Abstract
The betaherpesvirus human cytomegalovirus (HCMV) encodes several molecules that block antigen presentation by the major histocompatibility complex (MHC) proteins. Humans also possess one other family of antigen-presenting molecules, the CD1 family; however, the effect of HCMV on CD1 expression is unknown. The majority of CD1 molecules are classified on the basis of homology as group 1 CD1 and are present almost exclusively on professional antigen-presenting cells such as dendritic cells, which are a major target for HCMV infection and latency. We have determined that HCMV encodes multiple blocking strategies targeting group 1 CD1 molecules. CD1 transcription is strongly inhibited by the HCMV interleukin-10 homologue cmvIL-10. HCMV also blocks CD1 antigen presentation posttranscriptionally by the inhibition of CD1 localization to the cell surface. This function is not performed by a known HCMV MHC class I-blocking molecule and is substantially stronger than the blockage induced by herpes simplex virus type 1. Antigen presentation by CD1 is important for the development of the antiviral immune response and the generation of mature antigen-presenting cells. HCMV present in antigen-presenting cells thus blunts the immune response by the blockage of CD1 molecules.
The members of the CD1 family are nonclassical major histocompatibility complex class I (MHC-I) molecules which present primarily hydrophobic antigens such as lipids, in contrast to classical MHC-I molecules, which present peptides. They bind beta-2-microglobulin, require some of the same chaperones as classical MHC-I molecules, and have a structure similar to that of classical MHC-I molecules. However, they also bind the invariant chain and recycle through endocytic compartments, as do MHC-II molecules (reviewed in reference 4). They can present antigens derived from either the endoplasmic reticulum or endocytic compartments which are loaded onto CD1 molecules by a specialized antigen-loading complex (reviewed in reference 23). The possession of diverse features of the MHC-I and MHC-II systems has lead to the hypothesis that CD1 molecules are evolutionarily ancient and were present in the primordial MHC (reviewed in reference 31). This hypothesis was recently supported by the discovery of CD1 molecules in birds (24, 27, 37). CD1 molecules also have fundamental differences from the classical MHC-I system, originating partly in the hydrophobic nature of the antigens presented. CD1 genes, unlike either MHC-I or MHC-II genes, are also nonpolymorphic. Thus, the CD1 system is best considered as an ancient, unique antigen-presenting system with features common to other, better known systems.
Although CD1 genes are monoallelic or have a very restricted range of alleles, there is a high degree of divergence in the size of the CD1 family among species. For example, mice have only one functional CD1 molecule, CD1d. In contrast, humans have five CD1 molecules, CD1a to CD1e, which can be separated into two groups: group 1 consists of CD1a to CD1c, and group 2 consists of CD1d. CD1e is often classified as a group 1 molecule, although it shows significant differences from other group 1 molecules. The CD1 molecules recirculate to different intracellular compartments as dictated by tyrosine motifs within their cytoplasmic domains. The reasons behind the variation in the number of CD1 genes in contrast to the multiple alleles seen with classical MHC-I molecules are obscure but may be associated with the fact that the nature of the antigen loaded depends on the subcellular compartment to which the CD1 molecule localizes. The “traffic hypothesis” proposes that the deletion of individual CD1 genes during evolution is compensated for by the expansion of the recirculation pattern of the remaining CD1 molecules and that the recirculation patterns of CD1 molecules, like the structure of the presenting cleft, are under evolutionary pressure (9). The limited range of ligands known to be presented by CD1 molecules (4) also supports the idea that components other than the presenting cleft are important for antigen presentation.
CD1 molecules have a well-established role in antimicrobial immunity, particularly against mycobacteria. The best-studied molecule, CD1d, presents antigens to a subset of T cells called natural killer T (NKT) cells, so called due to the coexpression of NK markers and T-cell receptors on their surfaces. These cells undergo a unique development pathway during T-cell generation (reviewed in reference 3). Their role includes the modulation of both the adaptive and innate immune responses by rapid cytokine release.
Viral evasion of the immune system is a common phenomenon (reviewed in reference 1). The herpesviruses typically establish a life-long infection and thus have a high burden of genes that interfere with antigen presentation. Human cytomegalovirus (HCMV) is exceptional among the herpesviruses for having the highest number of known molecules that block MHC-I antigen presentation. These viral molecules form an overlapping and complementary shield in order to inhibit multiple pathways, reduce antigen presentation to a minimum, and thus enable HCMV to evade and persist in spite of a competent immune system.
Unlike MHC-I, which is ubiquitously expressed, the CD1 molecules have limited distributions, CD1a to CD1c and CD1e being restricted primarily to antigen-presenting cells such as dendritic cells (DC) and some B cells. Although it is well-known that HCMV interferes with antigen presentation by MHC-I molecules, little is known about the interplay between HCMV and the CD1 antigen presentation system. We and others have previously observed that CD1a is downregulated on HCMV-infected DC (12, 17, 32), and an impaired CD1a phenotype of DC isolated from immunocompetent individuals with active HCMV infections has been reported previously (11). Recent work has shown that the herpesviruses herpes simplex virus type 1 (HSV-1) and Kaposi's sarcoma-associated herpesvirus (KSHV) both inhibit CD1 presentation (34, 38, 43). CD1 molecules have also been implicated in immunity against several other viruses (2, 10, 13, 18, 19), including murine cytomegalovirus (MCMV) (42), although their precise function is a matter of debate. HCMV is known to infect DC (29, 32, 36), is latent within promonocytic cells (14, 22, 25), and can be regarded as having a strong tropism for antigen-presenting cells and DC in particular. With these characteristics in mind, we speculated that there was a high probability of finding CD1-blocking genes expressed by HCMV.
We demonstrate here the existence of multiple HCMV-encoded mechanisms for CD1 blockage that affect both the transcription and the cell surface expression of CD1 molecules. We identify one HCMV gene, the gene for the interleukin-10 (IL-10) homologue cmvIL-10, that has as part of its function the downregulation of CD1. We furthermore demonstrate that an HCMV early gene prevents the relocalization of CD1 molecules to the cell surface and thus inhibits the CD1-specific T-cell response.
MATERIALS AND METHODS
Cells and virus.
U373 cells were maintained in Eagle's minimal essential medium supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 IU of penicillin, 100 μg of streptomycin/ml, and 4.5 mM glutamine. HCMV was propagated in MRC5 cells, and HSV-1 strain F was propagated in Vero cells, the supernatant from lytic cells being cleared of debris by centrifugation and subsequently concentrated by centrifugation at 100,000 × g before being frozen in liquid nitrogen. Titers of virus stocks on MRC5 cells were determined. The infection of cells was performed by incubating the cells with virus for 1 h, and cells were then washed thrice with phosphate-buffered saline (PBS) before the addition of normal medium. The HCMV low-passage-number clinical strain NEWT was used for DC experiments, and strain AD169 was used for all U373 cell experiments except where otherwise stated. In order to generate supernatants from infected cells with or without cmvIL-10, fibroblasts were infected with AD169 or RVAdIL10C at a multiplicity of infection (MOI) of 1 (30). Medium was changed after 72 h to normal DC medium, and supernatants were collected at 96 h, filtered through 0.1-μm pores, and stored at −20°C.
Transfection of cells.
For the generation of CD1a, CD1b, and CD1c gene-transfected cell lines, cDNA derived from immature DC was amplified by PCR using appropriate primer sets and cloned into pEF6 (Invitrogen, Karlsruhe, Germany) or pEGFP-N1 (BD Biosciences Clontech, Heidelberg, Germany). CD1b in pEF6 was also mutated to remove the last eight cytoplasmic amino acid residues by PCR-based mutation. HCMV US2, US3, US6, and US11 genes were cloned from HCMV strain AD169 into the plasmid pIRES2 (BD Biosciences Clontech, Heidelberg, Germany). U373 cells were transfected with 5 μg of plasmid by using an Eppendorf multiporator at 410 V and 100 ms with hypoosmotic buffer and a 4-mm cuvette. Cotransfection with the plasmids pef-CD1a and pef-CD1b was achieved with a 1:1 mix of plasmids, 5 μg being the total volume used.
