Abstract
APOBEC3 proteins are mammal-specific cytidine deaminases that can restrict retroviral infection. The exact mechanism of the restriction remains unresolved, but one model envisions that uracilated retroviral cDNA, generated by cytidine deamination, is the target of cellular glycosylases. While restriction is unaffected by UNG deficiency, it has been suggested that the SMUG1 glycosylase might provide a backup. We found that retroviral restriction can be achieved by introducing human APOBEC3G into chicken cells (consistent with the components necessary for APOBEC3-mediated restriction predating mammalian evolution) and used this assay to show that APOBEC3G-mediated restriction can occur in cells deficient in both UNG and SMUG1.
APOBEC3 proteins can protect cells from a vast range of viral invaders, but most notably, they have been shown to participate in the restriction of retroviral and retrotransposable elements that could be a threat to genome stability through their random integration. Human APOBEC3G is packaged into assembling viral particles through its interaction with the nucleocapsid region of the Gag protein (possibly with the help of RNA molecules) (1, 6, 7, 26, 27, 30, 48, 52) and exerts its antiviral effect during reverse transcription (19, 31-33, 53).
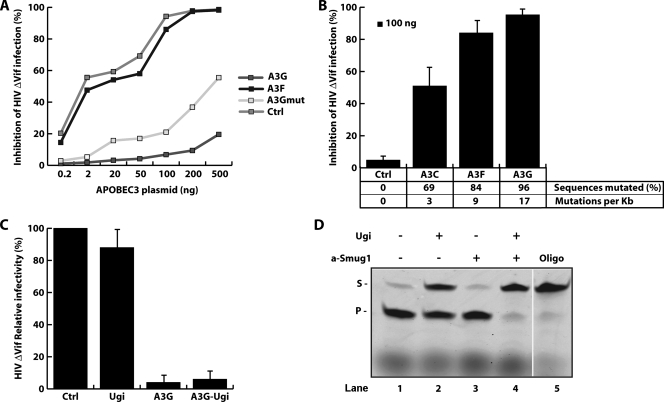
The exact mechanism of the retroviral restriction remains elusive. Although some degree of retroviral restriction can be obtained in transfection assays using active-site mutants of APOBEC3G, suggesting the existence of a deamination-independent restriction pathway (4, 8, 17, 22, 23, 29, 38, 46, 51), APOBEC3G mutants which lack deoxycytidine deaminase activity give a substantial reduction in the efficiency of restriction (5, 13, 22, 32, 34, 35, 37, 40, 45). This major deaminase-dependent pathway of restriction is associated with G-to-A hypermutation of the retroviral genome (19, 32, 33, 53). However, the restriction is unlikely to simply be the consequence of the accumulation of a high mutation load since, although dependent on the integrity of APOBEC3G's catalytic site (Fig. 1A), efficient restriction of human immunodeficiency virus (HIV) (ΔVif) particles is evident even with low mutation loads, such as those produced in presence of APOBEC3C (Fig. 1B).
FIG. 1.
APOBEC3G-mediated restriction of HIV (ΔVif) in human cells is independent of UNG. (A) Titration of transiently transfected Flag-tagged APOBEC3G (A3G), APOBEC3F (A3F), and a catalytically inactivated mutant of APOBEC3G (A3Gmut) in 293T cells, comparing their potencies in restricting HIV (ΔVif) infection. APOBEC3 expression plasmids (28), the HIV (ΔVif) vector p8.91 expressing the eGFP reporter (36), and viral infectivity assays in 293T cells are described elsewhere (28). A3Gmut has a cysteine-to-serine substitution at amino acid 288 that has been previously shown to compromise both deaminase and antiviral activities (13, 32, 34, 35, 40, 45, 53). (B) Infection assay showing that mutation load does not directly correlate with the level of inhibition of HIV (ΔVif) infection. Ctrl, control plasmid. Error bars indicate standard deviations. (C) Infection assay in 293T cells, showing that transient transfection of Ugi in A3G-expressing cells has no significant effect on HIV (ΔVif) restriction. (D) Oligonucleotide assay showing glycosylase activities of UNG and SMUG1 in 293T cell extracts. S indicates the uncleaved oligonucleotide substrate and P the cleaved product. Detailed procedures for the oligonucleotide assays were described previously (11). The amount of extract was such that the UNG activity in the control sample (lane 1) was sufficient to saturate the assay, shifting all the substrate to product.
