Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) nsp1 protein has unique biological functions that have not been described in the viral proteins of any RNA viruses; expressed SARS-CoV nsp1 protein has been found to suppress host gene expression by promoting host mRNA degradation and inhibiting translation. We generated an nsp1 mutant (nsp1-mt) that neither promoted host mRNA degradation nor suppressed host protein synthesis in expressing cells. Both a SARS-CoV mutant virus, encoding the nsp1-mt protein (SARS-CoV-mt), and a wild-type virus (SARS-CoV-WT) replicated efficiently and exhibited similar one-step growth kinetics in susceptible cells. Both viruses accumulated similar amounts of virus-specific mRNAs and nsp1 protein in infected cells, whereas the amounts of endogenous host mRNAs were clearly higher in SARS-CoV-mt-infected cells than in SARS-CoV-WT-infected cells, in both the presence and absence of actinomycin D. Further, SARS-CoV-WT replication strongly inhibited host protein synthesis, whereas host protein synthesis inhibition in SARS-CoV-mt-infected cells was not as efficient as in SARS-CoV-WT-infected cells. These data revealed that nsp1 indeed promoted host mRNA degradation and contributed to host protein translation inhibition in infected cells. Notably, SARS-CoV-mt infection, but not SARS-CoV-WT infection, induced high levels of beta interferon (IFN) mRNA accumulation and high titers of type I IFN production. These data demonstrated that SARS-CoV nsp1 suppressed host innate immune functions, including type I IFN expression, in infected cells and suggested that SARS-CoV nsp1 most probably plays a critical role in SARS-CoV virulence.
Severe acute respiratory syndrome coronavirus (SARS-CoV) is the etiological agent of a newly emerged disease, SARS, which originated in southern China in 2002 and spread to various areas of the world in a 2003 epidemic (9, 21, 30, 47). The 5′-end two-thirds of the single-stranded, positive-sense RNA genome of SARS-CoV constitutes gene 1, which contains two large, partly overlapping open reading frames (ORFs), ORF1a and ORF1b. Upon infection, the incoming viral genomic RNA is translated to produce two large precursor polyproteins from gene 1 (31, 45). These precursors are proteolytically processed by two virally encoded proteinases to generate 16 mature proteins, nsp1 to nsp16 (45). In addition to the essential role of the gene 1 proteins in viral RNA synthesis (3, 11, 17, 27, 36, 55), some of the gene 1 proteins could have multiple functions besides viral RNA synthesis (2, 13, 25, 41).
Host innate immune defense mechanisms, such as the production of alpha/beta interferon (IFN-α/β), are the first responders against animal virus infections, and many viruses have developed strategies to actively suppress and/or evade the innate immune response (14). Some viruses encode proteins that selectively suppress the host innate antiviral functions (4, 14), while some have evolved to target the general host gene expression machinery to block the innate immune functions as well (24, 35, 51). Viral genes that inhibit host innate defenses are often identified as major viral virulence factors (5, 35). It appears that activation of type I IFN responses is inhibited in SARS-CoV-infected cells (7, 22, 28, 38, 39, 44, 56); for example, IFN regulatory factor 3 (IRF-3) activation, which is essential for IFN-β mRNA transcription and IFN-β mRNA accumulation, does not occur in SARS-CoV-infected human 293 cells (37). We previously reported that expressed SARS-CoV nsp1 induces degradation of host mRNAs, suppresses host translation, and suppresses accumulation of IFN-β mRNA without inhibiting IRF-3 dimerization (18). These observations led us to hypothesize that SARS-CoV nsp1 suppresses general host gene expression and thereby also blocks host antiviral immune responses in infected cells. The present study tested this hypothesis and demonstrated that SARS-CoV nsp1 indeed suppressed host gene expression, including type I IFN production, by promoting host mRNA degradation and contributing to host translation suppression in infected cells.
MATERIALS AND METHODS
Cells and viruses.
293 cells stably expressing the SARS-CoV receptor protein, angiotensin-converting enzyme 2 (ACE2) (293/ACE2 cells), and Vero E6 cells were maintained as described previously (18). Wild-type SARS-CoV (SARS-CoV-WT) and mutant SARS-CoV (SARS-CoV-mt), both of which were rescued by using a reverse genetics system (53), were passaged once in Vero E6 cells and used for infection studies. For virus growth analysis, 293/ACE2 and Vero E6 cells were infected at a multiplicity of infection (MOI) of 1 or 0.01 for 1 h at 37°C. After virus adsorption, cells were washed twice with phosphate-buffered saline and incubated with the appropriate medium. The infectious virus titers were determined by 50% tissue culture infectious dose (TCID50) analysis on Vero E6 cells and are expressed in log10 TCID50 units per ml. All experiments with infectious SARS-CoV were performed in an approved biosafety level 3 laboratory at The University of Texas Medical Branch at Galveston (UTMB). Sendai virus (SeV; Cantell strain), obtained from Charles River Laboratory (Wilmington, MA), was used to infect cells at 100 hemagglutination (HA) units/ml.
Plasmid construction and generation of infectious recombinant SARS-CoV.
A recombinant PCR procedure, using pCAGGS-Nsp1-WT (18) as the template, was employed to generate both pCAGGS-Nsp1Δ160-173 and pCAGGS-Nsp1-mt (K164A and H165A). For the generation of in vitro-synthesized RNA transcripts, pcD-CAT, pcD-Nsp1-WT, and pcD-Nsp1-mt were constructed by inserting the chloramphenicol acetyltransferase (CAT) ORFs Nsp1-WT ORF and Nsp1-mt ORF, respectively, into pcDNA 3.1 HisA-myc. A QuikChange site-directed mutagenesis kit (Stratagene) was used to generate mutations (K164A and H165A) in fragment A of the SARS-CoV reverse genetics system (53). The SARS-CoV full-length cDNA was assembled from fragments A through F, the full-length RNA transcripts were synthesized, and the recombinant viruses were recovered according to established protocols, as described previously (53).
Reporter gene assays.
293 cells in 24-well plates were cotransfected in triplicate with IFN-β promoter-driven luciferase (pIFNβ-Luc) reporter plasmid (0.1 μg) and the indicated pCAGGS-based nsp1 expression plasmids (0.4 μg) using TransIT-293 reagent (Mirus). At 24 h posttransfection, the cells were mock infected or infected with SeV. At 16 h postinfection (p.i.), the cells were lysed in reporter lysis buffer (Promega), and luciferase assays were performed using the Promega luciferase assay system. For transfections with the reporter plasmid pRL-SV40 (0.1 μg), the cells were lysed in Renilla luciferase (rLuc) lysis buffer, and rLuc activities were measured.
