Abstract
Enveloped virus entry into host cells is typically initiated by an interaction between a viral envelope glycoprotein and a host cell receptor. For budded virions of the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, the envelope glycoprotein GP64 is involved in host cell receptor binding, and GP64 is sufficient to mediate low-pH-triggered membrane fusion. To better define the role of GP64 in receptor binding, we generated and characterized a panel of antisera against subdomains of GP64. Eight subdomain-specific antisera were generated, and their reactivities with GP64 proteins and neutralization of virus infectivity and binding were examined. Antibodies directed against the N-terminal region of GP64 (amino acids 21 to 159) showed strong neutralization of infectivity and effectively inhibited binding of 35S-labeled budded virions to Sf9 cells. In addition, we generated virions displaying truncated GP64 constructs. A construct displaying the N-terminal 274 amino acids (residues 21 to 294) of the ectodomain was sufficient to mediate virion binding. Additional studies of antisera directed against small subdomains revealed that an antiserum against a 40-amino-acid region (residues 121 to 160) neutralized virus infectivity. Site-directed mutagenesis was subsequently used for functional analysis of that region. Recombinant viruses expressing GP64 proteins with single amino acid substitutions within amino acids 120 to 124 and 142 to 148 replicated to high titers, suggesting that those amino acids were not critical for receptor binding or other important GP64 functions. In contrast, GP64 proteins with single amino acid substitutions of residues 153 and 156 were unable to substitute for wild-type GP64 and did not rescue a gp64 knockout virus. Further analysis showed that these substitutions substantially reduced binding of recombinant virus to Sf9 cells. Thus, the amino acid region from positions 21 to 159 was identified as a putative receptor binding domain, and amino acids 153 and 156 appear to be important for receptor binding.
The Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is a large double-stranded DNA virus (approximately 134 kbp) that produces two structurally and functionally distinct virion phenotypes, the occlusion-derived virion (ODV) and the budded virion (BV), during the infection cycle (7, 26). The ODV is assembled within the nucleus and embedded within large proteinaceous occlusion bodies that are produced in the very late phase of the infection cycle. The ODV is adapted for propagation of infection from animal to animal through oral transmission and infection of the midgut epithelial cells (6, 14, 15, 28, 46, 48). In contrast to ODV, BV are produced when nucleocapsids bud from the plasma membrane at the cell surface. Thus, the BV is surrounded by a lipid bilayer derived from the plasma membrane (6, 14, 15, 28, 46, 48). The BV mediates movement of the viral infection from the midgut to other tissues and propagation of the infection from cell to cell within the infected animal. BV enter cells via receptor-mediated endocytosis (48), while the ODV appear to fuse directly with the plasma membrane at the cell surface (6, 11).
The entry of enveloped viruses into cells is typically initiated by an interaction between a viral envelope glycoprotein and a host cell receptor. For baculoviruses of the group I NPVs, such as AcMNPV, this function is mediated by the major envelope protein of the BV known as GP64 (10, 46, 48). AcMNPV also encodes and expresses a baculovirus F protein called Ac23 (23, 36). In group II NPVs, such as Spodoptera exigua MNPV and Lymantria dispar MNPV, F proteins serve as essential membrane fusion proteins (12, 21, 34-36, 51, 52) and are functional homologs of AcMNPV GP64 (22). However, the F protein (Ac23) of AcMNPV does not appear to be a functional fusion protein, and unlike F proteins of group II NPVs (21), Ac23 is nonessential and can be deleted from the AcMNPV genome with no substantial effect on virus production or infectivity in insect cell culture (23, 36). In contrast, GP64 is essential for cell-to-cell transmission of the virus in cell culture and in infected animals (28). GP64 serves two major roles during virus entry. First, GP64 is involved in host cell receptor binding (8). Second, GP64 mediates the low-pH-triggered membrane fusion activity necessary for release of the nucleocapsid into the cytosol during entry by endocytosis (2, 16, 25, 27, 37, 47).
The cellular receptor for AcMNPV BV attachment has not yet been identified, although a prior study suggested that a cellular protein may serve as the receptor (50). In addition, it was reported that baculovirus BV bind to insect cells in a saturable and competitive manner (53). The most direct data implicating GP64 as a viral attachment protein derive from studies showing that a soluble form of Orgyia pseudotsugata MNPV (OpMNPV) GP64 competes for BV binding (8). In addition, GP64 expressed transiently in insect cells is sufficient to mediate low-pH-triggered membrane fusion, suggesting that the initial target membrane attachment is provided by GP64 alone (2). A prior study of GP64 glycosylation reported that when certain glycosylation sites of GP64 were eliminated, virion binding to insect cells was decreased (13), suggesting that the glycosylation state of GP64 may be a factor in virion binding. Although few anti-GP64 monoclonal antibodies (MAbs) have been reported, several early studies demonstrated that MAbs directed against GP64 were capable of neutralizing the infectivity of BV (10, 15, 40, 47). However, no anti-GP64 antibodies that inhibit virion attachment to cells have been reported.
To better define the role of GP64 in receptor binding, we produced and characterized a panel of antisera raised against overlapping subdomains of GP64. Eight antisera were produced, and their reactivities with GP64 proteins were examined in neutralization and receptor binding assays. An antiserum against the N-terminal region of GP64, including amino acids 21 to 159, exhibited significant virus neutralization and inhibition of virion binding. We examined this region further by a combination of additional antibody neutralization studies, peptide display and direct virion binding studies, and site-directed mutagenesis and functional analysis. We identified the region of amino acids 121 to 160 as important, and substitutions at amino acid positions 153 and 156 were shown to substantially reduce virus binding to Sf9 cells, confirming an important role for this N-terminal domain in receptor binding.
MATERIALS AND METHODS
Protein expression and purification.
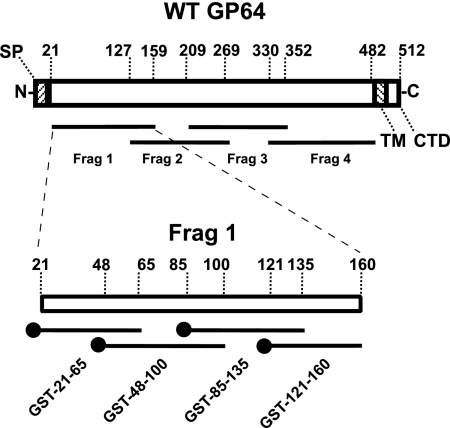
The ectodomain of the mature AcMNPV GP64 protein consists of approximately 462 amino acids. The GP64 ectodomain was subdivided into four partially overlapping subdomains of approximately 140 to 150 amino acids each (the subdomains were designated GP64 Frag1 [residues 21 to 159], GP64 Frag2 [residues 127 to 269], GP64 Frag3 [residues 209 to 352], and GP64 Frag4 [residues 330 to 482]). DNA segments encoding the above GP64 subdomains were PCR amplified from wild-type (wt) AcMNPV DNA by using primers that contained either an NheI site (upstream primer) or a HindIII site (downstream primer). The resulting PCR products were digested with NheI and HindIII, subcloned into the NheI/HindIII sites of a pET28a expression vector (Invitrogen), and used to generate a fusion protein containing an N-terminal six-histidine tag, a T7 epitope tag, and approximately 140 to 150 amino acids of the GP64 subdomain. The resulting plasmids, pET28a-GP64 Frag1, pET28a-GP64 Frag2, pET28a-GP64 Frag3, and pET28a-GP64 Frag4, were confirmed by DNA sequencing. The recombinant plasmids were transformed into Escherichia coli BL21(DE3) (Invitrogen) bacterial cells for protein expression. E. coli BL21(DE3) cells containing plasmids expressing the above GP64 subdomains were grown at 37°C in Luria-Bertani (LB) medium supplemented with 50 μg/ml kanamycin and 50 μg/ml chloramphenicol to an optical density at 600 nm of 0.5 to 0.7, induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM, and grown for an additional 3 h before being harvested. Bacterial cultures were pelleted and then lysed by sonication in lysis buffer (phosphate-buffered saline [PBS], pH 7.4, 100 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 mM dithiothreitol, and protease inhibitor cocktail [Complete; Roche Applied Science]). Insoluble cell debris was removed by centrifugation. Purification of recombinant proteins was achieved by absorption to nickel-nitrilotriacetic acid resin (Qiagen) as outlined in the manufacturer's instructions. The purity of the recovered affinity-purified recombinant proteins and polypeptides was determined by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue staining (data not shown). Rabbit polyclonal antibodies were generated from the purified proteins by standard protocols (Cocalico Biologicals Inc., Reamstown, PA). The polyclonal antibodies were designated anti-Frag1, anti-Frag2, anti-Frag3, and anti-Frag4, corresponding to the GP64 subdomains in the purified proteins. The immunoglobulin G (IgG) fraction from each antiserum was isolated using protein A agarose (Pharmacia). Antibody titers were determined by Western blotting using GP64 proteins.
Expression and purification of GST fusion proteins.
