Abstract
We performed an extensive two-dimensional differential in-gel electrophoresis proteomic analysis of the cellular changes in human T cells upon human immunodeficiency virus type 1 (HIV-1) infection. We detected 2,000 protein spots, 15% of which were differentially expressed at peak infection. A total of 93 proteins that changed in relative abundance were identified. Of these, 27 were found to be significantly downregulated and 66 were upregulated at peak HIV infection. Early in infection, only a small group of proteins was changed. A clear and consistent program of metabolic rerouting could be seen, in which glycolysis was downregulated and mitochondrial oxidation enhanced. Proteins that participate in apoptotic signaling were also significantly influenced. Apart from these changes, the virus also strongly influenced levels of proteins involved in intracellular transport. These and other results are discussed in light of previous microarray and proteomic studies regarding the impact of HIV-1 infection on cellular mRNA and protein content.
Human immunodeficiency virus type 1 (HIV-1) infection influences many physiological processes in the infected individual, ultimately leading to the development of AIDS. In order to replicate efficiently, the virus has to adapt the intracellular metabolism of the host cell. With the help of its accessory proteins, HIV-1 is able to alter many physiological aspects of both the infected cells and the organism as a whole (17, 39, 47). HIV-1 disturbs immune responses, changes infected cells so that it can survive and persist, and prepares the cells for production and release of new viral particles. The intracellular processes influenced by HIV infection have been the subject of intense research, including studies that focus mainly on the role of HIV-1 accessory proteins in these processes and more general studies using microarrays to detect mRNA changes in the cell (11, 16, 42, 51). The proteome changes, however, have only recently begun to be studied in detail (9).
Coordinated gene expression patterns characterize cell types and specific cellular responses. Changes are often accompanied by coordinated protein modifications. A gene only encodes the amino acid sequence of a protein product, which can exist in many different isoforms with different functions. Thus, a relatively limited number of genes can give rise to an enormous number of functionally distinct proteins (23). This is for the most part achieved by posttranslational protein modification (PTM). PTM provides cells with very agile systems to adapt swiftly to changing environments, e.g., as exemplified by the key role of protein phosphorylation in signal transduction or intracellular transport. Such PTM responses, however, cannot be directly detected using DNA technologies. Indeed, there often seems to be a discrepancy between experimental results based on analysis of products of transcription versus translation (40). Therefore, proteomic studies of many proteins using techniques that can detect subtle differences in protein expression and discriminate between different protein isoforms are crucial for the characterization of the multitude of intracellular responses to HIV infection (51). Proteomic studies can thus give rise to new insights that can eventually lead to new intervention strategies.
In this study, we used the fluorescence two-dimensional differential in-gel electrophoresis (2D-DIGE) technique to compare the differences between uninfected and HIV-1-infected T cells. The DIGE technique is based on protein labeling with cyanine-based fluorescent probes and subsequent 2D gel electrophoresis to quantify changes in protein expression (1, 50). Identities of differentially expressed proteins were subsequently determined by matrix-assisted laser desorption mass spectrometry. We found general correlations at the protein level with results from previous studies using microarray-based techniques (11, 16, 42, 51). Using annotated databases, we identified classes of proteins and general pathways that are altered upon HIV infection. This proteomic approach confirmed at the protein level several HIV-1 effects on pathways and processes that previously have been described using other techniques (9). Furthermore, it allowed us to reinterpret and extend earlier findings, identifying additional processes hitherto only suspected to be involved in HIV infection.
MATERIALS AND METHODS
Cells and viruses.
C33A cervical carcinoma cells (ATCC HTB) (31, 2) were grown as a monolayer in Dulbecco's minimal essential medium supplemented with 10% (vol/vol) fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin, 20 mM glucose, and minimal essential medium nonessential amino acids at 37°C and 5% CO2. The cells were transfected by the calcium phosphate method as described previously (13).
The human T-lymphocyte cell line PM1 (31) was cultured in advanced RPMI 1640 (Gibco BRL, Gaithersburg, MD) supplemented with 1% (vol/vol) FCS, 20 U/ml penicillin, and 20 μg/ml streptomycin at 37°C and 5% CO2.
HIV-1 infections were performed with C33A-produced virus stocks of the HIV-1 LAI molecular clone (38). For peak-infection analysis, infection was started with virus corresponding to 500 ng CA-p24 in a 20-ml cell culture. Cells were maintained and increased to 250-ml cultures with around 50 to 90 million cells. The infection grade was monitored by fluorescence-activated cell sorter (FACS) analysis, and the cells were harvested if at least 85% of the cells were positive for intracellular CA-p24 (7 to 10 days). For the massive 42-h infections, around 50 to 90 million uninfected cells were collected by centrifugation (5 min, 400 × g) and resuspended in the supernatant from the peak-infection culture. These infections were maintained for 42 h before harvesting of the cells. Aliquots were taken for intracellular CA-p24 FACS staining and supernatant CA-p24 enzyme-linked immunosorbent assay (ELISA).
Sample preparation.
Cells were harvested by centrifugation (5 min, 400 × g), washed once with 50 ml phosphate-buffered saline (PBS) (Gibco), and resuspended in 1 ml PBS. The cell suspension was transferred to a 2-ml Eppendorf tube and the cells collected by centrifugation (4 min, 4,000 rpm, Eppendorf centrifuge). The cell pellets were lysed at a ratio of 107 cells in 50 μl of lysis buffer (30 mM Tris buffer [pH 8.5] containing 7.7 M urea, 2.2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate; Sigma}, and 2 mM dithiothreitol [DTT]). Protein concentrations were determined using the 2-D Quant kit according to the manufacturer's instructions (GE Healthcare), and protein concentrations were adjusted in lysis buffer to a final concentration of 1.5 mg/ml.
CA-p24 internal staining and FACS analysis.
Flow cytometry was performed with RD1 or fluorescein isothiocyanate-conjugated mouse monoclonal anti-CA-p24 (clone KC57; Coulter). Cells from a 1-ml culture sample were collected (4 min, 4,000 rpm, Eppendorf centrifuge) and fixed in 250 μl 4% formaldehyde for 5 min at room temperature. The cells were washed with 500 μl BD Perm/Wash buffer (BD Pharmingen) and stained for 30 min at 4°C in 20 μl of BD Perm/Wash buffer and 5 μl of the appropriate antibody (1 in 100 diluted). Excess antibody was removed by washing the cells with 500 μl BD Perm/Wash buffer. The cells were collected, and 750 μl FACS buffer (PBS with 2% FCS) was added. The cells were analyzed on a FACSCalibur flow cytometer with CellQuest Pro software (BD Biosciences, San Jose, CA). Cell populations were defined based on forward/sideward scattering and isotype controls (clone MsIgG-RD1; Coulter) were used to set markers.
CA-p24 ELISA.
Culture supernatant was heat inactivated at 56°C for 30 min in the presence of 0.05% Empigen-BB (Calbiochem, La Jolla, CA). The CA-p24 concentration was determined by a twin-site ELISA with D7320 (Biochrom, Berlin, Germany) as the capture antibody and alkaline phosphatase-conjugated anti-p24 monoclonal antibody (EH12-AP) as the detection antibody. Detection was done with the Lumiphos Plus system (Lumigen, MI) in a LUMIstar Galaxy (BMG Lab Technologies, Offenburg, Germany) luminescence reader. Recombinant CA-p24 expressed in a baculovirus system was used as the reference standard.
Protein labeling with DIGE fluorophores.
