Abstract
During coronavirus replication, viral proteins induce the formation of endoplasmic reticulum (ER)-derived double-membrane vesicles for RNA synthesis, and viral structural proteins assemble virions at the ER-Golgi intermediate compartment. We hypothesized that the association and intense utilization of the ER during viral replication would induce the cellular unfolded protein response (UPR), a signal transduction cascade that acts to modulate translation, membrane biosynthesis, and the levels of ER chaperones. Here, we report that infection by the murine coronavirus mouse hepatitis virus (MHV) triggers the proximal UPR transducers, as revealed by monitoring the IRE1-mediated splicing of XBP-1 mRNA and the cleavage of ATF6α. However, we detected minimal downstream induction of UPR target genes, including ERdj4, ER degradation-enhancing α-mannosidase-like protein, and p58IPK, or expression of UPR reporter constructs. Translation initiation factor eIF2α is highly phosphorylated during MHV infection, and translation of cellular mRNAs is attenuated. Furthermore, we found that the critical homeostasis regulator GADD34, which recruits protein phosphatase 1 to dephosphorylate eIF2α during the recovery phase of the UPR, is not expressed during MHV infection. These results suggest that MHV modifies the UPR by impeding the induction of UPR-responsive genes, thereby favoring a sustained shutdown of the synthesis of host cell proteins while the translation of viral proteins escalates. The role of this modified response and its potential relevance to viral mechanisms for the evasion of innate defense signaling pathways during coronavirus replication are discussed.
A hallmark of viral replication is the rapid modification of the host cell environment to facilitate the replication of the viral genome and the assembly of virus particles. For positive-strand RNA viruses, sites of viral RNA synthesis are composed of viral replicase proteins and host cell membranes, which together assemble into novel structures in the cytoplasm of infected cells. For example, the picornavirus poliovirus generates rosette-like structures (2), the flavivirus hepatitis C virus generates membranous webs (14), and coronaviruses generate double-membrane vesicles (13, 15, 43) using membranes likely derived from the endoplasmic reticulum (ER). In addition, enveloped viruses must modify and perturb membranes to generate new virus particles. Often these modifications and perturbations trigger host cell stress responses, such as the unfolded protein response (UPR). We investigated how the murine coronavirus mouse hepatitis virus (MHV) activates and modulates the UPR during viral replication.
MHV is a prototype virus in the family Coronaviridae. Coronaviruses are associated with a variety of human and animal diseases, including severe acute respiratory syndrome (SARS) (9, 23, 33). The replication of MHV takes place in the cytoplasm of the infected cell. The 5′ end of the 31.5-kb genomic RNA is translated to generate two polyproteins, pp1a and pp1ab, that are proteolytically processed and assembled with intracellular membranes to generate double-membrane vesicles. Double-membrane vesicles have been demonstrated to be the sites of viral genomic and subgenomic RNA synthesis (13, 15, 43). The subgenomic mRNAs all contain the identical 5′ leader sequence which is derived via a discontinuous mechanism from the 5′ end of the genomic RNA (30, 40). The subgenomic mRNAs are translated to produce nonstructural proteins and the viral structural proteins nucleocapsid, membrane, envelope, and spike glycoprotein. The structural proteins, along with genomic RNA, assemble into viral particles, egress through the exocytic pathway, and are secreted at the cell surface (52).
This extensive use of intracellular membranes for the generation of replication complexes and for the assembly of virus particles likely overloads the ER during the acute infection. Previous studies have indicated that the expression of the coronavirus spike glycoprotein is sufficient to overload the ER and, therefore, to activate and modulate the UPR (6, 55), but the specific contributions of downstream regulators of the response have not been elucidated. The UPR is activated in response to a variety of ER stresses (calcium flux, unfolded proteins, and lipid depletion) and functions to upregulate the production of ER chaperones, to increase phospholipid biosynthesis, and to induce the transient attenuation of translation in order to reduce ER stress (37). Viruses, such as hepatitis C virus (49, 50), reovirus (42), rotavirus (31), herpes simplex virus (7), Borna disease virus (57), and cytomegalovirus (20), have been shown to activate and modulate the UPR to facilitate viral replication.
The UPR consists of three signaling pathways, mediated by the ER transmembrane proteins IRE1 (inositol-requiring protein 1), ATF6 (activating transcription factor 6α and -β), and PERK (protein kinase R [PKR]-like ER kinase) (25, 28, 37). These ER transmembrane proteins have lumenal sensor domains and cytosolic effector domains that mediate the activation of downstream genes. The activation of these sensors is proposed to be mediated by the disassociation of the ER chaperone BiP/GRP78 (immunoglobulin binding protein/glucose-regulated protein of 78 kDa) from the lumenal sensor domains to bind to unfolded proteins (37). The activation of IRE1 induces the autophosphorylation and dimerization of IRE1, which activates the endonuclease domain. This endonuclease cleaves the XBP-1u (X-box binding protein 1 unspliced) mRNA, removing 26 nucleotides from the center of the mRNA to generate XBP-1s (X-box binding protein 1 spliced) (4, 24). The XBP-1u mRNA is translated to produce a 33-kDa protein [XBP-1(u)] with a DNA binding domain but no activation domain. Following IRE1-mediated splicing, a frameshift is entered in the center of the mRNA, and when the XBP-1s mRNA is translated, a 54-kDa protein [XBP-1(S)] is produced. XBP-1(S) contains both a DNA binding domain and an activation domain and translocates to the nucleus to activate the transcription of UPR target genes, such as those associated with ER-associated degradation and lipid biosynthesis (24, 45). The activation of the UPR induces the translocation of ATF6α to the Golgi compartment, where site 1 and site 2 proteases process the 90-kDa ATF6α to generate the 50-kDa cleaved form, ATF6α(C), a soluble transcription factor that translocates to the nucleus and upregulates the expression of genes including many ER chaperones and folding enzymes. In addition, ATF6α(C) works in concert with another UPR transcription factor, ATF4, to activate further downstream genes, such as CHOP (C/EBP-homologous protein) which functions in the regulation of the recovery phase of the UPR as described below.
The third UPR ER transmembrane sensor is PERK, which is critical for the phosphorylation of eIF2α (eucaryotic initiation factor 2α) at serine 51 and translational attenuation. The phosphorylation of eIF2α inhibits the recycling of eIF2B to its active, GTP-bound form. The phosphorylation of eIF2α can also be mediated by at least three other kinases (PKR, heme-regulated inhibitor kinase, and general control non-derepressible 2) as part of an integrated stress response (16, 17). The overall impact of the phosphorylation of eIF2α is the rapid attenuation of translation. Translation attenuation is proposed to reduce the protein load in the ER. Interestingly, some viruses have evolved mechanisms to either dephosphorylate eIF2α (7) or to translate viral mRNAs in the presence of high levels of phosphorylated eIF2α (36, 53) and thereby avoid the stress-induced translational attenuation. Recovery from translational attenuation occurs through the expression of the transcription factors CHOP and ATF4 which are induced downstream of eIF2α phosphorylation and drive the expression of GADD34 (27, 29). GADD34 recruits type 1 protein phosphatase (PP1) and promotes the dephosphorylation of eIF2α, leading to translational recovery (3, 32). Recovery from translation attenuation is important for the optimal translation of mRNAs upregulated by XBP-1(S) and ATF6α(C) and the return to homeostasis. Recent studies indicate that the modulation of UPR induction requires factors such as XBP-1(S), ATF6α(C), ATF4, CHOP, and GADD34. These factors either allow the cell to recover from transient stress or, alternatively, to progress to apoptosis (25, 39, 58, 59).
