Abstract
Earlier studies have shown that ICP22 and the UL13 protein kinase but not the US3 kinase are required for optimal expression of a subset of late (γ2) genes exemplified by UL38, UL41, and US11. In primate cells, ICP22 mediates the disappearance of inactive isoforms of cdc2 and degradation of cyclins A and B1. Active cdc2 acquires a new partner, the viral DNA synthesis processivity factor UL42. The cdc2-UL42 complex recruits and phosphorylates topoisomerase IIα for efficient expression of the γ2 genes listed above. In uninfected cells, the cdc25C phosphatase activates cdc2 by removing two inhibitory phosphates. The accompanying report shows that in the absence of cdc25C, the rate of degradation of cyclin B1 is similar to that occurring in infected wild-type mouse embryo fibroblast cells but the levels of cdc2 increase, and the accumulation of a subset of late proteins and virus yields are reduced. This report links ICP22 with cdc25C. We show that in infected cells, ICP22 and US3 protein kinase mediate the phosphorylation of cdc25C at its C-terminal domain. In in vitro assays with purified components, both UL13 and US3 viral kinases phosphorylate cdc25C and ICP22. cdc25C also interacts with cdc2. However, in infected cells, the ability of cdc25C to activate cdc2 by dephosphorylation of the inactive cdc2 protein is reduced. Coupled with the phosphorylation of cdc25C by the US3 kinase, the results raise the possibility that herpes simplex virus 1 diverts cdc25C to perform functions other than those performed in uninfected cells.
Herpes simplex virus 1 (HSV-1) proteins are made in a sequential, orderly fashion (15, 16). The α proteins, made immediately after infection, regulate and enable all subsequent gene expression. The β proteins, made next, are largely concerned with viral nucleic acid synthesis. The γ1 and γ2 genes largely encode the structural proteins of the virus. Whereas γ1 proteins can be made in the absence of viral DNA synthesis, they accumulate in larger amounts once DNA synthesis is enabled (14). In contrast, γ2 genes require viral DNA synthesis for their expression (20). The list of γ2 genes includes UL38, UL41, UL44, US11, etc. Although all of these genes require viral DNA synthesis for their expression, UL38, UL41, and US11 require functional ICP22, the product of the α22 gene, and the protein kinase encoded by the UL13 gene for optimal expression, especially in primary human cells or rodent cell lines infected at low ratios of virus per cell (25, 29). The UL44 gene, in contrast, does not have such a requirement (25). The domain of ICP22 required for optimal expression of the subset of γ2 genes is at or near the carboxyl terminus of the protein. The same domain is the target of the UL13 and US3 protein kinases (22, 25). In reports on studies designed to unravel the mechanism by which ICP22 and UL13 regulate the subset of γ2 genes, it was noted that in infected primate cells, in an ICP22-dependent manner, the inactive forms of cdc2 disappear and its partners, cyclins A and B1, are degraded (1). cdc2 physically interacts with UL42, the viral DNA synthesis processivity factor (2). The cdc2-UL42 complex recruits and phosphorylates topoisomerase IIα to enable optimal expression of the ICP22-regulated subset of γ2 genes (3, 4).
A key step in this cascade of events is the mechanism of interaction between ICP22 and cdc2. In uninfected cells, the major activator of cdc2 is the cdc25C phosphatase. In the accompanying report, we have shown that cyclin B1 is degraded in both cdc25C+/+ and cdc25C−/− cells but the amount of cdc2 increases in infected cells lacking cdc25C (30). Furthermore, the expression of the ICP22-dependent subgroup of γ2 genes is decreased, and the yield of virus is at least 10-fold lower than in sibling, wild-type cells. These results indicate that the cdc25C phosphatase plays a role in viral replication. While this role could include activation of cdc2, it does not provide any clues as to the mechanisms by which viral proteins could in turn activate cdc25C phosphatase.
In this report, we show that cdc25C physically interacts in infected-cell lysates with ICP22 but only in the presence of the US3 protein kinase. cdc25C is phosphorylated in infected-cell lysates depending on the presence of US3, and the predominant site of phosphorylation of cdc25C is in the carboxyl-terminal domain, containing the catalytic site of the protein. We also show that in reaction mixtures containing purified proteins, both cdc25C and ICP22 are each independently phosphorylated by UL13 and US3 protein kinase. Finally, we demonstrate that in the course of infection, there is a reduction in the ability of cdc25C to activate cdc2 by dephosphorylation of the inactive cdc2 protein, suggesting that cdc25C may be diverted by HSV-1 to target novel substrates during infection.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 cells were initially obtained from the American Type Culture Collection and were grown in Dulbecco's modified Eagle medium supplemented with 5% newborn-calf serum. The insect cell line Sf9 was obtained from Pharmingen and was grown in TNM-FH (Pharmingen) insect cell medium. HSV-1 strain F [HSV-1(F)] is the prototype strain used in this laboratory (9). All recombinant viruses used in these studies contain mutations on an HSV-1(F) background. R325 (α22 with a deletion of the region encoding the C-terminal domain), R7041 (ΔUS3), R7353 (ΔUS3 ΔUL13), and R7356 (ΔUL13) have been previously described (25, 27, 29).
Plasmids.
The plasmid pGC52(cdc25Hs), a kind gift from H. Piwnica-Worms (Washington University, St. Louis, MO), contains the open reading frame (ORF) of human cdc25C inserted into the BamHI-XhoI site of pGC52, as previously described (19). The ORF was digested and inserted into the BamHI-XhoI site of the mammalian expression vector pcDNA3.1(+) (Invitrogen, Carlsbad, CA), creating the plasmid pcDNA-cdc25C. The plasmids pcDNA-UL13 and pcDNA-α22 contain the ORFs of the viral genes UL13 and α22, respectively, inserted into the vector pcDNA3.1(+). Site-directed mutagenesis was performed, in which complementary oligonucleotides containing a specific mutation in cdc25C or in UL13 were annealed to pcDNA-cdc25C or pcDNA-UL13 DNA, using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) to generate the single amino acid substitutions C377S in cdc25C and K176M in UL13. The C377S substitution abolishes phosphatase activity of cdc25C, and the K176M substitution abolishes kinase activity of UL13 (18, 31). The resulting plasmids were designated pcDNA-cdc25C(C377S) and pcDNA-UL13(K176M), respectively, to be used in further constructs.
The shuttle vector pRB5950 (MTS1) was derived from the pAcSG2 baculovirus transfer vector (Pharmingen, San Diego, CA) but contains a cytomegalovirus promoter, as described previously (28). pRB5915 contains the entire ORF of US3 inserted into the BglII site of pRB5950 (21). Plasmid pRB5914 is identical to pRB5915 except for a point mutation encoding the amino acid substitution K220N, shown to block kinase activity of US3 (17), inserted into the BglII site of pRB5950.
The ORFs from pcDNA-cdc25C and from pcDNA-cdc25C(C377S) were each amplified by PCR and inserted in frame into the pGEX4T-1 vector (Amersham Biosciences) between the BamHI and NotI restriction sites, resulting in pGEX4T1-cdc25C and pGEX4T1-cdc25C(C377S). An amino-terminal truncation of cdc25C was created by PCR amplification of the first 272 codons from pcDNA-cdc25C followed by insertion into pGEX4T-1 between BamHI and NotI to create pGEX4T1-cdc25C-NTD. Two carboxyl-terminal truncations of cdc25C were created by PCR amplification of the final 201 codons from pcDNA-cdc25C and pcDNA-cdc25C(C377S), followed by insertion into pGEX4T-1 in the BamHI and NotI sites to create pGEX4T1-cdc25C-CTD and pGEX4T1-cdc25C-CTD(C377S). These plasmids were used to generate glutathione S-transferase (GST) chimeric proteins in Escherichia coli.
The ORFs from pcDNA-cdc25C, pcDNA-cdc25C(C377S), and pcDNA-α22 were each amplified by PCR and inserted in frame into the pMal-c2 vector (New England Biolabs) in the BamHI and SalI sites or, in the case of α22, in the EcoRI and BamHI restriction sites, resulting in pMalc2-cdc25C, pMalc2-cdc25C(C377S), and pMalc2-α22. These plasmids were used to generate maltose-binding protein (MBP) chimeric proteins in E. coli.
The ORFs from pRB5915, pRB5914, pcDNA-UL13, and pcDNA-UL13(K176M) were each amplified by PCR and inserted in frame into the pAcGHLT-C baculovirus transfer vector (Pharmingen) in the EcoRI and NotI sites, resulting in pAcGHLTC-US3, pAcGHLTC-US3(K220N), pAcGHLTC-UL13, and pAcGHLTC-UL13(K176M), respectively. These plasmids were used to generate baculovirus encoding GST chimeric proteins for expression in Sf9 cells.