Sixty micrograms of protein sample was labeled according to the manufacturer's protocol with 3,200 pmol N-hydroxysuccinimide esters of cyanine dyes (Cy2, Cy3, or Cy5 Cy dye; GE Healthcare) for 30 min in the dark on ice. For each experimental time point, six biological replicates of infected and uninfected control cells were Cy dye labeled, including a dye swap so that three replicates of infected and control cells were labeled with Cy3 and Cy5, respectively, and three replicates with Cy5 and Cy3, respectively. An internal standard of 60 μg protein consisting of combined equimolar amounts of protein from all six biological replicates of infected and control cells was labeled with Cy2. After quenching the labeling reaction with 8 μl of 10 mM lysine solution (Sigma), the labeled proteins from control cells, infected cells, and the internal standard were mixed together and adjusted to end concentrations of 1% DTT and 1% IPG buffer in a total volume of 150 μl with lysis buffer.
2D gel electrophoresis.
First-dimension isoelectric focusing (IEF) of the samples was performed using 24-cm precast IPG strips (pH 3 to 11, nonlinear [NL]; GE Healthcare). Samples of 150 μl were applied by anodic cup loading onto the strips that had been rehydrated overnight with 450 μl rehydration buffer containing 30 mM Tris (pH 8.5), 7.7 M urea, 2.2 M thio-urea, 2 mM DTT, 4% CHAPS 0.5% IPG (pH 3 to 11, NL; GE Healthcare), and 1.2% Destreak reagent (GE Healthcare). IEF of all six strips of one experiment containing the six experimental replicates was carried out in parallel with the IPGPhor equipped with the Ettan Cup loading manifold (GE Healthcare). The strips were focused at 0.05 mA/IPG strip for approximately 55 kV·h at 20°C following a stepwise increase to 150 V for 3 h, a stepwise increase to 300 V for 3 h, a gradient to 1,000 V for 6 h, a gradient of 8,000 V for 5 h, a constant step of 8,000 V for 3 h, and a gradient decrease to 500 V for 0.5 h. After IEF, the strips were sequentially incubated in freshly prepared solutions of 1% DTT and 2% iodoacetamide in 50 mM Tris (pH 8.0), 6 M urea, 30% glycerol, and 2% sodium dodecyl sulfate (SDS) for 15 min. The second-dimension SDS-polyacrylamide gel electrophoresis was performed by mounting the IPG strips onto 12% polyacrylamide gels between low-fluorescent glass plates and running the gels in the Ettan Dalt-six electrophoresis system (GE Healthcare) for 30 min at 45 mA/gel and overnight at 13 mA/gel at 20°C.
Image acquisition and analysis.
After 2D electrophoresis, the gels were scanned on a Typhoon Trio image scanner (GE Healthcare) according to the manufacturer's protocol at 100-μm resolution to produce a Cy2, a Cy3, and a Cy5 image for each gel. After cropping with ImageQuant software (GE Healthcare), the images were subjected to automated difference-in-gel analysis and biological variation analysis (BVA) using Decyder version 6.5 software (GE Healthcare).
Protein identification.
After the gel images were acquired, the gels were removed from the glass plates, fixed with 50% methanol and 5% acetic acid, and stained with Coomassie brilliant blue R250 in 50% methanol and 2% acetic acid overnight to visualize the protein spots for isolation and subsequent protein identification. Protein-containing gel spots were digested with unmodified trypsin (Roche Molecular Biochemicals, sequencing grade) and extracted as described by Shevchenko et al. (46). Extracted peptides were concentrated using a Speedvac, and the pellet was taken up in 6 μl of 1% formic acid and 60% acetonitrile. A 0.6-μl portion of the peptide solution was mixed with an equal volume of 10 mg/ml α-cyano-4-hydroxycinnamic acid (Sigma Chemical Co.) solution in acetonitrile-ethanol (1:1, vol/vol) with 1% trifluoroacetic acid and 1 mM ammonium acetate. Before dissolving, the α-cyano-4-hydroxycinnamic acid was washed briefly with acetone. Then, 1.2 μl was spotted on target and allowed to dry at room temperature. Matrix-assisted laser desorption ionization-time-of-flight mass spectra were acquired on a Micromass M@LDI (Micromass, Wythenshawe, United Kingdom). The resulting peptide spectra were used to search a nonredundant protein sequence database (Swiss-Prot/TREMBL) using the Proteinprobe program or using the MASCOT search engine and database.
For annotation, GoMiner (http://discover.nci.nih.gov/gominer/), was used as well as searches in the Swiss-Prot (http://www.expasy.org) and Ensemble (http://www.ensembl.org) databases and PubMed literature (http://www.ncbi.nlm.nih.gov/Literature/).
RESULTS
Experimental setup.
To analyze the influence of HIV-1 infection on the cellular host, a 2D-DIGE proteomic approach was set up. As the host cells, we used a uniform cell population of the human T-cell line PM1 because these cells do not display syncytium formation upon virus infection, which could induce unwanted secondary effects. PM1 cells were infected with a virus stock of the molecular clone LAI (38).
For protein analysis, uniform samples contribute to sensitivity. This is particularly important for the analysis of virus infections in which asynchronous replication and multiple rounds of infection easily can lead to masking of protein changes. On the other hand, single-round infections with relative high virus titers will minimize such masking but will lead to rapid cytopathic effects and may reduce detection of long-term cellular responses. Furthermore, aspecific short-term stress responses can affect the results. Therefore, two different time points in an HIV-1 infection were analyzed: one at the peak of infection (around 7 to 10 days after inoculation) and one for which we used a massive amount of virus and harvested the cells after 42 h. Importantly, we did not observe significant differences in amounts of apoptotic cells between infected and uninfected cell populations (as judged by forward and side scatter plots in the FACS analyses) in either of these settings. The peak-infection experiment resembles a spreading infection and was designed to detect long-term effects of HIV-1 replication. The 42-h infection was used to detect acute infection changes resulting from a massive synchronous infection in a single round of replication. In principle, a comparison of the two experiments would allow us to monitor proteomic changes over time. FACS analyses of PM1 cells with anti-CA-p24 (as an intracellular indicator of viral production) before infection, after 42 h, and at the peak of regular infection are shown in Fig. 1A, B, and C, respectively. This demonstrates that all cells are producing the viral Gag protein.
FIG. 1.
FACS analysis of PM1 cells after intracellular staining against HIV-1 CA-p24 without infection (A), at the peak of a regular infection (B), and after 42 h of infection (C). Dot plot analysis with unstained FL1 (y axis) and anti-CA-p24 fluorescence (KC57-RD1) (x axis) is shown. For details, see Materials and Methods.
Proteome changes at peak HIV-1 infection of PM1 cells.
To determine accurately which proteins would be differentially expressed at peak infection of PM1 cells we performed DIGE analysis on six different batches of control cells and six different batches of cells at peak infection. Figure 2 shows a representative Cy2 image of one of the six gels (the master gel). Using the difference-in-gel analysis module of the De-cyder software (GE Healthcare), ratios between expression of proteins in control and infected cells were determined, whereas the biological variance analyses module was used on the six biological replicates of control and HIV-1-infected cells to determine which differential expression ratios are considered significant (i.e., to calculate the P value).
FIG. 2.
2D gel analysis of HIV-1-infected PM1 cells. A representative Cy2 image of a 2D gel of PM1 cells at the peak of a regular infection is shown. First dimension, IEF pH 3 to 11 NL (left to right); second dimension, SDS—12% polyacrylamide gel electrophoresis. For further details see Materials and Methods.