We have investigated the UPR during MHV infection. Here, we show that both the IRE1 and ATF6α pathways are activated during MHV infection; however, only minimal induction of UPR target genes occurs in MHV-infected cells. Furthermore, in agreement with the results of previous studies, we show that translation initiation factor eIF2α is phosphorylated during MHV infection and that the translation of most cellular mRNAs is inhibited. ATF4, which is translated via a translational shunting mechanism in the presence of phosphorylated eIF2α, is produced during MHV infection, but the expression of its downstream targets, such as CHOP and GADD34, is not realized. Thus, although MHV infection activates the proximal transducers of the UPR, this is not accompanied by the expression of factors necessary to mediate the dephosphorylation of eIF2α and promote eIF2α-dependent translation. Interestingly, the translation of MHV mRNA occurs in the presence of phosphorylated eIF2α. Thus, MHV modulates the UPR and circumvents cellular mechanisms of translational control. The implications of these results for understanding how coronaviruses optimize viral replication and potentially evade innate defense signaling pathways are discussed.
MATERIALS AND METHODS
Virus propagation, cell culture, and transfection.
MHV strain A59 stocks were propagated as previously described (47). The delayed brain tumor (DBT) cell line was cultured in minimal essential medium (Gibco) supplemented with 5% fetal calf serum (Atlas), 10% tryptose phosphate broth, 2% penicillin/streptomycin (Mediatech), and 2% l-glutamine (Mediatech). HeLa-MHV receptor (HeLa-MHVR) cells were cultured in Dulbecco's minimal essential medium (Gibco) supplemented with 10% fetal calf serum and 2% l-glutamine. As the positive control for UPR activation, cells were treated with 2 μg/ml of tunicamycin (MP Biochemicals). Cells were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
RNA isolation and RT-PCR analysis of XBP-1 mRNA.
Total RNA was isolated from tunicamycin-treated or MHV-infected cells by using a Qiagen RNeasy kit (Qiagen, Valencia, CA). Equivalent amounts of RNA (1 μg) from each sample were subjected to reverse transcription-PCR (RT-PCR) using an Access RT-PCR system (Promega, Madison, WI) according to the manufacturer's instructions. The primers used were forward primer, 5′-GTTGAGAACCAGGAGTTAAG-3′, and reverse primer, 5′-AGAGAAAGGGAGGCTGGTAAG-3′. For the analysis of PCR products, 10 μl of each reaction mixture was loaded on a 5% polyacrylamide-TBE (20 mM Tris-borate, 0.5 mM EDTA, pH 8.0) gel and subjected to electrophoresis to separate the products. The gel was stained in a solution of 5% ethidium bromide in TBE, and the bands were visualized on a Typhoon imager.
Preparation of cell lysates, SDS-polyacrylamide gel electrophoresis, and immunoblotting.
Whole-cell lysates were prepared from mock-treated, tunicamycin-treated, or MHV-infected cells by solubilization in lysis buffer A (LBA) (3% dithiothreitol, 2% sodium dodecyl sulfate [SDS], 40% glycerol, 0.065 M Tris, pH 6.8) followed by passage through a 20-gauge needle. Approximately 0.5 × 104 cell equivalents were loaded per lane and separated by electrophoresis on SDS-polyacrylamide gels. Kaleidoscope (Bio-Rad) prestained molecular-weight standards were used to monitor the separation and transfer. The proteins were transferred to nitrocellulose membranes (Schleicher & Schuell), and the probing protocol was adapted from Gass et al. (12). Briefly, the nitrocellulose membrane was blocked in blocking buffer (1× Tris-buffered saline [TS], 0.1% Tween 20, 5% milk) for 2 h at room temperature. The primary antibody was then mixed in probing buffer (1× TS, 0.1% Tween 20, 1% milk) and incubated overnight at 4°C. The primary antibodies were anti-nsp8 (also termed αD14 [15]); rabbit anti-XBP-1 and anti-ATF6α antibodies (kindly provided by K. Mori, Tokyo University); anticalnexin antibody (kindly provided by L. Hendershot, St. Jude's Children's Hospital, Memphis, TN); anti-GADD34, anti-CHOP, and anti-eIF2α antibodies (Santa Cruz Biotechnologies); and anti-phosphorylated eIF2α antibody (Cell Signaling). Following incubation with the primary antibody, the blot was washed in 1× TS and incubated with the appropriate secondary antibody conjugated to horseradish peroxidase (HRP) (goat anti-mouse HRP or donkey anti-rabbit HRP; Southern Biotech). The blot was washed three times in 1× TS and developed using Western lightening chemiluminescence reagent (PerkinElmer).
Preparation of nuclear and cytoplasmic fractions.
The nuclear and cytoplasmic fractions were prepared by using a protocol adapted from Reimers et al. (35). Briefly, approximately 4.5 × 106 cells were resuspended in 800 μl of lysis buffer (10 mM KCl, 10 mM HEPES, pH 7.9, 0.1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, and 0.2 mM phenylmethylsulfonyl fluoride) and incubated on ice for 20 min. After the incubation, the plasma membrane was permeabilized by the addition of 37.5 μl of 10% Nonidet P-40. The nuclear and cytosolic fractions were separated by centrifugation at 10,000 rpm for 5 min, and the cytosol-enriched supernatant was transferred to a new tube. The nuclear pellet was resuspended in 30 μl of extraction buffer B (0.4 M NaCl, 20 mM HEPES, pH 7.9, 1 mM EDTA, 1 mM EGTA) and rotated for 30 min at 4°C, followed by the addition of 750 μl of water. The cytoplasmic and nuclear fractions were extracted and precipitated by the addition of 800 μl of methanol and 200 μl of chloroform. The samples were mixed by vortexing and then centrifuged for 2 min at 14,000 rpm. The upper layer was removed, and 200 μl of methanol was added to precipitate the proteins. The proteins were pelleted at 14,000 rpm for 2 min, and the supernatant removed by aspiration. The pellet was dried for 5 min and resuspended in 80 μl of LBA. Approximately 1.1 × 106 cell equivalents were subjected to SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and probed as described above.
Detection of UPRE and ERSE activation using the dual-luciferase reporter assay.
DBT cells were transfected with reporter plasmids (Kazutoshi Mori, Kyoto University, Kyoto, Japan) carrying the UPR element (UPRE) (pUPRE-fluc) or the ER stress response element (ERSE) (pERSE-fluc) and a plasmid carrying Renilla internal control (pRL; Promega) and cultured for 24 h. The cells were treated with tunicamycin or infected with MHV, and whole-cell lysates were harvested in 1× passive lysis buffer (Promega) at the time points indicated (see Fig. 2). The luciferase activity was measured by using a Promega dual-luciferase reporter system and a TD-20/20 luminometer. To measure luciferase activity, 5 μl of lysates was added to 20 μl of LAR II buffer (Promega) and read in a luminometer with 2-s-delay and 10-s-measurement settings. Following the read, 20 μl of Stop and Glo buffer (Promega) was added, and the sample was read again. The samples were assayed in triplicate. For the XBP-1(S)-green fluorescent protein (GFP) and ATF6α(C)-GFP expression experiments, cells were transfected with expression constructs along with the pRL and reporter constructs.