All plasmids described above were sent to the University of Chicago Cancer Research Center DNA Sequencing Facility to confirm that the DNA sequence was correct and contained no unintended mutations.
Cell infections.
HEp-2 cells in 25-cm2 flasks were infected with the appropriate virus at the indicated multiplicity of infection in medium 199V (199 medium supplemented with 1% calf serum) on a rotary shaker at 37°C. After 2 h, the inoculum was replaced with fresh growth medium and culture flasks were incubated at 37°C until cells were harvested. Infection times delineated in the figures show time zero to indicate when infection was initiated. Cells were harvested by scraping into their own medium, pelleted by low-speed centrifugation, washed twice in phosphate-buffered saline A [PBS(A)] (0.14 M NaCl, 3 mM KCl, 10 mM Na2HPO4, 1.5 mM KH2PO4), and then lysed in the appropriate buffer.
Electrophoresis and immunoblotting.
Cell pellets were lysed and denatured in disruption buffer (50 mM Tris [pH 7.0], 2.75% sucrose, 5% β-mercaptoethanol, 2% sodium dodecyl sulfate). Protein samples were boiled for 5 min and then were electrophoretically separated in a 10% denaturing polyacrylamide gel and electrically transferred to a nitrocellulose sheet. The membrane was then blocked with 5% nonfat milk and reacted with primary antibody followed by appropriate secondary antibody conjugated to alkaline phosphatase (Bio-Rad Laboratories) or horseradish peroxidase (Sigma). Immunoblots were developed either with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (Sigma) or through enhanced chemiluminescence (ECL; Amersham Biosciences).
Antibodies.
The antibodies used in these studies were antiactin (catalog no. A4700; Sigma), anti-cdc2 (catalog no. sc-54; Santa Cruz), anti-cdc25C (catalog no. sc-13138; Santa Cruz), anti-cyclin B1 (catalog no. sc-245; Santa Cruz), anti-GST (catalog no. sc-138; Santa Cruz), anti-MBP (catalog no. E8032S; NEB), all monoclonal, and polyclonal antibody anti-ICP22 (Goodwin Cancer Research Institute). Anti-mouse immunoglobulin G (IgG)-peroxidase (catalog no. A4416; Sigma), anti-rabbit IgG-peroxidase (catalog no. A0545; Sigma), anti-mouse IgG-alkaline phosphatase (AP) conjugate (catalog no. 170-6520; Bio-Rad), and anti-rabbit IgG-AP conjugate (catalog no. 170-6518; Bio-Rad) were used as secondary antibodies for immunoblotting.
Expression and purification of GST chimeric proteins in E. coli.
GST chimeric proteins containing GST alone or GST fused to full-length cdc25C, a cdc25C construct carrying the C377S substitution (cdc25C-M), truncated cdc25C constructs containing the N-terminal 272 amino acids (NTD) or the C-terminal 201 amino acids (CTD), and a truncated cdc25C construct containing the C-terminal 201 amino acids and the C377S substitution (CTD-M) correspond to the plasmids pGEX4T-1, pGEX4T1-cdc25C, pGEX4T1-cdc25C(C377S), pGEX4T1-cdc25C-NTD, pGEX4T1-cdc25C-CTD, and pGEX4T1-cdc25C-CTD(C377S), respectively. They were produced as previously described.
GST-cdc25C kinase assay and pull-down assay.
HEp-2 cells were infected as indicated and harvested as described above. The rinsed cell pellet was lysed in high-salt lysis buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40, 400 mM NaCl, 0.1 mM Na orthovanadate, 10 mM NaF, 2 mM dithiothreitol [DTT]) containing a Complete protease mixture (Roche) and maintained for 1 h on ice, and then the insoluble material was cleared by centrifugation and protein concentrations were measured by Bradford assay (Bio-Rad). Purified GST or GST chimeric proteins on beads were incubated with 40 μg cell lysate in kinase buffer (50 mM Tris [pH 7.4], 10 mM MgCl2, 5 mM DTT, 10 μM ATP, and 20 μCi of [γ-32P]ATP) for a total volume of 40 μl per sample, incubated at 30°C for 20 min. The beads were rinsed five times with PBS(A) before addition of 50 μl of disruption buffer and heating for 5 min at 95°C. Alternatively, when specified, whole-kinase reactions were stopped with 13 μl of 4× disruption buffer in the absence of any washing steps. The samples were subjected to electrophoresis in 10% polyacrylamide gels, transferred to a nitrocellulose membrane, and subjected to autoradiography. Quantification of 32P phosphorylation of the substrate was done with the aid of a Molecular Dynamics PhosphorImager (Storm 860).
For the pull-down assay, cell pellets were lysed as described above in high-salt lysis buffer. Next, purified chimeric proteins on beads were incubated with 300 μg cell lysate in high-salt lysis buffer in a total volume of 1 ml with rotation overnight at 4°C for 20 min. The beads were rinsed five times with PBS(A) before addition of 50 μl of disruption buffer and heating for 5 min at 95°C. The samples were subjected to electrophoresis in 10% polyacrylamide gels, transferred to a nitrocellulose membrane, and immunoblotted as described above.
Expression and purification of MBP chimeric proteins in E. coli.
MBP chimeric proteins containing MBP alone or MBP fused to cdc25C, cdc25C-M, or ICP22 correspond to the plasmids pMal-c2, pMalc2-cdc25C, pMalc2-cdc25C(C377S), and pMalc2-α22, respectively. Preparation was identical to that for GST chimeric proteins in E. coli, with two notable exceptions: 1% Tween 20 was used instead of 1% Triton X-100, and MBP chimeric proteins were adsorbed to amylose resin (catalog no. E8021S; New England Biolabs) instead of glutathione-agarose, as described previously (18).
Expression and purification of GST chimeric proteins in Sf9 cells.
GST chimeric proteins expressed in Sf9 cells containing GST fused to US3, US3-M, UL13, and UL13-M correspond to the baculovirus transfer plasmids pAcGHLTC-US3, pAcGHLTC-US3(K220N), pAcGHLTC-UL13, and pAcGHLTC-UL13(K176M), respectively. Baculoviruses corresponding to each plasmid were generated using the Pharmingen (San Diego, CA) baculovirus expression vector system by cotransfecting each transfer plasmid along with Baculogold linearized baculovirus DNA (Pharmingen) into Sf9 cells according to the manufacturer's instructions. Baculoviruses were propagated in Sf9 cells grown in 150-cm2 flasks in TNM-FH insect cell medium. The supernatant containing virus was harvested and cleared by centrifugation at 1,000 rpm for 5 min at 4°C. The baculovirus was twice amplified by infecting fresh flasks of Sf9 cells.
Once baculoviruses were amplified, the baculovirus-induced expression of each GST chimeric protein was optimized in Sf9 cells and was determined to peak at 48 h after infection. The GST chimeric proteins were purified according to a protocol described previously (18). The eluted proteins (GST-US3, GST-US3-M, GST-UL13, and GST-UL13-M) were stored at 4°C and protected from light for use in further experiments.
In vitro kinase assay using purified viral kinases.
Similar kinase assays employing the GST-US3 and GST-UL13 protein kinases in reactions with substrates fused to MBP have been described previously (17, 18). Briefly, purified MBP chimeric proteins captured on amylose beads were rinsed twice with washing buffer (50 mM Tris-HCl [pH 8.0] and 1 mM DTT) and were subjected to in vitro kinase assays. The assays were performed to determine whether certain MBP chimeric proteins could serve as substrates for GST-US3 or GST-UL13. Kinase buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 15 mM MgCl2, 0.1% Nonidet P-40, and 1 mM DTT) containing 10 μM ATP, 10 μCi of [γ-32P]ATP, and purified GST chimeric protein was added to the beads that had captured MBP chimeric proteins, and samples were reacted for 30 min at 30°C. After incubation, the samples were extensively washed with TNE buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, and 1 mM EDTA), subjected to electrophoresis on 10% polyacrylamide denaturing gels, transferred to a nitrocellulose membrane, and subjected to autoradiography. The membrane was also used for immunoblotting, as described above.
Endogenous cdc25C phosphatase assay.