A series of plasmids encoding glutathione S-transferase (GST) fusion proteins was generated by the following methods. First, DNA fragments encoding GP64 amino acids 21 to 65, 48 to 100, 85 to 135, and 121 to 160 were PCR amplified from a wt AcMNPV DNA template. EcoRI and XhoI restriction sites were engineered into the 5′ and 3′ ends, respectively, of each PCR product. Each PCR product was then cloned into the EcoRI and XhoI sites of vector pGEX-4T-1 (Amersham Pharmacia Biosciences) to generate a GP64 subdomain fused in-frame with an N-terminal GST tag. The resulting plasmids were transformed into E. coli strain BL21(DE3) (Invitrogen), grown to an optical density at 600 nm of 0.7, and then induced for 3 h at 37°C by adding IPTG to a final concentration of 1 mM. Cells were pelleted and disrupted by sonication in lysis buffer, and then the lysates were cleared by centrifugation and filtered through a 0.45-μm filter. The cleared supernatants were incubated with immobilized glutathione agarose (Pierce) overnight at 4°C with rotation. The resins were transferred to a minicolumn, washed five times with ice-cold PBS (pH 7.4), and then eluted with 50 mM Tris-HCl (pH 8.0) containing 10 mM reduced glutathione at room temperature. Excess glutathione was removed by extensive dialysis against PBS (pH 7.4). The purified GST fusions were confirmed with an anti-GST antibody (Immunology Consultants Laboratory, Inc.) and used to generate polyclonal antibodies in guinea pigs (Cocalico Biologicals Inc., Reamstown, PA) according to standard protocols. The resulting antibodies were designated anti-GST (a control), anti-GST-21-65, anti-GST-48-100, anti-GST-85-135, and anti-GST-121-160, with numbers corresponding to the amino acid regions of the GP64 protein.
Neutralization assay.
For analysis of antibody neutralization, antibodies were purified from crude antisera with protein A agarose (Pharmacia). A series of antibody dilutions were prepared and used in neutralization experiments such that 0.1 to 60 μg of each antibody was incubated with virus in 50-μl reaction mixtures containing 10 μl of wt AcMNPV BV (1.49 × 106 IU/ml). Antibody-virus mixtures were incubated for 2 h at 27°C and then titrated on Sf9 cells to determine the 50% tissue culture infective dose.
For anti-GST-21-65, anti-GST-48-100, anti-GST-85-135, and anti-GST-121-160, neutralization assays were performed as follows. Dilutions of polyclonal antisera (1:1.6, 1:2, 1:2.5, 1:5, and 1:10) in a total volume of 40 μl TNMFH (8a) were incubated with 10 μl wt AcMNPV BV (1.49 × 106 IU/ml) for 2 h at 27°C. For each treatment, three replicates were performed. Titers of the treated virus mixtures were determined by end-point dilution assay as described previously (33, 47).
Assay of inhibition of neutralization.
The assay of inhibition of neutralization measures the ability of GST fusion proteins to block the neutralizing activity of an antibody preparation. Various quantities of GST fusion proteins (10 to 50 μg) and anti-GST-121-160 antibody (1.1.6 or 1:2 dilution) were mixed and incubated at 27°C for 2 h. The mixture was adjusted to a final volume of 40 μl with TNMFH. Each of the above dilutions was incubated with 10 μl wt AcMNPV BV (1.49 × 106 IU/ml) for 2 h at 27°C, and the titer of the virus was then determined by end-point dilution assay (33, 47).
[35S]methionine-labeled virions and binding inhibition assay.
wt AcMNPV or Ac23null AcMNPV (23, 36) BV were labeled with [35S]methionine in the following manner. Sf9 cells (2 × 106 cells) were plated in T-25 flasks (Corning Inc.), allowed to attach for 1 h, and then infected with wt or Ac23null AcMNPV BV at a multiplicity of infection (MOI) of 10. At 29 h postinfection (hpi), cells were starved by incubation in 3 ml methionine-free Grace's medium (Invitrogen) for 1 h, followed by the addition of 35S-EasyTag Express protein labeling mix (1,175.0 Ci/mmol; Perkin-Elmer) to a final concentration of 10 μCi/ml. At 37 hpi, unlabeled methionine was added to a final concentration of 10 mM and cells were incubated at 27°C for an additional 48 h. Virions were purified by centrifugation through a 25% sucrose cushion, and the purified labeled wt or Ac23null virus was titrated by end-point dilution assay.
For the binding inhibition assay, cells were seeded into 24-well plates (Corning Inc.) at 1 × 105 cells per well and allowed to attach for 1 h at 27°C. Ten microliters of [35S]methionine-labeled purified wt AcMNPV BV (1.93 × 109 IU/ml; 8,106 cpm/μl) or 10 μl of [35S]methionine-labeled purified Ac23null AcMNPV BV (1.53 × 109 IU/ml; 7,333.2 cpm/μl) was incubated with 100 μg purified antibodies (anti-Frag1, anti-Frag2, anti-Frag3, anti-Frag4, or MAb AcV1) separately in a total volume of 40 μl PBS (pH 6.2) or with 40 μl undiluted rabbit serum or 40 μl undiluted anti-Lef11 antiserum (a rabbit polyclonal antibody as a negative control) for 2 h at 27°C. The above antibody-virus mixtures were added to cells and incubated for 5 h at 4°C to permit virus binding. The cell culture medium was then removed, and the cells were rinsed three times with 1 ml PBS at 4°C (pH 6.2). The Sf9 cells were then lysed in 0.5 ml NET buffer (20 mM Tris, 150 mM NaCl, 0.5% deoxycholate, 1.0% Nonidet P-40, 1 mM EDTA, pH 7.5) at 27°C for 15 min, and lysates were added to vials containing 5 ml EcoLume liquid scintillation fluid. The tissue culture plates were rinsed twice with 200 μl lysis buffer (1% SDS, 0.2 N NaOH), and lysis buffer rinses were pooled with cell lysates before counting of radioactivity in an LS6500 liquid scintillation counter (Beckman).
Construction of plasmids and baculoviruses encoding constructs containing a GP64 stem region and receptor binding assay.
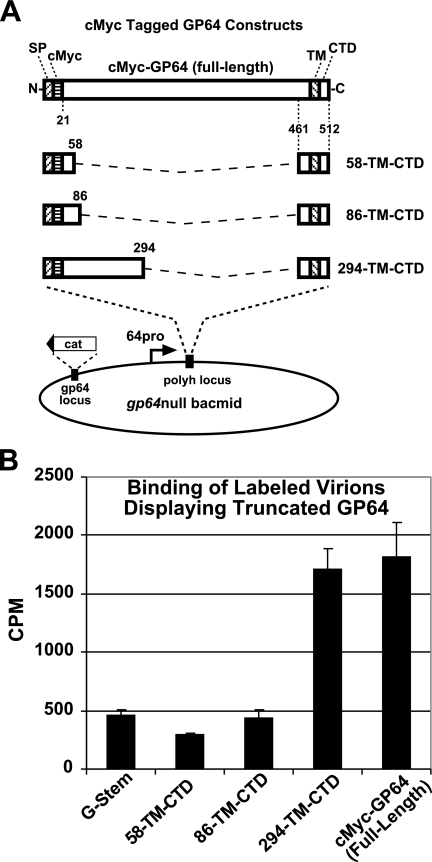
A truncated version of the human vesicular stomatitis virus envelope glycoprotein G (VSV G) was constructed previously (54). The truncated VSV G construct (G-stem) contains the AcMNPV gp64 promoter and signal peptide, a c-Myc epitope tag (at the N terminus of the mature protein), 42 amino acids (positions 421 to 463) from the VSV G ectodomain, and the predicted transmembrane (TM) domain and cytoplasmic tail domain (CTD) of VSV G. This G-stem construct was inserted into the polyhedrin locus of a gp64null AcMNPV bacmid. Cells stably expressing OpMNPV GP64 (cell line Sf9Op1D) (32, 37) were transfected with the bacmid DNA, and the resulting virus was designated vAc/G-stem.
A series of plasmids encoding various portions of the N terminus of the GP64 ectodomain combined with the C-terminal GP64 region were constructed previously (54, 55) (see Fig. 4). Sf9 cells were coinfected with the vAc/G-stem virus and each of the following viruses, at a total MOI of 10 (5 infectious units each of the vAc/G-stem virus and the virus expressing a truncated GP64 protein): vAc/58-TM-CTD (amino acids 21 to 58), vAc/86-TM-CTD (amino acids 21 to 86), and vAc/294-TM-CTD (amino acids 21 to 294). Progeny virus was then labeled with [35S]methionine as described previously, purified, and used for the receptor binding assay. Each [35S]methionine-labeled purified virus preparation (equal cpm quantities) was incubated with Sf9 cells for 15 h at 4°C. Cells were washed three times with PBS (pH 6.2) and then solubilized in 0.5 ml NET buffer for 15 min. The [35S]methionine associated with the lysed cells was counted as described above.
FIG. 4.