Western blot analysis.
Western blot analysis was performed as described previously (18). Anti-myc antibody, anti-IFN-stimulated gene 15 (anti-ISG15) polyclonal antibody (Cell Signaling Technology), and anti-nsp1 peptide antibody (16) were used as primary antibodies, and goat anti-mouse immunoglobulin G-horseradish peroxidase and goat anti-rabbit immunoglobulin G-horseradish peroxidase (Santa Cruz) were used as secondary antibodies.
Northern blot analysis.
Northern blot analyses using an rLuc probe, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe, and β-actin probe were performed using total intracellular RNAs as described previously (18). The 564-nucleotide (nt)-long, 293 cell-derived, digoxigenin (DIG)-labeled IFN-β riboprobe was used for IFN-β mRNA detection. A 492-nt-long, DIG-labeled ISG15 riboprobe and a 394-nt-long, DIG-labeled ISG56 riboprobe were used for the detection of ISG15 and ISG56 mRNAs, respectively. We used a DIG-labeled random-primed probe, corresponding to nt 29,084 to 29,608 of the SARS-CoV genome, to detect SARS-CoV mRNAs.
In vitro RNA synthesis and RNA transfection.
Capped and polyadenylated RNA transcripts were synthesized from linearized plasmids using the mMESSAGE mMACHINE T7 Ultra kit (Ambion). Subconfluent 293 cells were transfected with in vitro RNA transcripts using TransIT mRNA (Mirus) according to the manufacturer's instructions.
Radiolabeling of intracellular proteins.
293 cells were independently transfected with the indicated in vitro RNA transcripts. One hour after RNA transfection, the cells were incubated with medium containing 4 μg/ml actinomycin D (ActD) (Sigma). Eight hours after ActD addition, the cells were incubated in methionine-free medium for 30 min and then metabolically labeled with 20 μCi/ml of Tran35S-label for 1 h. For the radiolabeling of SARS-CoV-infected cells, replicate cultures of infected 293/ACE2 cells were radiolabeled for 1 h at different times, as indicated in the figures, starting from 5 h to 24 h p.i. in methionine-free medium containing 100 μCi/ml of Tran35S-label. Similarly, replicate cultures of infected Vero E6 cells were radiolabeled for 1 h at different times, starting from 5 h to 12 h p.i. Cell extracts were prepared in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (18), and equivalent amounts of extracts were applied on 12% SDS-PAGE gels. The gels were visualized by autoradiography, and the amounts of radioactivity present in selected regions of the gel were determined by using a Storm 860 PhosphorImager (Molecular Dynamics). The results were normalized to the amounts of radioactivity detected in the same region of the gel in mock-infected cells, and the data are represented as the percentage of radioactivity relative to mock-infected cells.
IFN bioassay.
293/ACE2 cells were independently infected with SARS-CoV-WT and SARS-CoV-mt at an MOI of 3 or with 100 HA units/ml of SeV. Supernatants were collected from SARS-CoV-infected cells at 24 and 48 h p.i. and from SeV-infected cells at 24 h p.i. After complete inactivation of viruses by 60Co irradiation (2 × 106 rads) (16), the samples were subjected to acid treatment. After neutralization of the samples, the human type I IFN activity was measured by a standard plaque reduction assay using Sindbis virus on Vero cells as described previously (12). The IFN levels are expressed as the reciprocal of the dilution that inhibited the formation of 50% of viral plaques. The data are averages of two independent experiments.
RESULTS
Generation of SARS-CoV nsp1 mutants that do not suppress host gene expression.
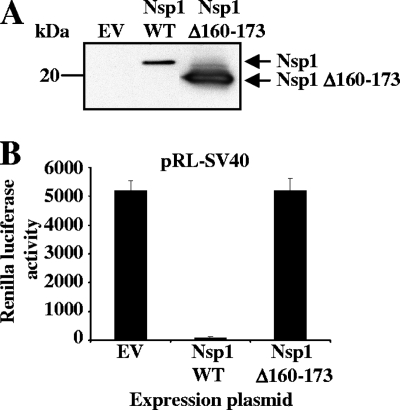
To know whether SARS-CoV nsp1 exerts host gene expression suppression in SARS-CoV-infected cells, we first constructed pCAGGS-Nsp1Δ160-173, in which the C-terminal amino acids 160 to 173 of nsp1 were removed from the 180-amino-acid-long, full-length nsp1 protein (Nsp1-WT). 293 cells were cotransfected with pCAGGS-Nsp1Δ160-173 and a reporter plasmid, pRL-SV40, in which the rLuc gene was cloned downstream of the simian virus 40 (SV40) promoter (18). As controls, the parental plasmid, pCAGGS, and pCAGGS-Nsp1-WT encoding the full-length nsp1 protein (Nsp1-WT) were used in place of pCAGGS-Nsp1Δ160-173. Both expressed Nsp1-WT, and the Nsp1Δ160-173 protein had a C-terminal myc tag. Western blot analysis using anti-myc antibody resulted in an efficient accumulation of the Nsp1Δ160-173 protein compared to low levels of accumulation found with the Nsp1-WT protein at 30 h posttransfection (Fig. 1A), which suggested that nsp1, but not Nsp1Δ160-173, suppressed its own gene expression (18). As expected, Nsp1-WT protein expression strongly suppressed rLuc activity, while Nsp1Δ160-173 protein expression did not inhibit rLuc activity (Fig. 1B), which led to the suggestion that the Nsp1Δ160-173 protein did not suppress host gene expression.
FIG. 1.
The Nsp1 C-terminal region is important for inhibition of reporter gene activity. (A) 293 cells were independently cotransfected with pRL-SV40 and pCAGGS (EV), pRL-SV40 and pCAGGS-Nsp1-WT (Nsp1 WT), or pRL-SV40 and pCAGGS-Nsp1Δ160-173 (Nsp1Δ160-173). (A) Total intracellular proteins were extracted at 30 h posttransfection, and Western blot analysis was performed using anti-myc antibody. (B) At 30 h after transfection, cells were lysed in Renilla luciferase lysis buffer, and aliquots of lysates were used to measure rLuc activities.