This assay was adapted from a previously described two-step assay for cdc25C phosphatase activity (12). Briefly, HEp-2 cells were mock or HSV-1(F) infected for the appropriate time or treated with 5 μg/ml nocodazole (Sigma) or 10 mM hydroxyurea (Sigma) in normal growth medium for 18 h. Cells were harvested by scraping into the medium, rinsed twice with PBS, lysed in eukaryotic lysis buffer (50 mM Tris-HCl [pH 7.4], 0.25 M NaCl, 50 mM NaF, 0.1% Triton X-100, 5 mM EDTA, 1 mM DTT, 1 mM Na3VO4) containing the Complete protease mixture (Roche), and kept on ice for 30 min. Insoluble material was cleared from the lysate by centrifugation for 10 min at 10,000 × g, and the total protein concentration was determined by Bradford assay (Bio-Rad).
A total of 500 μg of S-phase cell extracts (from hydroxyurea-treated cells) or 3 mg of sample extracts were brought up to 1 ml with eukaryotic lysis buffer and precleared with 25 μl protein A Sepharose beads, rotating 30 min at 4°C before centrifugation for 1 min at 10,000 × g. The supernatant was transferred to new reaction tubes containing 25 μl washed protein A Sepharose beads, and 5 μl of antibody for immunoprecipitation of cyclin B1 (for S-phase extracts) or 10 μl monoclonal antibody for the immunoprecipitation of cdc25C (sample extracts) was added, with rotation for 2 h at 4°C. After centrifugation, the supernatant was discarded and beads were washed three times with l ml eukaryotic lysis buffer, pelleting the beads after each wash by centrifugation for 1 min at 10,000 × g.
Phosphatase assay buffer (500 μl of 50 mM Tris-HCl [pH 8.0], 10 mM DTT) was added to the beads containing the immunoprecipitated cdc25C. These samples were individually resuspended by pipetting and combined with immunoprecipitated cyclin B1, mixed well, and centrifuged at 10,000 × g to remove the supernatant fluid. One hundred microliters phosphatase buffer was added for a reaction with incubation at 30°C for 15 min with constant shaking (200 rpm), followed by centrifugation at 10,000 × g to remove the supernatant fluid.
Immediately after the phosphatase reaction, 50 μl histone H1 kinase buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 1 mM DTT, 50 μM ATP, 5 μCi [γ-32P]ATP, 10 μg histone H1 [Roche] per reaction) was added to the pellet in a reaction incubated at 30°C for 15 min with constant shaking (200 rpm). The kinase reaction was stopped by adding 15 μl 4× disruption buffer and heating for 5 min at 95°C. The samples were subjected to electrophoresis in 10% polyacrylamide gels, transferred to a nitrocellulose membrane, and subjected to autoradiography.
RESULTS
GST-cdc25C chimeric protein is phosphorylated by lysates of HSV-1(F)-infected cells.
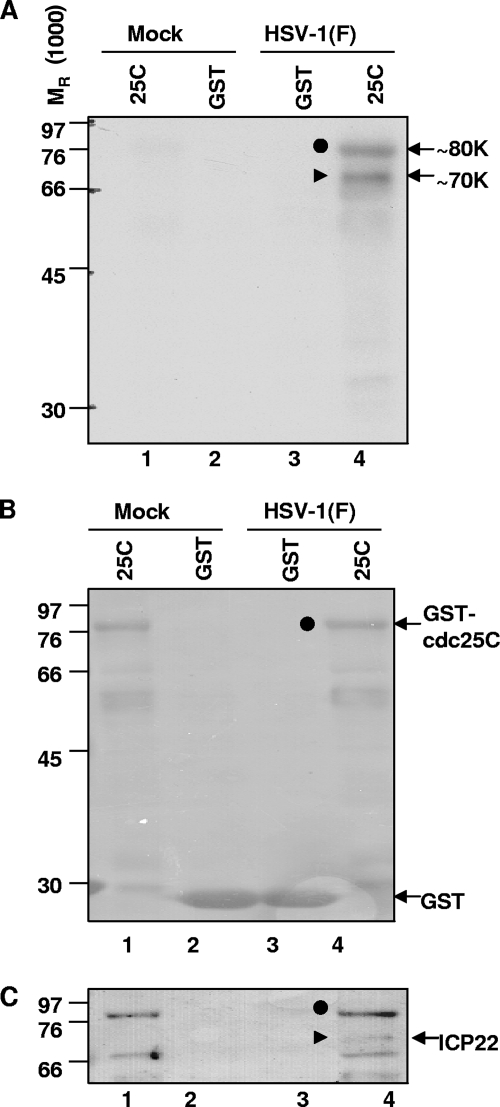
The purpose of this series of experiments was to determine whether cdc25C can be phosphorylated by lysates of wild-type-virus-infected cells. GST or GST-cdc25C chimeric protein was expressed in E. coli BL21 bacteria, bound to glutathione Sepharose beads as described in Materials and Methods, and reacted in kinase reaction buffer containing [γ-32P]ATP with 40 μg of lysate from mock- or HSV-1(F)-infected HEp-2 cells harvested 18 h postinfection. After incubation for 30 min at 30°C, the glutathione Sepharose beads were collected and rinsed five times with PBS and the bound proteins were electrophoretically separated in a denaturing 10% polyacrylamide gel, transferred to a membrane, and visualized by autoradiography. The autoradiogram (Fig. 1A) showed that the Sepharose bead-bound proteins in the reaction mixtures containing GST-cdc25C and infected-cell lysates formed two phosphorylated bands with molecular masses of 70 and 80 kDa, respectively (Fig. 1A, lane 4). These bands were not detected in any other reactions, indicating that the phosphorylation was specific to cdc25C and was dependent on HSV-1 infection. The blot was stained with Ponceau S to detect total protein, confirming that GST-cdc25C and GST were present in each lane (Fig. 1B). Of the two phosphorylated protein bands seen in the autoradiogram (Fig. 1A, lane 4), the 80-kDa band was detectable by Ponceau S staining of proteins bound to Sepharose beads in the reaction mixtures containing GST-cdc25C (Fig. 1B, lanes 1 and 4) whereas the 70-kDa protein was not detected by the Ponceau S stain. Thus, the 80-kDa band corresponded to GST-cdc25C, while the 70-kDa band represented a protein from the infected-cell lysate that was pulled down by cdc25C. This 70-kDa phosphorylated protein was consistent with ICP22, as demonstrated by immunoblotting with antibody specific for ICP22 (Fig. 1C). Nonspecific bands corresponding to the large amounts of GST-cdc25C can be seen in lanes 1 and 4. The results of this experiment showed that GST-cdc25C was phosphorylated by HSV-1(F)-infected cell lysate, consistent with an interaction with phosphorylated ICP22.
FIG. 1.
GST-cdc25C chimeric protein is phosphorylated by lysates of HSV-1(F)-infected cells. GST-cdc25C chimeric protein, on beads, was incubated with 40 μg of lysate from mock- or HSV-1(F)-infected HEp-2 cells (18 h postinfection) in kinase reaction buffer containing [γ-32P]ATP for 30 min at 30°C. Glutathione Sepharose beads were rinsed, and bound proteins were electrophoretically separated in 10% polyacrylamide gel, transferred to a membrane, and visualized by autoradiography. (A) Autoradiogram reveals two major phosphorylated protein bands (70 and 80 kDa). (B) Ponceau S staining to detect total protein reveals 80-kDa protein in large quantity. (C) ICP22 immunoblot reveals a 70-kDa band, marked by a filled triangle. GST alone and GST-cdc25C are abbreviated as GST and 25C, respectively.
Interaction of GST-cdc25C chimeric protein with ICP22 from infected-cell lysate depends on viral kinases US3 and UL13.
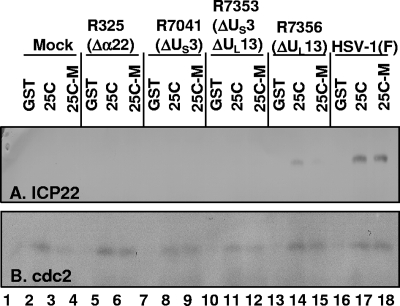
To further examine the interaction of ICP22 with cdc25C, aliquots of 400 μg of lysate from HEp-2 cells infected with wild-type or mutant viruses were reacted overnight with GST, GST-cdc25C, or GST-cdc25C-M (containing a serine in place of cysteine at position 377 which eliminates phosphatase activity [10, 31]) chimeric proteins bound to beads. The beads were collected and extensively rinsed, and the bound proteins were electrophoretically separated on a denaturing 10% polyacrylamide gel and analyzed by immunoblotting for the presence of ICP22 and cdc2. cdc2 was pulled down from all lysates with similar efficiencies by GST-cdc25C or GST-cdc25C-M but not by GST alone (Fig. 2B). Similarly, ICP22 was pulled down from wild-type-virus-infected cell lysate by GST-cdc25C or GST-cdc25C-M but not by GST alone (Fig. 2A, lanes 16 to 18). As expected, ICP22 was not detected in reactions containing lysates of mock-infected or Δα22 mutant virus-infected cells (Fig. 2A, lanes 1 to 6). Interestingly, ICP22 was not pulled down from ΔUS3 virus- or ΔUS3 ΔUL13 virus-infected cell lysates (Fig. 2A, lanes 7 to 12). However, ICP22 was pulled down by GST-cdc25C but not by the GST-cdc25C-M mutant from ΔUL13 virus-infected cell lysate (Fig. 2A, lanes 14 and 15), although the amount was reduced compared to that for HSV-1(F)-infected lysate (Fig. 2A, compare lanes 14 and 17). In summary, US3 was required for the pull-down interaction between cdc25C and ICP22, and UL13 was necessary for maximal interaction and was absolutely required for the pull-down of ICP22 by cdc25C lacking cysteine 377.