It was initially suggested that restriction might result from the uracilation of the retroviral cDNA (16, 21, 32, 33). However, it is notable that although some reports have indicated a role for the UNG as well as possibly the SMUG1 uracil DNA glycosylase (42, 50), APOBEC3-mediated restriction is actually unaffected by a deficiency of UNG activity (24, 34). Consistent with this result, we find that expression of Ugi, a potent and irreversible inhibitor of UNG, in human 293T cells does not influence the antiviral properties of APOBEC3G on HIV ΔVif infection whether the inhibitor is expressed in the producer cells and/or in the target cells (Fig. 1C). Sequence analysis of integrated HIV DNA also revealed no significant difference in the accumulation of G→A mutations in viruses produced in presence or absence of Ugi (Table 1). However, although UNG is an evolutionarily ancient uracil-DNA glycosylase and is implicated in excising uracils that arise in DNA through dUTP misincorporation (as well as during antibody gene diversification), SMUG1 appears to be the major uracil DNA glycosylase responsible for excising uracils that arise in cellular DNA from cytidine deamination (2). Indeed, SMUG1 could be a strong contender, since it is capable of excising uracils from both single-stranded and double-stranded uracilated DNA (2, 39). Thus, the lack of effect of UNG deficiency on APOBEC3G-mediated HIV restriction does not exclude the possibility that uracil excision is important to the process. Indeed, Fig. 1D depicts a biochemical assay in which a double-stranded oligonucleotide (5′-ATTATTATTATTCCUGGATTTATTTATTTATTTATTTATTT-fluorescein-3′ and 5′-AAATAAATAAATAAATAAATAAATCCAGGAATAATAATAAT-3′) containing a central U·A pair and a fluorescein marker at its 3′ end was incubated with whole-cell extracts of Ugi-expressing 293T transfectants. The results reveal the presence of substantial residual uracil excision activity; incubation with the SMUG1-inhibiting antibody Pms-1 (25) shows this residual activity to be due to SMUG1.
TABLE 1.
Editing of HIV (ΔVif) and RSV DNA by human APOBEC3Ga
| Virus and protein | Total no. of:
|
G-to-A mutation rate (mutations/kb) | |||
|---|---|---|---|---|---|
| Sequences | Bases sequenced | Mutations | G-to-A mutations | ||
| HIV (ΔVif) | |||||
| A3G | 20 | 14,368 | 251 | 239 | 16.6 |
| A3G-Ugi | 20 | 14,216 | 226 | 212 | 14.9 |
| RSV | |||||
| A3G | 48 | 35,903 | 211 | 194 | 5.4 |
| A3G-Ugi | 48 | 35,837 | 163 | 152 | 4.2 |
Genomic DNA was extracted from target cells infected by either HIV (ΔVif) or RSV particles produced from cells expressing Flag-tagged APOBEC3G (A3G) or APOBEC3G-Ugi (A3G-Ugi). A segment of integrated viral DNA containing the eGFP reporter sequence was amplified by PCR and cloned. Independent clones were sequenced and mutations analyzed.
We were therefore interested in ascertaining whether APOBEC3G-mediated restriction would be affected by simultaneous deficiencies in both UNG and SMUG1. Although SMUG1-deficient mammalian lines have not been described, we have previously found that chicken cells do not exhibit SMUG1 activity or indeed any major UNG backup activity as judged by biochemical assays of cell extracts (9-11). We therefore asked whether human APOBEC3G could restrict an avian retrovirus in chicken cells.
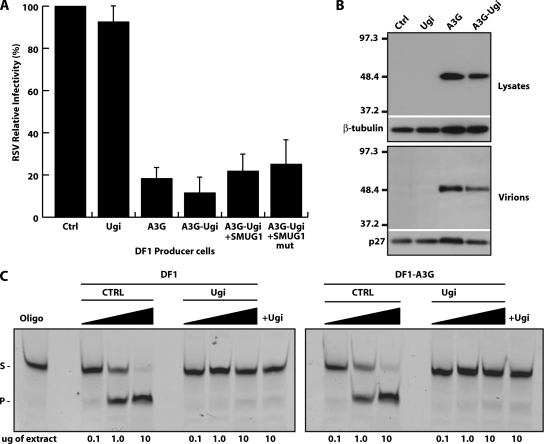
Stocks of replication-competent Rous sarcoma virus (RSV) in which the src gene had been replaced by an enhanced green fluorescent protein (eGFP) reporter (41) were used to infect either chicken fibroblast DF1 cells or DF1 transfectants that stably expressed human APOBEC3G. Supernatants from these infected cell populations were then used to infect either DF1 control cells or DF1[APOBEC3G] transfectants; eGFP expression was monitored by flow cytometry 24 h later (Fig. 2A). Expression of human APOBEC3G in the viral producer cells yielded viruses that exhibited an approximately 80% reduction in viral infection as judged by eGFP fluorescence. As with APOBEC3G-mediated restriction of HIV type 1 infectivity in human cells (44), the restriction depends upon APOBEC3G expression in the producer cells rather than in the targets. Similarly, viral encapsidation of human APOBEC3G was detected in purified virions produced from APOBEC3G-expressing DF1 cells (Fig. 2B). It is intriguing that a mammal-specific retroviral restriction factor is packaged even by an avian retrovirus, suggesting that it might exploit some ancestral pathway to force itself into the virus; binding to conserved cellular cofactors, such as the 7SL RNA, could provide such a mechanism (26, 47, 48). The restriction is also associated with retroviral deamination. Target DF1 cells were collected and integrated viral sequences analyzed for mutations. The vast majority of mutations were G-to-A transitions as read on the plus-strand viral DNA (Table 1), with a local sequence specificity characteristic of human APOBEC3G (preference for Cs at positions −1 and −2 relative to the target cytosine) (Table 2) (18- 20, 33).