Out of 1,921 detected protein spots, 288 were designated to be significantly differentially expressed at peak HIV infection using the following criteria.
A spot had to be present in at least 9 of the 18 Cy dye images of the peak-infection experiment and changed (up- or downregulated) at least 1.5-fold with a P value of <0.01 (which applied to 241 spots), changed at least 1.1-fold with a P value of <0.001 (adding a further 23 spots), or changed at least 2.0-fold with a P value of <0.2 (adding a final 24 spots).
Of these 288 differentially expressed protein spots, 106 contained enough protein to allow identification by peptide mass fingerprinting. One hundred two protein spots yielded 92 unique proteins, as several proteins were found more than once, and four gel spots contained more than one protein. Of the 92 identified proteins, 27 were significantly downregulated and 65 were upregulated at peak HIV infection. One of the upregulated proteins was identified as the HIV capsid protein CA-p24. In an earlier experiment, we were also able to identify the HIV accessory protein Nef (data not shown). A complete list of all differentially expressed proteins at HIV peak infection with their ratios and P values can be found in Table S1 in the supplemental material.
Differentially expressed proteins after single-cycle HIV-1 infection of PM1 cells.
At 42 h after massive HIV infection, only 12 protein spots out of 1,955 were designated to be significantly differentially expressed using the following criteria.
A spot had to be present in at least 9 of the 18 Cy dye images of the 42-h experiment and changed (up- or downregulated) at least 1.5-fold with a P value of <0.01 (which applied to 6 spots), changed at least 1.1-fold with a P value of <0.001 (adding a further 2 spots), or changed at least 2.0-fold with a P value of <0.2 (adding a final 4 spots).
We were able to determine the protein identities of nine protein spots by peptide mass fingerprinting, yielding nine unique proteins. Of these nine identified proteins, only one protein was found to be significantly downregulated and eight were upregulated at 42 h after infection. Among the upregulated proteins were the HIV CA-p24 and Nef proteins. Of the six upregulated cellular proteins, five were also identified at peak virus infection (not only the tubulin alpha but also the tubulin beta subunit was found to be upregulated at 42 h). However, the one downregulated protein (DDX3X) was observed only in this single-cycle experiment, and the 14-3-3 epsilon protein was upregulated (instead of downregulated as at peak infection). Only 0.6% of proteins that were detected changed in expression level. This further corroborates the absence of apoptosis, as confirmed by FACS analysis. A complete list of all differentially expressed proteins at 42 h of HIV infection with their ratios and P values can be found in Table S2 in the supplemental material.
Protein annotation, location, and function.
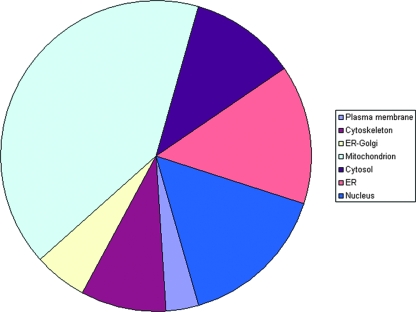
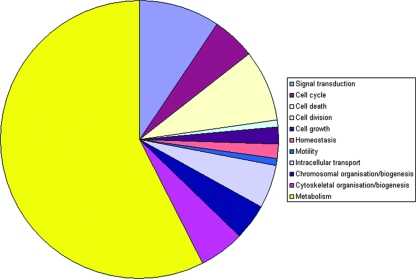
The complete list of proteins was evaluated against the literature and public databases. We were able to assign database accession numbers to all human proteins identified so that all human protein identifications were recognized by GoMiner to perform a gene ontology analysis (57). An overview of the differentially expressed proteins at peak infection, grouped according to gene ontology categories, is given in Table S3 in the supplemental material. This allowed us to rapidly analyze their annotated subcellular location, biological function, and molecular interactions. Figure 3 gives an overview of the distribution of the identified proteins over different cellular locations. Figure 4 illustrates the distribution over several main categories of cellular processes represented by many of the proteins identified.
FIG. 3.
Locations of differentially expressed PM1 proteins. The overall contribution of different cellular locations to the differentially expressed PM1 proteins upon HIV-1 peak infection, as determined using GoMiner, is displayed.
FIG. 4.
Gene ontology analyses of differentially expressed PM1 proteins. The contributions of the main cellular process categories represented by differentially expressed PM1 proteins upon HIV-1 peak infection as determined, using GoMiner, are displayed.
As can be seen in Fig. 3 and 4, there is a very high proportion of differentially expressed mitochondrial (mt) proteins and a high proportion of differentially expressed metabolic pathway proteins, suggesting that metabolic reprogramming occurs upon HIV infection of the T cell. Table 1 lists the upregulated mt proteins with a short description of their function, which is mainly in the tricarboxylic acid (TCA) cycle, aerobic respiration, and catabolism of fatty acids and amino acids. Table 2 shows a list of differentially regulated proteins categorized according to major metabolic pathways. Interestingly, all four differentially regulated proteins that are involved in glycolysis are downregulated, whereas all that are involved in other major metabolic pathways are upregulated. The upregulation of pyruvate dehydrogenase E1 beta (P11177) might seem to contradict the downregulation of the glycolytic enzymes, but pyruvate can also result from amino acid breakdown. One of the specific enzymes leading to the production of pyruvate from these precursors, serine hydroxymethyltransferase, was indeed found to be upregulated (Table 1). Several other enzymes involved in fatty acid oxidation and amino acid catabolism producing substrates for the TCA cycle are upregulated, as well as some of the TCA cycle enzymes themselves. These, of course, provide the substrates for oxidative phosphorylation, and there is a concerted upregulation of proteins involved in this process and thus in the generation of ATP. Many other proteins were found to be differentially expressed upon HIV infection of T cells; the most important ones are highlighted below.
TABLE 1.