Analysis of Sf9 cell binding by gp64null AcMNPV BV displaying truncated forms of GP64. (A) Strategy for construction of GP64 proteins with C-terminal ectodomain deletions and insertion into a gp64null bacmid. Each construct contains a C-terminal GP64-stem domain and various portions of the GP64 ectodomain, as illustrated. The GP64-stem domain consists of amino acids 461 to 512 from the C terminus of the AcMNPV GP64 protein (22 residues from the predicted GP64 ectodomain, 23 residues from the predicted GP64 TM domain, and the 7-residue CTD). Various portions of the GP64 ectodomain were fused to the GP64-stem such that in the mature protein, the protein construct contained a c-Myc epitope tag and 38 (residues 21 to 58), 66 (residues 21 to 86), 138 (residues 21 to 158), or 274 (residues 21 to 294) amino acids from the N terminus of the GP64 ectodomain. (B) Analysis of virion binding to Sf9 cells. Labeled virions displaying truncated GP64 proteins were examined for binding to Sf9 cells. Production of virions displaying truncated GP64 proteins was augmented by coexpression of a VSV G-stem construct (vAc/G-stem) that mediates virion budding (54). Progeny virions were labeled with [35S]methionine, purified, and bound to Sf9 cells at 4°C. Labeled virus bound to Sf9 cells was quantified, and cpm values are indicated. Each bar represents the average value from three independent experiments with three replicates.
Western blot analysis and syncytium formation assays.
Western blots were performed as described previously (2, 55). In brief, 10 μl of the cell lysate (approximately 1 × 104 cells) was mixed with 10 μl of 2× Laemmli buffer (125 mM Tris, 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue, pH 6.8) and heated to 100°C for 5 min prior to analysis by SDS-10% PAGE. Proteins were transferred to Immobilon-P membranes (Millipore) and blocked overnight at 4°C in TBST (25 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween 20, 5% powdered milk). Blots were incubated for 1 h at room temperature with one of the following primary antibodies diluted in TBST: AcV5 MAb (which recognizes a linear epitope within AcMNPV GP64) diluted 1:100, anti-cMyc MAb (ATCC CRL-1729; Myc 1-9E10.2) diluted 1:100, anti-His polyclonal antibody (MBL International) diluted 1:4,000, or anti-GST polyclonal antibody (MBL International) diluted 1:1,000. After being washed three times in TBST, blots were incubated with secondary antibodies consisting of alkaline phosphatase-conjugated goat anti-mouse, goat anti-rabbit, or goat anti-guinea pig IgG (Promega) at a dilution of 1:10,000. Detection of bound antibody was performed as described earlier (2).
Syncytium formation assays were performed with the mutant viruses listed in Table 1. Sf9 cells were infected at an MOI of 10. At 48 hpi, the cell culture medium was removed, replaced with PBS (pH 5.0) for 5 min, exchanged with fresh PBS (pH 6.2), and incubated for 2 h prior to scoring for syncytium formation. For this assay, three or more nuclei in a syncytial mass were scored as a syncytium.
TABLE 1.
Summary of results from analysis of BV displaying full-length GP64 constructs with single amino acid substitution mutationsa
| GP64 mutant | Virus titer (IU/ml)
|
% Syncytium formation | |
|---|---|---|---|
| Sf9 cells | Sf9Op1D cells | ||
| R120E | 1.78 × 108 | ND | |
| W121A | 1.61 × 107 | ND | |
| G122A | 1.43 × 108 | ND | |
| S123A | 1.43 × 108 | ND | |
| D124K | 7.20 × 107 | ND | |
| N132A | 2.74 × 108 | ND | |
| K142L | 1.87 × 108 | ND | >60 |
| E143K | 2.74 × 108 | ND | |
| L144K | 6.49 × 107 | ND | |
| V145T | 2.80 × 108 | ND | |
| K146L | 1.43 × 108 | ND | |
| R147L | 2.74 × 108 | ND | |
| Q148L | 2.80 × 108 | ND | |
| N149A | 3.51 × 105 | ND | >60 |
| N150A | 3.51 × 106 | ND | >60 |
| N151V | 3.16 × 105 | ND | <21 |
| H152V | 3.98 × 106 | ND | >60 |
| H152A | NA | 3.51 × 106 | <5 |
| F153A | NA | 1.43 × 107 | <10 |
| A154S | NA | 1.43 × 106 | <10 |
| H155V | 7.20 × 107 | ND | >60 |
| H155A | 2.31 × 108 | ND | >60 |
| H156V | NA | 3.51 × 106 | |
| H156A | NA | 3.51 × 106 | |
| T157V | 2.15 × 108 | ND | |
Viruses were titrated on Sf9 or Sf9Op1D cells by end-point dilution assay. Syncytium formation assays were performed with mutant viruses by infecting Sf9 cells at an MOI of 10. At 48 hpi, medium was removed, replaced with PBS (pH 5.0) for 5 minutes, exchanged with fresh PBS (pH 6.2), and incubated for 2 h prior to scoring for syncytium formation. Masses were scored as syncytia only if three or more nuclei were present. The percentage of cells found in syncytia, as calculated from representative fields of cells, is listed in the table. ND, not done; NA, not available.
Site-directed mutagenesis and analysis of virion binding.
Site-directed mutagenesis was performed as described previously (17). Briefly, a DNA fragment containing the AcMNPV gp64 promoter and open reading frame was PCR amplified from a wt AcMNPV DNA template as two separate subdomains. Each PCR of the 5′ fragment was performed with a primer pair that included a forward primer (KpnI gp64-pro forward [5′-AA GGTACC CGG CAT GTC GAC TGA GCG T-3′] [restriction site is underlined]) and a reverse mutagenic primer. For example, the mutagenic primer for substitution of alanine for phenylalanine at amino acid position 153 was 5′-AA GGTCTCN gca CTG CTC ATT AAA GAT GAC AAA AAT AAC C-3′ (N stands for any nucleotide, lowercase letters identify the alanine codon, and a BsaI recognition sequence is underlined). For PCR of the 3′ fragment of the target sequence, we used a primer pair that included a forward mutagenic primer (for the Phe-to-Ala substitution at amino acid position 153, the primer was 5′-AA GGTCTCN G tgc CGC AGCTTT TAT TTG GCG CGT-3′) and a reverse primer (HindIII gp64-TAA reverse [5′-GG AAGCTT TTA ATA TTG TCT ATT ACG GTT TC-3′]). The two amplified fragments were digested with BsaI, ligated together, digested with KpnI and HindIII, and subcloned into the KpnI and HindIII sites of pFastBac1-lacZ (23). The resulting plasmid DNA contained the gp64 promoter and the gp64 open reading frame with a specific substitution. All constructs were confirmed by DNA sequencing and then used for transposition into a gp64null AcMNPV genome (gp64− bacmid-DH10B+pMON7124) (22). Sf9 cells or cells stably expressing OpMNPV GP64 (cell line Sf9Op1D) (37) were transfected with DNA of each bacmid construct, and the resulting viruses were harvested from cell supernatants and titrated on Sf9 or Sf9Op1D cells by end-point dilution assay (33). For labeling and cell binding assays, virions displaying each substitution mutation construct were labeled and purified as described above. Labeled BV preparations were initially examined and confirmed by SDS-PAGE and PhosphorImager analysis of BV proteins, and the total label incorporated into each BV preparation was determined by liquid scintillation counting. BV preparations of AcMNPVs that displayed substituted GP64 proteins were adjusted such that equal cpm values were added to cells and were incubated for 5 h at 4°C to permit virus binding. Cells were then rinsed and lysed, and lysates were counted as described above.
RESULTS
N-terminal subdomain-specific antibodies neutralize infectivity of AcMNPV BV.
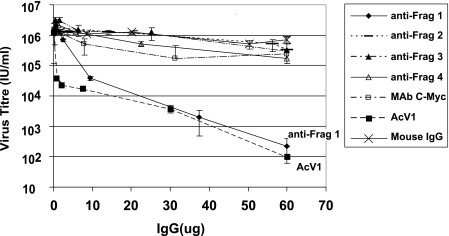
To investigate the role of the GP64 glycoprotein in receptor binding, we first generated antisera against several subdomains of GP64. Four overlapping subdomains covering the entire ectodomain of the GP64 protein (Fig. 1, Frag1 to Frag4) were expressed in E. coli and used to generate polyclonal antibodies, designated anti-Frag1, anti-Frag2, anti-Frag3, and anti-Frag4. High-antibody titers were observed, as effective dilutions for detection of GP64 ranged from 1:10,000 to 1:20,000 (data not shown). To determine whether the antibodies could reduce or neutralize the infectivity of AcMNPV BV, we performed neutralization assays. BV (1.49 × 104 IU) were incubated with various concentrations of each anti-GP64 subdomain antibody preparation, and viral infectivity was then determined by end-point dilution assays. Control antibodies included MAb AcV1, mouse IgG, or an anti-c-Myc antibody. In prior studies, it was demonstrated that the MAb AcV1 effectively neutralized AcMNPV BV infectivity (10, 48, 55). Therefore, we used AcV1 as a positive control for neutralization. We found that one subdomain antibody, anti-Frag1, was able to substantially reduce virus infectivity (Fig. 2), whereas parallel assays of anti-Frag2, anti-Frag3, and anti-Frag4 antibodies did not show similar reductions in titer, even at higher antibody concentrations. Treatment with negative control antibodies, such as mouse IgG and an anti-c-Myc antibody, showed no substantial reduction in BV titer when similarly increasing antibody concentrations were used.
