Abstract
Disruption of pRB-E2F interactions by E1A is a key event in the adenoviral life cycle that drives expression of early viral transcription and induces cell cycle progression. This function of E1A is complicated by E2F1, an E2F family member that controls multiple processes besides proliferation, including apoptosis and DNA repair. Recently, a second interaction site in pRB that only contacts E2F1 has been discovered, allowing pRB to control proliferation separately from other E2F1-dependent activities. Based on this new insight into pRB-E2F1 regulation, we investigated how E1A affects control of E2F1 by pRB. Our data reveal that pRB-E2F1 interactions are resistant to E1A-mediated disruption. Using mutant forms of pRB that selectively force E2F1 to bind through only one of the two binding sites on pRB, we determined that E1A is unable to disrupt E2F1's unique interaction with pRB. Furthermore, analysis of pRB-E2F complexes during adenoviral infection reveals the selective maintenance of pRB-E2F1 interactions despite the presence of E1A. Our experiments also demonstrate that E2F1 functions to maintain cell viability in response to E1A expression. This suggests that adenovirus E1A's seemingly complex mechanism of disrupting pRB-E2F interactions provides selectivity in promoting viral transcription and cell cycle advancement, while maintaining cell viability.
The retinoblastoma protein (pRB) regulates cell cycle progression by interacting with a number of cellular proteins. Binding to E2F transcription factors inhibits activation of S-phase transcriptional target genes, such as cyclin E, B-myb, and PCNA, among others (20, 47). Recruitment of LXCXE-containing chromatin-remodeling proteins, such as histone methyltransferases (Suv39h1 and -2) and histone deacetylases (HDAC1 and -2) to these promoters by pRB serves to actively control the accessibility of target gene promoters and further regulates transcription (6, 27, 30, 31, 36). Recently, it has also been shown that pRB can regulate cyclin/cdk activity directly by using APCcdh1 to antagonize Skp2's ability to degrade p27 (5, 22). Under normal proliferative conditions, mitogen stimulation leads to inactivation of pRB through G1 cyclin/cdk complexes that phosphorylate pRB and disrupt its ability to bind to its regulatory targets (13).
In a remarkably similar fashion, advancement of the cell cycle into S phase can arise from expression of small DNA tumor virus proteins, such as adenovirus E1A (25, 43). E1A drives transcription of early region viral genes and through interaction with pRB and other proteins induces S-phase entry (34). A key activity in E1A's ability to stimulate transcription and viral propagation is the disruption of E2F transcription factor regulation by the retinoblastoma family of proteins. Collectively, E2Fs were originally described as cellular transcription factors required for the transcriptional activation of the adenovirus E2 promoter (26). Currently, there are eight distinct E2F family proteins, and each is known to be capable of binding to the E2F consensus DNA sequence element (23). However, not all of these are potent activators of transcription or interact with RB family proteins. Since E2Fs 1 to 5 interact with pRB family proteins (46), the disruption of these interactions is of particular interest for understanding how E1A functions in directing viral gene transcription.
Two conserved regions (CR) within E1A have been shown to be necessary for disruption of pRB-E2F transcription factor complexes (39). E1A CR1 and CR2 show high protein sequence similarity among numerous E1A serotypes (1), as well as to regions in viral oncogenes from other DNA tumor viruses (18, 38). The LXCXE motif within CR2 of E1A forms a strong interaction with the pRB pocket domain (16). Once tethered to pRB, the CR1 region of E1A mediates disruption of E2F binding (16). CR1 can make a relatively weak interaction with pRB on its own because it resembles the portion of E2Fs that contact pocket proteins (14, 29). This suggests that E2Fs are ultimately removed from pRB by competition with CR1 (16). At present there is little indication why E1A uses a multistep mechanism rather than direct competition to disrupt E2F regulation.