FIG. 2.
UNG-independent inhibition of RSV by human APOBEC3G. (A) DF1 stable transfectants expressing A3G and/or Ugi were infected with replicative RSV-eGFP as described previously (41). Viral supernatants were then used to infect DF1 cells, and infectivity was assessed according to the percentage of cells expressing eGFP relative to the control after 24 h. eGFP expression was detectable in 95% of control (Ctrl) cells after 24 h (data not shown). APOBEC3G had no effect on viral infectivity when expressed in the target cells (data not shown). SMUG1mut contains an inactivating mutation in the catalytic site (12). Error bars indicate standard deviations. (B) Top, Western blotting performed on cell lysates, depicting expression of empty vector (Ctrl), Ugi, Flag-tagged APOBEC3G (A3G), and Flag-tagged APOBEC3G and Ugi (A3G-Ugi) stably transfected in DF1 cells. Bottom, encapsidation of Flag-tagged APOBEC3G into RSV particles. Encapsidation assays were performed as previously described (28). (C) Oligonucleotide assay performed on DF1-Ugi whole-cell extracts. The methods and oligonucleotide pair used were identical to those for Fig. 1D. UNG activity was tested on a double-stranded oligonucleotide with a central U·A pair and a fluorescein marker on the 3′ end of the U-containing oligonucleotide. S indicates the uncleaved oligonucleotide substrate and P the cleaved product.
TABLE 2.
DNA target specificity of APOBEC3G on RSVa
| Base in DNA | % at the indicated position from the deaminated cytosine
|
|||||
|---|---|---|---|---|---|---|
| APOBEC3G
|
APOBEC3G-Ugi
|
|||||
| −2 | −1 | 0 | −2 | −1 | 0 | |
| A | 12 | 5 | 14 | 4 | ||
| C | 71 | 82 | 100 | 78 | 87 | 100 |
| G | 6 | 3 | 3 | 4 | ||
| T | 11 | 9 | 5 | 5 | ||
Genomic DNA was extracted from DF1 target cells infected by RSV particles produced from DF1 cells stably expressing Flag-tagged APOBEC3G or APOBEC3G-Ugi. A segment of integrated viral DNA containing the eGFP reporter sequence was amplified by PCR and cloned. G-to-A mutations induced by APOBEC3G were compiled, and the deamination target specificity in the presence or absence of Ugi was analyzed.
We then investigated whether inhibition of uracil excision activity in DF1 cells affected APOBEC3G restriction of RSV. Introduction of a Ugi-expressing vector into DF1[APOBEC3G] cells yielded stable transfectants that exhibited no detectable uracil excision activity as judged by biochemical analysis of cell extracts using an oligonucleotide cleavage assay (Fig. 2C). Viral infectivity carried out using these cells showed that the Ugi-mediated neutralization of UNG activity in the viral producer cells had no effect on APOBEC3G-mediated viral restriction (Fig. 2A). We also did not observe any significant difference in viral restriction if Ugi was expressed in the target (as opposed to producer) cells (data not shown). The Ugi-mediated inhibition of uracil excision in DF1 also had no effect on the extent or nature of the retroviral mutations that accompanied the restriction (Tables 1 and 2). Furthermore, not only is APOBEC3G-mediated restriction in DF1 cells not diminished by inhibiting UNG activity, the restriction also is not enhanced by ectopic expression of SMUG1 (Fig. 2A).