mt proteins upregulated upon HIV-1 infection
| Ratio | t test value | Description | Function | Swiss-Prot accession no. |
|---|---|---|---|---|
| 1.88 | 4.10E−04 | Aconitate hydratase | TCA cycle | Q99798 |
| 2.51 | 2.60E−04 | ATP synthase alpha chain | Oxidative phosphorylation/ATP-synthase | P25705 |
| 2.76 | 2.20E−04 | ATP synthase beta chain | Oxidative phosphorylation/ATP-synthase | P06576 |
| 2.29 | 9.60E−04 | ATP synthase D chain | Oxidative phosphorylation/ATP-synthase | O75947 |
| 2.03 | 1.60E−03 | d-Beta-hydroxybutyrate dehydrogenase | Ketone and butanoate metabolism | Q02338 |
| 2.14 | 1.50E−03 | Complement 1 Q subcomponent-binding protein | Q07021 | |
| 1.96 | 1.70E−05 | Citrate synthase | TCA cycle/acetyl transfer to cytosol | O75390 |
| 2.98 | 4.10E−04 | Citrate synthase | O75390 | |
| 2.20 | 3.40E−04 | Dihydrolipoyl dehydrogenase | Glycolysis/TCA/amino acid metabolism | P09622 |
| 1.94 | 1.50E−04 | Succinyltransferase of oxoglutarate dehydrogenase | TCA/amino acid (lysine) catabolism | P36957 |
| 2.92 | 9.80E−05 | Enoyl-coenzyme A hydratase | Fatty acid metabolism/amino acid catabolism | P30084 |
| 2.66 | 8.50E−05 | Enoyl-coenzyme A hydratase | P30084 | |
| 1.87 | 9.90E−05 | Electron transfer flavoprotein beta (beta-ETF) | Oxidative phosphorylation/electron transfer | P38117 |
| 2.30 | 1.60E−04 | Fumarate hydratase | TCA cycle/cholesterol metabolism | P07954 |
| 1.66 | 2.60E−03 | Aspartate aminotransferase | Amino acid catabolism | P00505 |
| 2.38 | 1.40E−05 | 3-Hydroxyacyl-coenzyme A dehydrogenase type 2 | Fatty acid catabolism/amino acid catabolism | Q5H927 |
| 2.05 | 4.90E−04 | Long chain 3-hydroxyacyl-coenzyme A dehydrogenase | Fatty acid catabolism/amino acid catabolism | P40939 |
| 1.68 | 4.60E−04 | Stress 70 protein | Mitogen-activated protein kinase signaling | P38646 |
| 2.47 | 1.50E−03 | Stress 70 protein | P38646 | |
| 3.47 | 8.10E−04 | 60-kDa heat shock protein | P10809 | |
| 2.37 | 3.70E−04 | 60-kDa heat shock protein | P10809 | |
| 2.32 | 9.30E−04 | 60-kDa heat shock protein | P10809 | |
| 2.09 | 3.30E−05 | 60-kDa heat shock protein | P10809 | |
| 2.01 | 3.80E−05 | Isocitrate dehydrogenase [NAD] subunit alpha | TCA cycle | P50213 |
| 2.05 | 1.30E−03 | Methylcrotonoyl-coenzyme A carboxylase beta chain | Amino acid catabolism | Q9HCC0 |
| 2.24 | 5.40E−04 | Malate dehydrogenase | TCA cycle/acetyl transfer to cytosol | P40926 |
| 2.49 | 3.50E−05 | NADH-ubiquinone oxidoreductase 75-kDa subunit | Oxidative phosphorylation/ubiquinone synthesis | P28331 |
| 2.67 | 7.50E−04 | NADH-ubiquinone oxidoreductase 30-kDa subunit | O75489 | |
| 1.59 | 4.70E−04 | Ornithine aminotransferase | Amino acid catabolism | P04181 |
| 2.06 | 3.30E−04 | Succinyl-coenzyme A:3-ketoacid-coenzyme A transferase 1 | Ketone metabolism/amino acid catabolism | P55809 |
| 1.73 | 1.10E−04 | Pyruvate dehydrogenase E1 component subunit beta | Glycolysis/amino acid metabolism | P11177 |
| 2.75 | 4.70E−05 | Prohibitin | Regulation of mt response and protein synthesis | P35232 |
| 1.88 | 1.80E−04 | Thioredoxin-dependent peroxide reductase | Redox Regulation/NF-κB regulation | P30048 |
| 1.51 | 8.50E−03 | Succinate dehydrogenase flavoprotein subunit | TCA cycle/oxidative phosphorylation | P31040 |
| 2.33 | 8.10E−05 | Serine hydroxymethyltransferase | Amino acid metabolism | P34897 |
| 2.87 | 2.10E−05 | Superoxide dismutase [Mn] | Destroys radicals | P04179 |
| 2.96 | 1.50E−06 | Single-stranded DNA binding protein | mt DNA replication | Q04837 |
| 1.86 | 1.90E−04 | Heat shock protein, 75 kDa | Tumor necrosis factor receptor associated | Q12931 |
| 1.90 | 1.50E−03 | Elongation factor Tu (EF-Tu) (P43) | Protein synthesis | P49411 |
| 2.62 | 1.40E−04 | Ubiquinol-cytochrome c reductase core protein I | Oxidative phosphorylation/electron transfer | P31930 |
| 2.40 | 7.00E−05 | Voltage-dependent anion-selective channel protein 1 | Anion channel/Ca signaling | P21796 |
| 2.62 | 2.20E−05 | Voltage-dependent anion-selective channel protein 1 | P21796 | |
| 2.12 | 2.40E−05 | Voltage-dependent anion-selective channel protein 2 | Anion channel/Ca signaling | P45880 |
TABLE 2.
Differentially expressed proteins grouped by metabolic pathway
| Pathway (GoMiner term) | Ratio | P value | Protein name | Swiss-Prot accession no. |
|---|---|---|---|---|
| Glycolysis | −1.51 | 2.40E−03 | Fructose-bisphosphate aldolase A | P04075 |
| −1.67 | 1.60E−04 | Glucose-6-phosphate isomerase | P06744 | |
| −1.60 | 9.40E−03 | Phosphoglycerate mutase 1 | P18669 | |
| −1.57 | 6.10E−03 | Triosephosphate isomerase | P60174 | |
| TCA cycle | 1.88 | 4.10E−04 | Aconitate hydratase | Q99798 |
| 1.96 | 1.70E−05 | Citrate synthase | O75390 | |
| 2.20 | 3.40E−04 | Dihydrolipoyl dehydrogenase | P09622 | |
| 2.30 | 1.60E−04 | Fumarate hydratase | P07954 | |
| 2.01 | 3.80E−05 | Isocitrate dehydrogenase [NAD] subunit alpha | P50213 | |
| 2.24 | 5.40E−04 | Malate dehydrogenase | P40926 | |
| 1.73 | 1.10E−04 | Pyruvate dehydrogenase E1 component subunit beta | P11177 | |
| 1.51 | 8.50E−03 | Succinate dehydrogenase flavoprotein subunit | P31040 | |
| Amino acid metabolism | 2.15 | 6.90E−04 | Protein disulfide-isomerase precursor | P07237 |
| 1.66 | 2.60E−03 | Aspartate aminotransferase | P00505 | |
| 2.05 | 1.30E−03 | Methylcrotonoyl-coenzyme A carboxylase beta chain | Q9HCC0 | |
| 1.59 | 4.70E−04 | Ornithine aminotransferase | P04181 | |
| 2.33 | 8.10E−05 | Serine hydroxymethyltransferase | P34897 | |
| Oxidative phosphorylation | 2.62 | 1.40E−04 | Ubiquinol-cytochrome c reductase core protein I | P31930 |
| 2.49 | 3.50E−05 | NADH-ubiquinone oxidoreductase 75-kDa subunit | P28331 | |
| 2.67 | 7.50E−04 | NADH-ubiquinone oxidoreductase 30-kDa subunit | O75489 | |
| 2.29 | 9.60E−04 | ATP synthase D chain | O75947 | |
| 2.51 | 2.60E−04 | ATP synthase alpha chain | P25705 | |
| 2.76 | 2.20E−04 | ATP synthase beta chain | P06576 | |
| Fatty acid metabolism | 2.92 | 9.80E−05 | Enoyl-coenzyme A hydratase | P30084 |
| 2.05 | 4.90E−04 | Trifunctional enzyme subunit alpha | P40939 | |
| 1.65 | 7.20E−04 | Trifunctional enzyme subunit beta | P55084 | |
| Glucose metabolism | 1.73 | 1.10E−04 | Pyruvate dehydrogenase E1 component subunit beta | P11177 |
| UDP glucoses metabolism | 1.35 | 5.50E−04 | UTP-glucose-1-phosphate uridylyltransferase 2 | Q16851 |
DISCUSSION
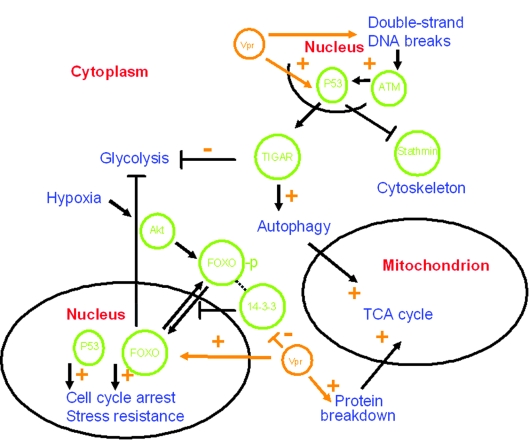
Our proteomic analysis demonstrates that infection of the T-cell line PM1 by HIV-1 gives rise to coordinated changes in protein expression patterns, together representing reprogramming of the T cell. In absolute amounts, these changes are less strong than one might have naively expected. However, this result is in agreement with microarray analyses and a recent proteomic analysis of HIV T-cell interactions (9, 34, 52). Such relatively subtle changes in protein levels can be reliably identified by our approach using multiple independent replicates, and this analysis becomes even more convincing given the concerted changes of many proteins within certain metabolic pathways. Metabolic rerouting as shown in Fig. 5 is apparent, as well as a “tug of war” between host cell and virus with regard to apoptotic signals.