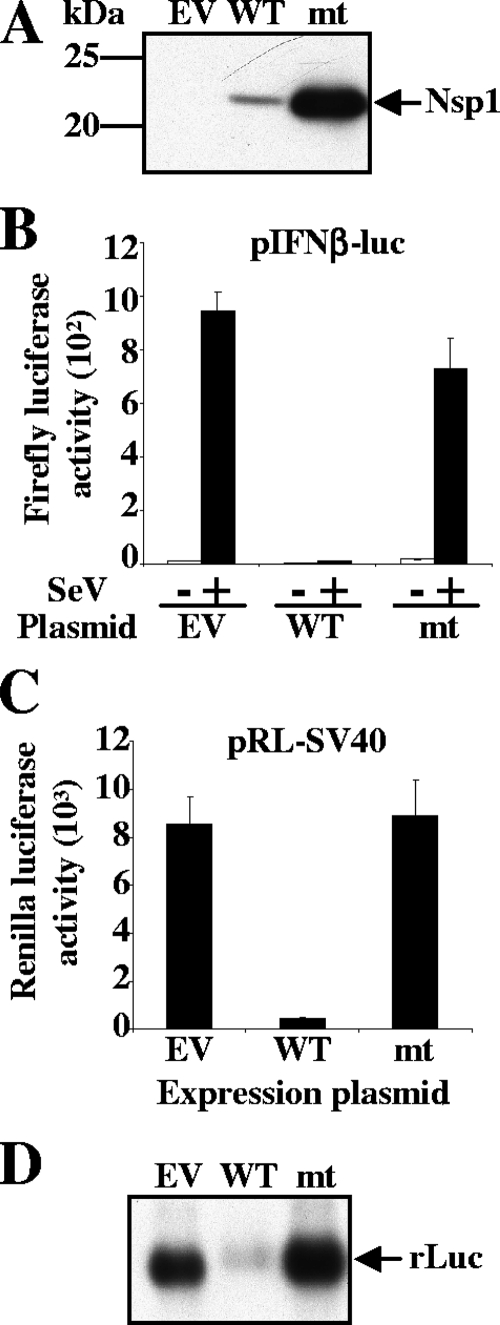
Because the above data indicated that an alteration of amino acids 160 to 173 of nsp1 may abolish the biological functions of nsp1, we next generated pCAGGS-Nsp1-mt, in which the positively charged amino acids K164 and H165 in nsp1's C-terminal region were replaced with alanines and the myc tag was added to the C-terminal end. Like Nsp1Δ160-173, the Nsp1-mt protein accumulated efficiently in transfected 293 cells (Fig. 2A). To determine the effect of Nsp1-mt expression on reporter gene expression, we cotransfected 293 cells with an IFN-β-promoter-driven luciferase reporter plasmid (pIFNβ-luc) and pCAGGS-Nsp1-mt. pCAGGS and pCAGGS-Nsp1-WT were used as controls in place of pCAGGS-Nsp1-mt. At 24 h posttransfection, cells were mock infected or infected with SeV, and cell extracts were prepared at 16 h p.i. As we reported previously (18), SeV infection significantly increased Luc activity in pCAGGS-transfected cells, while Nsp1-WT expression strongly inhibited the SeV-induced Luc activity (Fig. 2B). In contrast, the Nsp1-mt protein expression did not inhibit the SeV-induced Luc activity (Fig. 2B). Next, 293 cells were cotransfected with pCAGGS-Nsp1-mt and pRL-SV40. The controls used were pCAGGS and pCAGGS-Nsp1-WT, in place of pCAGGS-Nsp1-mt. As shown in Fig. 2C, Nsp1-WT expression strongly reduced Luc activity, whereas Nsp1-mt protein did not affect the expression of Luc activity. Northern blot analysis clearly demonstrated that the amounts of rLuc mRNA in pCAGGS-transfected cells and in cells expressing Nsp1-mt protein were significantly higher than in those expressing Nsp1-WT protein (Fig. 2D), while the amounts of 28S and 18S rRNAs were similar in all of these samples (data not shown). These data demonstrated that the Nsp1-mt protein had lost its ability to suppress reporter gene mRNA accumulation.
FIG. 2.
Mutations in the C-terminal region of Nsp1 abrogate its ability to inhibit reporter gene expression. (A) 293 cells were transfected with pCAGGS (EV), pCAGGS-Nsp1-WT (WT), or pCAGGS-Nsp1-mt (mt). At 30 h posttransfection, total intracellular proteins were extracted and Western blot analysis was performed using anti-myc antibody. (B) 293 cells were cotransfected with pIFNβ-luc and pCAGGS (EV), pIFNβ-luc and pCAGGS-Nsp1-WT (WT), or pIFNβ-luc and pCAGGS-Nsp1-mt (mt). At 24 h posttransfection, cells were infected with SeV (+) or mock infected (-). At 16 h p.i., firefly luciferase (Luc) activities were measured. (C and D) 293 cells were independently cotransfected with pRL-SV40 and pCAGGS (EV), pRL-SV40 and pCAGGS-Nsp1-WT (WT), or pRL-SV40 and pCAGGS-Nsp1-mt (mt). At 30 h posttransfection, cell extracts were prepared and used to measure rLuc activities (C), or total RNAs were extracted and Northern blot analysis was performed using a riboprobe specific for rLuc (D).
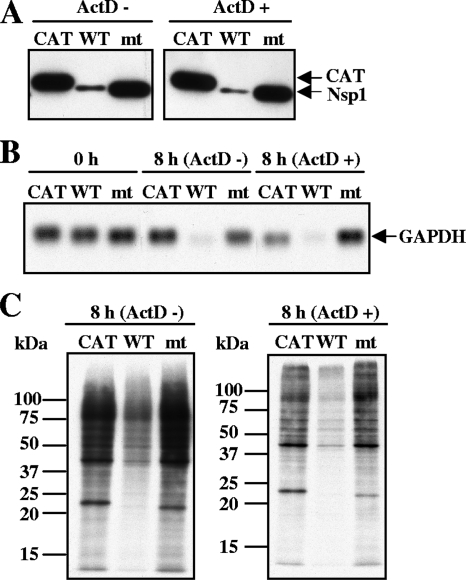
To know whether the Nsp1-mt protein expression induced endogenous host mRNA degradation, 293 cells were transfected independently with one of three in vitro-synthesized, capped, and polyadenylated RNA transcripts: RNA transcripts encoding Nsp1-WT containing a C-terminal myc tag (Nsp1-WT), Nsp1-mt containing a C-terminal myc tag (Nsp1-mt), and CAT protein containing a C-terminal myc tag. At 1 hour posttransfection, the cells were incubated in the absence or presence of 4 μg/ml of ActD; ActD treatment blocks new RNA synthesis and hence allows the analysis of the fate of preexisting cellular mRNAs. Intracellular proteins were extracted at 8 h after ActD addition (Fig. 3A). Intracellular RNAs were extracted at 1 h (Fig. 3B, 0 h) after RNA transfection or at 8 h (Fig. 3B, 8 h) after ActD addition. The expression level of Nsp1-WT was significantly lower than that of CAT and Nsp1-mt proteins (Fig. 3A), a finding that was similar to those in the plasmid transfection experiment. Northern blot analysis showed that the levels of endogenous GAPDH mRNA in the 8-h samples were significantly reduced in Nsp1-WT-expressing cells, whereas the levels of these mRNAs were not affected in the CAT- and Nsp1-mt-expressing cells in repeated experiments (Fig. 3B). A somewhat higher level of GAPDH mRNA in Nsp1-mt-expressing cells in the presence of ActD was not reproducible. These data strongly suggested that the Nsp1-mt protein lost the biological activity to promote host mRNA degradation.