These results reveal that restriction by APOBEC3G requires neither UNG nor SMUG1. Therefore, the dependence on the integrity of the catalytic site for the major pathway of APOBEC3-mediated restriction reflects either (i) a need for APOBEC3 to recognize cytosine but not necessarily for it to deaminate cytosine, (ii) that the deamination-induced alteration of particular nucleotide sequences within the retroviral genome compromises specific interactions necessary for retroviral replication and/or genomic integration (an explanation we think unlikely in view both of the low mutation load that accompanies restriction as well as the fact that different restricting APOBEC3 family members exhibit different local sequence preferences), or (iii) that restriction does occur through recognition of the APOBEC3-generated uracil but this recognition is not attributable to UNG or SMUG1 and might or might not involve base excision. Although thymine-DNA glycosylase is another glycosylase capable of excising uracil from DNA (14), it seems to us an unlikely candidate since it excises uracil only from U·G mispairs, which would arise only from cytidine deamination occurring following retroviral second-strand DNA synthesis. However, apart from the possibility of an involvement of an as-yet-unidentified mammalian uracil excision enzyme, it is also conceivable that restriction could be mediated by uracil recognition without excision. For example, it has recently been shown that archaebacteria harbor an endonuclease (which displays regions of homology to mammalian apurinic/apyrimidinic endonuclease 1) that cleaves DNA adjacent to uracil (15). A similar activity has been identified in flies (3). It will obviously be interesting to ascertain whether such activities can be found in mammalian cells.
Finally, it is notable that while high-level expression of APOBEC3G has been shown to be capable of inhibiting TY element transposition in yeast (13, 43), the restriction of retroviral infection achieved by expressing human APOBEC3G in chicken DF1 cells is strikingly efficient. APOBEC3 proteins appear to be restricted to mammals, and so, consistent with a recent report by Wiegand and Cullen (49), it appears that the evolution of APOBEC3 proteins was sufficient to create an effective pathway of retroviral restriction with all the factors necessary for incorporating APOBEC3 into the retrovirus as well as for the mediation of the downstream pathway of restriction already being in existence prior to mammalian evolution.
Acknowledgments
We are especially grateful to Javier Di Noia for technical assistance and valuable discussions throughout this project.
M.-A.L. is supported by a fellowship from the Canadian Institutes of Health Research and by an MRC Career Development Fellowship.
Footnotes
Published ahead of print on 13 February 2008.
REFERENCES
- 1.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 27934083-34086. [DOI] [PubMed] [Google Scholar]
- 2.An, Q., P. Robins, T. Lindahl, and D. E. Barnes. 2005. C→T mutagenesis and gamma-radiation sensitivity due to deficiency in the Smug1 and Ung DNA glycosylases. EMBO J. 242205-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bekesi, A., M. Pukancsik, V. Muha, I. Zagyva, I. Leveles, E. Hunyadi-Gulyas, E. Klement, K. F. Medzihradszky, Z. Kele, A. Erdei, F. Felfoldi, E. Konya, and B. G. Vertessy. 2007. A novel fruitfly protein under developmental control degrades uracil-DNA. Biochem. Biophys. Res. Commun. 355643-648. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 808450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, and B. R. Cullen. 2007. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology 364486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett, A., and P. Spearman. 2007. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 815000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 27933177-33184. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435108-114. [DOI] [PubMed] [Google Scholar]
- 9.Di Noia, J., and M. S. Neuberger. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 41943-48. [DOI] [PubMed] [Google Scholar]
- 10.Di Noia, J. M., and M. S. Neuberger. 2004. Immunoglobulin gene conversion in chicken DT40 cells largely proceeds through an abasic site intermediate generated by excision of the uracil produced by AID-mediated deoxycytidine deamination. Eur. J. Immunol. 34504-508. [DOI] [PubMed] [Google Scholar]
- 11.Di Noia, J. M., C. Rada, and M. S. Neuberger. 2006. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 25585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Noia, J. M., G. T. Williams, D. T. Y. Chan, J. M. Buerstedde, G. S. Baldwin, and M. S. Neuberger. 2007. Dependence of antibody gene diversification on uracil excision. J. Exp. Med. 2043209-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallinari, P., and J. Jiricny. 1996. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature 383735-738. [DOI] [PubMed] [Google Scholar]
- 15.Georg, J., L. Schomacher, J. P. Chong, A. I. Majernik, M. Raabe, H. Urlaub, S. Muller, E. Ciirdaeva, W. Kramer, and H. J. Fritz. 2006. The Methanothermobacter thermautotrophicus ExoIII homologue Mth212 is a DNA uridine endonuclease. Nucleic Acids Res. 345325-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goff, S. P. 2003. Death by deamination: a novel host restriction system for HIV-1. Cell 114281-283. [DOI] [PubMed] [Google Scholar]