FIG. 5.
Metabolic rerouting of T cells upon HIV-1 infection. A highly schematic overview of pathways and proteins possibly involved in metabolic rerouting of T cells as a result of HIV-1 infection is shown. Cellular proteins are in green, cellular locations in red, and cellular processes in blue. Some of the overall effects of Vpr on the given cellular pathways are indicated (orange). For details, see the text.
Metabolic rerouting.
We observed upregulation of proteins involved in fatty acid oxidation and amino acid catabolism, the TCA cycle, and oxidative phosphorylation. Thus, enzymes involved in the generation of ATP from fatty acids (e.g., the beta oxidation trifunctional enzyme alpha and beta subunits) and amino acids are strongly upregulated, which would lead to increased energy production. In striking contrast, all the differentially regulated proteins that catalyze steps in glycolysis are downregulated: glucose-6-phosphate isomerase, aldolase A, phosphoglycerate mutase 1, and triosephosphate isomerase. HIV apparently induces a metabolic change in the infected cells to decrease energy production from glucose but increases the energy production from amino acids and fatty acids (Tables 1 and 2). That this rerouting represents a highly specific metabolic program is clear not only from the downregulation of glycolytic enzymes but also from the downregulation of E-FABP, the immune cell-specific fatty acid transport protein that is consistently upregulated in activated immune cells (20). Apart from the specific downregulation of the glycolytic enzymes, our results agree with the very recent proteomic studies of Chan et al. (9). They describe one enzyme that is (also) involved in glycolysis (pyruvate kinase), which is upregulated. Although they used a totally different technique, similar effects with regard to oxidative phosphorylation, the TCA cycle, and valine, leucine, and isoleucine degradation were observed. However, their findings appear less consistent: indications for a “dysregulated” TCA cycle are presented, and some proteins involved in respiration (such as cytochrome c oxidase II) are downregulated.
Several studies have shown effects of HIV on cell metabolism. An effect of HIV on glucose metabolism of human intestinal epithelial cells was measured (32). HIV infection resulted in a disturbance of glycolytic and oxidative activities. The results indicated an increase in intracellular glucose concentration due to either increased uptake or decreased glycolysis. Glycolytic impairment was detected in neuronal cells upon incubation with HIV gp120 protein only (53). How could “metabolic rerouting” by the virus be induced at the molecular level? It is known that the retroviral Vpr protein interferes with the suppressive effects of insulin on FOXO transcription factors, which normally stimulate gluconeogenesis by upregulating glucose-6-phosphatase and phosphoenolpyruvate carboxykinase (26). FOXO transcription factors are negatively regulated through phosphorylation by the protein kinase Akt in response to insulin and growth factors and subsequent relocalization from the nucleus into the cytoplasm via interaction with 14-3-3 proteins (25, 26). Vpr is known to interact with 14-3-3 proteins and thus influence their binding specificities. FOXO factors could thus be positively regulated by Vpr, which binds 14-3-3 proteins, allowing FOXO to go to the nucleus. Another way to interfere with nuclear relocalization of certain transcription factors would be downregulation of the 14-3-3 proteins themselves. We indeed find the 14-3-3 gamma, epsilon, tau, and zeta proteins to be significantly (all about 1.6-fold) downregulated. The 14-3-3 pathway is also found to be strongly influenced upon HIV-1 infection (9). Besides their role in glucose metabolism, FOXO transcription factors are involved in cell cycle arrest (as are 14-3-3 proteins, mediating Vpr's cell cycle-arresting activity, e.g., by enhanced binding of Cdc25C [25, 28]), DNA repair, apoptosis, and stress resistance. For instance, FOXO factors stimulate Mn superoxide dismutase transcription, and we indeed find it to be upregulated after HIV infection (as would be appropriate given the increase in mt oxidation). In muscle cells, repression of FOXO activity by Akt signaling induces a decrease in protein degradation. Thus, a Vpr-mediated indirect increase in FOXO activity could lead to enhanced protein degradation, generating substrates for the increased amino acid catabolism that we seem to observe upon HIV infection.
The role of FOXO transcription factors in mediating cell cycle arrest, DNA repair, apoptosis, and stress resistance mimics the functions of the tumor suppressor protein p53, and FOXO and p53 seem to be part of a complex regulatory network (19, 41). HIV infection indeed activates p53 (15). Recently, p53 has also been shown to regulate glucose metabolism and autophagy via TIGAR (4, 18). An important role of TIGAR is to redirect glucose from catabolism (energy production) to anabolism (including synthesis of nucleotides) by blocking glycolysis and activating the pentose phosphate shunt (18). The observed downregulation of glycolysis, leading to an increase in glucose available for ribose precursors, should induce the pentose phosphate pathway. This would lead to a concomitant increase in NADPH, which can also be used for the synthesis of new viral particles and which is needed for the stress response. Indeed, we find a twofold upregulation of the purine nucleoside phosphorylase (also called inosine phosphorylase), a central player in nucleoside metabolism of blood cells, inactivation of which results in severe combined immunodeficiency (37). In this context, the slight upregulation of UDPglucose pyrophosphorylase, which plays a central role as a glucosyl donor, should also be mentioned. This is in agreement with studies of HIV infection of human intestinal epithelial cells (32), where fructose-1,6-phosphate was found to be decreased. Apart from these possibly indirect p53 effects on metabolism, p53 also downregulates stathmin (the central regulator of proliferative activation of microtubules), which is strongly reduced (2.3-fold) upon HIV infection, in agreement with p53-mediated cell cycle arrest.
The activation of p53 might again be mediated by Vpr. p53 has been shown to be activated by Vpr (16). This is possibly a consequence of Vpr-induced DNA double-strand breaks activating ataxia telangiectasia mutated protein-dependent signals such as p53 (36, 48). HIV-1-induced p53 signaling is also in agreement with our observations concerning RuvBL1 and RuvBL2, which are part of a large nuclear protein complex containing TIP60. This multifunctional complex can act as a transcriptional coregulator for several factors, including p53 (44). It can be recruited to double-strand breaks and activate ataxia telangiectasia mutated protein by acetylation and, like FOXO and p53, is also involved in cell cycle regulation and apoptosis (44). The HIV Tat protein, however, is known to suppress TIP60's apoptotic function, either via inhibition of its acetylation activity or by TIP60 degradation via polyubiquitination (44). The upregulated RuvBL1 and downregulated RuvBL2 proteins (ATP-dependent DNA helicases with 3′-to-5′ and 5′-to-3′ activity, respectively) indeed play antagonistic roles, thus underlining the coordinated changes that we observe (24, 43).