In addition to regulating proliferation, E2F1 has distinguished itself from other E2Fs by its unique roles in apoptosis and DNA repair (23). Endogenous E2F1 is activated by DNA damage signaling and can induce apoptosis (28, 37, 44). Thymocytes from E2F1 knockout mice are resistant to apoptosis induced by DNA damage and during T-cell development (17, 28). In addition, these mice frequently succumb to lymphomas, suggesting E2F1 may use this activity to function as a tumor suppressor gene (50). Conversely, endogenous E2F1 has also been demonstrated to regulate expression of genes necessary for DNA repair and maintenance of cell viability (40). In this regard E2f1−/− mice are more prone to apoptosis in the epidermis following DNA damage, and this correlates with an inability to engage DNA repair mechanisms (4, 49). This background emphasizes that E2F1 has additional functions that modulate cell viability to cause cell death or to promote survival, and for this reason it is different from other E2F family proteins. How the cellular context controls which E2F1 activity is activated by DNA damage or other stimuli remains speculative (23). Following infection of a host cell, E1A is the first adenoviral protein expressed. The need to maintain cell viability and induce S-phase entry by adenoviruses at this stage of infection is underscored by the observation that other early viral proteins are potent apoptotic inhibitors (3). Given the necessity to induce cell cycle advancement while maintaining viability early after viral infection, it seems counterintuitive that E1A would deregulate E2F1 along with the other E2Fs. It seems more logical that E1A would want to manipulate E2F1 and its specialized roles to help maintain cell viability.
It has previously been demonstrated that pRB contains two independent binding sites for E2F transcription factors (7, 10, 24). One type of pRB-E2F interaction is termed the general interaction, because it is unselective for its E2F binding partner and appears to function mainly to regulate cell cycle advancement (10). The second, specific, E2F binding site on pRB is used exclusively by E2F1, and this interaction has previously been shown to provide negative regulation of apoptosis while having almost no impact on proliferative control (24). Based on this division of function between different E2F binding configurations with pRB, we wondered if E1A was also selective in its ability to regulate E2F1. In this report we demonstrate that E1A is unable to disrupt E2F1 binding to pRB. In transfection experiments, E1A disrupted only the proliferative control, or general, interaction between pRB and E2F1. During a productive adenoviral infection where viruses infect quiescent cells, induce advancement of the cell cycle, and replicate, endogenous E2F1-pRB complexes are selectively retained. Furthermore, our analysis reveals that E2F1 functions to maintain viability following expression of E1A. These data strongly argue that E1A has developed a mechanism of selectively disrupting proliferative control mechanisms by pRB while leaving a specialized form of E2F1 regulation intact to maintain cell viability.
MATERIALS AND METHODS
Cell culture.
RB-1-deficient C33A cervical carcinoma cells, IMR90 human diploid fibroblasts, and 293 human embryonic kidney cells were obtained from the American Type Culture Collection. Mouse embryonic fibroblasts (MEFs) were generated from the appropriate genotypes of mice using previously published methods (21). All cells were cultured in Dulbecco's modified Eagle's medium supplemented with penicillin (100 U/ml), l-glutamine (200 mM), streptomycin (10 μg/ml), and 10% fetal bovine serum. Cells were maintained in a humidified chamber at 37°C with 5% CO2.
Plasmids and recombinant proteins.
CMV-β-Gal, CMV-HA-DP1, CMV-HA-E2F1, CMV-HA-E2F4, CMV-RB, and CMV-RBΔE2F-G have all been previously described (10). CMV-RBΔE2F-S was constructed similarly to other pRB mutants and has been described by Julian et al. (24). GST-E1A12S, GST-E1A13S, and mutant pEGFP-E1A constructs have been previously reported (16, 19). GST-E1A12S dl1107 was created by ligating an EcoRI-SalI fragment of pEGFP-E1A12S dl1107 into pGEX-4T1 cut with the same enzymes. GST-E1A12S.Y47H was constructed by digesting pEGFP-E1A12S.Y47H with EcoRI and XhoI and ligating into a pGEX-based GST-E1A12S plasmid as above. Glutathione S-transferase (GST)-fusion proteins were expressed and purified by standard techniques and quantified by the Bio-Rad protein assay. The retroviral pLPC-E1A12S construct was obtained from S. Lowe (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) (42).
In vitro competition assays.
