate early promoter. J. Biol. Chem. 27916111-16120. [DOI] [PubMed] [Google Scholar]
- 9.Chao, S. H., J. R. Walker, S. K. Chanda, N. S. Gray, and J. S. Caldwell. 2003. Identification of homeodomain proteins, PBX1 and PREP1, involved in the transcription of murine leukemia virus. Mol. Cell. Biol. 23831-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clauss, I. M., M. Chu, J. L. Zhao, and L. H. Glimcher. 1996. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 241855-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duyao, M. P., D. J. Kessler, D. B. Spicer, C. Bartholomew, J. L. Cleveland, M. Siekevitz, and G. E. Sonenshein. 1992. Transactivation of the c-myc promoter by human T cell leukemia virus type 1 tax is mediated by NFκB. J. Biol. Chem. 26716288-16291. [PubMed] [Google Scholar]
- 12.Franklin, A. A., M. F. Kubik, M. N. Uittenbogaard, A. Brauweiler, P. Utaisincharoen, M. A. Matthews, W. S. Dynan, J. P. Hoeffler, and J. K. Nyborg. 1993. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 26821225-21231. [PubMed] [Google Scholar]
- 13.Gachon, F., S. Thebault, A. Peleraux, C. Devaux, and J. M. Mesnard. 2000. Molecular interactions involved in the transactivation of the human T-cell leukemia virus type 1 promoter mediated by Tax and CREB-2 (ATF-4). Mol. Cell. Biol. 203470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii407-410. [DOI] [PubMed] [Google Scholar]
- 15.Hai, T., and M. G. Hartman. 2001. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 2731-11. [DOI] [PubMed] [Google Scholar]
- 16.Harrod, R., Y.-L. Kuo, Y. Tang, Y. Yao, A. Vassilev, Y. Nakatani, and C.-Z. Giam. 2000. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 27511852-11857. [DOI] [PubMed] [Google Scholar]
- 17.Isler, J. A., A. H. Skalet, and J. C. Alwine. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 796890-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwakoshi, N. N., A. H. Lee, and L. H. Glimcher. 2003. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 19429-38. [DOI] [PubMed] [Google Scholar]
- 19.Iwakoshi, N. N., A. H. Lee, P. Vallabhajosyula, K. L. Otipoby, K. Rajewsky, and L. H. Glimcher. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4321-329. [DOI] [PubMed] [Google Scholar]
- 20.Jeang, K. T., I. Boros, J. Brady, M. Radonovich, and G. Khoury. 1988. Characterization of cellular factors that interact with the human T-cell leukemia virus type I p40x-responsive 21-base-pair sequence. J. Virol. 624499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenner, R. G., K. Maillard, N. Cattini, R. A. Weiss, C. Boshoff, R. Wooster, and P. Kellam. 2003. Kaposi's sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc. Natl. Acad. Sci. USA 10010399-10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with Tax and stimulates Tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 198136-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 131211-1233. [DOI] [PubMed] [Google Scholar]
- 24.Kohno, K., K. Normington, J. Sambrook, M.-J. Gething, and K. Mori. 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13877-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ku, S. C., D. T. Ng, M. G. Yap, and S. H. Chao. 2008. Effects of overexpression of X-box binding protein 1 on recombinant protein production in Chinese hamster ovary and NS0 myeloma cells. Biotechnol. Bioeng. 99155-164. [DOI] [PubMed] [Google Scholar]
- 26.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380642-646. [DOI] [PubMed] [Google Scholar]
- 27.Lee, A. H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 237448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, J., Z. Klase, X. Gao, J. S. Caldwell, M. F. Stinski, F. Kashanchi, and S.-H. Chao. 2007. Cellular homeoproteins, SATB1 and CDP, bind to the unique region between the human cytomegalovirus UL127 and major immediate-early genes. Virology 366117-125. [DOI] [PubMed] [Google Scholar]
- 29.Lin, H.-C., M. Hickey, L. Hsu, D. Medina, and A. B. Rabson. 2005. Activation of human T cell leukemia virus type 1 LTR promoter and cellular promoter elements by T cell receptor signaling and HTLV-1 Tax expression. Virology 3391-11. [DOI] [PubMed] [Google Scholar]
- 30.Matsuzaki, Y., J. Fujisawa, and M. Yoshida. 1995. Identification of transcriptional activation domain of TREB5, a CREB/ATF family protein that binds to HTLV-1 enhancer. J. Biochem. (Tokyo) 117303-308. [DOI] [PubMed] [Google Scholar]