FIG. 2.
Wild-type and C377S mutant GST-cdc25C chimeric proteins pull down ICP22 from infected-cell lysate in the presence of US3. Four hundred micrograms of lysate from HEp-2 cells infected with wild-type or Δα22, ΔUS3, ΔUS3 ΔUL13, or ΔUL13 mutant virus was incubated with GST, GST-cdc25C, or GST-cdc25C-M protein bound to beads. The beads were rinsed after 12 h, and the bound proteins were electrophoretically separated on a 10% polyacrylamide gel and analyzed by immunoblotting. (A) Immunoblot detecting ICP22 that was pulled down by GST-cdc25C or GST-cdc25C-M. (B) Immunoblot detecting cdc2 that was pulled down by GST-cdc25C and GST-cdc25C-M. GST alone, GST-cdc25C, and GST-cdc25C-M are abbreviated as GST, 25C, and 25C-M, respectively.
US3 is required for phosphorylation of GST-cdc25C.
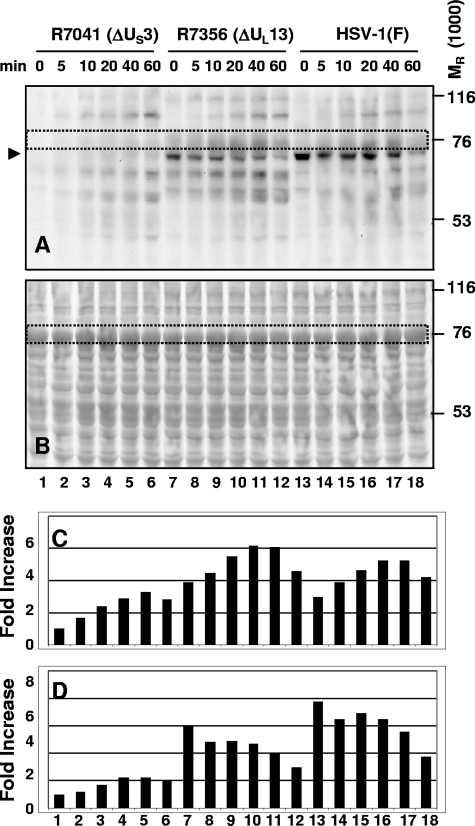
GST-cdc25C was shown to be phosphorylated by HSV-1(F)-infected cell lysate and to interact with ICP22. In light of the evidence that the interaction between GST-cdc25C and ICP22 was dependent on the presence of the viral protein kinase US3 and to a lesser extent on that of the viral protein kinase UL13, we next examined the role of the viral protein kinases in the phosphorylation of cdc25C. The GST-cdc25C-M protein, containing a serine in place of a cysteine residue at position 377 which eliminates phosphatase activity (10, 31), was used because it was phosphorylated by HSV-1(F) in a manner similar to that of active phosphatase (GST-cdc25C) but retained the phosphate modification for a longer period of time (data not shown). Lysates of HEp-2 cells harvested 12 h after infection with HSV-1(F) or with ΔUS3 or ΔUL13 mutant virus were reacted with the GST-cdc25C-M chimeric protein for a total of 60 min in kinase reaction buffer containing [γ-32P]ATP, spending 0, 5, 10, 20, 40, or 60 min in incubation at 30°C following incubation on ice for 60, 55, 50, 40, 20 or 0 min, respectively. The mixtures were electrophoretically separated on a denaturing 10% gel, transferred to a membrane, and visualized by autoradiography (Fig. 3A). The blot was stained with Ponceau S to detect total protein levels (Fig. 3B). The GST-cdc25C-M chimeric protein migrated as an 80-kDa protein (enclosed within a rectangle in Fig. 3A and B for identification). phosphorylation of this chimeric protein was readily apparent in reaction mixtures containing ΔUL13 mutant- or wild-type HSV-1(F)-infected lysates but not ΔUS3 mutant-infected cell lysates. We conclude that phosphorylation of GST-cdc25C-M was dependent on the presence of US3. Interestingly, a slight increase in the intensity of the phosphorylation was seen in reactions containing ΔUL13-infected lysates compared to those with wild-type HSV-1(F)-infected lysates. The amount of radioactivity of the 80-kDa GST-cdc25C-M band, as measured with the aid of a Molecular Dynamics 860 PhosphorImager, is shown in Fig. 3C.
FIG. 3.
GST-cdc25C-M chimeric protein is phosphorylated by infected-cell lysate in the presence of US3. Lysates of HEp-2 cells harvested 12 h after infection with ΔUS3, ΔUL13, or wild-type HSV-1(F) virus were incubated with GST-cdc25C-M chimeric protein for a total of 60 min in kinase reaction buffer containing [γ-32P]ATP, spending the indicated time of 0, 5, 10, 20, 40, or 60 min at 30°C after being incubated on ice for 60, 55, 50, 40, 20, or 0 min, respectively. The mixtures were electrophoretically separated on a 10% denaturing gel, transferred to a membrane, and visualized by autoradiography (A). The blot was then stained with Ponceau S to detect total protein levels (B). The GST-cdc25C-M band is outlined by a dotted rectangle and was quantified as shown in panel C. The 70-kDa band consistent with ICP22 is marked by an arrowhead and was quantified as shown in panel D. Quantification of 32P phosphorylation of the substrate was done using a Molecular Dynamics PhosphorImager. The quantification of the amounts of radioactivity in each band was normalized with respect to the amount of radioactivity present in the R7041-infected lysate at 0 min (lane 1).
A strong phosphorylated protein band (consistent with ICP22 and running just below the GST-cdc25C-M chimeric protein at around 70 kDa) can be seen in kinase reactions containing ΔUL13 mutant-infected or wild-type HSV-1(F)-infected lysates (Fig. 3A, lanes 7 to 18), marked by an arrowhead. This band is present and strong at 0 min, indicating that the phosphorylation occurred while on ice, in contrast to the GST-cdc25C-M band. This 70-kDa band was not detected in reactions with ΔUS3 virus-infected lysates, and it was stronger in reactions with wild type HSV-1(F)-infected than with ΔUL13 virus-infected lysates. The amount of radioactivity of the 70-kDa band was measured with the aid of a Molecular Dynamics 860 PhosphorImager as shown in Fig. 3D. phosphorylation of GST-cdc25C-M on beads and of putative ICP22 in infected-cell lysates was each dependent on US3. This result mirrors the US3 dependence of the interaction between cdc25C and ICP22 observed in the experiments described above (Fig. 2A).
US3 specifically targets phosphorylation of the carboxyl-terminal domain of cdc25C.