Generation of DC.
Human DC were generated from monocytes isolated from buffy coat preparations supplied by the German Red Cross, Berlin, Germany. Monocytes were isolated from peripheral blood mononuclear cells (PBMC) by negative selection with antibody-coupled magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany) and cultured in RPMI 1640 supplemented with 10% heat-inactivated FCS, 100 IU of penicillin, 100 μg of streptomycin/ml, 4.5 mM glutamine, 500 U of granulocyte-macrophage colony-stimulating factor (GM-CSF)/ml, and 200 U of IL-4/ml. Except where stated, DC were used for experiments after 2 days of culture.
Antibodies.
For phenotypic analyses of cell markers by flow cytometry, the following reagents were used: human CD1a monoclonal antibody (MAb) clone HI149 was obtained from Immunotools (Friesoythe, Germany), human CD1b MAb clone 4A7.6 and human CD1c MAb clone L161 were purchased from Immunotech (Marseille, France), and human MHC-I MAb clone W6/32 was a kind gift from G. Moldenhauer (German Cancer Research Center, Heidelberg, Germany). Human CD63 MAb clone H5C6 and isotype-matched control antibodies were purchased from BD Biosciences Pharmingen (Heidelberg, Germany). Polyclonal rabbit anti-HCMV was obtained from Chemicon (Ochsenhausen, Germany). MAb against HCMV glycoprotein B (gB) clone 1-M-12 was purchased from Dunn Labortechnik (Asbach, Germany), and MAb against HCMV IE1 clone NEA9221 was supplied by NEN Life Sciences (Heidelberg, Germany). Secondary reagents were purchased from Dianova (Hamburg, Germany).
Cytokines, Toll-like receptor (TLR) agonists, and reagents.
GM-CSF and IL-4 were supplied from Immunotools (Friesoythe, Germany). Viral and cellular IL-10 compounds were supplied from R & D Systems (Bad Nauheim, Germany). Lipoteichoic acid from Staphylococcus aureus was supplied by InvivoGen (Toulouse, France). Cycloheximide, actinomycin D, and phosphonoacetic acid were obtained from Sigma-Aldrich (Hannover, Germany). Cycloheximide was used at 200 μg/ml from 30 min preinfection to 6 h postinfection, when cells were washed thrice with PBS before being incubated in normal medium with actinomycin D at 10 μg/ml. Phosphonoacetic acid was used at 150 μg/ml.
Flow cytometry.
Cells in suspension were washed once with ice-cold FACSwash solution (PBS with 1% FCS and 0.02% sodium azide) before being resuspended with the first antibody in ice-cold FACSblock (PBS with 10% heat-inactivated FCS and 0.2% sodium azide) for 1 h. The cells were then washed in ice-cold FACSwash solution, and the staining was repeated with phycoerythrin (PE)-coupled anti-mouse secondary antibody. Subsequently, cells were washed in ice-cold FACSwash and then resuspended in 100 μl of PBS with 0.3% formaldehyde. Flow cytometry was performed on a FACSCalibur flow cytometer (Becton Dickinson, Heidelberg, Germany). Intracellular flow cytometry was performed in a similar fashion with the following differences. Cells were fixed with 4% paraformaldehyde for 5 min before being washed thrice with PBS. Cells were permeabilized by the addition of 1% saponin to FACSblock and 0.1% saponin to FACSwash.
Antigen-specific mean fluorescence index (MFI) scores were calculated by subtracting the intensity of nonspecific staining by an isotype-matched control primary antibody from the intensity of antigen-specific primary antibody staining. Enhanced intracellular background staining due to HCMV-encoded Fc binding proteins was thus excluded.
qRT-PCR.
Quantitative reverse transcriptase PCR (qRT-PCR) for CD1 genes was performed using 106 cells collected in 300 μl of lysis buffer from the MagnaPure mRNA isolation kit I (Roche Diagnostics, Mannheim, Germany). mRNA was isolated with the MagnaPure-LC device by using the mRNA kit I standard protocol. The elution volume was set to 50 μl. An aliquot of 8.2 μl of RNA was reverse transcribed in a thermocycler by using avian myeloblastosis virus RT and oligo(dT) as the primer, both from a first-strand cDNA synthesis kit, according to the protocol of the kit manufacturer (Roche). After the termination of the cDNA synthesis, the reaction mix was diluted to a final volume of 500 μl and stored at −20°C until PCR analysis. Primer sets specific for the sequences of CD1a to CD1e genes and optimized for the LightCycler (RAS, Mannheim, Germany) were developed and provided by SEARCH-LC GmbH (Heidelberg, Germany). PCR was performed with the LightCycler FastStart DNA Sybr green I kit (RAS) according to the protocol provided in the parameter-specific kits. To correct for differences in the contents of mRNA, the calculated copy numbers were normalized according to the average expression of two housekeeping genes, the cyclophilin B and beta-actin genes.
Endocytosis and recirculation assays.
For the analysis of endocytosis, cells were stained with the relevant MAb in normal culture medium on ice for 1 h. After being washed once, they were incubated for 4 h at either 4 or 37°C. Cells were then stained for remaining cell surface-bound MAb with a PE-labeled secondary antibody. The reduction in specific staining at 37°C compared to the specific staining at 4°C represents the level of endocytosis.
For the analysis of the CD1 molecules returning to the cell surface, cells were incubated with the relevant MAb for 3 h in normal culture medium at 37°C. Cells were then washed, and remaining cell surface antibody was removed by incubation in a solution of 300 mM glycine (pH 3) and 1% FCS for 2 min. Cells were washed twice in PBS and then incubated in normal medium at either 4 or 37°C for 3 h. Cells were then stained for remaining cell surface-bound MAb with a PE-labeled secondary antibody. The increase in specific staining at 37°C compared to the specific staining at 4°C represents the level of recirculation to the cell surface.
Confocal microscopy.
U373 transfectants grown on coverslips were fixed and permeabilized by acetone-methanol (1:1) at −20°C for 5 min. Thereafter, slides were washed three times before being blocked with FACSblock containing nonspecific human immunoglobulin G (Sigma-Aldrich, Germany) to block nonspecific staining. Cells were subsequently stained with MAbs against CD1a to CD1c at a 1:200 dilution and rabbit polyclonal antiserum against HCMV (Chemicon, Ochsenhausen, Germany) at a dilution of 1:400 or anti-CD63 at a dilution of 1:200. After three washes in PBS, primary antibody staining was followed by secondary antibody staining using isotype- and species-specific fluorescein isothiocyanate-, Alexa 568-, or Cy5-coupled secondary reagents at a dilution of 1:500. After a further three washes in PBS, coverslips were mounted and analyzed using a Leica DM 2500 confocal microscope and LCS software.
T-cell assays.
For proliferation assays, 5 × 104 CD1b gene-transfected U373 cells (U373-CD1b cells) were irradiated with 100 Gy and pulsed with 10 μg of lipoarabinomannan (LAM)/ml for 30 min before 5 × 104 antigen-specific T cells/well were added. The LAM-reactive T-cell line LCD4.7 was derived from lesions of a leprosy patient. Cells were cultured in triplicate in complete medium in 96-well round-bottom plates for 3 days at 37°C and 5% CO2 before [3H]thymidine (1 μCi/well; Amersham) was added over 18 h. Thereafter, cells were harvested and liquid scintillation measurements were performed using a Top Count scintillation counter (Packard, Darmstadt, Germany). The specific T-cell response was calculated by subtracting the level of baseline proliferation (that in cultures of U373-CD1b and T cells) from the level of proliferation in the presence of antigen (that in cultures of U373-CD1b and T cells with LAM).
In order to generate CD1b-specific and nonspecific CD4+ T-cell lines, 1.5 × 106 PBMC/ml were cultivated either with a 4 × 104/ml concentration of CD1b gene-transfected LCL 721.174 cells previously irradiated with 100 Gy and pulsed with 10 μM sulfatide (Sigma-Aldrich, Hannover, Germany) for 2 h or with the same quantity of untransfected, unloaded LCL 721.174 cells. LCL 721.174 is an MHC-I- and MHC-II-negative B-cell line, and sulfatide is a promiscuous antigen that is presented by group 1 CD1 molecules. PBMC were cultured in medium containing 25 U of IL-2/ml for 7 days before CD4+ cells were selected by a MACS system (Miltenyi Biotec, Bergisch Gladbach, Germany). CD4+ T cells were allowed to rest in medium with IL-2 for 2 days before use.