FIG. 3.
Nsp1 mutant protein expression does not promote host endogenous mRNA degradation and host protein synthesis inhibition. 293 cells were transfected independently with in vitro-synthesized CAT RNA transcripts (CAT), Nsp1-WT RNA transcripts (WT), or Nsp1-mt RNA transcripts (mt). One hour after RNA transfection, cells were incubated in the absence of ActD (ActD-) or presence of ActD (ActD+). (A) Total proteins were extracted at 8 h after ActD addition, and Western blot analysis was performed using anti-myc antibody to demonstrate the expression of CAT, Nsp1 WT, and Nsp1 mt proteins. (B) Total RNAs were also extracted at 0 or 8 h after ActD addition. The abundance of endogenous GAPDH mRNA was examined by Northern blot analysis using a riboprobe specific for GAPDH. (C) Cells were radiolabeled with 20 μCi of Tran35S-label from 8.5 to 9.5 h after ActD addition. Equivalent amounts of intracellular proteins were analyzed by 12% SDS-PAGE and visualized by autoradiography. The molecular masses of marker proteins (in kilodaltons) are shown to the left of the gel.
Next, the effect of Nsp1-mt protein expression on host protein synthesis was tested. 293 cells were independently transfected with the CAT RNA transcripts, Nsp1-WT RNA transcripts, and Nsp1-mt RNA transcripts, and at 1 h after transfection, the cells were either treated with ActD or left untreated. The cells were radiolabeled with Tran35S-label from 7 h to 8 h after ActD addition, and cell extracts were prepared and then analyzed by SDS-PAGE. Consistent with our previous report (18), Nsp-WT protein expression strongly suppressed host protein synthesis (Fig. 3C). In contrast, Nsp1-mt protein expression had little effect on host protein synthesis. The results following colloid Coomassie blue staining of the gel demonstrated the loading amounts of proteins in these samples to be similar (data not shown). These data clearly showed that the Nsp1-mt protein did not inhibit host protein synthesis.
Characterization of a SARS-CoV mutant expressing Nsp1-mt protein.
To determine the biological function of nsp1 in infected cells, we generated SARS-CoV-mt carrying K164A and H165A mutations in the nsp1 gene by using a SARS-CoV reverse genetics system (53); the nsp1 protein in SARS-CoV-mt had the same amino acid substitutions as did the Nsp1-mt protein in expression studies, but it lacked the C-terminal myc tag. The rescued SARS-CoV-mt retained the introduced sequence alterations with no additional nucleotide alterations within the nsp1 gene and had no nucleotide sequence alterations in nsp3, -3b, -6, and -N genes, all of which have been reported to affect host innate immune functions in expression studies (8, 19). Three SARS-CoV-mt isolates were recovered from three independent rescue experiments, and all showed similar biological phenotypes. Hence, representative data from one isolate are described below. SARS-CoV-WT was also rescued by using the reverse genetics system (53) and is presented as a control. SARS-CoV-WT and SARS-CoV-mt showed similar virus replication kinetics after infection with an MOI of 1 in Vero E6 cells and in 293/ACE2 cells, which are 293 cells that stably express the SARS-CoV receptor protein ACE2 (Fig. 4A) (18). Analysis of the virus growth kinetics after infection at an MOI of 0.01 in 293/ACE2 and Vero E6 cells showed that the SARS-CoV-mt virus had slightly higher titers than did SARS-CoV-WT in both cell types, except at day 4 p.i. in 293/ACE2 cells (Fig. 4B). Both viruses accumulated similar amounts of virus-specific mRNAs in both cell lines (Fig. 4C) and showed similar levels of nsp1 protein accumulation in infected cells (Fig. 4D). The SARS-CoV-mt nsp1 protein migrated slightly more slowly than did the SARS-CoV-WT nsp1 (Fig. 4D). Similarly, in expression studies, the untagged Nsp1-mt protein also migrated slightly more slowly than the untagged Nsp1-WT protein (data not shown), which suggested that the substitutions of two amino acids affected the nsp1 protein migration in SDS-PAGE. Confocal microscopic analysis using anti-SARS-CoV N protein antibody showed that 80 to 90% of the cells were infected with SARS-CoV-WT or SARS-CoV-mt. Experiments using anti-nsp1 antibody showed that the majority of SARS-CoV-WT nsp1 protein and SARS-CoV-mt nsp1 protein accumulated in the cytoplasm in infected cells (data not shown). Overall, there were no significant differences between the replication of SARS-CoV-WT and SARS-CoV-mt in infected cells.
FIG. 4.
Characterization of the SARS-CoV Nsp1 mutant. 293/ACE2 (A to D) and Vero E6 (A to C) cells were infected with SARS-CoV-WT (WT) or SARS-CoV-mt (mt) at an MOI of 1 (A), 0.01 (B), or 3 (C and D). (A and B) Culture supernatants were collected at the indicated times, and virus titers were determined by TCID50 analysis in Vero E6 cells. The results represent the averages of three independent experiments. (C) At 16 h p.i., total RNAs were extracted. The viral mRNAs were detected by Northern blot analysis using a probe that binds to the 3′-end of the SARS-CoV genome. (D) At 16 h p.i., total proteins were extracted, and Western blot analysis was performed to detect the Nsp1 protein by using an anti-nsp1 peptide antibody.