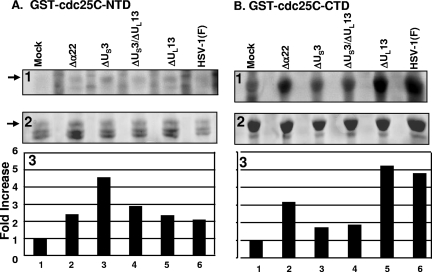
In light of the evidence that US3 mediates the phosphorylation of cdc25C, it was of interest to determine more precisely the region of cdc25C that is targeted for phosphorylation by US3. Two truncated cdc25C constructs, NTD and CTD, were each fused to GST. The GST-NTD and GST-CTD chimeric proteins on glutathione beads were reacted with lysates of HEp-2 cells harvested 12 h after infection with Δα22, ΔUS3, ΔUS3 ΔUL13, ΔUL13, or wild-type HSV-1(F) or mock infection. The beads were then rinsed extensively, and the bound proteins were electrophoretically separated, transferred to membranes, and visualized by autoradiography (Fig. 4A and B, panels 1). The membranes were stained with Ponceau S to detect total protein levels (Fig. 4A and B, panels 2). The GST-NTD chimeric protein migrated at ∼60 kDa (Fig. 4A), while the GST-CTD chimeric protein migrated at ∼50 kDa (Fig. 4B). The amount of radioactivity in each of the GST-NTD and GST-CTD bands was measured with the aid of a Molecular Dynamics 860 PhosphorImager as shown in panels 3 of Fig. 4A and B. The amount of phosphorylation of the GST-NTD construct was increased in the presence of wild-type or mutant virus-infected-cell lysates compared to that with mock infection, although there was not much difference among the various mutant virus-infected-cell lysates (Fig. 4A, panels 1 and 3, compare lane 1 with lanes 2 to 6, noting that the ΔUS3 mutant virus lysate in lane 3 showed a slightly greater increase than the others). GST-CTD phosphorylation was similarly increased in the presence of wild-type HSV-1(F)-infected lysate compared to results with mock-infected lysate (Fig. 4B, panels 1 and 3, compare lane 1 with lane 6). On the other hand, whereas the ΔUS3 and ΔUS3 ΔUL13 mutant virus-infected lysates did not significantly increase the phosphorylation of GST-CTD compared to results with mock-infected lysates (Fig. 4B, panels 1 and 3, compare lane 1 with lanes 3 and 4), the ΔUL13 mutant virus-infected lysate sharply increased the amount of GST-CTD phosphorylation compared to results for mock-infected lysate in a manner similar to that with wild-type HSV-1(F)-infected lysate (Fig. 4B, panels 1 and 3, compare lane 1 with lanes 5 and 6). The Δα22 mutant virus-infected lysate also increased the amount of GST-CTD phosphorylation compared to that with mock lysate but to an intermediate extent (Fig. 4B, panels 1 and 3, compare lane 1 with lanes 2 and 6). In summary, GST-NTD phosphorylation was increased by viral infection independently of the α22, US3, and UL13 genes, whereas GST-CTD phosphorylation was sharply increased in a manner dependent upon the viral gene US3 and to a lesser extent α22. We conclude that HSV-1 infection results in phosphorylation of both the amino- and carboxyl-terminal domains of cdc25C but the viral gene US3, and to some extent α22, specifically targets the carboxyl-terminal domain for phosphorylation.
FIG. 4.
phosphorylation of carboxyl-terminal domain of cdc25C requires α22 and US3. The GST-NTD (A) and GST-CTD (B) chimeric proteins on beads were incubated in the presence of [γ-32P]ATP with lysates of HEp-2 cells harvested 12 h after mock infection or infection with Δα22, ΔUS3, ΔUS3 ΔUL13, ΔUL13, or wild-type HSV-1(F). Beads were rinsed, and bound proteins were electrophoretically separated, transferred to membranes, and visualized by autoradiography (panel 1). The membrane was stained with Ponceau S to detect total protein levels (panel 2). Quantification of 32P phosphorylation was done using a Molecular Dynamics PhosphorImager. For each band measured, the background radioactivity for the lane was subtracted from the total value and this background-adjusted value was normalized with respect to the amount of radioactivity present in reaction mixtures containing lysates of mock-infected cells (panel 3).
It is noteworthy that a phosphorylated protein band (consistent with ICP22 and running at ∼70 kDa) was detected in kinase reactions containing GST-CTD incubated with ΔUL13 or wild-type HSV-1(F) lysates (data not shown). This band was not visible in reactions with mock- or Δα22 virus-, ΔUS3 virus-, or ΔUS3 ΔUL13 virus-infected-cell lysates. Since the beads containing GST-CTD were rinsed following the kinase reaction, this phosphorylated protein band was pulled down by the GST-CTD chimeric protein specifically in the presence of both α22 and US3. We conclude from this finding that the carboxyl-terminal domain of cdc25C likely interacts with ICP22 in a manner dependent on US3.
Phosphorylation of the carboxyl-terminal domain of cdc25C requires a cysteine residue at position 377.
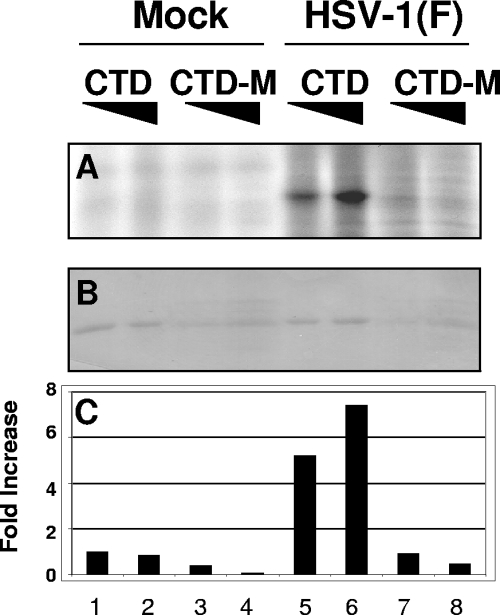
The domain of cdc25C phosphorylated by infected-cell lysates was narrowed down to the carboxyl-terminal 201 amino acids and more specifically the domain containing the catalytic phosphatase activity of the protein. The objective of this series of experiments was to investigate whether phosphatase activity was required for the phosphorylation of the carboxyl-terminal domain of cdc25C. Like other dual-specificity protein phosphatases, cdc25C depends on an essential cysteine residue in its active site for enzymatic activity (10). Thus, cdc25C loses its phosphatase activity when cysteine is replaced with serine at residue 377 (31). To investigate whether this residue, necessary for the phosphatase activity, is important for the phosphorylation of the carboxyl-terminal domain of cdc25C, we constructed chimeric proteins (GST-CTD and GST-CTD-M) in which the native carboxyl-terminal 201 amino acids or corresponding structure bearing the substitution C377S was fused to GST. The chimeric proteins were used in kinase assays in which lysates of HEp-2 cells mock infected or infected with wild-type HSV-1(F) were mixed with increasing amounts of GST-CTD or GST-CTD-M. The beads containing the chimeric proteins were collected and rinsed extensively, and the bound proteins were separated by electrophoresis on a denaturing gel, transferred, and analyzed by autoradiography (Fig. 5A). The membrane was stained with Ponceau S (Fig. 5B), and the amount of radioactivity in each band was measured with the aid of a Molecular Dynamics 860 PhosphorImager as shown in Fig. 5C. The results were as follows. Neither GST-CTD nor GST-CTD-M was phosphorylated in reaction mixtures containing lysates of uninfected cells (Fig. 5A and C, lanes 1 to 4). The GST-CTD chimeric protein was phosphorylated in a dose-dependent fashion in reaction mixtures containing lysates of wild-type-virus-infected cells (Fig. 5A and C, lanes 5 and 6). In stark contrast, the GST-CTD-M construct was not phosphorylated by the lysates of wild-type-virus-infected cells (Fig. 5A and C, lanes 7 and 8). From this we conclude that the active cysteine residue 377 of cdc25C is required in cis for the phosphorylation of the carboxyl-terminal 201 amino acids.
FIG. 5.
HSV-1(F) phosphorylation of the carboxyl-terminal domain of cdc25C requires intact cysteine residue 377. Lysate of HEp-2 cells mock infected or infected with wild-type HSV-1(F) was mixed with increasing amounts of the GST-CTD or GST-CTD-M chimeric protein on beads in the presence of [γ-32P]ATP for 30 min. The beads were rinsed, and bound proteins were eluted, separated by electrophoresis, transferred to a membrane, and analyzed by autoradiography (A). The membrane was then stained with Ponceau S (B). The amount of radioactivity in each band was quantified (C) as described in the legend to Fig. 4. GST-CTD and GST-CTD-M are abbreviated as CTD and CTD-M, respectively.
US3 and UL13 are each able to phosphorylate cdc25C and ICP22.
In earlier studies, whole-cell lysates were used to characterize the phosphorylation of cdc25C. In this set of experiments, purified viral kinases were used. The HSV-1 protein kinases US3 and UL13 and their kinase-dead point mutants US3-M and UL13-M were prepared as GST chimeras expressed in Sf9 insect cells and purified on glutathione Sepharose beads as described in Materials and Methods. The GST-US3 and GST-UL13 chimeric proteins were eluted from the beads with glutathione prior to addition to the kinase reaction. In order to avoid nonspecific GST-GST interaction between kinase and substrate, cdc25C or ICP22 was expressed as an MBP chimeric protein in E. coli and purified on amylose beads.