For cytokine release assays, autologous DC were generated by incubation with GM-CSF and IL-4 for 6 days. DC were infected with HCMV at a MOI of 3 for 3 days before use. Infected or uninfected DC (2 × 104) were mixed with 2 × 105 autologous CD4+ T cells, and the mixtures were incubated for 2 days before the collection of supernatants. Gamma interferon (IFN-γ) release was measured by an enzyme-linked immunosorbent assay (Immunotools, Friesoythe, Germany). The level of sulfatide-specific IFN-γ released by CD1b-specific CD4+ T cells was determined, as was the level of sulfatide-specific IFN-γ released from nonspecific CD4+ T-cell lines (background).
For cytotoxicity assays, 3 × 104 U373-CD1b cells were loaded with 15 mM calcein for 30 min before being pulsed with 10 μM sulfatide (Sigma-Aldrich, Hannover, Germany) for 1 h. Cells were washed and added to 3 × 105 CD1b-specific CD4+ T cells. Supernatant was collected after 5 h and evaluated on a Mithras LB940 instrument (Berthold Technologies, Barsinghausen, Germany). HCMV-specific cytotoxicity was measured by subtracting UV-inactivated virus-infected cells from virus-infected cells. The percentage of lysis was determined by comparing results with the same cells not mixed with T cells (null lysis) and cells exposed to 2% sodium dodecyl sulfate (maximum lysis).
RESULTS
Group 1 CD1 molecules on DC are downregulated by HCMV infection.
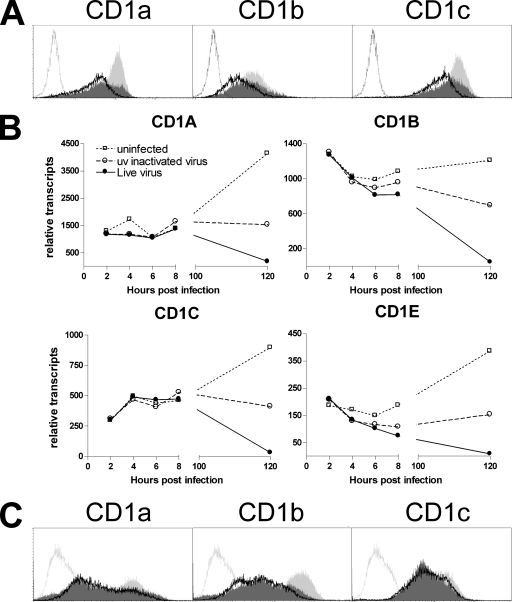
In order to investigate if HCMV affects the expression of CD1 molecules, we infected primary human cells expressing high levels of endogenous group 1 CD1 molecules with a low-passage-number clinical strain of HCMV capable of productively infecting human antigen-presenting cells (32). Flow cytometric analysis of human DC infected with HCMV strain NEWT at a MOI of 3 revealed the downregulation of all group 1 CD1 molecules 3 days postinfection (Fig. 1A and C). Transcriptional analysis by qRT-PCR showed strongly reduced transcript levels postexposure to live virus and, to a lesser extent, to UV-inactivated HCMV (Fig. 1B). These observations demonstrate that HCMV downregulates CD1 molecules on the transcriptional level.
FIG. 1.
Reduced CD1 expression on DC post-HCMV infection. (A and B) Human DC generated from monocytes were infected with HCMV strain NEWT at a MOI of 3, mock infected with UV-inactivated HCMV, or not infected before incubation for 3 days and analysis for group 1 CD1 molecules by flow cytometry (A) or qRT-PCR (B). The unfilled solid-line curves represent data for live HCMV-infected DC, the dark gray filled curves represent data for UV-inactivated HCMV-treated DC, and the light gray filled curves represent data for uninfected DC. The unfilled stippled-line curves represent data for isotype-matched control antibody. One typical example from three is given. (C) Human DC were generated by 7 days of incubation with cytokines before being infected as described in the legend to panels A and B. One typical example of two is given.
HCMV-encoded IL-10 interferes with CD1 antigen presentation on the transcriptional level.
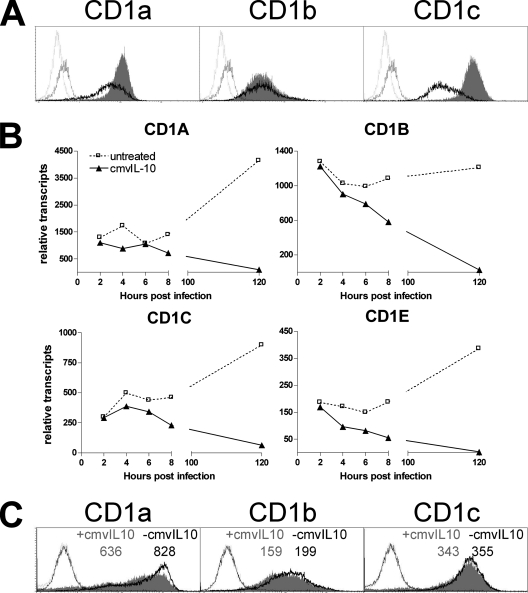
Since in DC, transcriptional downregulation postinfection was apparent, we decided to test viral factors that may regulate CD1 transcription. HCMV encodes an IL-10 homologue (cmvIL-10) which shapes the phenotype, function, and survival of DC (20, 33). We investigated whether cmvIL-10 also interferes with CD1 antigen presentation pathways. Both cell surface expression and relative transcript numbers of group 1 CD1 molecules on DC were decreased after exposure to cmvIL-10 (Fig. 2A and B). Similar observations were made using human IL-10 and an IL-10 homologue encoded by Epstein-Barr virus (data not shown). DC cultivated in supernatant from fibroblasts infected with either AD169 or the cmvIL-10 gene deletion virus RVAdIL10C (30) also showed stronger downregulation of CD1 molecules in the presence of cmvIL-10 than in the absence of cmvIL-10 (Fig. 2C). Thus, virus-encoded IL-10 mimics endogenous IL-10 by downregulating CD1 transcripts and represents a novel mechanism by which HCMV may evade CD1-restricted T-cell responses.
FIG. 2.
cmvIL-10 downregulates the transcription of CD1 molecules. (A and B) Human DC were exposed to cmvIL-10 at 25 ng/ml or left untreated for 3 days before analysis for group 1 CD1 molecules by flow cytometry (A) or qRT-PCR (B). The unfilled solid-line curves represent data for cmvIL-10-treated DC, the gray filled curves represent data for untreated DC, and the unfilled black-, stippled-line curves and unfilled gray-, stippled-line curves represent data for isotype-matched control antibodies corresponding to cmvIL-10-treated DC and untreated DC, respectively. One typical example of three is given. (C) DC were incubated for 3 days in medium supplemented with 10% supernatant from fibroblasts infected with AD169 or RVAdIL10C before analysis by flow cytometry. RVAdIL10C is a mutant of AD169 which lacks cmvIL-10. Data for DC treated with RVAdIL10C supernatant are represented by unfilled black-, solid-line curves, those for DC treated with AD169 supernatant are represented by light gray filled curves, and unfilled black-, stippled-line curves and unfilled gray-, stippled-line curves represent data for isotype-matched control antibodies corresponding to DC treated with RVAdIL10C supernatant and DC treated with AD169 supernatant, respectively. MFIs are shown above the respective curves. One typical example of two is given.
Posttranslational HCMV-induced downregulation of cell surface CD1.