FIG. 1.
Strategy for mapping the receptor binding domain by generation and analysis of subdomain-specific antisera. Lines below the GP64 diagram represent peptides used for generation of antisera in sequential mapping experiments. Amino acid positions are numbered according to the full-length GP64 coding region. The subdomains contained the following amino acids from the GP64 ectodomain: Frag 1, amino acids 20 to 159; Frag 2, amino acids 127 to 269; Frag 3, amino acids 209 to 352; Frag 4, amino acids 330 to 482. Various domains of AcMNPV GP64 are also shown, including SP (signal peptide), TM (transmembrane domain), and CTD (cytoplasmic tail domain). The closed circle at the end of each Frag 1 subdomain construct represents the N-terminal position of a GST tag.
FIG. 2.
Analysis of subdomain-specific antibodies for neutralization of AcMNPV BV. wt AcMNPV BV (1.49 × 104 IU) was incubated with various quantities of anti-GP64 subdomain antibodies (anti-Frag1, anti-Frag2, anti-Frag3, and anti-Frag4) or with control antibodies (anti-c-Myc, AcV1, or mouse IgG) for 2 h at 27°C (see Materials and Methods). Viral infectivity was subsequently determined in triplicate by end-point dilution assays on Sf9 cells. The average values for three independent experiments are presented.
Anti-Frag1 antibodies inhibit binding of Ac23null BV to Sf9 cells.
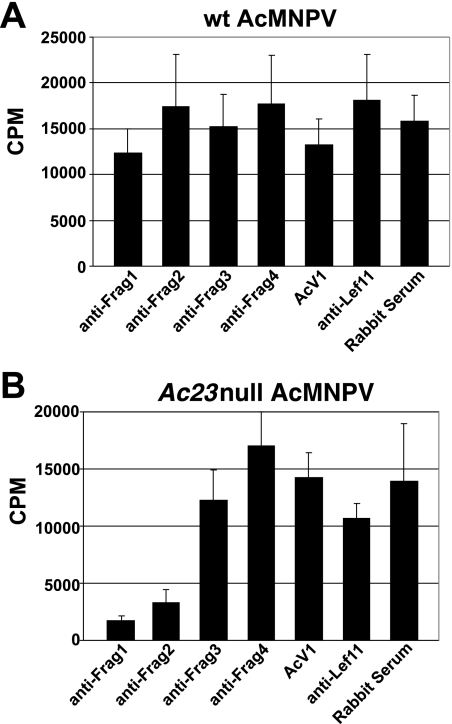
To determine whether neutralization by anti-Frag1 was caused by inhibition of receptor binding, we conducted binding inhibition assays. Effects on virus binding were measured by examining binding of radiolabeled BV in the presence of each subdomain-specific antibody. After treating labeled BV with each antibody preparation and then allowing BV to bind to cells at 4°C, the cells were washed and lysed and the cell-associated label was quantified. First, we used metabolically labeled wt AcMNPV BV in the binding inhibition assay. The results showed no significant reduction in binding when each antisubdomain antibody preparation was compared with the negative control antibodies (AcV1, anti-Lef11 polyclonal antibodies, or rabbit serum) (Fig. 3A).
FIG. 3.
Inhibition of labeled virus binding to Sf9 cells by antibodies directed against subdomains of AcMNPV GP64. (A) Antibodies raised against the indicated GP64 subdomains were analyzed for the capacity to inhibit binding of [35S]methionine-labeled wt AcMNPV BV to Sf9 cells (see Materials and Methods). (B) The same panel of subdomain-specific antibodies was used in a parallel experiment to examine their capacity to inhibit binding of [35S]methionine-labeled Ac23null AcMNPV BV to Sf9 cells. Each bar represents the average value obtained from three independent experiments with three replicates.
Because AcMNPV BV have two envelope proteins (GP64 and Ac23), which may both contribute to virus binding, we used labeled BV from an Ac23null virus (23) in the same assay to eliminate any possible effects from Ac23. When [35S]methionine-labeled Ac23null AcMNPV BV (23) were used in the binding inhibition assay, we observed significant reductions in binding after treatment with two of the antibody preparations (Fig. 3B). Binding of viruses incubated with the anti-Frag1 and anti-Frag2 polyclonal antibodies was reduced to 12 and 23%, respectively, of that for virus incubated with the negative control MAb AcV1 (14,301 cpm). Previous studies of BV attachment to host cells in the presence of AcV1 indicated that AcV1 neutralization does not result from inhibition of BV attachment but rather from a stage after initial viral adsorption (47). Additional control antibodies used in parallel confirmed that the binding inhibition by the antisubdomain antibodies was specific (Fig. 3B). These results provide indirect evidence that in addition to the role of GP64 in virion attachment, the Ac23 protein may also augment virion binding to host cells. Overall, the results from studies of both (i) neutralization of infectivity and (ii) inhibition of binding identified the Frag1 region as playing an important role in receptor binding. Therefore, we focused further studies on the region containing amino acids 21 to 159.
Virions displaying a truncated GP64 protein bind to Sf9 cells.
Antibody neutralization and binding inhibition studies indicated that the GP64 receptor binding domain was located in the N-terminal portion of the GP64 ectodomain. If this was correct, the N-terminal portion of GP64 may be capable of binding independently to receptors on the host cell. Therefore, we asked whether AcMNPV BV expressing only the N-terminal portion of the GP64 ectodomain could bind to Sf9 cells. To generate virions displaying only truncated forms of GP64, we used the strategy outlined below. Although deletion of gp64 (and most truncations of GP64) results in a severe defect in virion budding (32), we recently found that this problem could be overcome by expressing a truncated form of the VSV G protein, a construct that contains only 42 amino acids from the C-terminal portion of the ectodomain plus the TM domain and CTD. This so-called G-stem construct is sufficient to rescue the budding defect in a gp64null virus (54). Therefore, in the current study, we coinfected Sf9 cells with (i) a gp64null virus encoding the VSV G-stem construct (vAc/G-stem) and (ii) a series of gp64null viruses expressing portions of the N terminus of GP64 (Fig. 4A). Thus, defects in virion budding should be overcome by coexpression of the G-stem construct. We previously found that the selected truncated GP64 constructs were stably expressed and displayed on virions (see Fig. 3D to F in reference 54). After coinfection with vAc/G-stem and each truncated GP64 construct, virions were labeled with [35S]methionine. Labeled virions were purified and used for direct receptor binding assays. In each case, the virions displayed no wt GP64, but only truncated GP64 and the small G-stem construct. As negative and positive controls, we used gp64null virions (generated by coinfection with G-stem viruses and thus displaying G-stem only) and a virus displaying an epitope-tagged full-length GP64 construct, respectively. The results from the direct receptor binding studies (Fig. 4B) show that one truncated GP64 construct displaying 274 amino acids from the N terminus of the ectodomain (residues 21 to 294; construct 294-TM-CTD) bound to Sf9 cells in a manner similar to that of the full-length GP64 construct. In contrast, two shorter constructs, displaying 38 and 66 N-terminal amino acids of GP64 (construct 58-TM-CTD and 86-TM-CTD, respectively), bound to cells at much reduced levels, similar to that of the negative control virions displaying only the G-stem construct and no GP64 (Fig. 4B). These results implicate the N-terminal 274 amino acids (residues 21 to 294) of GP64 as the putative receptor binding region.
Anti-GST-121-160 antibodies neutralize infectivity of AcMNPV BV.
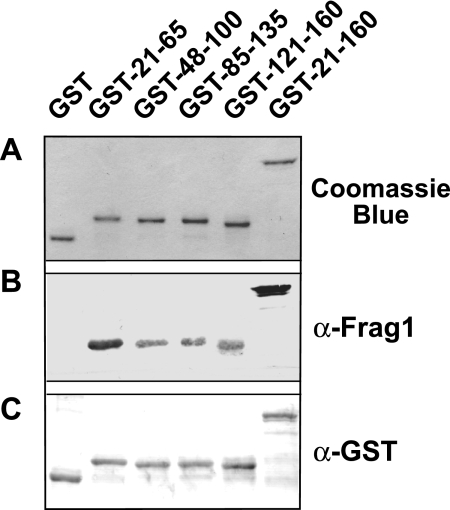
Direct binding studies showed that the ectodomain 21-274 region was sufficient to mediate virion binding (Fig. 4B). In addition, antibodies directed against the region of amino acids 21 to 160 inhibited virion binding (Fig. 3) and infectivity (Fig. 2). To further examine the role of the GP64 region of amino acids 21 to 160 in receptor binding, we generated a second panel of antibodies against this region. The region of amino acids 21 to 160 was subdivided into four partially overlapping regions, comprised of amino acids 21 to 65, 48 to 100, 85 to 135, and 121 to 160 (Fig. 1, lower diagram). Each subdomain was expressed in E. coli as a GST fusion protein, purified using affinity chromatography (Fig. 5A), and used to generate polyclonal antisera. Using Western blot analysis, we found that the rabbit polyclonal anti-Frag1 antibody preparation and polyclonal anti-GST reacted strongly with each fusion protein (Fig. 5B and C), as expected. The purified fusion proteins (GST-21-65, GST-48-100, GST-85-135, and GST-121-160) and a GST control were used to generate antibodies in guinea pigs. Assessment of antibody titers by Western blotting using the GP64 proteins demonstrated relatively high antibody titers, with effective antibody dilutions ranging from 1:5,000 to 1:10,000 (data not shown).