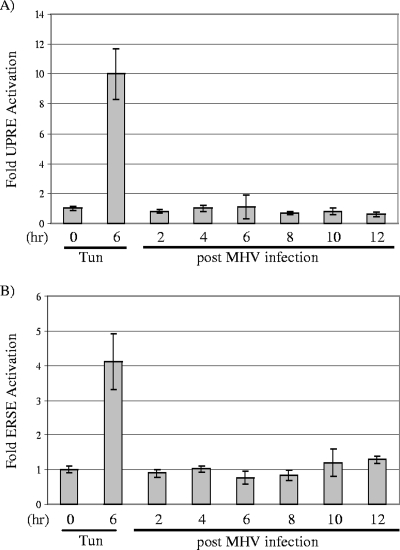
FIG. 2.
Analysis of induction of UPRE and ERSE promoters during MHV infection. DBT cells were transfected with either UPR reporter plasmid pUPRE-GL3 or ERSE reporter plasmid pERSE-GL3 DNA and, 24 h later, infected with MHV or treated with tunicamycin (Tun; 2 μg/ml). Cell lysates were prepared at the times indicated and assayed in triplicate using a dual-luciferase assay as described in Materials and Methods. Error bars show standard deviations.
Quantitative RT-PCR analysis of XBP-1 mRNA.
TaqMan quantitative real-time RT-PCR analysis was performed with reagents from Applied Biosystems according to the manufacturer's instructions, using an ABI PRISM 7700 sequence detection system. The probes used were XBP-1 forward, 5′ GCC ATT GTC TGA GAC CAC CTT 3′; XBP-1 reverse, 5′ TCT GTA CCA AGT GGA GAA GAC ATG 3′; and Taqman internal probe, 5′ FAM-TGC CTG CTG GAC GCT CAC AGT GAC-TAMRA 3′. TaqMan rRNA control reagents were used to generate the standard curve.
Northern blot analysis.
Northern blotting was performed as previously described in Gass et al. (12). Briefly, total RNA was prepared as described above and 15 μg of each RNA sample was combined with RNA sample buffer (0.12 M sodium acetate, 10 mM Tris, pH 7.4, 1 mM EDTA, 0.05% SDS, and 50 mg/ml Escherichia coli tRNA) and resolved by electrophoresis on a 1.1% agarose-formaldehyde gel. The samples were then transferred to Duralose-UV membrane (Stratagene) and hybridized overnight at 42°C in hybridization buffer (50% formamide, 6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA, pH 7.7], 5× Denhardt's solution, 0.5% SDS, 100 mg/ml salmon sperm DNA). Probes were prepared by random primer labeling using a Prime-It II kit (Stratagene) and [α-32P]dCTP (GE Healthcare). The probes used were mouse ERdj4, an 1,800-bp EcoRI/NotI insert from pME18FL3-mERdj4 (Linda Hendershot, St. Jude Children's Research Hospital, Memphis, TN); mouse XBP-1, a 1,900-bp SalI/NotI insert from pCMV-SPORT6 msXBP-1; mouse ATF6α, a 660-bp PCR product encoding the cytosolic region; mouse p58IPK, a 1,600-bp EcoRI/XhoI insert from pBS-mouse p58IPK (provided by Michael Katze, University of Washington, Seattle, WA); and MHV nucleocapsid gene, a 1,100-bp XhoI/BamHI insert from pcDNA 3.1-MHV-N. Approximately 500,000 cpm/ml of probe was added to the hybridization buffer and incubated overnight at 42°C. The following day, the blot was removed and washed three times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS at 65°C. The blot was dried and imaged on a Typhoon imager.
XBP-1-GFP and ATF6α-GFP expression constructs.
The XBP-1(S) and ATF6α(C) domains were amplified from human expression plasmids pcDNA-XBP1(spliced) and pcDNA-ATF6α(373) (provided by Kazutoshi Mori, Kyoto University, Kyoto, Japan) using the following primer pairs: XBP-1 forward, 5′-TAAAGCTTATGATGGTGGTGGTGGCAG-3′; XBP-1 reverse, 5′-TATGGATCCCGGACACTAATCAGCTG-3′; ATF6 forward, 5′-TACTCGAGTGTGAAATGGGGGAGCCGGCTG-3′; and ATF6 reverse, 5′-TGGAACTTACTAGGGACTTTAAGC-3′. The XBP-1(S) PCR product was digested with HindIII and BamHI and ligated into the corresponding sites of pCMV-GFP (Clontech) to generate pCMV-XBP1-GFP. The ATF6α(C) PCR product was digested with XhoI and HindIII and ligated into the corresponding sites of pCMV-GFP to generate pCMV-ATF6α(C)-GFP. These constructs were transfected into DBT cells, the cells were treated with tunicamycin, and the localization of GFP was monitored by microscopy. GFP fluorescence was detected in the nucleus of tunicamycin-treated cells, indicating that the expressed proteins translocated to the nucleus as a consequence of a stress response.
Analysis of radiolabeled proteins.
DBT cells were treated with 2 μg/ml tunicamycin or MHV at a multiplicity of infection (MOI) of 10 for the indicated periods of time (see Fig. 6) and then radiolabeled with 50 μCi/ml of 35S-translabel (MP Biochemicals) for 15 min. Following radiolabeling, cells were washed with phosphate-buffered saline and harvested in 200 μl LBA, and a 10-μl amount was resolved on a 12% gel. The gel was dried and assessed by using a Typhoon phosphorimager (Applied Biosystems).
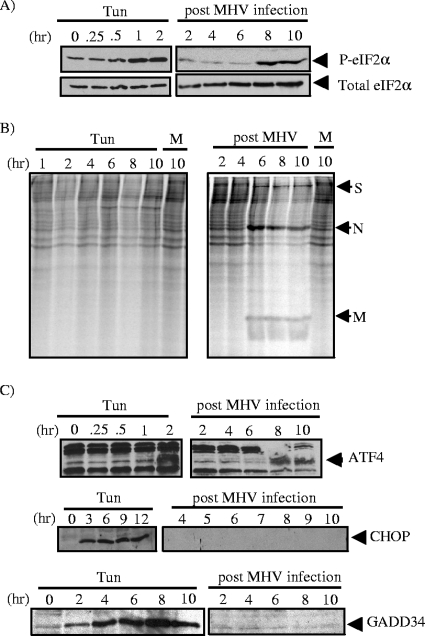
FIG. 6.
Analysis of the eIF2α pathway in MHV-infected cells. (A) Lysates were generated from either MHV-infected cells or cells treated with tunicamycin (Tun) (2 μg/ml), and Western blot analysis was performed using antibodies directed against phosphorylated eIF2α (P-eIF2α) and eIF2α. (B) Analysis of protein synthesis in MHV-infected cells. DBT cells were either treated with tunicamycin or infected with MHV at an MOI of 10 and incubated for the indicated periods of time. A half hour before harvest, cells were starved in medium without methionine for 15 min and then pulsed with [35S]methionine for 15 min. Cell lysates were separated by electrophoresis on a 12% polyacrylamide gel and imaged using a phosphoimager. MHV structural proteins are indicated: S, spike; N, nucleocapsid; M, membrane. (C) Western blot analysis of lysates prepared from MHV-infected or tunicamycin-treated cells probed with antibodies specific to ATF4, CHOP, and GADD34.