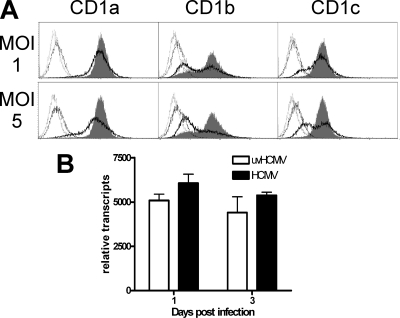
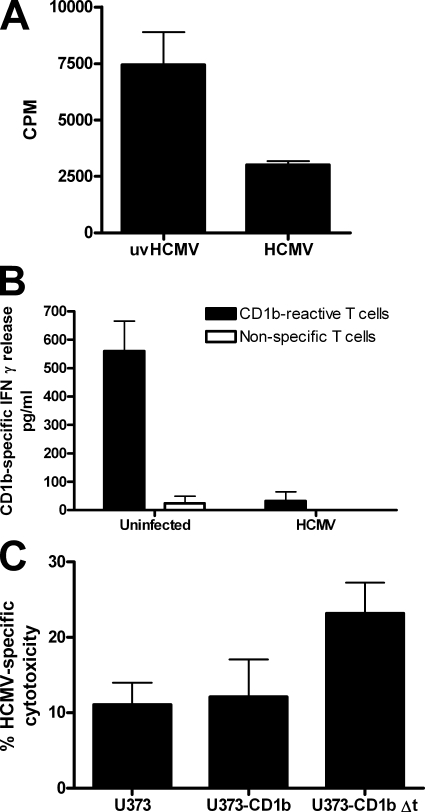
As both alpha- and gammaherpesviruses (HSV-1 and KSHV) have been shown to possess posttranscriptional CD1-blocking mechanisms (34, 38, 43), the possibility of a posttranscriptional component of HCMV-encoded CD1 interference was investigated. U373 astroglioma cells expressing group 1 CD1 molecules driven by the constitutively active human EF-1α promoter were infected with HCMV strain NEWT. Infection downregulated all group 1 CD1 molecules, although CD1b was especially sensitive to this effect (Fig. 3A). This downregulation was posttranscriptional, as qRT-PCR analysis showed no significant inhibition (Fig. 3B). HCMV reduction of cell surface CD1b resulted in the functional inhibition of CD1b-reactive T cells (Fig. 4). Effective blocking of CD1b-specific cytotoxicity was demonstrated in cell lines in which the CD1b cytoplasmic domain was intact, whereas increased cytotoxicity was shown when this domain was removed (Fig. 4C). This blocking mechanism was furthermore conserved in the fibroblast-adapted HCMV strain AD169 (Fig. 5A), which has fewer open reading frames than clinical strains of HCMV (6). Taken together, these data indicate that HCMV interferes with antigen presentation through group 1 CD1 molecules by a posttranslational mechanism.
FIG. 3.
Antigen presentation by CD1 molecules is blocked by HCMV via a posttranscriptional mechanism. U373 cell lines permanently transfected with group 1 CD1 molecules were infected with HCMV strain NEWT at a MOI of 1 or 5 or mock infected with the same quantity of UV-inactivated virus (uvHCMV). Cells were analyzed by flow cytometry after 3 days (A) or by qRT-PCR analysis of CD1b after 1 and 3 days (B). The unfilled black-, solid-line curves represent data for HCMV-infected cells, the gray filled curves represent data for mock-infected cells, and the unfilled black-, stippled-line curves and unfilled gray-, stippled-line curves represent data for isotype-matched control antibodies corresponding to HCMV-infected cells and mock-infected cells, respectively. One typical example of three is given. n ≥ 3; error bars show standard deviations (SD) of the means.
FIG. 4.
HCMV interferes with the stimulation of CD1b-specific T cells. (A) U373-CD1b cells were infected with HCMV strain NEWT at a MOI of 2 or mock infected with the same quantity of UV-inactivated virus (uvHCMV) and used at 3 days postinfection to stimulate a CD1b-specific T-cell line. The levels of antigen-specific activation of T cells were measured by evaluating [3H]thymidine uptake during proliferation and are shown as counts per minute, with error bars showing SD of the means. (B) Autologous DC were infected with HCMV strain NEWT at a MOI of 3 for 3 days before the addition of CD1b-reactive autologous CD4+ T cells or nonspecific T cells. IFN-γ release was measured by an enzyme-linked immunosorbent assay 2 days after stimulation. (C) U373-CD1b cells or U373-CD1b cells with the CD1b cytoplasmic domain deleted (U373-CD1b Δt) were infected as described for panel A and then loaded with calcein and mixed with CD1b-specific T cells at a ratio of 1:10. Supernatant was collected after 5 h, and the quantity of calcein released was determined by fluorimetry. Results reflect experiments with cells from two different donors.
FIG. 5.
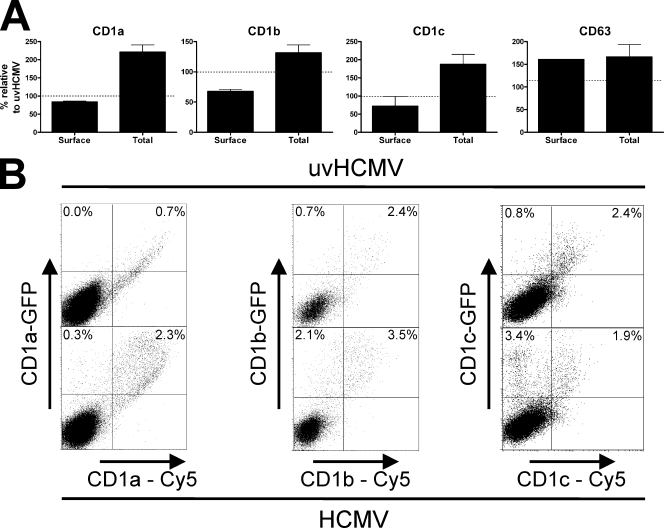
HCMV-induced cell surface downregulation of group 1 CD1 molecules is accompanied by intracellular accumulation. (A) U373 cell lines permanently transfected with group 1 CD1 molecules were infected with strain AD169 or UV-inactivated virus (uvHCMV) at a MOI of 5 for 3 days. Cells were then harvested and either analyzed by flow cytometry for cell surface antigens or fixed, permeabilized, and analyzed for whole-cell antigens. (B) U373 cells were infected with strain AD169 or UV-inactivated virus at a MOI of 5 for 1 day. Cells were then transiently transfected with plasmids containing group 1 CD1 molecules fused to GFP. Two days later, cells were analyzed by flow cytometry for total CD1 expression (GFP) and cell surface CD1 expression (Cy5). n ≥ 3; error bars show SD of the means.
Accumulation of intracellular group 1 CD1 molecules post-HCMV infection.
In order to investigate how this reduction takes place, we used intracellular flow cytometry to measure the total level of CD1 in infected cells. A comparison of cell surface group 1 CD1 staining with total CD1 expression from permeabilized cells revealed that HCMV-infected cells upregulated the total quantity of CD1 while downregulating cell surface CD1 expression (Fig. 5A). Among the CD1 molecules, cell surface CD1b was the most sensitive to HCMV infection. The apparently weak reduction of cell surface CD1a, however, reflects an efficient block with regard to the large increase in CD1a present in infected cells. As transcription was not affected, the conclusion was that group 1 CD1 molecules accumulated postinfection. The transient transfection of infected cells with genes for group 1 CD1 molecules expressing green fluorescent protein (GFP) in the cytoplasmic domain clearly demonstrated that HCMV infection resulted in the retention of CD1 molecules intracellularly (Fig. 5B), with a high percentage of cells expressing reduced cell surface CD1 in comparison to total CD1 (GFP). These findings demonstrate that the posttranslational HCMV inhibition of CD1 cell surface expression is accompanied by the intracellular accumulation of CD1 molecules.
HCMV-induced CD1 blocking is rapid and efficient.