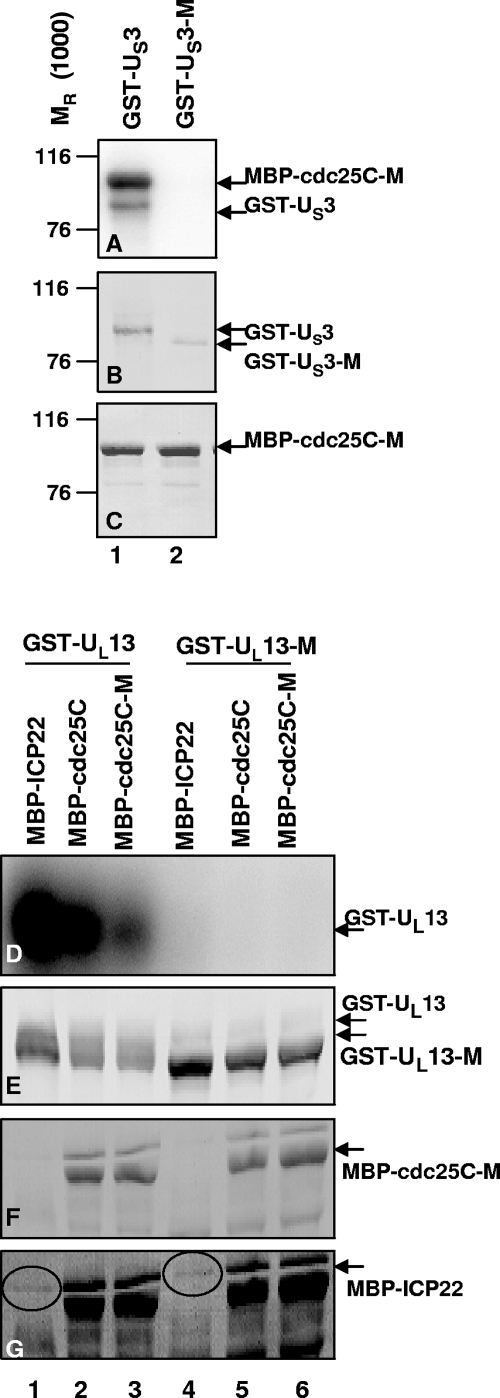
GST-US3 or GST-US3-M was reacted with MBP-cdc25C or MBP-cdc25C-M in a kinase reaction, after which the amylose beads containing the substrate were rinsed extensively and the bound proteins were eluted, electrophoretically separated on denaturing gels, and subjected to autoradiography (Fig. 6A). The membrane was stained with Ponceau S to detect total protein (Fig. 6C) and then immunoblotted for GST (Fig. 6B). GST-US3 retained interaction with the amylose beads, autophosphorylated, and also phosphorylated both MBP-cdc25C-M (Fig. 6A, lane 1) and MBP-cdc25C (data not shown) in an equivalent manner. In contrast, GST-US3-M showed no kinase activity under similar conditions (Fig. 6A, lane 2). Thus, the kinase activity associated with GST-US3 is specific to functional US3.
FIG. 6.
Purified US3 and UL13 protein kinases are active. GST-US3 or GST-US3-M was reacted with the substrates MBP-cdc25C and MBP-cdc25C-M in the presence of [γ-32P]ATP for 30 min. Beads containing the substrate were rinsed, and proteins separated by electrophoresis, transferred, and analyzed by autoradiography (A). The membrane was stained with Ponceau S to detect total protein (C) and then immunoblotted for GST (B). GST-UL13 and GST-UL13-M were each reacted with MBP-ICP22, MBP-cdc25C, or MBP-cdc25C-M on beads in the presence of [γ-32P]ATP for 30 min. The entire reaction was separated by electrophoresis, transferred to a membrane, and subjected to autoradiography (D). The membrane was stained with Ponceau S to detect total protein (F) and immunoblotted for GST (E). The Ponceau S stain from panel F was darkened (G) to highlight the MBP-ICP22 band, circled.
GST-UL13 and GST-UL13-M were each reacted with MBP-ICP22, MBP-cdc25C, or MBP-cdc25C-M on beads in a kinase reaction, after which proteins from the entire reaction were processed as described above and subjected to autoradiography (Fig. 6D). The membrane was stained with Ponceau S to detect total protein (Fig. 6F) and immunoblotted for GST (Fig. 6E). The Ponceau S stain image was darkened (Fig. 6G) to highlight the MBP-ICP22 band. GST-UL13 autophosphorylated and seemed to phosphorylate all three MBP-chimeric substrates (Fig. 6D, lanes 1 to 3), while GST-UL13-M exhibited no kinase activity (Fig. 6D, lanes 4 to 6). Thus, the kinase activity associated with GST-UL13 was specific to functional UL13. In a separate control experiment (data not shown), the bacterially expressed MBP protein by itself was not phosphorylated by GST-US3 or GST-UL13, suggesting that ICP22 and cdc25C of the MBP chimeric proteins are specific substrates of the US3 and UL13 kinases. MBP-cdc25C-M, which as a substrate behaved similarly to wild-type MBP-cdc25C, was used in subsequent kinase assays to limit dephosphorylation during the reaction.
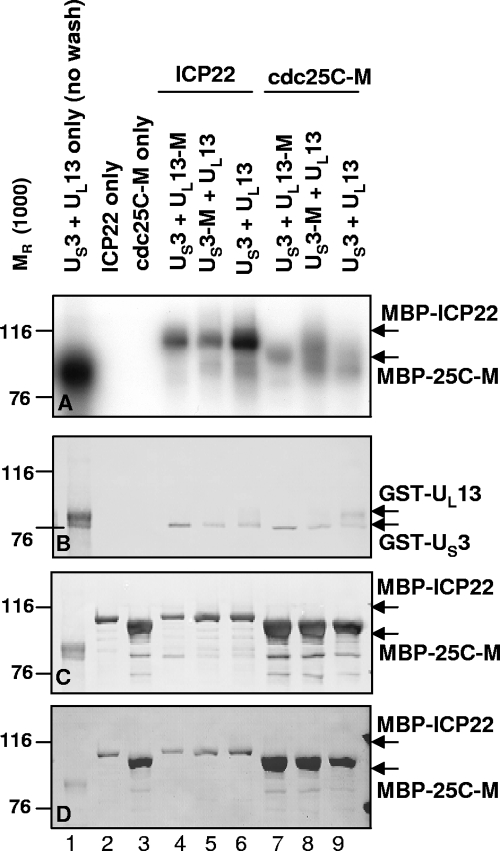
To verify that either US3 or UL13 alone is able to phosphorylate ICP22 and cdc25C, GST-US3 or GST-US3-M was mixed with GST-UL13 or GST-UL13-M in various combinations to react with MBP-ICP22 or MBP-cdc25C-M bound to beads in kinase assays. After 30 min at 30°C, the beads were rinsed extensively and the amylose bead-bound proteins separated by electrophoresis, transferred, and subjected to autoradiography (Fig. 7A). The membrane was stained with Ponceau S to detect total protein (Fig. 7D), immunoblotted first for GST (Fig. 7B), and then reprobed for MBP (Fig. 7C). In the absence of a substrate and without any washing of beads, GST-US3 and GST-UL13 each are phosphorylated (Fig. 7A, lane 1, 80- to 90-kDa bands; compare to Fig. 7B, lane 1). Substrate MBP-ICP22 or MBP-cdc25C-M did not phosphorylate itself (Fig. 7A, lanes 2 and 3), indicating that each of the substrates was free of contaminating kinase activity. The MBP-ICP22 substrate was phosphorylated by the kinase pairs GST-US3 plus GST-UL13-M or GST-US3-M plus GST-UL13 (Fig. 7A, lanes 4 and 5) but was further phosphorylated by the kinase pair GST-US3 plus GST-UL13 (Fig. 7A, lane 6), suggesting an additive effect of two active kinases on the phosphorylation of the MBP-ICP22 substrate. The MBP-cdc25C-M substrate was also strongly phosphorylated by the kinase pair GST-US3 plus GST-UL13-M, GST-US3-M plus GST-UL13, or GST-US3 plus GST-UL13 (Fig. 7A, lanes 7 to 9), showing no further increase in phosphorylation in the presence of both active kinases.
FIG. 7.
Purified US3 and UL13 kinases phosphorylate ICP22 and cdc25C. GST-US3 or GST-US3-M was mixed with GST-UL13 or GST-UL13-M in various combinations and added to MBP-ICP22 or MBP-cdc25C-M bound to beads in the presence of [γ-32P]ATP for 30 min. Beads were rinsed and proteins separated by electrophoresis, transferred, and analyzed by autoradiography (A). The membrane was stained with Ponceau S to detect total protein (D), immunoblotted first for GST (B), and then reprobed for MBP (C).