RESULTS
Analysis of the IRE1/XBP-1 pathway in MHV-infected cells.
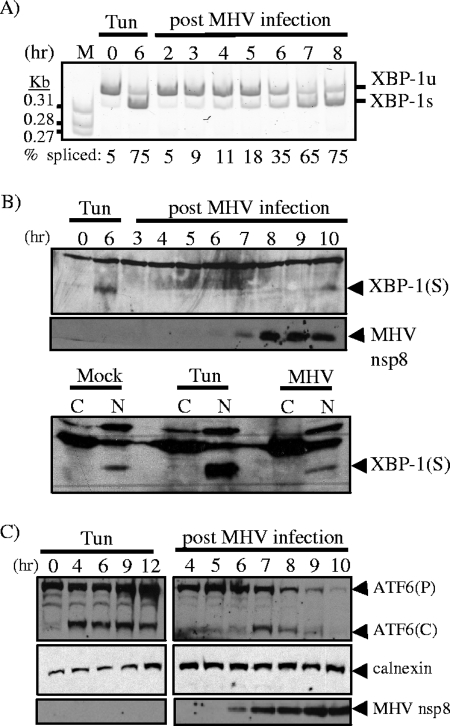
The results of previous studies showed that the expression of the coronavirus spike glycoprotein is sufficient to activate the UPR (6, 55); however, the translation of UPR transcription factors and the potential modulation of the response was not assessed in these studies. Here, we examined both XBP-1 mRNA splicing and the production of the XBP-1(S) protein product during MHV infection. A pharmacologic inducer of ER stress, tunicamycin, was used as a positive control in all experiments. To determine if replication of the murine coronavirus MHV strain A59 activates the IRE1/XBP-1 arm of the UPR, DBT cells were infected with MHV and mRNA was isolated at various time points postinfection. RT-PCR analysis was then used to determine the extent of XBP-1 mRNA splicing. RT-PCR was performed by using primers flanking the XBP-1 splice site, and PCR products were resolved on a 5% polyacrylamide gel and visualized by staining with ethidium bromide (Fig. 1A). The predominant RT-PCR product generated from mock-infected cells represents the unspliced form of XBP-1 mRNA (95%), compared to only 5% of the spliced form. In tunicamycin-treated cells, the predominant form of the XBP-1 mRNA was the spliced form, with 75% of the RT-PCR products generated from spliced mRNA. During MHV infection, there was a gradual increase in the amount of spliced XBP-1 mRNA, and by 7 h postinfection (hpi), the spliced form of the mRNA was predominant (65%). These results are in agreement with those of a previous study (55) and demonstrate that MHV infection activates IRE1-mediated splicing of the XBP-1 mRNA in all cell lines studied to date. To extend this analysis, we asked if the XBP-1(S) protein is produced during MHV infection. Whole-cell lysates were prepared from MHV-infected cells and analyzed by Western blotting using antibodies which recognize the XBP-1(S) protein and the MHV replicase product (nsp8). We found that the XBP-1(S) protein was detectable at 6 h posttunicamycin treatment. In sharp contrast, very little XBP-1(S) protein was present in MHV-infected cells, with the greatest amount apparent at late intervals (10 hpi) when cells are experiencing the cytopathic effects (syncytium formation) of infection (Fig. 1B). To further probe for XBP-1(S), we isolated the cytosolic and nuclear fractions from mock-infected, tunicamycin-treated, or MHV-infected cells at 7 hpi and analyzed these fractions by Western blotting. In agreement with the results of the time-course experiment, we detected a substantial increase in XBP-1(S) in the nuclear fraction from tunicamycin-treated cells, but the levels of XBP-1(S) in mock-infected and MHV-infected cells were similar. Thus, these results demonstrate that although the IRE1/XBP-1 pathway is activated early during MHV infection, as evidenced by splicing of the XBP-1 mRNA, the XBP-1(S) protein does not accumulate during MHV infection to the levels observed under conditions of tunicamycin-induced ER stress.
FIG. 1.
Analysis of the activation status of the IRE1/XBP-1 and ATF6 pathways during MHV infection. Cells were incubated in the absence or presence of 2 μg/ml of tunicamycin (Tun) or infected with MHV. (A) RT-PCR analysis of XBP-1u and XBP-1s mRNAs during MHV infection. RNA was extracted from cells at the times indicated and subjected to RT-PCR analysis using primers flanking the XBP-1 splice site. The PCR products were resolved on a 5% polyacrylamide gel and visualized by staining with ethidium bromide. The PCR products generated from the XBP-1u and XBP-1s mRNAs are indicated. M, molecular-size markers. (B and C) Western blot analysis of XBP-1(S) and ATF6α proteins from MHV-infected cells. Whole-cell lysates or nuclear (N) and cytosolic (C) fractions were prepared from untreated (Mock), tunicamycin-treated, or MHV-infected cells and subjected to Western blot analysis using antisera specific to XBP-1 or ATF6α. For ATF6α, both precursor (P) and cleaved (C) products are indicated in panel C. Detection of the resident ER protein calnexin and MHV nonstructural protein 8 (nsp8) was used as a loading control and a marker for MHV infection, respectively.
Analysis of the ATF6α pathway in MHV-infected cells.
Next, we investigated the activation of the ATF6α pathway as an indicator of UPR activation. We prepared whole-cell lysates from MHV-infected cells and monitored the extent of processing of ATF6α by Western blotting. Both ATF6α(P) and ATF6α(C) were detected during MHV infection, with the peak level of ATF6α(C) occurring at 7 hpi (Fig. 1C). However, we noted a steady decline in both the ATF6α precursor and cleavage product over the time course of MHV infection. This is in contrast to the results for tunicamycin-induced ER stress, where the ATF6α precursor and cleavage product increased over the entire time course of treatment. Together, these data indicate that the ATF6α pathway is activated during MHV infection but the level of ATF6α(C) diminishes at later intervals of the infection process.
Analysis of UPR-regulated gene expression in MHV-infected cells.
As part of the ER stress response, the XBP-1(S) and ATF6α(C) transcription factors translocate to the nucleus and activate the transcription of specific target genes. To determine if ER stress response promoters are upregulated during MHV infection, we monitored their activities by using UPRE- and ERSE-luciferase reporters. The UPRE contains a consensus motif that is recognized by XBP-1(S), and the ERSE contains a consensus motif that is recognized by both XBP-1(S) and ATF6α(C); these reporters have been used to characterize stress responses (59). DBT cells were transfected with p5xUPRE-GL3 or -ERSE-GL3 DNA and, 24 h later, infected with MHV. Cell lysates were prepared at the indicated time points after MHV infection and tested for luciferase activity (Fig. 2). As expected, treatment with tunicamycin strongly induced UPRE (10-fold) and ERSE (fourfold) reporter activity. In contrast, during MHV infection the UPRE reporter activity was unchanged over the time course of infection, and the ERSE reporter activity was only slightly elevated (less than twofold) at 10 and 12 hpi, the same time as virus particles are released from infected cells. These results indicate that although the IRE1/XBP-1 and ATF6α branches of the UPR are activated during MHV infection, ER stress-responsive promoters exhibit very little activity under these conditions.