Many HIV-1 effects could be mediated by Vpr through FOXO, p53, and TIP60. These three all share regulation by the p300/CBP nuclear coactivator (19, 25, 41, 44), which contains acetyltransferase and ubiquitin ligase activity (41). p300/CBP is targeted by several viruses, and both HIV Vpr and Tat can bind and influence this coactivator (7, 12, 22, 27).
An apoptotic “tug of war.”
Although p53 activation by HIV-1 could trigger apoptosis by a mt pathway (8, 15), viral proteins can actively suppress apoptotic programs (12, 35). In fact, many proteins that are (in)directly involved in apoptosis are differentially expressed upon infection. Chaperonins, mostly associated with antiapoptotic activity, are upregulated, including mt HSP60 and GRP75 (Mortalin) and endoplasmic reticulum (ER)-located GRP94 and GRP78, the latter being involved in virion binding (14). Some mt regulators of apoptopsis are upregulated, including BAG-2 and prohibitin, which is associated with cell cycle arrest. On the other hand, the proapoptotic mt VDAC1 protein is strongly upregulated (as is VDAC2), while antiapoptotic 14-3-3 gamma, Cofilin-1, and high-mobility-group protein B1 are all 1.5-fold downregulated. Clearly, the virus does not suppress all apoptotic signals. Many proteins involved in cell cycle regulation and apoptosis do so via ubiquitination and subsequent proteolysis. Complex changes in the amounts of such proteins are described in this study (see Table S1 in the supplemental material) as well as in the liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based proteomic study (9).
hnRNPs and stress responses.
The heterogeneous nuclear ribonucleoproteins (hnRNPs) are a group of proteins sharing common structural domains, which seem to have roles in DNA repair, telomere biogenesis, cell signaling, RNA export and splicing, and regulation of gene expression. We find hnRNPs K and A1 to be downregulated. The hnRNP A1 protein regulates HIV-1 Tat mRNA splicing and seems to be involved in HIV-1 mRNA export from the nucleus (49). However, hnRNP H, which is also involved in HIV-1 mRNA splicing (6), is upregulated. Another “anomaly” is obtained with the regulation of protein breakdown, in which we find the general ubiquitin-activating enzyme E1 to be downregulated, whereas the proteasome beta type 4 subunit is upregulated nearly fourfold. Its regulatory subunit 13 (PSMD13) is also increased. The latter two proteins are involved in the HIV-1 Vpu-mediated degradation of CD4 (33).
Proteins involved in stress responses are also strongly influenced by HIV infection and virus replication. The first one detected, in the “single” 42-h infection, is the cytoplasmic beta subunit of HSP90. Not surprisingly, many chaperonins (mostly ER located) are upregulated in the peak infection, including GRP 78 (BIP), ERp44, GRP 94 (endoplasmin), ERp 29 and mt HSP60. Many ER protein disulfide isomerases (such as A3, A4, and A6) are also upregulated, as are the alpha subunit of glucosidase II, heme oxygenase 2, and the 150-kDa hypoxia-upregulated protein 1 (HYOU1). The last two could be upregulated because of the increased mt oxidation of fatty acids and amino acids described above. This could explain the upregulation of the mt [Mn] superoxide dismutase as well. Interestingly, hypoxia can signal via Akt (already mentioned above as the protein kinase that can negatively regulate FOXO transcription factors; see also Fig. 5), and indications for the activation of this pathway have been found as well (9). Surprisingly, AHSA1, the 38-kDa activator of the ER-located 90-kDa heat shock protein, is slightly downregulated. It is known to interact with the cytoplasmic tail of the vesicular stomatitis virus glycoprotein (45), but interactions with HIV proteins have not been found so far. However, such interactions have been documented for two of our upregulated ER stress proteins, endoplasmin and cyclophilin B (a peptidyl-prolyl cis-trans-isomerase). Endoplasmin, which can also be induced by hypoxia, seems to be bound by the virion (10, 55), although the specific interaction is still unknown. Cyclophilins interact with Vpr; although this has been shown much more convincingly for cyclophilin A (21, 56). These interactions probably are important for virus routing and packaging, as are the interactions of the virus with cytoskeletal (associated) proteins.
Our observations confirm known interactions with HIV-1 proteins in the case of p21-activated kinase 2, which is bound by Nef to inhibit Bad-mediated apoptotic death and increase virus production (30, 54), and of eIF-5A, the eukaryotic translation initiation factor bound by Rev, which is involved in the nuclear export of Rev and HIV-1 replication (5). Both p21-activated kinase 2 and eIF-5A are downregulated.
Proteins involved in cellular transport.
Some of the proteins involved in cellular transport are strongly influenced by the virus. The most strongly upregulated (almost fivefold) cellular protein that we find is beta-actin, while alpha-tubulin, one of the major constituents of microtubules, is also 2.5-fold upregulated. Ezrin, which is probably involved in connecting cytoskeletal structures to the plasma membrane; microtubule-associated protein RP/EB family member 1 (a negative regulator of microtubular polymerization); and Rho GDP-dissociation inhibitor 2, which is involved in actin cytoskeleton organization, are all downregulated (as is stathmin, the regulator of microtubular activation, mentioned above). Also downregulated are adenylyl cyclase-associated protein 1, which binds actin monomers and is involved in filament dynamics; the actin binding lymphocyte cytosolic protein 1; and alpha actinine 1, an actin cross-linking protein. Interestingly, Rab GDP dissociation inhibitor beta, a protein involved in the regulation of vesicle-mediated cellular transport, is almost fourfold upregulated (3). In agreement with our findings, the previous proteomic study of (9) found the RAN regulation pathway, involved in nucleocytoplasmic transport, to be influenced by the infection process. How all these changes fit in with cell cycle arrest, viral production, and/or transport remains to be established.
Concluding remarks.
More than 30 mt proteins are differentially expressed upon HIV-1 infection, and all of these are upregulated. We do not think this stems from a systematic artifact, as we find an upregulation not of all mt proteins but of only a small subclass and because the magnitude of upregulation observed varies among proteins. Of these upregulated mt proteins, two remain to be mentioned: the glycoprotein gC1qBP/P33, of unknown function, which can interact with rubella virus capsid protein, and SSB, a single-stranded DNA binding protein (which is known to interact with viral proteins). The production of HIV proteins is seen more strongly (CA-p24 is more upregulated and Nef can also be detected) after 42 h (see Table S2 in the supplemental material). The six upregulated cellular proteins at 42 h are all also detected at peak infection, but the downregulated DDX3X was detected only in this experiment. Recent studies showed that DEAD box protein RNA helicases, such as DDX3 and DDX1, are important for HIV infection by facilitating the export of singly spliced or even unspliced HIV RNAs from the nucleus via the CRM1-Rev pathway. Of the DEAD box protein family, DDX3 showed the strongest mRNA downregulation upon HIV-1 replication (at 24 h after induction of replication), in agreement with our observation at the protein level (29).