In summary, purified GST-US3 and GST-UL13 were active kinases, while GST-US3-M and GST-UL13-M were inactive, suggesting that the kinase preparations were free of contaminating kinase activity. Either GST-US3 alone or GST-UL13 alone was able to phosphorylate MBP-ICP22 or MBP-cdc25C-M but not MBP. Thus, ICP22 and cdc25C were true substrates of the kinases.
Endogenous cdc25C phosphatase activity decreases following infection with HSV-1(F) virus.
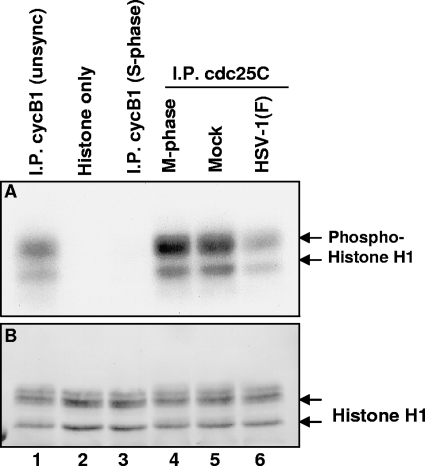
The objective of the experiments described here was to assess the activity of endogenous cdc25C in the course of HSV-1 replication using a two-step assay: first, immunoprecipitated cdc25C is used to activate cdc2 in a phosphatase reaction; next, activated cdc2 phosphorylates histone H1 in a kinase reaction. cdc25C was immunoprecipitated from lysates of HEp-2 cells harvested 18 h after infection with HSV-1(F) or mock infection. As a positive control, cdc25C was also immunoprecipitated from M-phase lysates prepared from HEp-2 cells that were treated with the microtubule inhibitor nocodazole for 18 h. For use as a substrate for cdc25C activity, inactive cyclin B1-cdc2 complex was prepared by immunoprecipitation from S-phase lysates prepared from HEp-2 cells that were treated with the DNA synthesis inhibitor hydroxyurea for 18 h. The immunoprecipitated complexes were rinsed extensively, and each cdc25C complex was mixed with inactive cyclin B1-cdc2 complex in phosphatase reaction buffer for 15 min at 30°C—the first step of this two-step assay. After the phosphatase reaction, the supernatant was removed and the combined beads were reacted with histone H1 kinase buffer containing [γ-32P]ATP for 15 min at 30°C—the second step of this two-step assay. Proteins were electrophoretically separated, transferred, and subjected to autoradiography (Fig. 8A). The blots were stained with Ponceau S to detect total protein (Fig. 8B). The results were as follows.
FIG. 8.
Endogenous cdc25C phosphatase activity decreases after HSV-1(F) infection. We examined the phosphatase activity of endogenous cdc25C using a two-step assay. cdc25C was immunoprecipitated from lysates of HEp-2 cells harvested 18 h after infection with HSV-1(F) or mock infection. As a positive control, cdc25C was also immunoprecipitated from M-phase lysates prepared from HEp-2 cells treated for 18 h with nocodazole. Simultaneously, cyclin B1-cdc2 complex was immunoprecipitated from S-phase lysates prepared from HEp-2 cells that were treated for 18 h with hydroxyurea. The immunoprecipitated (I.P.) complexes were rinsed, and each cdc25C complex was mixed with cyclin B1-cdc2 complex from S-phase cells in phosphatase reaction buffer for 15 min at 30°C—the first step of this two-step assay. After the phosphatase reaction, the supernatant was removed and the combined complex beads were incubated with histone H1 kinase buffer containing [γ-32P]ATP for 15 min at 30°C—the second step of this two-step assay. Proteins were electrophoretically separated, transferred, and analyzed by autoradiography (A). The blots were stained with Ponceau S to detect total protein (B). unsync, unsynchronized.
The histone buffer by itself showed no kinase activity (Fig. 8A, lane 2), suggesting it was free of contaminating kinases. The cyclin B1 antibody used for immune precipitation worked successfully, since it pulled down active cdc2-cyclin B1 complex from unsynchronized cell lysate (Fig. 8A, lane 1), a positive control, but very little activity from S-phase lysate (Fig. 8A, lane 3), a negative control. The inactive cyclin B1-cdc2 complex was used to determine the phosphatase activity of cdc25C complex immunoprecipitated from lysates of mock-infected or HSV-1(F)-infected cells (Fig. 8A, lanes 4 to 6). By 18 h after HSV-1(F) infection, cdc25C activity had noticeably decreased compared to mock levels of cdc25C activity (Fig. 8A, lanes 5 and 6). A similar decrease in cdc25C activity was observed at 7 h as well (data not shown). In summary, endogenous cdc25C activity decreased after HSV-1(F) infection as measured by this two-step assay.
DISCUSSION
The focus of this article and its companion article is on the function of cdc25C in infected cells. The chain of reports that led to the studies presented here may be summarized as follows.
(i) The α protein ICP22 and the viral kinase UL13 are required at least in primary cell lines but also in rodent cell lines infected at low ratios of PFU/cell for the expression of a subset of γ2 genes exemplified by UL38, UL41, and US11 (24, 25, 29). The domain of ICP22 crucial for this function maps at or near the carboxyl terminus of the protein (6, 22).
(ii) Both the US3 and UL13 protein kinases have been shown to mediate the phosphorylation of ICP22 (17, 25, 26). Analyses based on amino acid substitutions or deletions suggested that the sites of phosphorylation were in the carboxyl-terminal domain of ICP22 (22, 23).
(iii) ICP22 and UL13 induce the degradation of cyclins A and B1 and a disappearance or failure of replenishment of the inactive form of their partner, the cdc2 kinase (1).
(iii) The residual cdc2 kinase is enzymatically highly active, possibly because it acquires a new interactive partner—the UL42 viral DNA synthesis processivity factor (2).
(iv) The cdc2-UL42 complex recruits and phosphorylates topoisomerase IIα in an ICP22-dependent fashion to perform a function associated with upregulation of the γ2 gene listed above (3, 4). The question we have posed is whether cdc25C plays a role in this process.
In the accompanying report, we examined viral replication in murine cdc25C−/− cells (30). Those studies indicated that in murine cdc25C−/− cells infected at low ratios of PFU/cell, the yield of HSV-1 was at least 10-fold lower that that in sibling, wild-type cells. We also noted that in these cells cyclin B1 was degraded at the same rate as in infected wild-type MEFs and that there was an increase in the level of cdc2 over that in mock-infected cells. To resolve the role of cdc25C, we investigated the interactions of cd25C with viral proteins. The salient features of our results may be summarized as follows.
(i) cdc25C physically interacts with ICP22 but only in lysates of cells containing at least the US3 protein kinase; the interaction is greater when both the US3 and UL13 kinases are present. cdc25C also physically interacts with cdc2. (Fig. 2).
(ii) Both the US3 and UL13 protein kinases can independently phosphorylate ICP22 and cdc25C phosphatase as well as an inactive cdc25C construct carrying the C377S substitution (Fig. 3). An interesting observation is that the small amount of ICP22 in the reaction mixture with the UL13 protein kinase appeared to accept far more phosphate than the much larger amount of cdc25C (Fig. 6). One hypothesis that could explain the disparity in the amount of phosphate is that UL13 phosphorylates many more sites in ICP22 than in cdc25C.
(iii) The predominant but probably not unique site of phosphorylation of cdc25C is in the catalytic domain contained in the carboxyl-terminal half of the protein (Fig. 4). phosphorylation of the carboxyl-terminal fragment of cdc25C is abolished in a mutant carrying the C377S substitution (Fig. 5). It is not clear whether the failure to phosphorylate the carboxyl terminus is due to a lack of phosphatase activity or the change in the secondary structure of the protein.
(iv) cdc25C remains active until late in the replicative cycle, but the activity is lower than that seen in uninfected cells (Fig. 8). The loss of activity with time is not surprising inasmuch as the shutoff of host protein synthesis would preclude replenishment of the cdc25C protein.
In essence, earlier studies have identified several of the components of the regulatory cascade that optimizes the expression of the subset of late genes exemplified by UL38, UL41, and US11. These studies established that the key proteins required for expression of these genes were ICP22, UL13, cdc2, UL42, and topoisomerase IIα. The missing link was the connection between ICP22 and cdc2. In this report, we have established that cdc25C physically interacts with ICP22 and, as expected, it also interacts with cdc2. We have also established that both UL13 and US3 can phosphorylate both ICP22 and cdc25C. Coupled with the accompanying report, these results indicate that cdc25C phosphatase is recruited by HSV-1 for optimal viral expression of its genes and that at least one of its functions may be to maintain active cdc2.