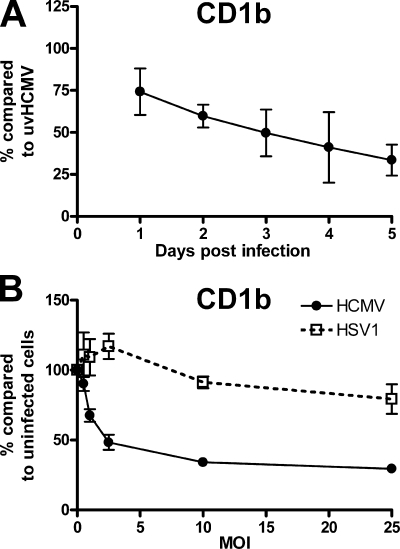
In order to better quantify the strength of blockage, we further analyzed cell surface CD1b expression postinfection. The analysis of U373-CD1b cells infected with HCMV at a MOI of 2 over time showed increasing blockage up to day 5 postinfection (Fig. 6A). Blockage was also dependent on the viral titer (Fig. 6B). The alphaherpesvirus HSV-1 has been shown previously to upregulate CD1b at a MOI of 1 and block expression at a MOI of 100 (34). HSV-1 infection at low MOIs (1-5) could be seen to result in an increase in cell surface CD1b (Fig. 6B). HCMV, in contrast, showed robust CD1b blockage with no sign of upregulation at low MOIs. Although tests showed that U373 cells were fully infected with both viruses at all viral titers above 1, the titers of HSV-1 and HCMV were determined on MRC5 and not U373 cells. Thus, the titers given refer to the input virus, and a higher level of infection with one or the other virus cannot be excluded. Collectively, these results show that HCMV induced more rapid, longer-lasting, and more efficient CD1 blocking than HSV-1.
FIG. 6.
Time course and titer dependency of HCMV-encoded CD1 blockade. U373-CD1b cells were infected with strain AD169 at a MOI of 2 for 1 to 5 days for a comparison of CD1b levels to those in cells infected with UV-inactivated virus (uvHCMV) (A) or a MOI of 0.5 to 25 for 3 days for a comparison of CD1b levels to those in uninfected cells (B). Cells were subsequently harvested, and cell surface CD1b was analyzed by flow cytometry. For comparison, cells were infected with HSV-1 strain F at a MOI of 0.5 to 25 for 1 day before analysis by flow cytometry. n = 3; error bars show SD of the means.
HCMV-encoded MHC-I-blocking proteins do not block group 1 CD1 molecules.
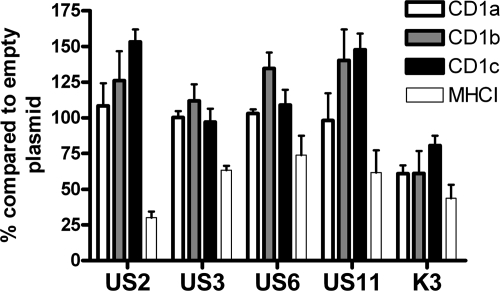
HCMV encodes several MHC-I-blocking molecules (US2, US3, US6, and US11) which may also interact with the MHC-I-related CD1 molecules. We transfected U373 cells expressing group 1 CD1 molecules with plasmids which expressed GFP in tandem with the viral immune evasion gene (Fig. 7). Neither US2, US3, US6, nor US11 blocked the cell surface expression of group 1 CD1 molecules. In contrast, KSHV K3, a molecule known to interfere with CD1d expression (38), could be shown to decrease the expression of all group 1 CD1 molecules. Furthermore, instead of being downregulated, CD1b and CD1c were upregulated by HCMV MHC-I-blocking molecules in a fashion similar to that of the upregulation of these molecules by HSV-1 ICP47 (34). Thus, the HCMV-encoded CD1b block is sufficiently strong to overcome CD1b upregulation induced by viral blockage of MHC-I surface expression. The KSHV K3-induced CD1 block of 25% was also significantly weaker than the CD1b block induced by HCMV infection, which at a MOI of 25 was 70% (Fig. 6B). Thus, the overexpression of the multifunctional KSHV K3 molecule was not as efficient as active HCMV infection in reducing CD1b surface expression. These results demonstrate that HCMV-encoded CD1 blockage is efficient and not a result of a viral MHC-I evasion protein.
FIG. 7.
HCMV-encoded MHC-I-blocking molecules do not inhibit group 1 CD1 molecule surface expression. U373 cells expressing group 1 CD1 molecules were transfected with plasmids expressing viral MHC-I-blocking molecules under the control of the cytomegalovirus immediate-early promoter in tandem with GFP under the control of an internal ribosome entry site sequence. Thirty-six hours posttransfection, cells were analyzed by flow cytometry. The CD1 or MHC-I on GFP+ cells was compared to that on GFP− cells, and the resulting ratio was compared to that given by control (empty) vector transfection. The decrease in this ratio denotes the efficiency of the blockage. n = 3; error bars show SD of the means.
Group 1 CD1 blockage by HCMV is caused by an early viral gene.
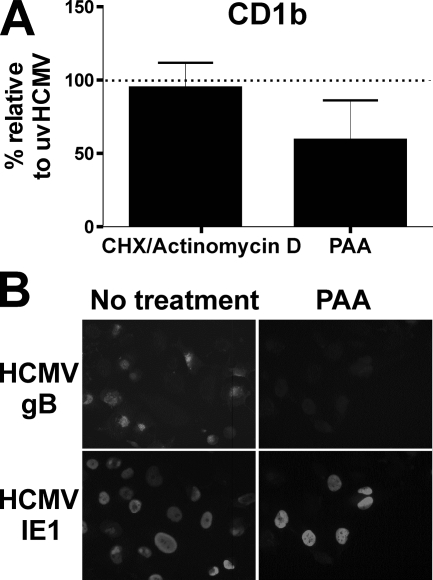
In order to identify which class of viral gene was responsible for CD1 blockage, we infected cells under conditions in which only immediate-early or immediate-early and early genes were expressed. The analysis of CD1b molecule expression 3 days post-HCMV infection in the presence of actinomycin D after an initial short phase of transcription demonstrated that an immediate-early gene was not responsible (Fig. 8A). When viral DNA replication was inhibited by phosphonoacetic acid, thus preventing late gene expression, there was only a slight alleviation of blockage (Fig. 8). Similar results were seen for CD1a and CD1c (data not shown). The responsible function is therefore encoded by an early viral gene.
FIG. 8.
The HCMV-induced group 1 CD1 block is caused by an early viral protein. U373 cells expressing group 1 CD1 molecules were infected with strain AD169 or UV-inactivated virus (uvHCMV) at a MOI of 5 for 3 days. Cells were treated with 150 μg of phosphonoacetic acid (PAA)/ml or cycloheximide (CHX)-actinomycin D before being analyzed by flow cytometry (A) and immunocytometry staining (B) for the expression of the HCMV immediate-early IE1 gene and the late gB gene. n = 3; error bars show SD of the means.
Group 1 CD1 blockage by HCMV is dependent on the cytoplasmic domain of the CD1 molecule.
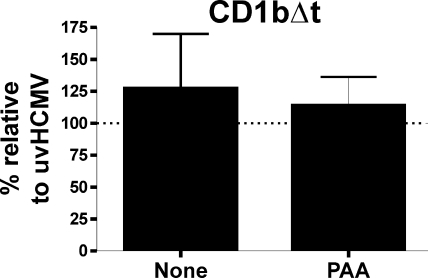
The analysis of a mutant CD1b molecule in which the cytoplasmic tyrosine motif was missing (CD1bΔt) and which was thus not endocytosed showed no significant downregulation in response to HCMV infection (Fig. 9). These results show that the cytoplasmic domain of group 1 CD1 molecules is necessary for HCMV-encoded CD1 blockage.
FIG. 9.
The HCMV-induced group 1 CD1 block is dependent on the cytoplasmic domain of the CD1 molecule. U373 cells expressing a mutant CD1b which lacks the last eight amino acids (CD1bΔt) were infected with strain AD169 or UV-inactivated virus (uvHCMV) at a MOI of 5 for 3 days. Cells were treated with 150 μg of phosphonoacetic acid (PAA)/ml or mock treated before being analyzed by flow cytometry. n = 3; error bars show SD of the means.
Confocal analysis of CD1 expression in HCMV-infected cells.