To study the T-cell-virus interaction, we used transformed PM1 cells cultured in vitro, which likely differ in several ways from primary T cells found in vivo. It is currently not clear how the observed PM1 changes compare to cellular changes in vivo or whether direct cytopathicity of HIV-1 is enhanced in vitro, although large-scale apoptosis was not observed. How the observed changes relate to pathogenicity for the infected individual is also unclear. Though superficially the cellular proteome seems relatively stable, PM1 cells are reprogrammed upon HIV-1 infection. This reprogramming begins early on in infection and has functions, among others, at the level of general metabolism, cytoskeletal organization, and primary suppression of apoptotic responses. Many of these processes are known to be affected by the viral Vpr protein, subverting cellular pathways involved in their regulation at many different levels (Fig. 5). Our DIGE analysis is, for the most part, in agreement with the recent findings (9) in a HIV-T-cell interaction study using LC-MS/MS. There are even many instances in which the findings perfectly complement each other (e.g., upregulation of components of the TCA cycle or the pyruvate dehydrogenase complex in both studies, but with different sets of proteins). There are three notable differences. We observed downregulation of glycolytic enzymes, upregulation of proteins involved in fatty acid breakdown (whereas the previous study [9], e.g., found a strong increase in fatty acid synthase), and a much more consistent increase in TCA cycle enzymes. Possibly, these variations result from differences in the cell types used, infection procedures, and harvest times. More likely the differences are due to the different methods used, with LC-MS/MS yielding more candidate proteins but at the cost of more false positives. This first DIGE analysis of changes in the T-cell proteome upon HIV-1 infection not only uncovered specific reprogramming of T cells but also allowed us to identify (and verify) many proteins potentially involved in the virus-host interactions. Upon verification of these new cofactors of HIV-1 replication, they may become future drug targets.
Supplementary Material
Acknowledgments
We thank Stef Heynen for performing the CA-p24 ELISA experiments and Tony de Ronde for critical reading of the manuscript.
This research was supported by grants from the Dutch Cancer Society (KWF Kankerbestrijding, AMC 2000-210), the EU project Hidden HIV Challenge (FP6-012182), and the AIDS Fund (2007028). This study was further supported by the Academic Medical Center Anton Meelmeijer Fund.
Footnotes
Published ahead of print on 20 February 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alban, A., S. O. David, L. Bjorkesten, C. Andersson, E. Sloge, S. Lewis, and I. Currie. 2003. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 336-44. [DOI] [PubMed] [Google Scholar]
- 2.Auersperg, N. 1964. Long-term cultivation of hypodiploid human tumor cells. J. Natl. Cancer Inst. 32135-163. [PubMed] [Google Scholar]
- 3.Bachner, D., Z. Sedlacek, B. Korn, H. Hameister, and A. Poustka. 1995. Expression patterns of two human genes coding for different rab GDP-dissociation inhibitors (GDIs), extremely conserved proteins involved in cellular transport. Hum. Mol. Genet. 4701-708. [DOI] [PubMed] [Google Scholar]
- 4.Bensaad, K., A. Tsuruta, M. A. Selak, M. N. Vidal, K. Nakano, R. Bartrons, E. Gottlieb, and K. H. Vousden. 2006. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126107-120. [DOI] [PubMed] [Google Scholar]
- 5.Bevec, D., H. Jaksche, M. Oft, T. Wohl, M. Himmelspach, A. Pacher, M. Schebesta, K. Koettnitz, M. Dobrovnik, R. Csonga, F. Lottspeich, and J. Hauber. 1996. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science 2711858-1860. [DOI] [PubMed] [Google Scholar]
- 6.Caputi, M., and A. M. Zahler. 2002. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 21845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caron, C., E. Col, and S. Khochbin. 2003. The viral control of cellular acetylation signaling. Bioessays 2558-65. [DOI] [PubMed] [Google Scholar]
- 8.Castedo, M., J. L. Perfettini, M. Piacentini, and G. Kroemer. 2005. p53—a pro-apoptotic signal transducer involved in AIDS. Biochem. Biophys. Res. Commun. 331701-706. [DOI] [PubMed] [Google Scholar]
- 9.Chan, E. Y., W. J. Qian, D. L. Diamond, T. Liu, M. A. Gritsenko, M. E. Monroe, D. G. Camp, R. D. Smith, and M. G. Katze. 2007. Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J. Virol. 817571-7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chertova, E., O. Chertov, L. V. Coren, J. D. Roser, C. M. Trubey, J. W. Bess, Jr., R. C. Sowder, E. Barsov, B. L. Hood, R. J. Fisher, K. Nagashima, T. P. Conrads, T. D. Veenstra, J. D. Lifson, and D. E. Ott. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 809039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coiras, M., E. Camafeita, T. Urena, J. A. Lopez, F. Caballero, B. Fernandez, M. R. Lopez-Huertas, M. Perez-Olmeda, and J. Alcami. 2006. Modifications in the human T cell proteome induced by intracellular HIV-1 Tat protein expression. Proteomics. 6(Suppl. 1)S63-S73. [DOI] [PubMed] [Google Scholar]
- 12.Col, E., C. Caron, C. Chable-Bessia, G. Legube, S. Gazzeri, Y. Komatsu, M. Yoshida, M. Benkirane, D. Trouche, and S. Khochbin. 2005. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 242634-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, A. T., B. Klaver, and B. Berkhout. 1999. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 7381-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earl, P. L., B. Moss, and R. W. Doms. 1991. Folding, interaction with GRP78-BiP, assembly, and transport of the human immunodeficiency virus type 1 envelope protein. J. Virol. 652047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genini, D., D. Sheeter, S. Rought, J. J. Zaunders, S. A. Susin, G. Kroemer, D. D. Richman, D. A. Carson, J. Corbeil, and L. M. Leoni. 2001. HIV induces lymphocyte apoptosis by a p53-initiated, mitochondrial-mediated mechanism. FASEB J. 155-6. [DOI] [PubMed] [Google Scholar]
- 16.Giri, M. S., M. Nebozhyn, L. Showe, and L. J. Montaner. 2006. Microarray data on gene modulation by HIV-1 in immune cells: 2000-2006. J. Leukoc. Biol. 801031-1043. [DOI] [PubMed] [Google Scholar]
- 17.Gomez, C., and T. J. Hope. 2005. The ins and outs of HIV replication. Cell. Microbiol. 7621-626. [DOI] [PubMed] [Google Scholar]
- 18.Green, D. R., and J. E. Chipuk. 2006. p53 and metabolism: inside the TIGAR. Cell 12630-32. [DOI] [PubMed] [Google Scholar]
- 19.Greer, E. L., and A. Brunet. 2005. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 247410-7425. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto, S., T. Suzuki, H. Y. Dong, S. Nagai, N. Yamazaki, and K. Matsushima. 1999. Serial analysis of gene expression in human monocyte-derived dendritic cells. Blood 94845-852. [PubMed] [Google Scholar]
- 21.Hatziioannou, T., D. Perez-Caballero, S. Cowan, and P. D. Bieniasz. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hottiger, M. O., and G. J. Nabel. 1998. Interaction of the human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J. Virol. 728252-8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen, O. N. 2006. Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 7391-403. [DOI] [PubMed] [Google Scholar]
- 24.Kanemaki, M., Y. Kurokawa, T. Matsu-ura, Y. Makino, A. Masani, K. Okazaki, T. Morishita, and T. A. Tamura. 1999. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 27422437-22444. [DOI] [PubMed] [Google Scholar]