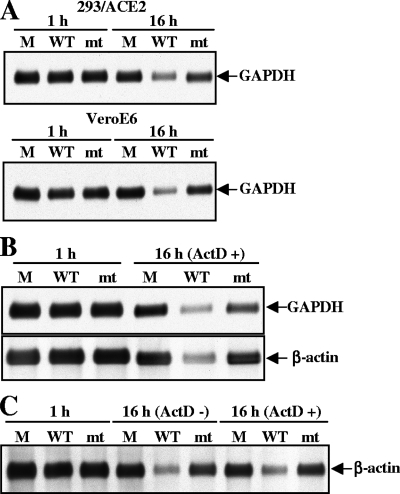
Next, we examined whether SARS-CoV-mt replication induced host mRNA degradation. The same amount of intracellular RNA from infected cells and mock-infected cells was examined by Northern blot analysis (Fig. 5). Consistent with our previous report (18), the abundance of GAPDH mRNA in SARS-CoV-WT-infected cells was clearly lower than that in mock-infected cells at 16 h p.i. (Fig. 5A). The abundance of GAPDH mRNA was clearly higher in SARS-CoV-mt-infected cells than in SARS-CoV-WT-infected cells and was only slightly lower than that in mock-infected cells (Fig. 5A). To find whether SARS-CoV-mt failed to promote efficient host mRNA degradation, 293/ACE2 or VeroE6 cells were mock infected or independently infected with SARS-CoV-WT or SARS-CoV-mt. After virus adsorption, intracellular RNAs were extracted immediately or cells were incubated in the presence of ActD, and intracellular RNAs were extracted after 16 h p.i. (Fig. 5B and C); ActD treatment does not affect SARS-CoV replication (18). As expected (18), a reduction in the abundance of GAPDH and β-actin mRNAs occurred in SARS-CoV-WT-infected cells, which demonstrated that SARS-CoV-WT replication induced the degradation of these host mRNAs (Fig. 5B). In contrast, the amounts of β-actin mRNA in SARS-CoV-mt-infected cells and mock-infected cells were similar, and the abundance of GAPDH mRNA in SARS-CoV-mt-infected cells was clearly higher than that in SARS-CoV-WT-infected cells (Fig. 5B and C). In all cases, the replication of SARS-CoV-WT and SARS-CoV-mt did not alter the abundance of 28S and 18S rRNAs (data not shown). These data revealed that SARS-CoV-mt replication failed to promote efficient host endogenous mRNA degradation.
FIG. 5.
Effect of SARS-CoV-mt replication on abundance of host endogenous mRNAs. 293/ACE2 cells (A and B) and Vero E6 cells (A and C) were either mock infected or infected with SARS-CoV-WT (WT) or SARS-CoV-mt (mt) at an MOI of 3. (A) Intracellular RNAs were extracted at 1 and 16 h p.i. The amount of endogenous GAPDH mRNA was determined by Northern blot analysis. (B and C) At 1 h p.i., intracellular RNAs were extracted (1 h) or ActD was added to the culture medium. Intracellular RNAs were extracted at 15 h after ActD addition (16 h). The abundance of GAPDH (B, upper panel) and β-actin (B, lower panel, and C) mRNAs was determined using Northern blot analysis.
Role of nsp1 on host translational inhibition and type I IFN production in infected cells.
Replication of mouse hepatitis virus (MHV), a group 2 coronavirus, suppresses host protein synthesis (15), whereas it is unknown whether replication of SARS-CoV, which is also a group 2 coronavirus (36), induces host protein synthesis inhibition. We examined whether SARS-CoV replication suppresses host translation and whether nsp1 contributes to the putative SARS-CoV-mediated host translational suppression. 293/ACE2 cells and Vero E6 cells were mock infected or independently infected with SARS-CoV-WT and SARS-CoV-mt. At different times postinfection as indicated in Fig. 6A, the cells were incubated with Tran35S-label for 1 h, and cell extracts were prepared. The cell extracts were heated at 100°C for 5 min in the presence of sample buffer and then analyzed by SDS-PAGE. Colloid Coomassie blue staining confirmed the loading of similar amounts of cell extracts in the gels (data not shown). In 293/ACE2 cells, SARS-CoV-WT replication induced efficient host protein synthesis inhibition that developed around 9 h p.i. and proceeded throughout the course of infection up to 24 h p.i. (Fig. 6A, top panel), while the synthesis of virus-specific S and N proteins was evident (Fig. 6A, top panel). Absence of a major viral M protein in the gels was probably due to the heat-induced M protein aggregation (23, 40). While SARS-CoV-mt replication also induced host protein synthesis inhibition, it was less prominent and developed more slowly than in SARS-CoV-WT infection (Fig. 6A, top panel). Phosphorimager analysis of the marked areas of the gels showed that the amounts of radiolabeled host proteins in SARS-CoV-WT-infected cells were lower than those in SARS-CoV-mt-infected cells from 9 h to 24 h p.i. (Fig. 6A, top panel). A similar time course of radiolabeling experiments using Vero E6 cells showed that protein synthesis inhibition induced by SARS-CoV-mt replication was less prominent than that induced by SARS-CoV-WT infection (Fig. 6A, bottom panel). Western blot analysis showed similar levels of SARS-CoV-WT and SARS-CoV-mt nsp1 protein accumulation during the course of infection in both 293/ACE2 and Vero E6 cells (Fig. 6B). We concluded that SARS-CoV-WT replication induced strong host protein synthesis inhibition and nsp1 contributed to the virus-induced host protein synthesis suppression.
FIG. 6.
Effect of SARS-CoV-mt replication on host protein synthesis. 293/ACE2 and Vero E6 cells were either mock infected (M) or infected with SARS-CoV-WT (WT) or SARS-CoV-mt (mt) at an MOI of 3. (A) Cells were radiolabeled for 1 h with 100 μCi of Tran35S-label, and extracts were prepared at the indicated times postinfection. Equivalent amounts of the intracellular proteins were analyzed on a 12% SDS-PAGE gel and visualized by autoradiography (top panel, 293/ACE2 cells; bottom panel, Vero E6 cells). Phosphorimager analysis was used to determine the level of host protein synthesis, and the numbers below the lanes represent the percent radioactivity relative to mock-infected cells. The boxes represent the region of the gel used for phosphorimager analysis. Representative data from two independent experiments are shown. (B) Western blot analysis was performed with extracts prepared at the indicated times postinfection to detect Nsp1 protein accumulation using anti-nsp1 peptide antibody.