- 17.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 8011710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hache, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 28010920-10924. [DOI] [PubMed] [Google Scholar]
- 19.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113803-809. [DOI] [PubMed] [Google Scholar]
- 20.Harris, R. S., S. K. Petersen-Mahrt, and M. S. Neuberger. 2002. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 101247-1253. [DOI] [PubMed] [Google Scholar]
- 21.Harris, R. S., A. M. Sheehy, H. M. Craig, M. H. Malim, and M. S. Neuberger. 2003. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat. Immunol. 4641-643. [DOI] [PubMed] [Google Scholar]
- 22.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 2822587-2595. [DOI] [PubMed] [Google Scholar]
- 23.Iwatani, Y., D. S. Chan, F. Wang, K. S. Maynard, W. Sugiura, A. M. Gronenborn, I. Rouzina, M. C. Williams, K. Musier-Forsyth, and J. G. Levin. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 327096-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser, S. M., and M. Emerman. 2006. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 80875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavli, B., O. Sundheim, M. Akbari, M. Otterlei, H. Nilsen, F. Skorpen, P. A. Aas, L. Hagen, H. E. Krokan, and G. Slupphaug. 2002. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 27739926-39936. [DOI] [PubMed] [Google Scholar]
- 26.Khan, M. A., R. Goila-Gaur, S. Opi, E. Miyagi, H. Takeuchi, S. Kao, and K. Strebel. 2007. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan, M. A., S. Kao, E. Miyagi, H. Takeuchi, R. Goila-Gaur, S. Opi, C. L. Gipson, T. G. Parslow, H. Ly, and K. Strebel. 2005. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 795870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langlois, M. A., R. C. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 331913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, X. Y., F. Guo, L. Zhang, L. Kleiman, and S. Cen. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J. Biol. Chem. 28232065-32074. [DOI] [PubMed] [Google Scholar]
- 30.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X. F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 7811841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X. F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 817238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 42499-103. [DOI] [PubMed] [Google Scholar]
- 33.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 11421-31. [DOI] [PubMed] [Google Scholar]
- 34.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 817099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyagi, E., S. Opi, H. Takeuchi, M. Khan, R. Goila-Gaur, S. Kao, and K. Strebel. 2007. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 8113346-13353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272263-267. [DOI] [PubMed] [Google Scholar]
- 37.Navarro, F., B. Bollman, H. Chen, R. Konig, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333374-386. [DOI] [PubMed] [Google Scholar]
- 38.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15166-170. [DOI] [PubMed] [Google Scholar]
- 39.Nilsen, H., K. A. Haushalter, P. Robins, D. E. Barnes, G. L. Verdine, and T. Lindahl. 2001. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 204278-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opi, S., H. Takeuchi, S. Kao, M. A. Khan, E. Miyagi, R. Goila-Gaur, Y. Iwatani, J. G. Levin, and K. Strebel. 2006. Monomeric APOBEC3G is catalytically active and has antiviral activity. J. Virol. 804673-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248305-311. [DOI] [PubMed] [Google Scholar]
- 42.Schrofelbauer, B., Q. Yu, S. G. Zeitlin, and N. R. Landau. 2005. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 7910978-10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 1029854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 45.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 27844412-44416. [DOI] [PubMed] [Google Scholar]
- 46.Strebel, K. 2005. APOBEC3G & HTLV-1: inhibition without deamination. Retrovirology 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian, C., T. Wang, W. Zhang, and X. F. Yu. 2007. Virion packaging determinants and reverse transcription of SRP RNA in HIV-1 particles. Nucleic Acids Res. 357288-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, T., C. Tian, W. Zhang, K. Luo, P. T. Sarkis, L. Yu, B. Liu, Y. Yu, and X. F. Yu. 2007. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 8113112-13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiegand, H. L., and B. R. Cullen. 2007. Inhibition of alpharetrovirus replication by a range of human APOBEC3 proteins. J. Virol. 8113694-13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, B., K. Chen, C. Zhang, S. Huang, and H. Zhang. 2007. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 28211667-11675. [DOI] [PubMed] [Google Scholar]
- 51.Yang, Y., F. Guo, S. Cen, and L. Kleiman. 2007. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology 36592-100. [DOI] [PubMed] [Google Scholar]
- 52.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 7812058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 42494-98. [DOI] [PMC free article] [PubMed] [Google Scholar]