FIG. 5.
Analysis of purified GP64 subdomains fused with GST. (A) Purified GST fusion proteins were separated by SDS-PAGE and stained with Coomassie brilliant blue. Purified GST fusion proteins were examined by Western blot analysis with anti-Frag1 antibodies (B) and an anti-GST polyclonal antibody (C).
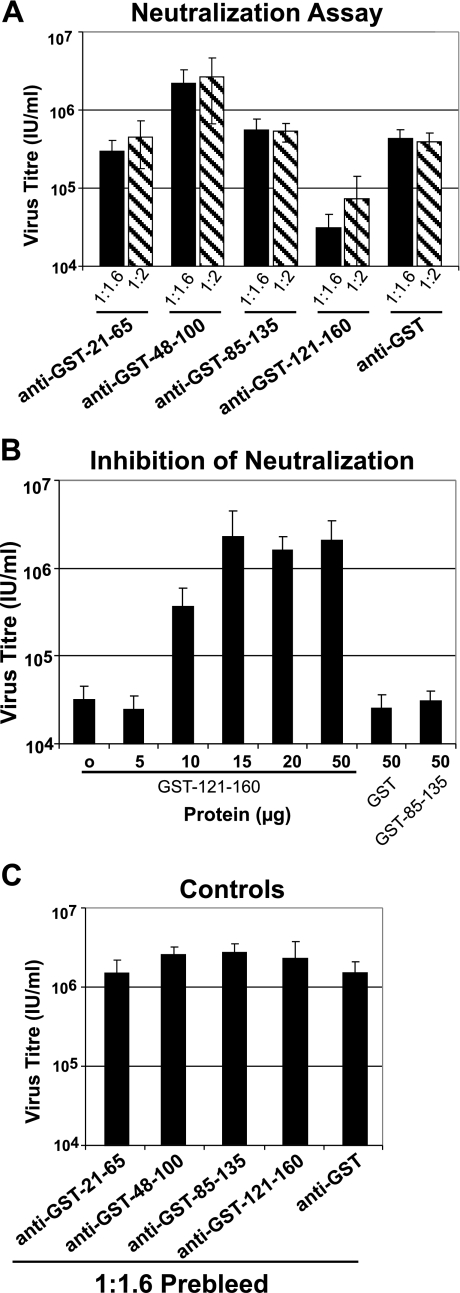
To determine whether the antibodies generated against the small-subdomain GST fusion proteins could reduce virus infectivity, we performed a neutralization assay. Using an antibody dilution of 1:1.6 or 1:2, we found that anti-GST-121-160 antibodies neutralized infectivity, while no significant reduction was detected with anti-GST-21-65, anti-GST-48-100, or anti-GST-85-135 examined at similar dilutions (Fig. 6A). These antibody dilutions of anti-GST-121-160 reduced the AcMNPV BV titer from approximately 2.08 × 106 IU/ml (as observed with the control anti-GST antibodies) to 2.98 × 104 or 6.85 × 104 IU/ml. To confirm the specificity of the neutralization by the anti-GST-121-160 antibodies, the anti-GST-121-160 antibody preparation was incubated with increasing concentrations of the GST-121-160 protein prior to incubation with virus in an assay of neutralization inhibition. While incubation with increasing concentrations of the specific protein (GST-121-160) resulted in an inhibition of neutralization (Fig. 6B), incubation with other proteins as negative controls failed to inhibit neutralization by the anti-GST-121-160 antibody preparation (Fig. 6B, GST and GST-85-135). These data confirmed that neutralization by anti-GST-121-160 antibodies resulted from recognition of the region of amino acids 121 to 160.
FIG. 6.
Analysis of AcMNPV BV neutralization by antisubdomain antibodies. (A) Neutralization assay. wt AcMNPV BV were incubated with a 1:1.6 or 1:2 dilution of the indicated subdomain-specific or control antibodies (anti-GST-21-65, anti-GST-85-135, anti-GST-48-100, anti-GST-121-160, and anti-GST) and then examined for infectivity in a neutralization assay. (B) Inhibition of neutralization. The neutralizing antibody preparation (anti-GST-121-160) was mixed with increasing quantities of the purified GST-121-160 protein or control proteins (GST and GST-85-135 [50 μg of each]), added to AcMNPV BV (1.49 × 106 IU/ml), and used to infect Sf9 cells. The titer of each BV preparation was then determined by end-point dilution assay. Each bar represents the average titer obtained for three separate experiments. (C) Controls. A neutralization assay was performed with wt AcMNPV BV and a 1:1.6 dilution of preimmune serum from each guinea pig used for the neutralization assays in panel A.
Analysis of single amino acid substitutions in the GP64 region spanning amino acids 121 to 160.
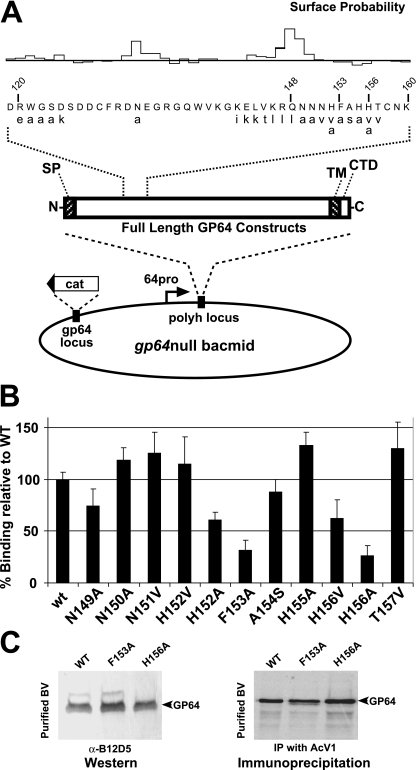
Our neutralization studies suggested that the region of the GP64 ectodomain spanning amino acids 121 to 160 may be important for GP64 interactions with the host cell receptor. Therefore, we next generated a series of single amino acid substitutions in a variety of positions in the region spanning amino acids 121 to 160 of a full-length GP64 protein construct. Each construct was used to replace wt GP64 in AcMNPV BV, and we examined BV from each recombinant virus for rescue of AcMNPV infectivity. Selected constructs were then examined for binding to host cells. For GP64 substitution mutations, we either changed the character of the amino acid (e.g., hydrophobic amino acids were replaced with hydrophilic amino acids or vice versa) or used a neutral amino acid (alanine) as a substitute. Many or most residues targeted for site-directed mutagenesis were predicted to be surface exposed (Fig. 7A). The resulting AcMNPV constructs that contained the GP64 substitution mutations were first assayed for the ability to form viral particles and to amplify infectious virus in Sf9 cells. Some substitution mutations appeared to have little or no effect on critical GP64 functions in binding, entry, or egress: GP64 proteins that contained single amino acid substitutions at positions 120 to 124, 132, 142 to 148, 155, and 157 rescued the gp64null AcMNPV, and the viruses replicated in Sf9 cells to high titers (107 to 108 IU/ml) (Fig. 7A; Table 1). These data suggested that those residues are not critical for the function of GP64. In a second group of substitution mutations (region spanning amino acids 149 to 152), the GP64 proteins were able to rescue the gp64null AcMNPV to replicate in Sf9 cells, but replication appeared to be less robust and lower titers were achieved (105 to 106 IU/ml). Finally, for substitutions at positions 152 to 154 and 156, we detected no replication in Sf9 cells. For position 152, one GP64 substitution construct (H152V) was able to rescue the gp64null virus (replicating to only moderate to low titers), while another construct (H152A) was not able to rescue the gp64null virus.
FIG. 7.