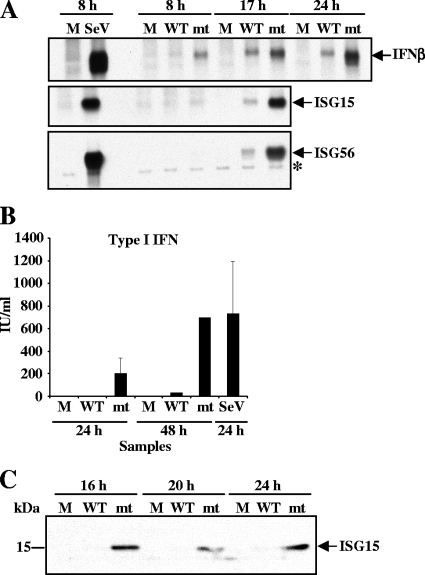
The data showing that SARS-CoV-mt replication induced a less efficient suppression of host gene expression, compared with SARS-CoV-WT replication, led us to investigate a possibility that SARS-CoV-mt replication, but not SARS-CoV-WT replication, induces the expression of IFN-β and ISGs, like ISG15 and ISG56, because these genes have been identified as direct IRF-3 target genes that are induced in response to virus infection. 293/ACE2 cells were mock infected or independently infected with SARS-CoV-WT or SARS-CoV-mt. As a control, 293/ACE2 cells were infected with 100 HA units of SeV. Northern blot analysis showed no accumulation of IFN-β, ISG15, and ISG56 mRNAs in mock-infected cells and efficient accumulation of these mRNAs in SeV-infected cells at 8 h p.i. (Fig. 7A). As expected, the accumulation of these mRNAs in SARS-CoV-WT-infected cells was very low throughout infection (Fig. 7A). In contrast, IFN-β mRNA accumulation was detected as early as 8 h p.i. in SARS-CoV-mt-infected cells, and the abundance of this mRNA increased during the time course of infection (Fig. 7A, top panel). Likewise, the efficient accumulation of ISG15 and ISG56 mRNAs occurred in SARS-CoV-mt-infected cells, but not in SARS-CoV-WT-infected cells (Fig. 7A, middle and bottom panels). A biological assay of type I IFN production showed significant type I IFN production from SARS-CoV-mt-infected cells at both 24 and 48 h p.i. (Fig. 7B); the titer of type I IFN produced from SARS-CoV-mt-infected cells at 48 h p.i. was comparable to that produced from SeV-infected cells at 24 h p.i. In contrast, very low levels of type I IFN were detected in SARS-CoV-WT-infected cells at both time points (Fig. 7B). Western blot analysis showed an increase in the level of ISG15 protein in SARS-CoV-mt-infected 293/ACE2 cells, but not in SARS-CoV-WT-infected cells, further confirming the SARS-CoV-mt-induced increase in ISG15 mRNA levels (Fig. 7C). These data clearly demonstrated that SARS-CoV nsp1 protein suppressed the accumulation of IFN-β and other ISG mRNAs and type I IFN production in infected cells.
FIG. 7.
Effect of SARS-CoV-mt replication on type I IFN and ISG induction. 293/ACE2 cells were either mock infected (M) or infected with SARS-CoV-WT (WT) or SARS-CoV-mt (mt) at an MOI of 3. SeV infection (100 HA units) was used as a positive control (A and B). (A) Total intracellular RNAs were extracted at the indicated times postinfection, and the amounts of endogenous IFN-β (top panel), ISG15 (middle panel), and ISG56 (bottom panel) mRNAs were determined by Northern blot analysis using riboprobes specific for the IFN-β, ISG15, and ISG56 genes, respectively. The identity of the band indicated by an asterisk in the bottom panel is unknown. (B) Culture supernatants were collected from mock-infected cells (M) and SARS-CoV-infected cells at 24 and 48 h p.i. and from SeV-infected cells at 24 h p.i. A type I IFN bioassay was performed to measure the concentration of type I IFN protein (in IU/ml) produced by infected cells. The data represent the averages of two independent experiments. (C) At the indicated times postinfection, total proteins were extracted and Western blot analysis was performed to detect ISG15 protein using anti-ISG15 antibody.
DISCUSSION
In expression studies, SARS-CoV nsp1 induces degradation of host mRNAs and suppresses host translation (18). The present study examined whether SARS-CoV nsp1 could suppress host gene expression in infected cells. We found that, unlike Nsp1-WT, expressed Nsp1-mt failed to suppress gene expression from cotransfected plasmids, had no impact on GAPDH mRNA stability, and did not inhibit host translation. Further, we found that the Nsp1-mt protein accumulated substantially better than did the Nsp1-WT protein in expressing cells, which suggested to us that Nsp1-WT, but not Nsp1-mt, suppressed its own expression. These data revealed that K164A and H165A substitutions in the nsp1 protein completely abolished the host gene suppression activities of the nsp1 protein. Analysis of the structure of SARS-CoV nsp1 by nuclear magnetic resonance showed that the C-terminal region, where we introduced two amino acid substitutions, is flexibly disordered (1); the effects of the K164A and H165A mutations on SARS-CoV nsp1 structure are unclear. Both SARS-CoV-WT and SARS-CoV-mt, the latter of which carried K164A and H165A mutations in the nsp1 gene, replicated efficiently in 293/ACE2 and Vero E6 cells, yet SARS-CoV-mt replication did not induce efficient host mRNA degradation and induced a less efficient inhibition of host protein synthesis, which led us to believe that nsp1 indeed promoted host mRNA degradation and contributed to host translation inhibition in SARS-CoV-infected cells. SARS-CoV nsp1 is the first viral protein among any RNA viruses that has been shown to promote host mRNA degradation in infected cells. Coronavirus replication suppresses host translation (15), and SARS-CoV 7a protein expression suppresses host translation in transfected cells (20); however, the viral protein(s) that contributes to host translational suppression in infected cells has not been identified. To our knowledge, SARS-CoV nsp1 is the first coronavirus protein that has been shown to contribute toward virus-mediated host translation suppression in infected cells.