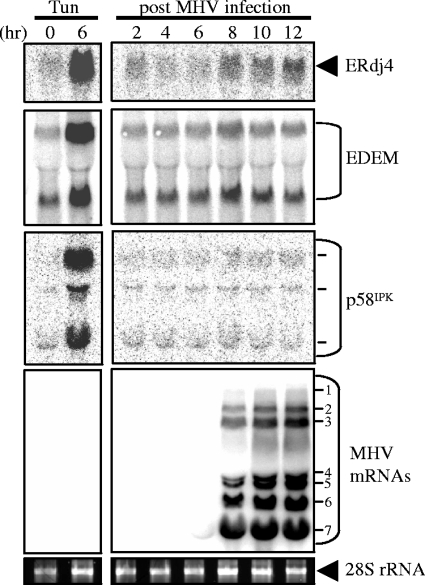
To further investigate if XBP-1(S)-responsive genes are activated during coronavirus replication, RNA was isolated from MHV-infected cells and analyzed by Northern blotting (Fig. 3). Northern blot analysis of the UPR-responsive genes ERdj4, EDEM (ER degradation-enhancing α-mannosidase-like protein), and p58IPK (24) revealed minimal induction during MHV infection. We detected an approximately twofold induction of ERdj4 transcripts at 8, 10, and 12 hpi but essentially no change in the levels of EDEM and p58IPK mRNAs. In contrast, tunicamycin treatment resulted in a 6- to 10-fold induction of these XBP-1(S)-responsive genes. Similar results were recently reported in a microarray analysis of MHV-infected mouse fibroblast cells, which revealed low-level activation of genes such as ERdj4 (2.5-fold induction), CHOP (3.6-fold induction), and HERP (fourfold induction) (54). This minimal induction of UPR genes during MHV infection is in stark contrast to the levels of induction detected in a similar microarray study of mouse embryo fibroblasts treated with tunicamycin, which resulted in a 9.6-fold induction of ERdj4, a 27.1-fold induction of CHOP, and an 11-fold induction of HERP (24). Collectively, these data indicate that UPR-regulated gene expression is very weak during MHV infection in comparison to the UPR profile characteristic of pharmacologically induced ER stress.
FIG. 3.
Northern blot analysis of UPR target genes during MHV infection. DBT cells were infected with MHV or treated with tunicamycin (Tun; 2 μg/ml), and RNA was extracted and subjected to Northern blot analysis using probes against the ERdj4, EDEM, p58IPK, and MHV nucleocapsid gene sequences. The seven MHV mRNAs and multiple p58IPK mRNAs are indicated. Ethidium bromide stain of 28S rRNA is visualized as a loading control.
Analysis of XBP-1 and ATF6α mRNAs during MHV infection.
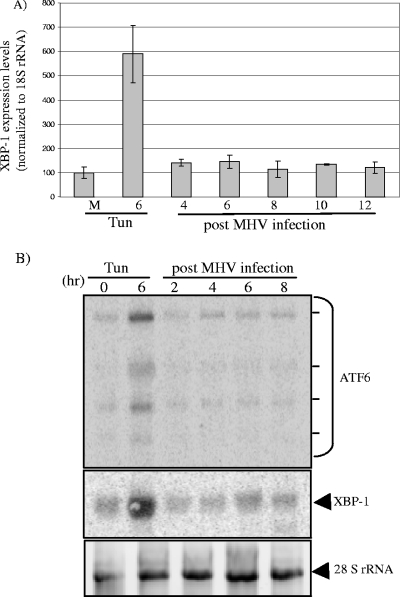
During homeostatic conditions, the XBP-1 and ATF6α mRNA levels are low, whereas during ER stress, these genes are upregulated and their mRNA levels are dramatically increased. In the case of XBP-1(S) production, the transcriptional induction may be necessary to compensate for the short half-life of the protein (4). To investigate transcriptional induction of XBP-1 and ATF6α mRNAs, we performed real-time RT-PCR and Northern blot analysis to measure the relative levels of the mRNAs. As expected, the level of XBP-1 mRNA increased substantially (sixfold) after treatment with tunicamycin (Fig. 4A). In contrast, the abundance of XBP-1 mRNA increased less than twofold over the time course of MHV infection. Similar results were obtained by Northern blot analysis of ATF6α and XBP-1 mRNAs isolated from MHV-infected cells (Fig. 4B). Therefore, while both ATF6α and XBP-1 mRNAs are significantly induced during the ER stress response induced by tunicamycin, MHV infection results in minimal changes in the levels of these transcripts.
FIG. 4.
Analysis of XBP-1 and ATF6α mRNAs during MHV infection. (A) Results of quantitative RT-PCR analysis of XBP-1 mRNA. RNA was isolated from MHV-infected, untreated, or tunicamycin-treated cells and subjected to quantitative RT-PCR analysis using rRNA as a control, as described in Materials and Methods. Error bars show standard deviations. M, mock-treated cells. (B) Northern blot analysis of ATF6α and XBP-1 mRNA during MHV infection. RNA was extracted and subjected to Northern blot analysis using probes against XBP-1 or ATF6α. Dashes indicate the mRNAs detected with the probe to ATF6α. Ethidium bromide staining of 28S rRNA is shown as a loading control. Tun, tunicamycin.
Transcriptional activities of overexpressed XBP-1(S) and ATF6α(C) during MHV infection.
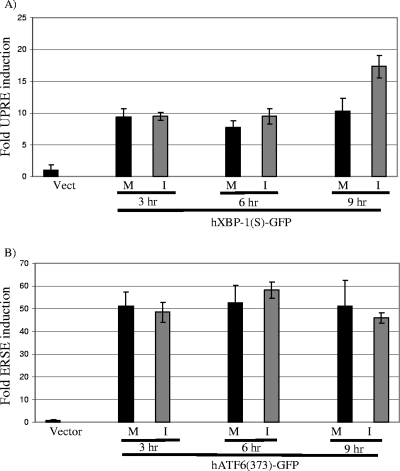
We reasoned that the lack of XBP-1(S)- and ATF6α(C)-mediated transcriptional activation during MHV infection might be due to the inactivation or degradation of these factors or to a block in their ability to efficiently localize to the nucleus. To test these possibilities, we established a dynamic system of monitoring transcriptional induction by XBP-1(S) and ATF6(C). We generated expression constructs of GFP fusions with XBP-1(S) and ATF6α(C) and transfected the DNA along with either UPRE or ERSE reporters. We then tested whether MHV infection inhibited the ability of these transcription factors to induce the reporters (Fig. 5). Since luciferase is rapidly turned over (2-h half-life), luciferase activity is a useful reporter for monitoring the dynamics of transcriptional induction (19). In addition, the transcription factors XBP-1(S) and ATF6(C) are rapidly turned over, with a half-life of 22 min for XBP-1(S) (4) and 40 min for ATF6(C) (51). Therefore, if MHV replication blocked the transport of XBP-1 or ATF6C to the nucleus or induced the degradation of these factors, we would expect to see a reduction in luciferase activity over the time course of MHV infection. However, we found that the overexpression of the XBP-1-GFP or ATF6α-GFP fusion protein resulted in the induction of both UPRE and ERSE reporter activities in both mock-infected and MHV-infected cells at 3, 6, and 9 hpi. We found no evidence for a block in transcriptional activation by these factors during MHV infection and even detected an increase in UPRE activity at 9 hpi. In addition, nuclear localization of both the XBP-1-GFP and ATF6α-GFP transcription factors was visualized by microscopy throughout MHV infection (data not shown). Therefore, when XBP-1(S) and ATF6α(C) were overexpressed, subsequent MHV infection neither disrupted the nuclear localization of these factors nor suppressed their ability to transactivate promoters. These findings suggest that the lack of strong UPR-regulated gene induction in MHV-infected cells is unlikely to result from virus-mediated inhibition of XBP-1(S) and ATF6α(C) activity.