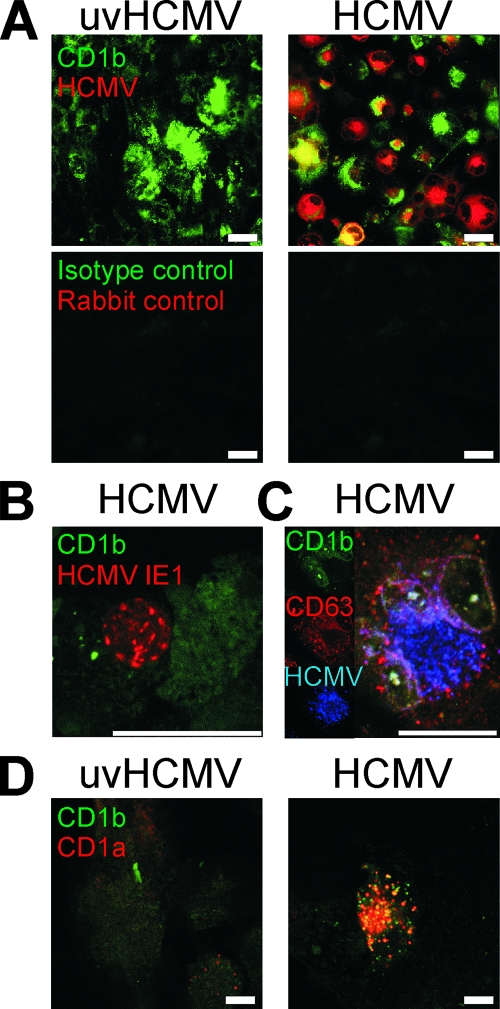
Confocal analysis was undertaken to visualize the subcellular changes shown by flow cytometry (Fig. 10). Confocal analysis of U373 transfectants infected with HCMV strain AD169 for 5 days demonstrated the principal features of HCMV infection and CD1 molecules. Group 1 CD1 molecules accumulated intracellularly postinfection (Fig. 10A). The partial colocalization of CD1 and HCMV proteins was often visible (Fig. 10A, C, and D). A central core of colocalization was often surrounded by HCMV-free CD1+ accumulation and an area of CD1-free HCMV staining. Colocalization was seen to increase later in infection, and large intracellular multivesicular CD1+ inclusions were a characteristic feature of late HCMV infection. The strong expression of viral proteins was associated with decreased CD1 expression in transfected cells and in primary DC (Fig. 10A and B). In order to analyze further the CD1+ compartment, we costained infected cells for CD1b and CD63, a lysosomal marker that colocalizes strongly with CD1b (41). CD1b in infected cells retained strong colocalization with CD63 while the cells gained positivity for HCMV proteins (Fig. 10C), thus demonstrating that CD1b remained primarily in the lysosome postinfection. In order to investigate if the recirculation and localization of all the CD1 molecules were normal postinfection, we cotransfected infected cells with genes for CD1a and CD1b, molecules which normally segregate into different subcellular compartments, the early endosome and the lysosome, respectively (Fig. 10D). As predicted, these molecules showed no colocalization in cells exposed to UV-inactivated virus. However, postinfection, strong colocalization could be seen, demonstrating that the normal recirculation of these molecules was altered by infection. The CD1+ compartment has some similarity to the previously reported virion assembly compartment (VAC) (39). Our data support the notion that viral infection disturbs the normal trafficking of CD1 molecules, which are trapped within the VAC or an associated compartment.
FIG. 10.
Confocal microscopy analyses of group 1 CD1 distribution and HCMV proteins postinfection. (A) U373 cells expressing CD1b were infected with AD169 or UV-inactivated virus (uvHCMV) at a MOI of 2 for 5 days before being fixed and permeabilized. Cells were stained for viral proteins (red) by using a polyclonal rabbit antiserum generated using preparations of HCMV virions and for CD1b (green). Normal rabbit serum and isotype-matched control antibodies were used to demonstrate specificity. (B) DC infected with HCMV strain NEWT at a MOI of 3 for 3 days were stained for HCMV IE1 (red) and CD1b (green). (C) U373-CD1b cells infected as described in the legend to panel A were stained for CD1b (green), CD63 (red), and HCMV proteins (blue). (D) U373 cells were infected with AD169 or UV-inactivated virus at a MOI of 2 and incubated for 24 h before being cotransfected with plasmids expressing CD1a and CD1b. After a further 4 days, cells were stained for CD1a (red) and CD1b (green). Scale bars show 25 μm.
CD1 recirculation affected in HCMV-infected cells.
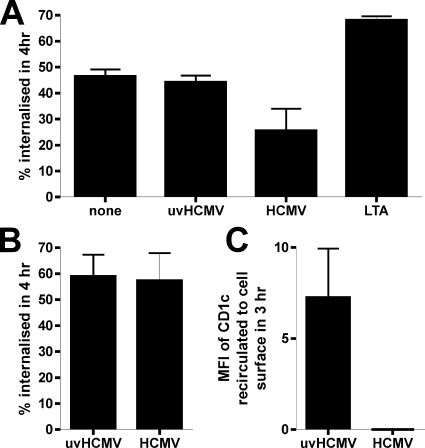
In order to clarify how the CD1 molecules were accumulating intracellularly, we measured the cellular uptake of cell surface CD1 molecules postinfection. CD1c was chosen in preference to CD1b, as the levels of CD1c expressed on infected DC were generally higher than those of CD1b and, thus, CD1c was more suited to this type of experiment. Furthermore, CD1c recirculates to the late endosome, a more benign environment for bound antibody than the acidic lysosome, where CD1b resides. DC infected with live HCMV strain NEWT at a MOI of 3 for 3 days showed a significant decrease in the uptake of CD1c (Fig. 11A), although this effect was less pronounced than the decrease in receptor-mediated phagocytosis (data not shown). This decrease in CD1c uptake was not due to DC maturation, because both UV-inactivated HCMV and lipoteichoic acid, a TLR-2 agonist, did not decrease CD1c endocytosis. This finding is also in line with previous reports that DC maturation does not affect CD1 molecule trafficking (5). U373-CD1c cells, in contrast, showed no alteration in CD1c internalization post-HCMV infection (Fig. 11B). However, an analysis of CD1c molecules reappearing at the surfaces of U373 cells showed the inhibition of CD1c recirculation post-HCMV infection (Fig. 11C). Thus, a major factor in HCMV-encoded group 1 CD1 molecule blockage is the inhibition of cell surface relocalization of endocytosed CD1 molecules. In DC, however, further mechanisms, such as the inhibition of endocytosis, also contribute to CD1 blockage. Taken together, these data demonstrate that HCMV prevents the correct recirculation of CD1 molecules.
FIG. 11.
The recirculation of CD1 molecules is altered post-HCMV infection. (A) DC were infected with HCMV (strain NEWT) or UV-inactivated HCMV (uvHCMV) at a MOI of 3 for 3 days. For comparison, DC were treated with lipoteichoic acid (LTA), a TLR-2 agonist. Thereafter, cells were stained with CD1c or isotype-matched control antibody, washed, and incubated for 4 h at 4 or 37°C in normal medium. Secondary antibody was then added, and the cells were analyzed by flow cytometry. The change in the staining intensity as a percentage of the staining intensity at 4°C was taken as the degree of endocytosis. (B) U373-CD1c cells infected with AD169 or UV-inactivated AD169 at a MOI of 2 for 5 days were stained with primary antibody or isotype-matched control antibody, washed, and incubated for 4 h at 4 or 37°C in normal medium. Secondary antibody was then added, and the degree of endocytosis was calculated as described for panel A. (C) Alternatively, the U373-CD1c cells were stained with primary antibody or isotype-matched control antibody for 3 h at 37°C. Cell surface antibody was removed by a 2-min wash in an acid buffer, and the cells were then incubated for 3 h at either 4 or 37°C in normal medium. Secondary antibody was then added, and the increase in the staining intensity (MFI) compared to the staining intensity at 4°C was taken as the level of recirculation to the cell surface within this time period. n = 3; error bars show SD of the means.