The innate antiviral response is initiated after the host detects virally encoded, pathogen-associated molecular patterns using cellular pattern recognition receptors (PRRs) (29, 43, 46). Several recent studies have explored how host cell sensors recognize coronavirus replication and activate host innate immune responses. Two studies reported that IRF-3 nuclear translocation and IFN-β mRNA accumulation do not occur in cells infected with MHV or SARS-CoV, and yet treatment of the infected cells with a synthetic, double-stranded RNA analog, poly(I·C), induced IFN-β mRNA accumulation (49, 54). These data led the authors to speculate that double-stranded RNAs of MHV and SARS-CoV, which presumably accumulate within double membrane vesicles in infected cells, are not accessible to PRRs (49, 54). In contrast to this speculation, SARS-CoV-mt replication in 293/ACE2 cells induced IFN-β and ISG mRNA accumulation and high titers of type I IFN production, which suggested to us that host PRRs indeed recognized replicating SARS-CoV-mt and triggered the signaling pathways that led to type I IFN and ISG induction. We showed that expressed SARS-CoV nsp1 does not suppress SeV-induced IRF-3 dimerization (18), which suggested that the nsp1 protein did not suppress signaling events upstream of IRF-3 activation, including the PRR functions. It has been shown that SARS-CoV replication in the human lung epithelial cell line Calu3 and in the rhesus monkey-derived fibroblast cell line MA104 triggers a weak induction of IFN-β mRNA (8). Furthermore, infection of MHV and SARS-CoV in plasmacytoid dendritic cells (pDCs) induced type I IFN production (6) and MHV replication in L2 cells induced IFN-β mRNA accumulation (34), suggesting that coronavirus replication is, in fact, recognized by these cells. Low levels of IFN-β mRNA accumulated in SARS-CoV-WT-infected 293/ACE2 cells (Fig. 7A), whereas Spiegel et al. failed to observe IFN-β-specific reverse transcription-PCR products in SARS-CoV-infected 293 cells (37), leading to the possibility that subtle differences between the two lineages of 293 cells influenced the efficiencies of PRR-mediated recognition of replicating SARS-CoV. We speculate that the expression and accessibility levels of the PRRs to coronavirus-derived, double-stranded RNAs or virus-specific protein(s) could account for the differences observed among cell types and cell lines.
Because host PRRs recognized replicating SARS-CoV-mt and induced type I IFN production in 293/ACE2 cells, it is not surprising that coronaviruses have developed strategies to actively suppress host innate immune functions. Expression studies showed that SARS-CoV nsp3, -3b, -6, and N proteins and MHV N protein suppress host innate immune functions (8, 19, 52). Thiel and his colleagues reported that the expression of MHV nsp1 also suppressed host gene expression and that an nsp1 mutant carrying a 99-nt deletion at the C-terminal region (nsp1Δ99) did not possess this activity (57). Replication of both the parental MHV and an MHV mutant carrying nsp1Δ99 in bone marrow-derived pDCs and primary splenic pDCs induced IFN-α production, which suggests that MHV nsp1 does not suppress IFN-α production in pDCs. It is unclear whether replication of this MHV nsp1 mutant in other MHV-susceptible cells would induce type I IFN production. Wathelet et al. also reported the generation of a SARS-CoV mutant virus carrying mutations in a different region of the nsp1 gene from that in our study (50). Their report showed that expressed SARS-CoV nsp1 suppressed the host antiviral signaling pathways. In their study, unlike Nsp1-mt, the mutations did not completely abolish the reporter gene suppression activity of the nsp1 protein. The mutant virus replicated poorly compared to the parental SARS-CoV in Calu3 cells after infection with a very low MOI. The authors speculated that IFN produced from the mutant virus-infected cells, along with the less efficient IFN-signaling inhibitory activity of the mutant nsp1 protein, could account for the poor growth of the mutant virus (50). However, no direct evidence of IFN production in mutant virus-infected cells was presented (50). Our finding that SARS-CoV-mt replication, but not SARS-CoV-WT replication, induced high titers of type I IFN from infected cells illuminated nsp1's vital role in suppressing host gene expression, including those involved in the host innate antiviral response, in infected cells. To our knowledge, the SARS-CoV nsp1 protein is the first identified coronavirus protein that suppresses IFN production in infected cells. We speculate that nsp1 induces degradation of IFN-β mRNA in SARS-CoV-WT-infected cells to block IFN-β production.
Viral proteins that suppress host gene expression often play critical roles in viral pathogenesis. For example, the herpes simplex virus type 1 vhs protein, a viral RNase (10, 42), suppresses host gene expression by promoting host and viral mRNA degradation and is a viral major virulence factor (35), as is Rift Valley fever virus NSs, which suppresses host transcription (24) and is a major viral virulence factor (5). Accordingly, it is conceivable that the SARS-CoV nsp1 is also a viral major virulence factor. Consistent with this notion, an MHV nsp1 deletion mutant carrying nsp1Δ99 is highly attenuated in the mouse (57). Because transgenic mice expressing human ACE2 are susceptible to SARS-CoV infection (26, 48) and alterations of the SARS-CoV S gene with the corresponding gene of the mouse-adapted SARS-CoV (32) and that of early isolates of SARS-CoV are highly virulent for aged mice (33), testing the role of SARS-CoV nsp1 in viral pathogenesis is now feasible using these mouse model systems.
Acknowledgments
We thank Ralph Baric (University of North Carolina at Chapel Hill) for providing us the SARS-CoV reverse genetics system. We thank Samuel Baron and Joyce Poast (UTMB at Galveston) for their advice and assistance with the IFN bioassay. We also thank Kui Li (UTMB at Galveston) for providing us the ISG15 and ISG56 probes.
This work was supported by Public Health Service grants AI29984 and AI72493 from NIH. C.H., W.K., and T.I. were supported by the James W. McLaughlin Fellowship Fund.
Footnotes
Published ahead of print on 27 February 2008.