FIG. 5.
Analysis of reporter induction by XBP-1-GFP and ATF6α-GFP during MHV infection. HeLa-MHVR cells were transfected with either pUPRE-GL3, pRL, and XBP-1(S)-GFP plasmid DNAs (A) or pERSE-GL3, pRL, and ATF6α(373)-GFP plasmid DNAs (B) using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, cells were either mock infected (M) or infected with MHV (I), and lysates were prepared at 3, 6, and 9 hpi. The pEGFP plasmid (Invitrogen) was used as the vector control (Vect). The lysates were analyzed in triplicate using a dual-luciferase assay. Error bars show standard deviations.
Translation during ER stress and MHV infection.
Another hallmark of ER stress is the phosphorylation of eIF2α, which results in a dramatic attenuation of translation. This translational attenuation is usually transient, and dephosphorylation of eIF2α mediated by GADD34 and PP1 is required to return to homeostasis. We hypothesized that the modulation of the XBP-1(S) and ATF6α pathways described above might be due, at least in part, to high levels of phosphorylated eIF2α. Such a condition would block the translation of most cellular mRNAs, including XBP-1(S) and ATF6α transcripts, but not MHV mRNAs (34). To explore this hypothesis, we prepared lysates from MHV-infected cells and probed for eIF2α and phosphorylated eIF2α by Western blotting. We found that the amount of phosphorylated eIF2α increased over the course of MHV infection, whereas the steady-state level of eIF2α was unchanged (Fig. 6A). Furthermore, in agreement with the results of previous studies (1, 18, 34, 41), we observed a marked decrease in the translation of cellular mRNAs, while MHV mRNAs resisted translational attenuation (Fig. 6B). These results demonstrate that MHV replication activates the phosphorylation of eIF2α and attenuates the translation of most cellular mRNAs.
To further examine translational attenuation and its potential relationship with the UPR during MHV infection, we asked if factors which are normally activated by high levels of phosphorylated eIF2α, such as ATF4, CHOP, and GADD34, are expressed under these conditions. ATF4 is translated using a translational shunting mechanism which functions most efficiently under conditions of limiting amounts of eIF2α. As expected, we detected the expression of ATF4 (Fig. 6C), indicating that phosphorylated eIF2α-mediated translational shunting occurs during MHV infection. However, ATF4 targets, such as CHOP and GADD34, were not detected during MHV infection (Fig. 6C). Importantly, CHOP has been implicated in the transcriptional induction of GADD34 (29) and GADD34 promotes the dephosphorylation of eIF2α, thereby facilitating translation (32). Indeed, the absence of GADD34 during MHV infection correlates with the persistent phosphorylation of eIF2α and sustained repression of most cellular protein synthesis (Fig. 6). In this state, only those mRNAs capable of being translated in the presence of phosphorylated eIF2α, such as ATF4 and MHV transcripts, are efficiently translated. Thus, the ability of MHV to minimize the induction of UPR target genes may have profound influences on how the virus alters and utilizes the cellular environment for optimal replication.
DISCUSSION
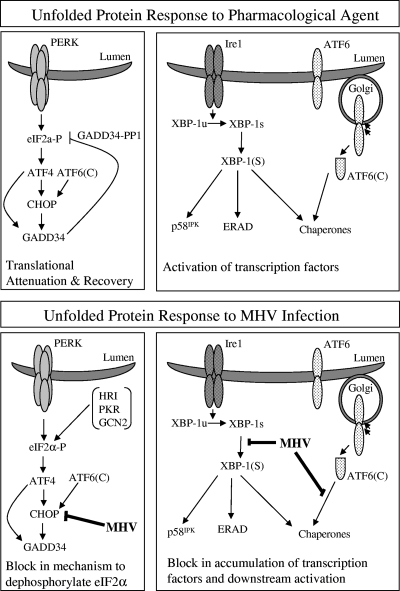
Viruses must rapidly modify the host cell environment to maximize viral genome replication and the assembly of virus particles. In addition, many viruses have evolved mechanisms to alter the expression of host cell proteins to modulate the activation of innate immune response pathways, and these can have important influences on viral pathogenesis. In this study, we investigated the activation and modulation of the UPR by murine coronavirus. We found that MHV-infected cells exhibit activation of the proximal transducers of the UPR, very weak induction of UPR target genes, and sustained phosphorylation of eIF2α. In keeping with these observations, the translation of most cellular mRNAs is reduced while the translation of viral mRNAs proceeds unabated during MHV infection. Unique to this study, we showed that the UPR transcription factor ATF4 is produced in MHV-infected cells but its downstream targets CHOP and GADD34 are not expressed. Since the GADD34-mediated dephosphorylation of eIF2α is required for the recovery of translation in cells subjected to pharmacologically induced ER stress (32), we propose that the absence of GADD34 in MHV-infected cells contributes to the sustained repression of the synthesis of most cellular proteins (Fig. 7). These findings have important implications for the identification of factors that facilitate the translation of viral mRNAs in the presence of phosphorylated eIF2α and for the understanding of potential mechanisms whereby viruses modulate the synthesis of cellular proteins involved in the innate immune response.
FIG. 7.
Models depicting the UPR during pharmacologically induced ER stress versus MHV infection. (Top) During pharmacological induction of the UPR, IRE1-mediated splicing of XBP-1 mRNA, cleavage of ATF6α, and PERK-mediated phosphorylation of eIF2α are triggered. Translational attenuation is induced, but translational recovery initiates with the production of ATF4 which, with ATF6α, activates the transcription of CHOP and GADD34. GADD34 recruits PP1 to the ER to dephosphorylate eIF2α and promote the return to homeostasis. XBP-1 and ATF6α activate the transcription of UPR targets, which are produced and further modulate recovery and homeostasis. (Bottom) During MHV infection, IRE1-mediated splicing of XBP-1 mRNA occurs and ATF6α is cleaved, but very little induction of UPR target genes is realized. eIF2α is phosphorylated and ATF4 is produced; however, CHOP and GADD34 are not expressed. Phosphorylation of eIF2α persists and this correlates with the ongoing repression of most cellular protein synthesis. eIF2α-P, phosphorylated eIF2α.
Our findings are in agreement with those of previous studies and shed new light on the interpretation of previous results. Chan and coworkers reported that the coronavirus spike protein can mediate the induction of the ER chaperone GRP78 and the phosphorylation of eIF2α (6). However, they detected minimal splicing of XBP-1 mRNA and no activation of ER stress-responsive reporters and therefore concluded that the IRE1/XBP-1 and ATF6α pathways are not activated during coronavirus infection. By direct analysis of the proximal transducers of these pathways, we show extensive IRE1-mediated splicing of XBP-1 mRNA and ATF6α cleavage during MHV infection. In agreement with the results of Chan and coworkers (6), we detected little or no activation of the UPRE and ERSE reporters. These results indicate the importance of analyzing both RNA and protein expression during MHV infection. For example, a previous study showed that tumor necrosis factor alpha and beta interferon (IFN-β) transcripts increase during MHV infection but that these cytokines are not made (46). In a recent study of intracellular chemokine production, Versteeg and coworkers found that Cxcl2 is upregulated at the mRNA level but the protein is not detected in the supernatant of coronavirus-infected cells (55). They also noted the reduced translation of cellular mRNAs during coronavirus infection, which prevented the synthesis of CXCL2 and ER stress response proteins despite increased intracellular mRNA concentrations. Finally, Raaben and coworkers recently reported that coronavirus replication induces host translational shutoff and mRNA decay, which likely results from the generation of stress granules and P bodies (34). Stress granules contain transcripts that have been stalled in translation, while P bodies are associated with both mRNA decay and mRNAs stalled in translation (10). Thus, the attenuation of translation and mRNA decay are both likely to alter the host cell environment during coronavirus replication.