DISCUSSION
The CD1 family is the premier antigen-presenting system for hydrophobic ligands in mammals. It is supported by a system of molecules responsible for the correct loading and processing of ligands, analogous to the transporter associated with antigen processing for MHC-I (reviewed in reference 23). HCMV is well-known to block MHC-I and MHC-II antigen presentation by both direct interference with the presenting molecule itself and alterations in the antigen-presenting machinery (reviewed in reference 21). We here show that HCMV infection also blocks CD1 antigen presentation. We have furthermore identified viral cmvIL-10 as being responsible for the transcriptional downregulation of CD1. The transcriptional downregulation of CD1 molecules as an immune evasion strategy has been demonstrated previously for Mycobacterium tuberculosis (40). Our data also clearly show that HCMV possesses at least one further CD1-blocking gene or mechanism which operates posttranscriptionally during the early phase of infection. This block is distinct from known HCMV-encoded MHC-I-blocking molecules and is associated with the intracellular accumulation of CD1 molecules in infected cells due to alterations in CD1 recirculation.
The potential advantage of CD1 blockage for the virus is substantial. CD1-reactive T cells are thought to jump-start the immune response to pathogens and help steer the subsequent development of the adaptive immune response. The most intensively researched system is that of the CD1d-reactive NKT cells. NKT cells react very rapidly to antigens, releasing high quantities of IFN-γ and potentiating NK activity and DC maturation. NKT cells also have a high cytotoxic capacity. The characteristics of group 1 CD1-restricted T cells are less well understood but include cytotoxicity and a high capacity to produce cytokines, particularly IFN-γ (reviewed in reference 4). The downregulation of CD1 molecules by viruses thus potentially impairs both adaptive and innate immunity, yielding for the virus a significant advantage. It is of interest that the expression of CD1 molecules is also frequently used as a marker for the positive identification of DC, and the active downregulation of CD1 by HCMV thus may impair the classification of infected cells.
It is well-known that MHC-I expression in HCMV-infected cells reflects a composite of cellular upregulation mechanisms and viral blocking mechanisms. Cellular IFN release and pattern recognition receptor signaling postinfection upregulate the transcription of MHC-I, and this upregulation is countered by HCMV with multiple mechanisms. These include blocks of IFN production, signaling, transcription, and posttranscriptional processing of the MHC-I heavy chain (reviewed in references 15, 16, 21, 26, 28, and 35). The CD1 phenotype of HCMV-infected cells is similarly a composite of cellular upregulation mechanisms and viral blocking mechanisms. Monocytes, in response to pathogen-induced inflammation and GM-CSF release, generate high levels of group 1 CD1 transcripts and molecules. The inhibition of MHC-I expression by viral infection further enhances CD1b expression on the cell surface. The most important site for the loading of ligands onto CD1 molecules, in contrast to MHC-I molecules, is thought to be the endosomal or lysosomal compartment (reviewed in references 4 and 23). Thus, unlike for MHC-I antigen presentation, not only fresh transcription but also recirculation is important for CD1 antigen presentation. We have shown that HCMV efficiently blocks both elements.
A number of parallels between HCMV and HSV-1 (34) with regard to group 1 CD1 molecules are apparent. Both viruses block CD1 molecule expression at the cell surface, both inhibit CD1 recirculation, and both show the colocalization of viral proteins and CD1 molecules. These parallels imply that the blocking mechanism is conserved between the viruses. The differences between the viruses within these broad parameters are also illuminating. The HSV-1 block of CD1b is relatively weak and requires a high MOI (34), whereas the HCMV block of CD1b is strong. This potent block suggests that CD1b may have a specific role in immunity against HCMV. This preference may be explained by the sites of latency of the two viruses, CD1b being found predominantly on DC. Other human herpesviruses have been shown previously to affect group 2 CD1 molecules (34, 38, 43), for which the inhibition of recirculation and the accumulation of CD1d could also be demonstrated. KSHV K3 and K5 both result in the accumulation of intracellular CD1d and the breakdown of MHC-I in the lysosome (38). A comparison with MCMV is also helpful. Investigations in vivo found that although CD1d is potentially capable of inhibiting MCMV infection, it does so only when exogenous ligand is added (42). Although the situation is not precisely the same, as mice lack group 1 CD1 molecules, it is plausible that MCMV encodes an effective CD1-blocking mechanism which prevents the presentation of endogenous ligand.
The nature of the posttranscriptional blocking molecule(s) remains, for the moment, a matter of speculation. The clinical strain used to infect DC did not have a significantly different phenotype when used to infect U373 cells expressing group 1 CD1 molecules, indicating that a blocking molecule was less likely to be present within the region deleted in strain AD169 than elsewhere. Furthermore, the fact that the viral gene was sensitive to actinomycin D but insensitive to phosphonoacetic acid treatment shows that an early gene is responsible for the blockage of cell surface CD1. This blockage requires the cytoplasmic domain of the CD1 molecule, either for the binding of the viral protein or for subsequent subcellular localization. KSHV K3 and K5 also require this domain in order to inhibit CD1d expression by promoting retention, suggesting that the HCMV-encoded blocker may work in a similar fashion.
It is also evident that the interaction between the host and HCMV with regard to CD1 molecules cannot be interpreted simply as a block induced exclusively by one viral molecule. The CD1+ intracellular inclusion and colocalization data suggest that a direct association between viral proteins and CD1 molecules may exist. However, colocalization takes place predominantly on day 4 postinfection in U373 cells, when the blockade is already well established. The CD1+ compartment may be the same as the VAC, where virions are assembled, and consequently a site of intense lipid trafficking. The trapping of CD1 molecules within this compartment may serve as a late blocking mechanism common to several herpesviruses. Furthermore, differences seen in recirculation between DC and U373 cells implicate additional viral genes which are active only within the environment of the DC. It has also not escaped our attention that the CD1 molecules most strongly affected by HCMV infection specifically and herpesviral infection generally are those that recirculate to the most central compartments. CD1b is localized almost exclusively in the lysosome, CD1c is localized in the late endosome, and CD1a recirculates only to the early endosome. CD1a is the least affected and CD1b is the most affected by HCMV infection. Such a block would most strongly affect the CD1 molecules that naturally come in closest contact with the VAC.
Bearing this situation in mind, one can postulate the contribution that CD1 evasion makes to HCMV infection. HCMV latent in myeloid progenitors reactivates as the progenitors mature into antigen-presenting cells such as DC. HCMV inhibits initial CD1 transcription via cmvIL-10 and can also prevent the subsequent presentation of antigen loaded onto CD1 molecules in endocytotic compartments as it reactivates from latent infection. The capacity to inhibit antigen presentation in DC, where group 1 CD1 molecules are predominantly located, would shield the reactivated virus during the vulnerable phase of transfer from latently infected cells into endothelial and other cell types where high viral titers can be generated and thus ultimately promote viral dissemination.
Acknowledgments
We thank T. Kaiser for assistance in flow cytometry, S. Schuhmann for assistance in confocal microscopy, U. Noack and A. Schorlies for excellent technical assistance, and G. Moldenhauer for providing helpful antibodies.
We have no conflicting financial interests.
This work was supported by the Deutsche Forschungsgemeinschaft (grants SFB421 to G.S. and S.H.E.K. and SFB490 to B.P.).
Footnotes
Published ahead of print on 20 February 2008.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashkar, A. A., and K. L. Rosenthal. 2003. Interleukin-15 and natural killer and NKT cells play a critical role in innate protection against genital herpes simplex virus type 2 infection. J. Virol. 7710168-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowski, C., and A. Bendelac. 2005. Signaling for NKT cell development: the SAP-FynT connection. J. Exp. Med. 201833-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brigl, M., and M. B. Brenner. 2004. CD1: antigen presentation and T cell function. Annu. Rev. Immunol. 22817-890. [DOI] [PubMed] [Google Scholar]
- 5.Cao, X., M. Sugita, N. van der Wel, J. Lai, R. A. Rogers, P. J. Peters, and M. B. Brenner. 2002. CD1 molecules efficiently present antigen in immature dendritic cells and traffic independently of MHC class II during dendritic cell maturation. J. Immunol. 1694770-4777. [DOI] [PubMed] [Google Scholar]
- 6.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 7078-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Reference deleted.