Effects of single amino acid substitutions on BV binding to Sf9 cells. (A) Strategy for generating single amino acid substitutions in full-length GP64 constructs and insertion of constructs into the polyhedrin locus of a gp64null AcMNPV bacmid. Substituted amino acids (lowercase) are shown below wt amino acids (uppercase), and numbers represent positions in the GP64 protein. (B) Analysis of binding to Sf9 cells of labeled AcMNPV BV displaying GP64 proteins with single amino acid substitution mutations. BV were labeled with [35S]methionine as described in Materials and Methods, and purified BV were analyzed for the ability to bind to Sf9 cells. Each bar represents the percent binding relative to binding by virus displaying wt GP64. The bars represent average values from three independent experiments with three replicates each. (C) (Left) Characterization of GP64 F153A and H156A mutants from purified BV by Western blot analysis with MAb B12D5. (Right) Immunoprecipitation from purified BV of [35S]methionine-labeled GP64 protein containing substitutions F153A and H156A by AcV1 antibody.
Because mutations with more severe effects on virus replication appeared to cluster in the 152-156 region, we examined viruses displaying GP64 with substitution mutations in this region in more detail. Each AcMNPV bacmid DNA with a modified gp64 gene encoding a single amino acid substitution (H152V, H152A, F153A, A154S, H155A, H156V, and H156A) in this region was transfected into Sf9Op1D cells, which express OpMNPV GP64. Each of these AcMNPV mutants was found to replicate to moderate levels in Sf9Op1D cells (Table 1, H152V, H152A, F153A, A154S, H155A, H156V, and H156A mutants). Because most of these constructs were unable to replicate in Sf9 cells yet replicated in Sf9Op1D cells, these data confirm that the observed defects were due to the substitution in the GP64 protein (Table 1).
We also asked whether low-pH-triggered membrane fusion was affected by the GP64 amino acid substitutions in the 121-160 region (Table 1). To address this, Sf9 cells were infected with each of the recombinant viruses and then assayed at 48 hpi for membrane fusion activity by triggering fusion at pH 5.0 and performing a syncytium formation assay after 2 h. wt virus-infected cells readily formed abundant and large syncytia, containing up to 10 nuclei, by 48 hpi. Substitutions at amino acid positions 120 to 124 and 142 to 150 retained >60% syncytium formation activity compared with wt GP64 (100%). In contrast, syncytium formation was reduced to <10% from GP64 substitution constructs H152A, F153A, A154S, H156V, and H156A (Table 1).
We next examined each of these constructs for incorporation of the modified GP64 protein into progeny virions. Each recombinant virus was amplified in Sf9Op1D cells, and then the resulting BV were used to infect Sf9 cells and progeny virions were labeled with [35S]methionine from 15 to 40 hpi. Labeled progeny virions were then purified, and relative BV quantities were examined by PhosphorImager analysis of virion structural protein abundance (32) and confirmed by Western blot analysis (data not shown). All of the modified GP64 proteins were incorporated into progeny BV particles, and BV were produced at levels similar to those from viruses expressing wt GP64 (data not shown).
Amino acid residues 153 and 156 are important for virus binding to Sf9 cells.
BV binding to host cells was compared among recombinant viruses displaying GP64 proteins with single amino acid substitutions in the 149-157 region. For these studies, viruses containing GP64 substitutions were amplified in Sf9Op1D cells (which constitutively express a wt GP64 protein), and virions produced in Sf9Op1D cells were used to infect Sf9 cells for the production of labeled virions displaying only the substituted GP64 construct. Labeled BV preparations were then bound to Sf9 cells, and the relative levels of BV binding were determined by liquid scintillation counting of cells (as described in Materials and Methods). The most dramatic effects on BV binding were observed for viruses expressing GP64 with single amino acid substitutions at positions 153 and 156. Comparison of relative binding revealed that binding was reduced 70% and 73% for BV displaying GP64 constructs F153A and H156A, respectively (Fig. 7B). The relative levels of GP64 in those virions showed no substantial differences from that of wt virus by Western blot or immunoprecipitation assays of purified BV (Fig. 7C). We also observed differences in binding when different amino acids were substituted at the same positions. Compared with wt AcMNPV BV binding, the H156V construct resulted in a 40% decrease in binding, while the H156A construct resulted in a more substantial 73% decrease in binding. In addition, while the H152V construct did not appear to affect binding, the H152A construct yielded an approximately 40% decrease in binding.
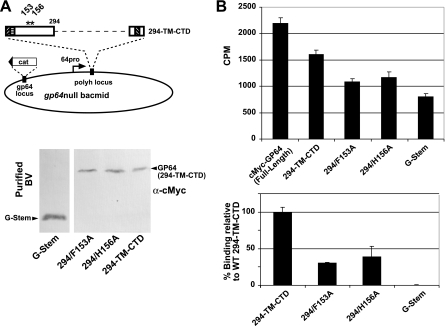
To further examine and confirm the effects of point mutations at positions 153 and 156 on virion binding, we also introduced each of the substitutions F153A and H156A into the truncated GP64 construct 294-TM-CTD (Fig. 8A, vAc/294-TM-CTD) and displayed each of these truncated GP64 constructs or BV, in the absence of wt GP64. Binding of each construct to Sf9 cells was examined as described earlier. The recombinant viruses were designated vAc294/F153A and vAc294/H156A (Fig. 8A). Examination of purified BV by Western blot analysis showed that the BV displayed the truncated GP64 constructs with substitution mutations (Fig. 8A, lower panel), and the modified truncated GP64 proteins were found on virions at levels similar to that of the unmodified truncated GP64 construct, 294-TM-CTD, indicating that no gross changes in expression or virion targeting had occurred as a result of the single amino acid substitutions. Labeled virions displaying the truncated GP64 constructs were used for direct cell binding assays as described above. Labeled BV from viruses vAc/294/F153A and vAc/294/H156A showed moderate reductions in binding compared with binding by vAc/294-TM-CTD (Fig. 8B, lower panel), which displays the unmodified truncated GP64 protein. As a negative control, we used BV containing no GP64 protein but instead displaying a stem domain derived from VSV G (virus vAc/G-stem) (54). BV from a virus (vAc/cMyc-GP64) expressing an epitope-tagged full-length GP64 protein construct was also included as a positive control, and this positive control bound to Sf9 cells at levels higher that that of the truncated GP64 construct (294-TM-CTD) (Fig. 8B, upper panel). These results further support prior data for substitution mutations in the full-length GP64 protein and indicate that amino acid residues 153 and 156 are important for receptor binding by GP64.
FIG. 8.
Analysis of the effects of single amino acid substitutions in a truncated GP64 construct on BV binding to Sf9 cells. (A) Strategy for inserting truncated GP64 constructs containing single amino acid substitutions at residues 153 and 156 into the polyhedrin locus of a gp64null AcMNPV bacmid. Truncated GP64 constructs were expressed under the control of an AcMNPV gp64 promoter. Purified BV displaying a truncated wt GP64 protein (294-TM-CTD) or similar constructs with single amino acid substitutions (294/F153A and 294/H156A) were examined by Western blot analysis with anti-c-Myc antibody. Purified BV from a control gp64null virus expressing a c-Myc-tagged G-stem construct were also included as a control (left lane). (B) Analysis of Sf9 cell binding by labeled purified BV displaying truncated GP64 constructs or controls, as shown in panel A. BV were labeled with [35S]methionine, purified, and used in binding assays as described in Materials and Methods. Each bar represents the average value for three independent experiments with three replicates. The top panel shows cpm values measured from BV bound to Sf9 cells and compares binding by each construct with control binding by a virus displaying a c-Myc-tagged full-length wt GP64 (cMyc-GP64, full-length). The bottom panel illustrates data from the same experiment but normalized to binding by the truncated wt GP64 construct (100%) and negative control BV displaying only the G-stem construct (0%).
For these studies, we examined the role of GP64 in receptor binding by a combination of approaches. Antisubdomain antibodies were used to examine (i) neutralization of virion infectivity and (ii) inhibition of virion binding and to map a putative receptor binding domain in the N-terminal region of GP64. To confirm the role of the GP64 N terminus in receptor binding, we displayed truncated forms of GP64 on the virion and examined binding directly. Finally, we used site-directed mutations in both full-length and truncated forms of GP64 to identify specific residues which are important for virion binding.
DISCUSSION
To identify GP64 domains that participate in the interaction of GP64 with the host cell receptor, our approaches included (i) antibody neutralization of infection and inhibition of binding, (ii) display of GP64 domains on virions and analysis of binding, and (iii) mutagenesis and functional analysis of a candidate receptor binding domain. In several prior studies, neutralizing MAbs that recognize GP64 were identified (10, 40, 47, 48). In the case of the best-studied anti-GP64 MAb, the neutralizing MAb (AcV1) inhibited virus entry, but at a step following virus attachment (10, 47, 48, 55). In other cases, the mechanism of neutralization was not determined, and those hybridoma lines are no longer available (40). To examine receptor binding in the current study, we subdivided the GP64 ectodomain into four partially overlapping subdomains (Fig. 1) and generated polyclonal antibodies against these major subdomains. The antibodies were examined for the capacity to neutralize AcMNPV BV infectivity and to inhibit virus binding to host cells. Polyclonal antibodies directed against one of the four subdomains (Frag1; amino acids 21 to 159) strongly neutralized AcMNPV BV infectivity (Fig. 2). Although the neutralizing efficiency of anti-Frag1 antibodies was similar to that of MAb AcV1, these antibodies recognize different regions of the GP64 ectodomain. We previously mapped the AcV1 epitope to a 24-amino-acid region in the central variable domain of GP64 (amino acids 273 to 294) (55), a region that is included in our current Frag3 subdomain (amino acids 209 to 352). However, anti-Frag3 antibodies did not neutralize infectivity. This may be explained by the fact that the AcV1 MAb recognizes a conformational epitope (55), whereas the anti-Frag3 antibodies were generated against a peptide expressed in E. coli and purified under denaturing conditions.