All of these studies point to the role of phosphorylated eIF2α in attenuating the translation of cellular mRNA and promoting the translation of coronavirus transcripts. The unique contribution of our work is the analysis of the normal recovery stage of the UPR, revealing that although ATF4 is generated, its downstream targets CHOP and GADD34 are not induced. One possibility is that a viral factor engages ATF4 and prevents the transactivation of its target genes. Alternatively, since ATF4 and ATF6α(C) work in concert to activate CHOP and GADD34 (58), it is possible that the levels of ATF6α(C) are too low to activate transcription during MHV infection. While multiple and complex mechanisms may be responsible for the persistent phosphorylation of eIF2α and the repression of host cell protein synthesis during MHV infection, it is reasonable to speculate that the absence of GADD34, at the very least, contributes to this phenomenon. In addition, we showed that a negative regulator of PERK and PKR, p58IPK, is not upregulated during MHV infection. The absence of p58IPK induction might facilitate persistent signaling from PERK and/or PKR and thus sustain eIF2α phosphorylation.
Several interesting questions arise from these studies. First, how are MHV mRNAs translated in the presence of phosphorylated eIF2α? Previous studies suggest that the presence of the leader RNA may facilitate translation under such conditions (48). For Sindbis virus, an alphavirus, Ventoso and coworkers identified a conserved structure in the 26S mRNA that facilitates translation in cells with high levels of phosphorylated eIF2α (53). This conserved structure, termed the downstream hairpin loop, promotes the translational resistance of Sindbis virus 26S mRNA to eIF2α phosphorylation, presumably by binding the ribosomes and then directing the initiation complex to the 26S mRNA start site AUG. Future studies will be needed to determine if sequences or RNA secondary structure in the leader or body sequence of coronavirus mRNAs functions in a similar fashion. Interestingly, coronavirus genomic RNAs encode small open reading frames upstream of the start site for the translation of ORF1a (44) and may utilize a more-classical shunting mechanism for the translation of pp1a and pp1ab in the presence of phosphorylated eIF2α. However, coronavirus subgenomic mRNAs do not contain the small, upstream open reading frame, but rather a leader sequence that is predicted to form a hairpin loop (26) which may shunt ribosomes through a distinct mechanism. We also note that the work of Raaben and coworkers revealed that MHV does not require phosphorylated eIF2α for efficient viral replication, as shown by efficient replication in the presence of an eIF2α-S51A mutant (34). However, while the ability to replicate in the presence of phosphorylated eIF2α may not be essential for MHV in cell culture, such a mechanism may play a role in vivo as a means to evade or modulate the innate immune response.
The results of recent studies indicate that coronaviruses have evolved multiple strategies to evade the innate immune response (11). These strategies include the production of viral proteins that block the signaling pathways used to induce the production of IFN and IFN-stimulated genes (8, 22, 56, 60). In addition, the results of a recent study suggest that MHV infection causes an inhibition of IFN-β at a posttranscriptional level, without altering RNA or protein stability (38). Our results provide further support for this concept and suggest that MHV causes sustained repression of most cellular protein synthesis, thereby impeding the translation of induced host cell mRNAs, including IFN-β. Further studies will be needed to determine if this attenuation of host cell mRNA translation is true of all coronaviruses, including the SARS coronavirus. However, the results of recent studies indicate that atypical innate and adaptive immune responses were associated with immunopathological events in SARS patients (5). The attenuation of translation of innate immune mediators may play a role in coronavirus pathogenesis, and further studies are needed to address this issue. In this regard, the results of our study argue that both RNA and protein levels must be monitored in coronavirus-infected cells and patients to obtain an accurate picture of the innate immune response to this virus.
Finally, what coronavirus protein(s) mediates the sustained repression of host cell protein synthesis? One candidate viral protein which may be involved in either instigating or modulating eIF2α phosphorylation or stress granule formation is nsp1. The expression of SARS coronavirus nsp1 has been demonstrated to decrease the half-life of reporter transcripts, suggesting that nsp1 either directly or indirectly instigates stress granule formation. It has also been demonstrated that exogenously expressed nsp1 from the MHV, 229E, and SARS coronaviruses blocks the induction of IFN, providing a possible rationale for virus-induced translational arrest and stress granule formation (21, 61). Thus, it is intriguing to speculate that nsp1 may somehow be involved in sustaining translational repression.
In summary, our data demonstrate that coronaviruses, like many viruses (7, 20, 31, 42, 49, 50, 57), activate and modulate the UPR. These findings underscore the remarkable variability in how the UPR pathway is utilized and/or modified under distinct conditions of physiological ER stress. The next step will be to delineate mechanisms by which viruses modulate UPR-mediated events. To this end, MHV offers a tractable model to further investigate how viruses might abrogate UPR-regulated gene expression and exploit cellular translational control mechanisms to promote viral replication.
Acknowledgments
We thank members of the Baker lab for helpful discussions.
This work was supported by funding from the NIH: grant AI45798 to S.C.B. and grant GM61970 to J.W.B. J.B. was supported by NIH training grant AI07508.
Footnotes
Published ahead of print on 27 February 2008.