- 9.Dascher, C. C., and M. B. Brenner. 2003. Evolutionary constraints on CD1 structure: insights from comparative genomic analysis. Trends Immunol. 24412-418. [DOI] [PubMed] [Google Scholar]
- 10.Exley, M. A., N. J. Bigley, O. Cheng, S. M. Tahir, S. T. Smiley, Q. L. Carter, H. F. Stills, M. J. Grusby, Y. Koezuka, M. Taniguchi, and S. P. Balk. 2001. CD1d-reactive T-cell activation leads to amelioration of disease caused by diabetogenic encephalomyocarditis virus. J. Leukoc. Biol. 69713-718. [PubMed] [Google Scholar]
- 11.Frascaroli, G., S. Varani, A. Mastroianni, S. Britton, D. Gibellini, G. Rossini, M. P. Landini, and C. Soderberg-Naucler. 2006. Dendritic cell function in cytomegalovirus-infected patients with mononucleosis. J. Leukoc. Biol. 79932-940. [DOI] [PubMed] [Google Scholar]
- 12.Gredmark, S., and C. Soderberg-Naucler. 2003. Human cytomegalovirus inhibits differentiation of monocytes into dendritic cells with the consequence of depressed immunological functions. J. Virol. 7710943-10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grubor-Bauk, B., A. Simmons, G. Mayrhofer, and P. G. Speck. 2003. Impaired clearance of herpes simplex virus type 1 from mice lacking CD1d or NKT cells expressing the semivariant V α14-Jα281 TCR. J. Immunol. 1701430-1434. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 953937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hengel, H., W. Brune, and U. H. Koszinowski. 1998. Immune evasion by cytomegalovirus: survival strategies of a highly adapted opportunist. Trends Microbiol. 6190-197. [DOI] [PubMed] [Google Scholar]
- 16.Hengel, H., U. H. Koszinowski, and K. K. Conzelmann. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol. 26396-401. [DOI] [PubMed] [Google Scholar]
- 17.Hertel, L., V. G. Lacaille, H. Strobl, E. D. Mellins, and E. S. Mocarski. 2003. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J. Virol. 777563-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, T. R., S. Hong, L. Van Kaer, Y. Koezuka, and B. S. Graham. 2002. NK T cells contribute to expansion of CD8+ T cells and amplification of antiviral immune responses to respiratory syncytial virus. J. Virol. 764294-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kakimi, K., L. G. Guidotti, Y. Koezuka, and F. V. Chisari. 2000. Natural killer T cell activation inhibits hepatitis B virus replication in vivo. J. Exp. Med. 192921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotenko, S. V., S. Saccani, L. S. Izotova, O. V. Mirochnitchenko, and S. Pestka. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA 971695-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lilley, B. N., and H. L. Ploegh. 2005. Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond. Immunol. Rev. 207126-144. [DOI] [PubMed] [Google Scholar]
- 22.Maciejewski, J. P., E. E. Bruening, R. E. Donahue, E. S. Mocarski, N. S. Young, and S. C. St. Jeor. 1992. Infection of hematopoietic progenitor cells by human cytomegalovirus. Blood 80170-178. [PubMed] [Google Scholar]
- 23.Major, A. S., S. Joyce, and L. Van Kaer. 2006. Lipid metabolism, atherogenesis and CD1-restricted antigen presentation. Trends Mol. Med. 12270-278. [DOI] [PubMed] [Google Scholar]
- 24.Maruoka, T., H. Tanabe, M. Chiba, and M. Kasahara. 2005. Chicken CD1 genes are located in the MHC: CD1 and endothelial protein C receptor genes constitute a distinct subfamily of class-I-like genes that predates the emergence of mammals. Immunogenetics 57590-600. [DOI] [PubMed] [Google Scholar]
- 25.Mendelson, M., S. Monard, P. Sissons, and J. Sinclair. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J. Gen. Virol. 773099-3102. [DOI] [PubMed] [Google Scholar]
- 26.Miller, D. M., C. M. Cebulla, and D. D. Sedmak. 2002. Human cytomegalovirus inhibition of major histocompatibility complex transcription and interferon signal transduction. Curr. Top. Microbiol. Immunol. 269153-170. [DOI] [PubMed] [Google Scholar]
- 27.Miller, M. M., C. Wang, E. Parisini, R. D. Coletta, R. M. Goto, S. Y. Lee, D. C. Barral, M. Townes, C. Roura-Mir, H. L. Ford, M. B. Brenner, and C. C. Dascher. 2005. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc. Natl. Acad. Sci. USA 1028674-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mocarski, E. S., Jr. 2004. Immune escape and exploitation strategies of cytomegaloviruses: impact on and imitation of the major histocompatibility system. Cell. Microbiol. 6707-717. [DOI] [PubMed] [Google Scholar]
- 29.Moutaftsi, M., A. M. Mehl, L. K. Borysiewicz, and Z. Tabi. 2002. Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood 992913-2921. [DOI] [PubMed] [Google Scholar]
- 30.Pepperl-Kindworth, S., K. Besold, N. Frankenberg, M. Farkas, J. Kuball, M. Theobald, and B. Plachter. 2006. Cytomegalovirus interleukin-10 expression in infected cells does not impair MHC class I restricted peptide presentation on bystanding antigen-presenting cells. Viral Immunol. 1992-101. [DOI] [PubMed] [Google Scholar]
- 31.Porcelli, S. A., and R. L. Modlin. 1999. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu. Rev. Immunol. 17297-329. [DOI] [PubMed] [Google Scholar]
- 32.Raftery, M. J., M. Schwab, S. M. Eibert, Y. Samstag, H. Walczak, and G. Schonrich. 2001. Targeting the function of mature dendritic cells by human cytomegalovirus: a multilayered viral defense strategy. Immunity 15997-1009. [DOI] [PubMed] [Google Scholar]
- 33.Raftery, M. J., D. Wieland, S. Gronewald, A. A. Kraus, T. Giese, and G. Schonrich. 2004. Shaping phenotype, function, and survival of dendritic cells by cytomegalovirus-encoded IL-10. J. Immunol. 1733383-3391. [DOI] [PubMed] [Google Scholar]
- 34.Raftery, M. J., F. Winau, S. H. Kaufmann, U. E. Schaible, and G. Schonrich. 2006. CD1 antigen presentation by human dendritic cells as a target for herpes simplex virus immune evasion. J. Immunol. 1776207-6214. [DOI] [PubMed] [Google Scholar]
- 35.Reddehase, M. J. 2002. Antigens and immunoevasins: opponents in cytomegalovirus immune surveillance. Nat. Rev. Immunol. 2831-844. [DOI] [PubMed] [Google Scholar]
- 36.Riegler, S., H. Hebart, H. Einsele, P. Brossart, G. Jahn, and C. Sinzger. 2000. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J. Gen. Virol. 81393-399. [DOI] [PubMed] [Google Scholar]
- 37.Salomonsen, J., M. R. Sorensen, D. A. Marston, S. L. Rogers, T. Collen, A. van Hateren, A. L. Smith, R. K. Beal, K. Skjodt, and J. Kaufman. 2005. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc. Natl. Acad. Sci. USA 1028668-8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez, D. J., J. E. Gumperz, and D. Ganem. 2005. Regulation of CD1d expression and function by a herpesvirus infection. J. Clin. Investig. 1151369-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stenger, S., K. R. Niazi, and R. L. Modlin. 1998. Down-regulation of CD1 on antigen-presenting cells by infection with Mycobacterium tuberculosis. J. Immunol. 1613582-3588. [PubMed] [Google Scholar]
- 41.Sugita, M., X. Cao, G. F. Watts, R. A. Rogers, J. S. Bonifacino, and M. B. Brenner. 2002. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity 16697-706. [DOI] [PubMed] [Google Scholar]
- 42.van Dommelen, S. L., H. A. Tabarias, M. J. Smyth, and M. A. Degli-Esposti. 2003. Activation of natural killer (NK) T cells during murine cytomegalovirus infection enhances the antiviral response mediated by NK cells. J. Virol. 771877-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan, W., A. Dasgupta, and P. Cresswell. 2006. Herpes simplex virus evades natural killer T cell recognition by suppressing CD1d recycling. Nat. Immunol. 7835-842. [DOI] [PubMed] [Google Scholar]