- 25.Kino, T., and G. P. Chrousos. 2004. Human immunodeficiency virus type-1 accessory protein Vpr: a causative agent of the AIDS-related insulin resistance/lipodystrophy syndrome? Ann. N. Y. Acad. Sci. 1024153-167. [DOI] [PubMed] [Google Scholar]
- 26.Kino, T., M. U. De Martino, E. Charmandari, T. Ichijo, T. Outas, and G. P. Chrousos. 2005. HIV-1 accessory protein Vpr inhibits the effect of insulin on the Foxo subfamily of forkhead transcription factors by interfering with their binding to 14-3-3 proteins: potential clinical implications regarding the insulin resistance of HIV-1-infected patients. Diabetes 5423-31. [DOI] [PubMed] [Google Scholar]
- 27.Kino, T., A. Gragerov, O. Slobodskaya, M. Tsopanomichalou, G. P. Chrousos, and G. N. Pavlakis. 2002. Human immunodeficiency virus type 1 (HIV-1) accessory protein Vpr induces transcription of the HIV-1 and glucocorticoid-responsive promoters by binding directly to p300/CBP coactivators. J. Virol. 769724-9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kino, T., A. Gragerov, A. Valentin, M. Tsopanomihalou, G. Ilyina-Gragerova, R. Erwin-Cohen, G. P. Chrousos, and G. N. Pavlakis. 2005. Vpr protein of human immunodeficiency virus type 1 binds to 14-3-3 proteins and facilitates complex formation with Cdc25C: implications for cell cycle arrest. J. Virol. 792780-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnan, V., and S. L. Zeichner. 2004. Alterations in the expression of DEAD-box and other RNA binding proteins during HIV-1 replication. Retrovirology 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linnemann, T., Y. H. Zheng, R. Mandic, and B. M. Peterlin. 2002. Interaction between Nef and phosphatidylinositol-3-kinase leads to activation of p21-activated kinase and increased production of HIV. Virology 294246-255. [DOI] [PubMed] [Google Scholar]
- 31.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. Reitz, Jr. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 693712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz, N. W., N. Yahi, J. Fantini, and P. J. Cozzone. 1997. Perturbations of glucose metabolism associated with HIV infection in human intestinal epithelial cells: a multinuclear magnetic resonance spectroscopy study. AIDS 11147-155. [DOI] [PubMed] [Google Scholar]
- 33.Margottin, F., S. P. Bour, H. Durand, L. Selig, S. Benichou, V. Richard, D. Thomas, K. Strebel, and R. Benarous. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1565-574. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell, R., C. Y. Chiang, C. Berry, and F. Bushman. 2003. Global analysis of cellular transcription following infection with an HIV-based vector. Mol. Ther. 8674-687. [DOI] [PubMed] [Google Scholar]
- 35.Moon, H. S., and J. S. Yang. 2006. Role of HIV Vpr as a regulator of apoptosis and an effector on bystander cells. Mol. Cell 217-20. [PubMed] [Google Scholar]
- 36.Nakai-Murakami, C., M. Shimura, M. Kinomoto, Y. Takizawa, K. Tokunaga, T. Taguchi, S. Hoshino, K. Miyagawa, T. Sata, H. Kurumizaka, A. Yuo, and Y. Ishizaka. 2007. HIV-1 Vpr induces ATM-dependent cellular signal with enhanced homologous recombination. Oncogene 26477-486. [DOI] [PubMed] [Google Scholar]
- 37.Pannicke, U., P. Tuchschmid, W. Friedrich, C. R. Bartram, and K. Schwarz. 1996. Two novel missense and frameshift mutations in exons 5 and 6 of the purine nucleoside phosphorylase (PNP) gene in a severe combined immunodeficiency (SCID) patient. Hum. Genet. 98706-709. [DOI] [PubMed] [Google Scholar]
- 38.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185661-672. [DOI] [PubMed] [Google Scholar]
- 39.Perez, O. D., and G. P. Nolan. 2001. Resistance is futile: assimilation of cellular machinery by HIV-1. Immunity 15687-690. [DOI] [PubMed] [Google Scholar]
- 40.Pradet-Balade, B., F. Boulme, H. Beug, E. W. Mullner, and J. A. Garcia-Sanz. 2001. Translation control: bridging the gap between genomics and proteomics? Trends Biochem. Sci. 26225-229. [DOI] [PubMed] [Google Scholar]
- 41.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187112-126. [DOI] [PubMed] [Google Scholar]
- 42.Roeth, J. F., and K. L. Collins. 2006. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol. Mol. Biol. Rev. 70548-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rottbauer, W., A. J. Saurin, H. Lickert, X. Shen, C. G. Burns, Z. G. Wo, R. Kemler, R. Kingston, C. Wu, and M. Fishman. 2002. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell 111661-672. [DOI] [PubMed] [Google Scholar]
- 44.Sapountzi, V., I. R. Logan, and C. N. Robson. 2006. Cellular functions of TIP60. Int. J. Biochem. Cell. Biol. 381496-1509. [DOI] [PubMed] [Google Scholar]
- 45.Sevier, C. S., and C. E. Machamer. 2001. p38: A novel protein that associates with the vesicular stomatitis virus glycoprotein. Biochem. Biophys. Res. Commun. 287574-582. [DOI] [PubMed] [Google Scholar]
- 46.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins on silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 47.Sierra, S., B. Kupfer, and R. Kaiser. 2005. Basics of the virology of HIV-1 and its replication. J. Clin. Virol. 34233-244. [DOI] [PubMed] [Google Scholar]
- 48.Tachiwana, H., M. Shimura, C. Nakai-Murakami, K. Tokunaga, Y. Takizawa, T. Sata, H. Kurumizaka, and Y. Ishizaka. 2006. HIV-1 Vpr induces DNA double-strand breaks. Cancer Res. 66627-631. [DOI] [PubMed] [Google Scholar]
- 49.Tange, T. O., C. K. Damgaard, S. Guth, J. Valcarcel, and J. Kjems. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 205748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unlu, M., M. E. Morgan, and J. S. Minden. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 182071-2077. [DOI] [PubMed] [Google Scholar]
- 51.Unwin, R. D., C. A. Evans, and A. D. Whetton. 2006. Relative quantification in proteomics: new approaches for biochemistry. Trends Biochem. Sci. 31473-484. [DOI] [PubMed] [Google Scholar]
- 52.Van't Wout, A., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+ T-cell lines. J. Virol. 771392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vignoli, A. L., I. Martini, K. G. Haglid, L. Silvestroni, G. Augusti-Tocco, and S. Biagioni. 2000. Neuronal glycolytic pathway impairment induced by HIV envelope glycoprotein gp120. Mol. Cell. Biochem. 21573-80. [DOI] [PubMed] [Google Scholar]
- 54.Wolf, D., V. Witte, B. Laffert, K. Blume, E. Stromer, S. Trapp, P. D'Aloja, A. Schurmann, and A. S. Baur. 2001. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat. Med. 71217-1224. [DOI] [PubMed] [Google Scholar]
- 55.Xu, A., A. R. Bellamy, and J. A. Taylor. 1998. BiP (GRP78) and endoplasmin (GRP94) are induced following rotavirus infection and bind transiently to an endoplasmic reticulum-localized virion component. J. Virol. 729865-9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zander, K., M. P. Sherman, U. Tessmer, K. Bruns, V. Wray, A. T. Prechtel, E. Schubert, P. Henklein, J. Luban, J. Neidleman, W. C. Greene, and U. Schubert. 2003. Cyclophilin A interacts with HIV-1 Vpr and is required for its functional expression. J. Biol. Chem. 27843202-43213. [DOI] [PubMed] [Google Scholar]
- 57.Zeeberg, B. R., W. Feng, G. Wang, M. D. Wang, A. T. Fojo, M. Sunshine, S. Narasimhan, D. W. Kane, W. C. Reinhold, S. Lababidi, K. J. Bussey, J. Riss, J. C. Barrett, and J. N. Weinstein. 2003. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 4R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.