REFERENCES
- 1.Banerjee, S., S. An, A. Zhou, R. H. Silverman, and S. Makino. 2000. RNase L-independent specific 28S rRNA cleavage in murine coronavirus-infected cells. J. Virol. 748793-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 662740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, M., K. F. Bryant, C. Jousse, K. Long, H. P. Harding, D. Scheuner, R. J. Kaufman, D. Ma, D. M. Coen, D. Ron, and J. Yuan. 2005. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science 307935-939. [DOI] [PubMed] [Google Scholar]
- 4.Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 41592-96. [DOI] [PubMed] [Google Scholar]
- 5.Cameron, M. J., J. F. Bermejo-Martin, A. Danesh, M. P. Muller, and D. J. Kelvin. 2008. Human immunopathogenesis of severe acute respiratory syndrome (SARS). Virus Res. 13313-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, C. P., K. L. Siu, K. T. Chin, K. Y. Yuen, B. Zheng, and D. Y. Jin. 2006. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 809279-9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, G., Z. Feng, and B. He. 2005. Herpes simplex virus 1 infection activates the endoplasmic reticulum resident kinase PERK and mediates eIF-2α dephosphorylation by the γ134.5 protein. J. Virol. 791379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devaraj, S. G., N. Wang, Z. Chen, M. Tseng, N. Barretto, R. Lin, C. J. Peters, C. T. Tseng, S. C. Baker, and K. Li. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 28232208-32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 3481967-1976. [DOI] [PubMed] [Google Scholar]
- 10.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 89-22. [DOI] [PubMed] [Google Scholar]
- 11.Frieman, M., M. Heise, and R. Baric. 2008. SARS coronavirus and innate immunity. Virus Res. 133101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gass, J. N., N. M. Gifford, and J. W. Brewer. 2002. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 27749047-49054. [DOI] [PubMed] [Google Scholar]
- 13.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 763697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 61099-1108. [DOI] [PubMed] [Google Scholar]
- 17.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11619-633. [DOI] [PubMed] [Google Scholar]
- 18.Hilton, A., L. Mizzen, G. MacIntyre, S. Cheley, and R. Anderson. 1986. Translational control in murine hepatitis virus infection. J. Gen. Virol. 67923-932. [DOI] [PubMed] [Google Scholar]
- 19.Ignowski, J. M., and D. V. Schaffer. 2004. Kinetic analysis and modeling of firefly luciferase as a quantitative reporter gene in live mammalian cells. Biotechnol. Bioeng. 86827-834. [DOI] [PubMed] [Google Scholar]
- 20.Isler, J. A., A. H. Skalet, and J. C. Alwine. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 796890-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamitani, W., K. Narayanan, C. Huang, K. Lokugamage, T. Ikegami, N. Ito, H. Kubo, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. USA 10312885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 3481953-1966. [DOI] [PubMed] [Google Scholar]
- 24.Lee, A. H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 237448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, J. H., H. Li, D. Yasumura, H. R. Cohen, C. Zhang, B. Panning, K. M. Shokat, M. M. Lavail, and P. Walter. 2007. IRE1 signaling affects cell fate during the unfolded protein response. Science 318944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, P., J. J. Millership, L. Li, D. P. Giedroc, and J. L. Leibowitz. 2006. A previously unrecognized UNR stem-loop structure in the coronavirus 5′ untranslated region plays a functional role in replication. Adv. Exp. Med. Biol. 58125-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma, Y., and L. M. Hendershot. 2003. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 27834864-34873. [DOI] [PubMed] [Google Scholar]
- 28.Malhotra, J. D., and R. J. Kaufman. 2007. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18716-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marciniak, S. J., C. Y. Yun, S. Oyadomari, I. Novoa, Y. Zhang, R. Jungreis, K. Nagata, H. P. Harding, and D. Ron. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 183066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montero, H., M. Rojas, C. F. Arias, and S. Lopez. 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J. Virol. 821496-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novoa, I., Y. Zhang, H. Zeng, R. Jungreis, H. P. Harding, and D. Ron. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 221180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 3611319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raaben, M., M. J. Groot Koerkamp, P. J. Rottier, and C. A. de Haan. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 92218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reimers, K., K. Buchholz, and H. Werchau. 2005. Respiratory syncytial virus M2-1 protein induces the activation of nuclear factor kappa B. Virology 331260-268. [DOI] [PubMed] [Google Scholar]
- 36.Robert, F., L. D. Kapp, S. N. Khan, M. G. Acker, S. Kolitz, S. Kazemi, R. J. Kaufman, W. C. Merrick, A. E. Koromilas, J. R. Lorsch, and J. Pelletier. 2006. Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2. GTP.Met-tRNA(i)(Met) ternary complex availability. Mol. Biol. Cell 174632-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ron, D., and P. Walter. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8519-529. [DOI] [PubMed] [Google Scholar]
- 38.Roth-Cross, J. K., L. Martinez-Sobrido, E. P. Scott, A. Garcia-Sastre, and S. R. Weiss. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 817189-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutkowski, D. T., S. M. Arnold, C. N. Miller, J. Wu, J. Li, K. M. Gunnison, K. Mori, A. A. Sadighi Akha, D. Raden, and R. J. Kaufman. 2006. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 8120-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siddell, S., H. Wege, A. Barthel, and V. ter Meulen. 1981. Coronavirus JHM: intracellular protein synthesis. J. Gen. Virol. 53145-155. [DOI] [PubMed] [Google Scholar]
- 42.Smith, J. A., S. C. Schmechel, A. Raghavan, M. Abelson, C. Reilly, M. G. Katze, R. J. Kaufman, P. R. Bohjanen, and L. A. Schiff. 2006. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 802019-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 805927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soe, L. H., C. K. Shieh, S. C. Baker, M. F. Chang, and M. M. Lai. 1987. Sequence and translation of the murine coronavirus 5′-end genomic RNA reveals the N-terminal structure of the putative RNA polymerase. J. Virol. 613968-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sriburi, R., S. Jackowski, K. Mori, and J. W. Brewer. 2004. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 16735-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stohlman, S. A., D. R. Hinton, D. Cua, E. Dimacali, J. Sensintaffar, F. M. Hofman, S. M. Tahara, and Q. Yao. 1995. Tumor necrosis factor expression during mouse hepatitis virus-induced demyelinating encephalomyelitis. J. Virol. 695898-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sturman, L. S., and K. K. Takemoto. 1972. Enhanced growth of a murine coronavirus in transformed mouse cells. Infect. Immun. 6501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tahara, S. M., T. A. Dietlin, C. C. Bergmann, G. W. Nelson, S. Kyuwa, R. P. Anthony, and S. A. Stohlman. 1994. Coronavirus translational regulation: leader affects mRNA efficiency. Virology 202621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tardif, K. D., K. Mori, R. J. Kaufman, and A. Siddiqui. 2004. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J. Biol. Chem. 27917158-17164. [DOI] [PubMed] [Google Scholar]
- 50.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 767453-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thuerauf, D. J., L. E. Morrison, H. Hoover, and C. C. Glembotski. 2002. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J. Biol. Chem. 27720734-20739. [DOI] [PubMed] [Google Scholar]
- 52.Tooze, J., and S. A. Tooze. 1985. Infection of AtT20 murine pituitary tumour cells by mouse hepatitis virus strain A59: virus budding is restricted to the Golgi region. Eur. J. Cell Biol. 37203-212. [PubMed] [Google Scholar]
- 53.Ventoso, I., M. A. Sanz, S. Molina, J. J. Berlanga, L. Carrasco, and M. Esteban. 2006. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2087-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Versteeg, G. A., O. Slobodskaya, and W. J. Spaan. 2006. Transcriptional profiling of acute cytopathic murine hepatitis virus infection in fibroblast-like cells. J. Gen. Virol. 871961-1975. [DOI] [PubMed] [Google Scholar]
- 55.Versteeg, G. A., P. S. van de Nes, P. J. Bredenbeek, and W. J. Spaan. 2007. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J. Virol. 8110981-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wathelet, M. G., M. Orr, M. B. Frieman, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J. Virol. 8111620-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams, B. L., and W. I. Lipkin. 2006. Endoplasmic reticulum stress and neurodegeneration in rats neonatally infected with Borna disease virus. J. Virol. 808613-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, J., D. T. Rutkowski, M. Dubois, J. Swathirajan, T. Saunders, J. Wang, B. Song, G. D. Yau, and R. J. Kaufman. 2007. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell 13351-364. [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto, K., T. Sato, T. Matsui, M. Sato, T. Okada, H. Yoshida, A. Harada, and K. Mori. 2007. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell 13365-376. [DOI] [PubMed] [Google Scholar]
- 60.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 812554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zust, R., L. Cervantes-Barragan, T. Kuri, G. Blakqori, F. Weber, B. Ludewig, and V. Thiel. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3e109. [DOI] [PMC free article] [PubMed] [Google Scholar]