To determine if neutralization by anti-Frag1 antibodies resulted from an inhibition of BV binding, [35S]methionine-labeled BV were preincubated with antibodies and then incubated with Sf9 cells in a binding assay. The initial binding assay performed with labeled wt AcMNPV BV showed no effect of the polyclonal antibodies on BV binding (Fig. 3A). However, when labeled Ac23null AcMNPV BV were used in the binding assay, anti-Frag1 and anti-Frag2 antibodies both inhibited BV binding (Fig. 3B). Previous studies showed that Ac23, an F protein homolog, is found in BV envelopes but is not essential and does not appear to mediate membrane fusion (23, 29). However, because F proteins from baculoviruses of the group II NPVs presumably have an attachment function, we reasoned that Ac23 may retain a receptor binding function. Our observation that these anti-GP64 antibodies inhibited binding of BV from the Ac23null virus but not from wt AcMNPV may suggest that Ac23 retains some cell binding activity. While this is the first study to provide data suggestive of a role for the Ac23 protein in host cell binding, additional, more-detailed studies will be necessary to confirm any possible role for Ac23 in virus binding and to determine what significance, if any, such a function may have in the AcMNPV infection cycle. Any such function would appear to be redundant and nonessential in AcMNPV, since Ac23null AcMNPV replicates to high titers in cell culture and provides a robust infection in larvae of the host Trichoplusia ni (23). In addition, GP64 alone has been used to pseudotype retroviruses and a paramyxovirus (19, 20, 30, 31, 41, 43) and therefore would clearly not require an additional receptor binding partner to mediate binding and entry.
To more directly examine virion binding and to confirm the role of the GP64 N-terminal domain in virion binding, portions of the GP64 ectodomain were displayed on virus particles and examined for the ability to mediate virus binding to Sf9 cells. We found that a construct containing the N-terminal 274 amino acids of the ectodomain (residues 21 to 294) mediated binding to Sf9 cells, whereas virions displaying smaller portions (38 and 66 amino acids) from the N terminus of the GP64 ectodomain did not efficiently bind to Sf9 cells (Fig. 4B). These data show directly that the N-terminal region of GP64 mediates binding to Sf9 cells. Combined with antibody neutralization and binding inhibition studies, these data strongly implicate the N terminus as the GP64 receptor binding domain.
Since anti-Frag1 antibodies neutralized infectivity and inhibited binding, we selected the region spanning amino acids 21 to 160 for further investigation. Antibodies were generated against four partially overlapping subdomains of this region, each of which was approximately 40 to 50 amino acids (Fig. 1B). Only antibodies directed against the 121-160 region of GP64 neutralized infectivity (Fig. 6A) compared with control anti-GST antibodies or other subdomain-specific antibodies. We also confirmed that neutralization was specific to the 121-160 region of GP64 by using an neutralization-of-inhibition assay (Fig. 6B). Thus, the effect of the anti-GST-121-160 antibody is indeed specific to the 121-160 region of the GP64 ectodomain.
We next examined individual amino acid positions in the 120-160 region of the GP64 ectodomain, using single amino acid substitutions at 22 selected positions (Fig. 7A). Full-length GP64 constructs, each containing a single amino acid substitution in 1 of the 22 positions, were inserted into a gp64null AcMNPV background such that only the modified GP64 was displayed on progeny virions. The effects of these mutations were examined by (i) determining whether the resulting virus replicated in Sf9 cells, (ii) measuring infectious virus titers for each, (iii) examining membrane fusion mediated by each GP64 construct, and (iv) examining virion binding to Sf9 cells (Table 1 and Fig. 7B). A large group of substitutions (positions 120 to 124, 132, 142 to 148, 155, and 157) had no substantial effect on virus replication in Sf9 cells and showed robust membrane fusion (Table 1). A second group (positions 149 to 152) replicated in Sf9 cells but produced lower titers of infectious virus. Fusion activity in this group was generally robust (>60% of wt fusion), although a substitution at position 151 resulted in less fusion (<20% of wt fusion). Viruses with a third group of substitutions (positions 152, 153, 154, and 156) were unable to replicate in Sf9 cells, and their fusion activity was very low (<10% of wt fusion). When viruses displaying proteins containing substitutions in this region (amino acids 149 to 157) were examined in virion binding assays, we found that substitutions at positions 153 and 156 had a substantial negative effect on virion binding to Sf9 cells (Fig. 7B). Thus, positions 153 and 156 were identified as important in virion binding. To further confirm this result, the same substitutions (F153A and H156A) were generated in the truncated GP64 construct that was previously found to contain the receptor binding domain (Fig. 4A, 294-TM-CTD), and the negative effects of these substitutions on binding were confirmed (Fig. 8). Thus, substitution mutations at positions 153 and 156 had a similar effect on virion binding in virions displaying either full-length GP64 or a truncated GP64 construct that contains the N-terminal receptor binding domain.
In the current study, we identified a portion of the N terminus of GP64 that is important for virion binding to host cells, thus implicating the N terminus of GP64 as the putative receptor binding domain. The identification of the receptor binding domain near the N terminus of the GP64 protein is not necessarily unexpected, since a number of well-studied viral envelope proteins (influenza virus hemagglutinin [HA], human immunodeficiency virus gp120, and coronavirus spike protein) contain receptor binding domains near their N termini. One of the best-studied virus-receptor interactions is the influenza virus HA protein and its interactions with sialic acid, the cellular receptor. In the case of HA, some single amino acid substitution mutations at or near the primary receptor binding pocket have little effect on virion binding to erythrocytes. Only three amino acid substitutions (at positions 98, 183, and 194) are known to abolish erythrocyte binding (44). HA also contains a second sialic acid binding site, located in the interface between monomers of the HA trimer, and that site dissociates upon exposure to low pH and conformational change. It is unclear whether the second binding site plays any significant role in the virus infection cycle. In the case of the two amino acid positions identified in the GP64 protein, substitutions resulted in reduced binding, but binding was not eliminated. Thus, it is unlikely that these changes completely removed the binding capacity of GP64. Indeed, binding by GP64 may be complex, and it is possible that the effects of these amino acid substitutions could be either direct (involving direct contacts between GP64 and the cellular ligand) or indirect (perhaps affecting conformational changes necessary for ligand binding at another site).
Although similar in activity and general structural aspects to other membrane fusion proteins that have a receptor binding function, GP64 also has several unusual characteristics. For example, GP64 monomers are disulfide linked to each other in the trimer, and the monomer does not require processing by cleavage for its function. GP64 appears to bind promiscuously to many cell types (3, 42), and this feature, combined with robust membrane fusion activity (16, 27, 37-39), has led to applications of baculoviruses as mammalian transduction vectors (4, 18). In addition, baculoviruses are being examined as possible gene therapy vectors, and GP64 has been used to pseudotype retrovirus vectors for human gene therapy (1, 5, 9, 19, 24, 43, 45, 49). Thus, a better understanding of GP64-mediated receptor binding and entry will be important not only for understanding its function in the infection of host insect cells but also for future development of a variety of applications.
Acknowledgments
We thank Joshua Huffer and Gerrit Heetderks for assistance with virus construction and for expert technical assistance.
This work was supported by a grant from the NIH (RO1 AI33657) and is Boyce Thompson Institute project 1255.
Footnotes
Published ahead of print on 20 February 2008.
REFERENCES
- 1.Airenne, K. J., M. O. Hiltunen, M. P. Turunen, A. M. Turunen, O. H. Laitinen, M. S. Kulomaa, and S. Yla-Herttuala. 2000. Baculovirus-mediated periadventitial gene transfer to rabbit carotid artery. Gene Ther. 71499-1504. [DOI] [PubMed] [Google Scholar]
- 2.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 666829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, F. M., and N. L. R. Bucher. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 932348-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condreay, J. P., S. M. Witherspoon, W. C. Clay, and T. A. Kost. 1999. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. USA 96127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facciabene, A., L. Aurisicchio, and N. La Monica. 2004. Baculovirus vectors elicit antigen-specific immune responses in mice. J. Virol. 788663-8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granados, R. R., and K. A. Lawler. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108297-308. [DOI] [PubMed] [Google Scholar]
- 7.Granados, R. R., and K. A. Williams. 1986. In vivo infection and replication of baculoviruses, vol. I. Biological properties and molecular biology. CRC Press, Boca Raton, FL.