A central question is the role of the US3 kinase. Earlier studies have shown that in infected cells, US3 is not required for optimal expression of the subset of γ2 genes regulated by ICP22 (24, 25, 29). The implication of this finding remains to be sorted out, particularly since the substrate of the US3 protein kinase is similar to that of protein kinase A (5). In infected cells, the interactions between ICP22 and cdk9 (8) and between ICP22 and cdc25C are dependent on or at least enhanced by the US3 kinase. It is conceivable that in the environment of uninfected cells, these interactions are facilitated by protein kinase A. A fundamental strategy of HSV-1 is to recruit and divert cellular proteins to perform novel functions, and in this instance, it is conceivable that ICP22 recruits cdc25C to perform a function in addition to or unrelated to the maintenance of cdc2 in an active state. A curious parallel is the recruitment of phosphatase 1α by the γ134.5 protein to dephosphorylate the α subunit of the translation initiation factor eIF-2 (13).
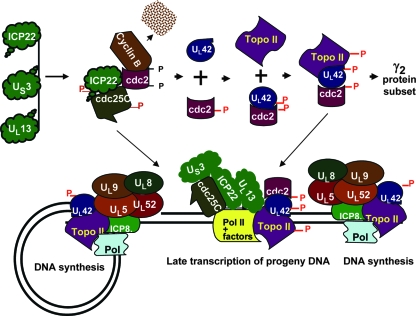
Recently Fraser and Rice (11) reported that ICP22 mediates the loss of serine-2-phosphorylated RNA polymerase 11 in the absence of other viral proteins. The data do not exclude the possibility that ICP22 is modified by cellular enzymes in the absence of infected-cell proteins. Earlier Dai-Ju et al. (7) reported that in the course of late protein synthesis, serine-2-phosphorylated RNA polymerase II is degraded in a proteasome-dependent fashion, and the authors suggested that this process is not related to dephosphorylation of the protein. Given the interaction of ICP22 with the transcriptional apparatus in the infected cell, one hypothesis that takes into account the interaction of ICP22 with cdc25C is that it brings the phosphatase to the transcriptional apparatus. The model we would like to propose is a modification of that proposed by Advani et al. (3), which describes the activation of cdc2 and its binding of UL42 to recruit topoisomerase IIα, leading to viral DNA synthesis and late transcription of progeny DNA (Fig. 9). Briefly, the new element presented in this model is that the interaction of ICP22 and cdc25C phosphatase has two objectives. One objective is to maintain cdc2 in an active state. The second objective is to recruit the complex to the transcriptional machinery, leading to dephosphorylation of serine 2.
FIG. 9.
A model of the interaction of ICP22 with cdc25C in the context of assembly of the cdc2/UL42/topoisomerase IIα complex and transcription of late genes. The model presented in this figure is an update of the model initially published by Advani et al. (4). The top part of the figure describes the formation of the UL42/cdc2/topoisomerase II complex essential for efficient transcription of the subset of γ2 proteins exemplified by UL38, UL41, and US11. The bottom panel illustrates at least in part the complexes of viral proteins involved in DNA synthesis on both strands of viral DNA and some of the proteins involved in the transcription of newly synthesized DNA. The studies presented in this and earlier reports indicate that several proteins in this complex (ICP22, UL13, and US3 cdc25C) play a role in enabling efficient transcription of late genes. The phosphates shown in red are attributed to viral kinases.
Acknowledgments
We thank Sunil Advani for invaluable discussions and H. Piwnica-Worms for providing the plasmid pGC52(cdc25Hs).
B.A.S.-D. was supported by UC MSTP. These studies were aided by National Cancer Institute grant CA83939.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Advani, S. J., R. Brandimarti, R. R. Weichselbaum, and B. Roizman. 2000. The disappearance of cyclins A and B and the increase in activity of the G2/M-phase cellular kinase cdc2 in herpes simplex virus 1-infected cells require expression of the α22/US1.5 and UL13 viral genes. J. Virol. 748-15. [PMC free article] [PubMed] [Google Scholar]
- 2.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2001. cdc2 cyclin-dependent kinase binds and phosphorylates herpes simplex virus 1 UL42 DNA synthesis processivity factor. J. Virol. 7510326-10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2003. Herpes simplex virus 1 activates cdc2 to recruit topoisomerase II alpha for post-DNA synthesis expression of late genes. Proc. Natl. Acad. Sci. USA 1004825-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2000. The role of cdc2 in the expression of herpes simplex virus genes. Proc. Natl. Acad. Sci. USA 9710996-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benetti, L., and B. Roizman. 2004. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. USA 1019411-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, K. L., and B. Roizman. 1996. The promoter and transcriptional unit of a novel herpes simplex virus 1 α gene are contained in, and encode a protein in frame with, the open reading frame of the α22 gene. J. Virol. 70172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai-Ju, J. Q., L. Li, L. A. Johnson, and R. M. Sandri-Goldin. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus 1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 803567-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand, L. O., S. J. Advani, A. P. Poon, and B. Roizman. 2005. The carboxyl-terminal domain of RNA polymerase II is phosphorylated by a complex containing cdk9 and infected-cell protein 22 of herpes simplex virus 1. J. Virol. 796757-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2357-364. [DOI] [PubMed] [Google Scholar]
- 10.Fauman, E. B., J. P. Cogswell, B. Lovejoy, W. J. Rocque, W. Holmes, V. G. Montana, H. Piwnica-Worms, M. J. Rink, and M. A. Saper. 1998. Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A. Cell 93617-625. [DOI] [PubMed] [Google Scholar]
- 11.Fraser, K. A., and S. A. Rice. 2007. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J. Virol. 815091-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassepass, I., and I. Hoffmann. 2004. Assaying Cdc25 phosphatase activity. Methods Mol. Biol. 281153-162. [DOI] [PubMed] [Google Scholar]
- 13.He, B., M. Gross, and B. Roizman. 1997. The gamma(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland, L. E., K. P. Anderson, C. Shipman, Jr., and E. K. Wagner. 1980. Viral DNA synthesis is required for the efficient expression of specific herpes simplex virus type 1 mRNA species. Virology 10110-24. [DOI] [PubMed] [Google Scholar]
- 15.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 148-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 721276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, A., M. Yamamoto, T. Ohno, H. Kodaira, Y. Nishiyama, and Y. Kawaguchi. 2005. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J. Virol. 799325-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi, Y., K. Kato, M. Tanaka, M. Kanamori, Y. Nishiyama, and Y. Yamanashi. 2003. Conserved protein kinases encoded by herpesviruses and cellular protein kinase cdc2 target the same phosphorylation site in eukaryotic elongation factor 1δ. J. Virol. 772359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, M. S., S. Ogg, M. Xu, L. L. Parker, D. J. Donoghue, J. L. Maller, and H. Piwnica-Worms. 1992. cdc25+ encodes a protein phosphatase that dephosphorylates p34cdc2. Mol. Biol. Cell 373-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavromara-Nazos, P., and B. Roizman. 1987. Activation of herpes simplex virus 1 γ2 genes by viral DNA replication. Virology 161593-598. [DOI] [PubMed] [Google Scholar]
- 21.Munger, J., A. V. Chee, and B. Roizman. 2001. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 755491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogle, W. O., and B. Roizman. 1999. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J. Virol. 734305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poon, A. P., W. O. Ogle, and B. Roizman. 2000. Posttranslational processing of infected cell protein 22 mediated by viral protein kinases is sensitive to amino acid substitutions at distant sites and can be cell-type specific. J. Virol. 7411210-11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Post, L. E., and B. Roizman. 1981. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25227-232. [DOI] [PubMed] [Google Scholar]
- 25.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 906701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc. Natl. Acad. Sci. USA 897310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 655757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sciortino, M. T., B. Taddeo, A. P. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 998318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sears, A. E., I. W. Halliburton, B. Meignier, S. Silver, and B. Roizman. 1985. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith-Donald, B. A., L. O. Durand, and B. Roizman. 2008. Role of cellular phosphatase cdc25C in herpes simplex virus 1 replication. J. Virol. 824527-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turowski, P., C. Franckhauser, M. C. Morris, P. Vaglio, A. Fernandez, and N. J. Lamb. 2003. Functional cdc25C dual-specificity phosphatase is required for S-phase entry in human cells. Mol. Biol. Cell 142984-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]