- 31.Medigeshi, G. R., A. M. Lancaster, A. J. Hirsch, T. Briese, W. I. Lipkin, V. Defilippis, K. Fruh, P. W. Mason, J. Nikolich-Zugich, and J. A. Nelson. 2007. West Nile virus infection activates the unfolded protein response leading to CHOP induction and apoptosis. J. Virol. 8110849-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen, D. G., K. C. Wolff, H. Yin, J. S. Caldwell, and K. L. Kuhen. 2006. “UnPAKing” human immunodeficiency virus (HIV) replication: using small interfering RNA screening to identify novel cofactors and elucidate the role of group I PAKs in HIV infection. J. Virol. 80130-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i1031-1032. [DOI] [PubMed] [Google Scholar]
- 34.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 777415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy, T. R., H. Tang, X. Li, and F. Wong-Staal. 1997. Functional interaction of the HTLV-1 transactivator Tax with activating transcription factor-4 (ATF4). Oncogene 142785-2792. [DOI] [PubMed] [Google Scholar]
- 36.Ressler, S., G. F. Morris, and S. J. Marriott. 1997. Human T-cell leukemia virus type 1 Tax transactivates the human proliferating cell nuclear antigen promoter. J. Virol. 711181-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rimsky, L., J. Hauber, M. Dukovich, M. H. Malim, A. Langlois, B. R. Cullen, and W. C. Greene. 1988. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature 335738-740. [DOI] [PubMed] [Google Scholar]
- 38.Shaffer, A. L., M. Shapiro-Shelef, N. N. Iwakoshi, A. H. Lee, S. B. Qian, H. Zhao, X. Yu, L. Yang, B. K. Tan, A. Rosenwald, E. M. Hurt, E. Petroulakis, N. Sonenberg, J. W. Yewdell, K. Calame, L. H. Glimcher, and L. M. Staudt. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 2181-93. [DOI] [PubMed] [Google Scholar]
- 39.Smith, M. R., and W. C. Greene. 1990. Identification of HTLV-I tax trans-activator mutants exhibiting novel transcriptional phenotypes. Genes Dev. 41875-1885. [DOI] [PubMed] [Google Scholar]
- 40.Su, H. L., C. L. Liao, and Y. L. Lin. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 764162-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tardif, K. D., K. Mori, R. J. Kaufman, and A. Siddiqui. 2004. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J. Biol. Chem. 27917158-17164. [DOI] [PubMed] [Google Scholar]
- 42.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 767453-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tirosh, B., N. N. Iwakoshi, B. N. Lilley, A. H. Lee, L. H. Glimcher, and H. L. Ploegh. 2005. Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J. Virol. 792768-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Twizere, J.-C., L. Lefebvre, D. Collete, C. Debacq, P. Urbain, H. Heremans, J.-C. Jauniaux, A. Burny, L. Willems, and R. Kettmann. 2005. The homeobox protein MSX2 interacts with Tax oncoproteins and represses their transactivation activity. J. Biol. Chem. 28029804-29811. [DOI] [PubMed] [Google Scholar]
- 45.Yin, M.-J., E. J. Paulssen, J.-S. Seeler, and R. B. Gaynor. 1995. Protein domains involved in both in vivo and in vitro interactions between human T-cell leukemia virus type I Tax and CREB. J. Virol. 693420-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107881-891. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 206755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida, H., M. Oku, M. Suzuki, and K. Mori. 2006. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172565-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 792031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshimura, T., J. Fujisawa, and M. Yoshida. 1990. Multiple cDNA clones encoding nuclear proteins that bind to the tax-dependent enhancer of HTLV-1: all contain a leucine zipper structure and basic amino acid domain. EMBO J. 92537-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, C. Y., Y. W. Hsu, C. L. Liao, and Y. L. Lin. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J. Virol. 8011868-11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, W., J. W. Nisbet, B. Albrecht, W. Ding, F. Kashanchi, J. T. Bartoe, and M. D. Lairmore. 2001. Human T-lymphotropic virus type 1 p30(II) regulates gene transcription by binding CREB binding protein/p300. J. Virol. 759885-9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao, L. J., and C. Z. Giam. 1992. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc. Natl. Acad. Sci. USA 897070-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]