REFERENCES
- 1.Almeida, M. S., M. A. Johnson, T. Herrmann, M. Geralt, and K. Wuthrich. 2007. Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 813151-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 7915189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 7812218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bode, J. G., E. D. Brenndorfer, and D. Haussinger. 2007. Subversion of innate host antiviral strategies by the hepatitis C virus. Arch. Biochem. Biophys. 462254-265. [DOI] [PubMed] [Google Scholar]
- 5.Bouloy, M., C. Janzen, P. Vialat, H. Khun, J. Pavlovic, M. Huerre, and O. Haller. 2001. Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J. Virol. 751371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervantes-Barragan, L., R. Zust, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 1091131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung, C. Y., L. L. Poon, I. H. Ng, W. Luk, S. F. Sia, M. H. Wu, K. H. Chan, K. Y. Yuen, S. Gordon, Y. Guan, and J. S. Peiris. 2005. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 797819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devaraj, S. G., N. Wang, Z. Chen, Z. Chen, M. Tseng, N. Barretto, R. Lin, C. J. Peters, C. T. Tseng, S. C. Baker, and K. Li. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 28232208-32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 3481967-1976. [DOI] [PubMed] [Google Scholar]
- 10.Everly, D. N., Jr., P. Feng, I. S. Mian, and G. S. Read. 2002. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 768560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, K., P. Wei, Q. Feng, S. Chen, C. Huang, L. Ma, B. Lai, J. Pei, Y. Liu, J. Chen, and L. Lai. 2004. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2791637-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira, P. C., M. L. Peixoto, M. A. Silva, and R. R. Golgher. 1979. Assay of human interferon in Vero cells by several methods. J. Clin. Microbiol. 9471-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham, R. L., A. C. Sims, S. M. Brockway, R. S. Baric, and M. R. Denison. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 7913399-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hengel, H., U. H. Koszinowski, and K. K. Conzelmann. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol. 26396-401. [DOI] [PubMed] [Google Scholar]
- 15.Hilton, A., L. Mizzen, G. MacIntyre, S. Cheley, and R. Anderson. 1986. Translational control in murine hepatitis virus infection. J. Gen. Virol. 67923-932. [DOI] [PubMed] [Google Scholar]
- 16.Ito, N., E. C. Mossel, K. Narayanan, V. L. Popov, C. Huang, T. Inoue, C. J. Peters, and S. Makino. 2005. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 793182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 785619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamitani, W., K. Narayanan, C. Huang, K. Lokugamage, T. Ikegami, N. Ito, H. Kubo, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 10312885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, and P. Palese. 2006. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 80785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 3481953-1966. [DOI] [PubMed] [Google Scholar]
- 22.Law, H. K., C. Y. Cheung, H. Y. Ng, S. F. Sia, Y. O. Chan, W. Luk, J. M. Nicholls, J. S. Peiris, and Y. L. Lau. 2005. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood 1062366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, Y. N., L. K. Chen, H. C. Ma, H. H. Yang, H. P. Li, and S. Y. Lo. 2005. Thermal aggregation of SARS-CoV membrane protein. J. Virol. Methods 129152-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le May, N., S. Dubaele, L. Proietti De Santis, A. Billecocq, M. Bouloy, and J. M. Egly. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116541-550. [DOI] [PubMed] [Google Scholar]
- 25.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 7915199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCray, P. B., Jr., L. Pewe, C. Wohlford-Lenane, M. Hickey, L. Manzel, L. Shi, J. Netland, H. P. Jia, C. Halabi, C. D. Sigmund, D. K. Meyerholz, P. Kirby, D. C. Look, and S. Perlman. 2007. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 1035108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng, L. F., M. L. Hibberd, E. E. Ooi, K. F. Tang, S. Y. Neo, J. Tan, K. R. Murthy, V. B. Vega, J. M. Chia, E. T. Liu, and E. C. Ren. 2004. A human in vitro model system for investigating genome-wide host responses to SARS coronavirus infection. BMC Infect. Dis. 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paun, A., and P. M. Pitha. 2007. The innate antiviral response: new insights into a continuing story. Adv. Virus Res. 691-66. [DOI] [PubMed] [Google Scholar]
- 30.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, and A. J. McGeer. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 3481995-2005. [DOI] [PubMed] [Google Scholar]
- 31.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 789977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts, A., D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, and K. Subbarao. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rockx, B., T. Sheahan, E. Donaldson, J. Harkema, A. Sims, M. Heise, R. Pickles, M. Cameron, D. Kelvin, and R. Baric. 2007. Synthetic reconstruction of zoonotic and early human severe acute respiratory syndrome coronavirus isolates that produce fatal disease in aged mice. J. Virol. 817410-7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth-Cross, J. K., L. Martinez-Sobrido, E. P. Scott, A. Garcia-Sastre, and S. R. Weiss. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 817189-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smiley, J. R. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J. Virol. 781063-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 792079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiegel, M., K. Schneider, F. Weber, M. Weidmann, and F. T. Hufert. 2006. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J. Gen. Virol. 871953-1960. [DOI] [PubMed] [Google Scholar]
- 39.Spiegel, M., and F. Weber. 2006. Inhibition of cytokine gene expression and induction of chemokine genes in non-lymphatic cells infected with SARS coronavirus. Virol. J. 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sulea, T., H. A. Lindner, E. O. Purisima, and R. Menard. 2005. Deubiquitination, a new function of the severe acute respiratory syndrome coronavirus papain-like protease? J. Virol. 794550-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taddeo, B., W. Zhang, and B. Roizman. 2006. The U(L)41 protein of herpes simplex virus 1 degrades RNA by endonucleolytic cleavage in absence of other cellular or viral proteins. Proc. Natl. Acad. Sci. USA 1032827-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi, O., and S. Akira. 2007. Signaling pathways activated by microorganisms. Curr. Opin. Cell Biol. 19185-191. [DOI] [PubMed] [Google Scholar]
- 44.Tang, B. S., K. H. Chan, V. C. Cheng, P. C. Woo, S. K. Lau, C. C. Lam, T. L. Chan, A. K. Wu, I. F. Hung, S. Y. Leung, and K. Y. Yuen. 2005. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J. Virol. 796180-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 842305-2315. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, A. J., and S. A. Locarnini. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol. Cell Biol. 85435-445. [DOI] [PubMed] [Google Scholar]
- 47.Tsang, K. W., P. L. Ho, G. C. Ooi, W. K. Yee, T. Wang, M. Chan-Yeung, W. K. Lam, W. H. Seto, L. Y. Yam, T. M. Cheung, P. C. Wong, B. Lam, M. S. Ip, J. Chan, K. Y. Yuen, and K. N. Lai. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 3481977-1985. [DOI] [PubMed] [Google Scholar]
- 48.Tseng, C. T., C. Huang, P. Newman, N. Wang, K. Narayanan, D. M. Watts, S. Makino, M. M. Packard, S. R. Zaki, T. S. Chan, and C. J. Peters. 2007. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human angiotensin-converting enzyme 2 virus receptor. J. Virol. 811162-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Versteeg, G. A., P. J. Bredenbeek, S. H. van den Worm, and W. J. Spaan. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 36118-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wathelet, M. G., M. Orr, M. B. Frieman, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 8111620-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weidman, M. K., R. Sharma, S. Raychaudhuri, P. Kundu, W. Tsai, and A. Dasgupta. 2003. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 9575-85. [DOI] [PubMed] [Google Scholar]
- 52.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 812554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 10012995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, H., and S. Perlman. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 81568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziebuhr, J. 2006. The coronavirus replicase: insights into a sophisticated enzyme machinery. Adv. Exp. Med. Biol. 5813-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ziegler, T., S. Matikainen, E. Ronkko, P. Osterlund, M. Sillanpaa, J. Siren, R. Fagerlund, M. Immonen, K. Melen, and I. Julkunen. 2005. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J. Virol. 7913800-13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zust, R., L. Cervantes-Barragan, T. Kuri, G. Blakqori, F. Weber, B. Ludewig, and V. Thiel. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3e109. [DOI] [PMC free article] [PubMed] [Google Scholar]