- 8.Hefferon, K., A. Oomens, S. Monsma, C. Finnerty, and G. Blissard. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258455-468. [DOI] [PubMed] [Google Scholar]
- 8a.Hink, W. F. 1970. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature 226466-467. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann, C., W. Lehnert, and M. Strauss. 1998. The baculovirus vector system for gene delivery into hepatocytes. Gene Ther. Mol. Biol. 1231-239. [Google Scholar]
- 10.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125432-444. [DOI] [PubMed] [Google Scholar]
- 11.Horton, H. M., and J. P. Burand. 1993. Saturable attachment sites for polyhedron-derived baculovirus on insect cells and evidence for entry via direct membrane fusion. J. Virol. 671860-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.IJkel, W. F. J., M. Westenberg, R. W. Goldbach, G. W. Blissard, J. M. Vlak, and D. Zuidema. 2000. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 27530-41. [DOI] [PubMed] [Google Scholar]
- 13.Jarvis, D. L., L. Wills, G. Burow, and D. A. Bohlmeyer. 1998. Mutational analysis of the N-linked glycans on Autographa californica nucleopolyhedrovirus gp64. J. Virol. 729459-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keddie, B. A., G. W. Aponte, and L. E. Volkman. 1989. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science 2431728-1730. [DOI] [PubMed] [Google Scholar]
- 15.Keddie, B. A., and L. E. Volkman. 1985. Infectivity difference between the two phenotypes of Autographa californica nuclear polyhedrosis virus: importance of the 64k envelope glycoprotein. J. Gen. Virol. 661195-1200. [Google Scholar]
- 16.Kingsley, D. H., A. Behbahani, A. Rashtian, G. W. Blissard, and J. Zimmerberg. 1999. A discrete stage of baculovirus GP64-mediated membrane fusion. Mol. Biol. Cell 104191-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko, J.-K., and J. Ma. 2005. A rapid and efficient PCR-based mutagenesis method applicable to cell physiology study. Am. J. Physiol. Cell Physiol. 2881273-1278. [DOI] [PubMed] [Google Scholar]
- 18.Kost, T. A., J. P. Condreay, and D. L. Jarvis. 2005. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 23567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kremer, K. L., K. R. Dunning, D. W. Parsons, and D. S. Anson. 2007. Gene delivery to airway epithelial cells in vivo: a direct comparison of apical and basolateral transduction strategies using pseudotyped lentivirus vectors. J. Gene Med. 9362-368. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, M., B. P. Bradow, and J. Zimmerberg. 2003. Large-scale production of pseudotyped lentiviral vectors using baculovirus GP64. Hum. Gene Ther. 1467-77. [DOI] [PubMed] [Google Scholar]
- 21.Long, G., X. Pan, M. Westenberg, and J. M. Vlak. 2006. Functional role of the cytoplasmic tail domain of the major envelope fusion protein of group II baculoviruses. J. Virol. 8011226-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lung, O., M. Westenberg, J. M. Vlak, D. Zuidema, and G. W. Blissard. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 765729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lung, O. Y., M. Cruz-Alvarez, and G. W. Blissard. 2003. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J. Virol. 77328-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makela, A. R., H. Matilainen, D. J. White, E. Ruoslahti, and C. Oker-Blom. 2006. Enhanced baculovirus-mediated transduction of human cancer cells by tumor-homing peptides. J. Virol. 806603-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markovic, I., H. Pulyaeva, A. Sokoloff, and L. V. Chernomordik. 1998. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J. Cell Biol. 1431155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, L. K. (ed.). 1997. The baculoviruses. Plenum Press, New York, NY.
- 27.Monsma, S. A., and G. W. Blissard. 1995. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 envelope fusion protein. J. Virol. 692583-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monsma, S. A., A. G. P. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 704607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okano, K., A. L. Vanarsdall, V. S. Mikhailov, and G. F. Rohrmann. 2006. Conserved molecular systems of the Baculoviridae. Virology 34477-87. [DOI] [PubMed] [Google Scholar]
- 30.Oomens, A. G., and G. W. Wertz. 2004. The baculovirus GP64 protein mediates highly stable infectivity of a human respiratory syncytial virus lacking its homologous transmembrane glycoproteins. J. Virol. 78124-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oomens, A. G., and G. W. Wertz. 2004. trans-Complementation allows recovery of human respiratory syncytial viruses that are infectious but deficient in cell-to-cell transmission. J. Virol. 789064-9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254297-314. [DOI] [PubMed] [Google Scholar]
- 33.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors, a laboratory manual. W. H. Freeman and Co., New York, NY.
- 34.Pearson, M. N., C. Groten, and G. F. Rohrmann. 2000. Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J. Virol. 746126-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, M. N., and G. F. Rohrmann. 2002. Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J. Virol. 765301-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson, M. N., R. L. Russell, and G. F. Rohrmann. 2001. Characterization of a baculovirus-encoded protein that is associated with infected-cell membranes and budded virions. Virology 29122-31. [DOI] [PubMed] [Google Scholar]
- 37.Plonsky, I., M. S. Cho, A. G. P. Oomens, G. W. Blissard, and J. Zimmerberg. 1999. An analysis of the role of the target membrane on the gp64-induced fusion pore. Virology 25365-76. [DOI] [PubMed] [Google Scholar]
- 38.Plonsky, I., and J. Zimmerberg. 1994. Direct measurement of fusion pore conductance during virus-induced cell-cell fusion. Biophys. J. 666-10. [Google Scholar]
- 39.Plonsky, I., and J. Zimmerberg. 1996. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. J. Cell Biol. 1351831-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts, S. R., and J. S. Manning. 1993. The major envelope glycoprotein of the extracellular virion of Autographa californica nuclear polyhedrosis virus possesses at least three distinct neutralizing epitopes. Virus Res. 28285-297. [Google Scholar]
- 41.Sastre, P., A. G. Oomens, and G. W. Wertz. 2007. The stability of human respiratory syncytial virus is enhanced by incorporation of the baculovirus GP64 protein. Vaccine 255025-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoji, I., H. Aizaki, H. Tani, K. Ishii, T. Chiba, I. Saito, T. Miyamura, and Y. Matsuura. 1997. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J. Gen. Virol. 782657-2664. [DOI] [PubMed] [Google Scholar]
- 43.Sinn, P. L., E. R. Burnight, M. A. Hickey, G. W. Blissard, and P. B. McCray, Jr. 2005. Persistent gene expression in mouse nasal epithelia following feline immunodeficiency virus-based vector gene transfer. J. Virol. 7912818-12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69531-569. [DOI] [PubMed] [Google Scholar]
- 45.Tani, H., C. K. Limn, C. C. Yap, M. Onishi, M. Nozaki, Y. Nishimune, N. Okahashi, Y. Kitagawa, R. Watanabe, R. Mochizuki, K. Moriishi, and Y. Matsuura. 2003. In vitro and in vivo gene delivery by recombinant baculoviruses. J. Virol. 779799-9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkman, L. E., and P. A. Goldsmith. 1984. Budded Autographa californica NPV 64K protein: further biochemical analysis and effects of postimmunoprecipitation sample preparation conditions. Virology 139295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volkman, L. E., and P. A. Goldsmith. 1985. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143185-195. [DOI] [PubMed] [Google Scholar]
- 48.Volkman, L. E., P. A. Goldsmith, R. T. Hess, and P. Faulkner. 1984. Neutralization of budded Autographa californica NPV by a monoclonal antibody: identification of the target antigen. Virology 133354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, C. Y., F. Li, Y. Yang, H. Y. Guo, C. X. Wu, and S. Wang. 2006. Recombinant baculovirus containing the diphtheria toxin A gene for malignant glioma therapy. Cancer Res. 665798-5806. [DOI] [PubMed] [Google Scholar]
- 50.Wang, P., D. A. Hammer, and R. R. Granados. 1997. Binding and fusion of Autographa californica nucleopolyhedrovirus to cultured insect cells. J. Gen. Virol. 783081-3089. [DOI] [PubMed] [Google Scholar]
- 51.Westenberg, M., F. Veenman, E. C. Roode, R. W. Goldbach, J. M. Vlak, and D. Zuidema. 2004. Functional analysis of the putative fusion domain of the baculovirus envelope fusion protein F. J. Virol. 786946-6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westenberg, M., H. Wang, W. F. IJkel, R. W. Goldbach, J. M. Vlak, and D. Zuidema. 2002. Furin is involved in baculovirus envelope fusion protein activation. J. Virol. 76178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickham, T. J., M. L. Shuler, D. A. Hammer, R. R. Granados, and H. A. Wood. 1992. Equilibrium and kinetic analysis of Autographa californica nuclear polyhedrosis virus attachment to different insect cell lines. J. Gen. Virol. 733185-3194. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, J., and G. W. Blissard. 2008. Display of heterologous proteins on gp64null baculovirus virions and enhanced budding mediated by a vesicular stomatitis virus G-stem construct. J. Virol. 821368-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, J., and G. W. Blissard. 2006. Mapping the conformational epitope of a neutralizing antibody (AcV1) directed against the AcMNPV GP64 protein. Virology 352427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]