C33A cells were transfected with pRB along with E2F and DP1 expression plasmids. Whole-cell lysates were prepared as described previously (11) in GSE buffer (20 mM HEPES [pH 7.5], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 25 mM dithiothreitol [DTT], with 0.1 mM Na3VO4,0.5 mM NaF, and 0.25 mM phenylmethylsulfonyl fluoride [PMSF]). In each immunoprecipitation (IP) competition reaction mixture, lysate (approximately 400 μg total) was diluted into 200 μl of GSE buffer to normalize for pRB. A 300-μl aliquot of wash buffer (20 mM HEPES [pH 7.5], 200 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25 mM DTT, and 0.1% NP-40) and 10-fold dilutions of GST-E1A from 10 μg to 0.01 μg were added. Reaction mixtures were incubated on ice for 30 min before addition of antihemagglutinin (anti-HA) antibodies and protein G-Sepharose beads. Beads were washed two times with wash buffer and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and pRB binding was analyzed by Western blotting. Competition assays using native 12S E1A were essentially the same as above except that transfected C33A nuclear extracts containing E1A were used. E1A present in these extracts was quantified by Western blotting alongside defined quantities of GST-E1A; 10-fold dilutions from 1 μg to 0.01 μg of E1A 12S were used.
GST binding assays.
GST binding assays were carried out essentially as described by Dick et al. (12). Briefly, C33A cells transfected with pRB expression constructs were harvested as above for competition assays. Extracts were normalized to pRB content by diluting into GSE buffer. Wash buffer was again added, along with 10-fold dilutions of GST-E1A proteins (from 10 μg to 0.01 μg) and glutathione-Sepharose beads. Binding reaction mixtures were mixed for 1 h at 4°C, and beads were pelleted and washed as described above. Bound pRB was detected by Western blotting.
Immunoprecipitations.
C33A cells were transfected with equal quantities of CMV-E1A12S or CMV-β-Gal in combination with plasmids encoding pRB, HA-E2Fs, and HA-DP1. Whole-cell extracts were prepared in GSE buffer and normalized to pRB content by diluting to a 200-μl final volume (approximately 400 μg per IP mixture). Wash buffer and anti-HA hybridoma supernatant were added as described above, and immune complexes were collected on protein G-Sepharose beads. Beads were washed, and bound pRB was detected by Western blotting.
Nuclear extracts were prepared from HEK293 cells by scraping cells into phosphate-buffered saline, and cells were then pelleted by centrifugation and resuspended in three cell volumes of hypotonic lysis buffer (10 mM HEPES [pH 7.5], 10 mM KCl, 3 mM MgCl2, 1 mM EDTA, 1 mM PMSF, 1 mM DTT, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 5 mM NaF, 0.1 mM Na3VO4). Cells were incubated on ice for 5 min before 0.05% NP-40 was added, and cells were lysed for 5 min. Nuclei were pelleted at 4,000 rpm and washed two times with hypotonic lysis buffer containing 0.05% NP-40. Nuclei were suspended in GSE buffer and stored frozen.
For each quantitative IP reaction, beads were prebound to antibodies by incubating 150 μl of 20% protein G-Sepharose (PGS) slurry with 15 μl C15 or preimmune serum and washing three to four times prior to resuspension as a 20% slurry in wash buffer. Each IP reaction mixture contained 600 μg nuclear extract and was diluted to 300 μl in GSE buffer before 450 μl of wash buffer was added. To this, 150 μl of 20% prebound PGS was added and incubated with gentle agitation for 1 hour at 4°C. After 1 hour, an additional aliquot of 20% prebound PGS slurry was added and allowed to adsorb soluble pRB. This was repeated one more time, before spinning out the PGS beads. The supernatant was transferred to a new tube, and beads were washed twice with wash buffer before being resuspended in a volume equal to the supernatant. Supernatant and bead fractions were each mixed with 5× sample buffer and processed for SDS-PAGE and Western blotting.
Sequential IP experiments were performed similarly to the quantitative IPs above except that twice as much extract was used, and after the first addition of beads bound complexes were collected by centrifugation and eluted with 1 mg/ml peptide corresponding to the C-terminal 15 amino acids of pRB. Eluted material was then precipitated as before with anti-E1A antibodies and processed for SDS-PAGE and Western blotting as described above.
Electrophoretic mobility shift assays.
All DNA probes used were as described by Hurford et al. (20). Double-stranded oligonucleotides were labeled with 50 μCi [α-32P]dCTP using Klenow fragment for 15 min at 25°C and purified on a G25 spin column. HEK293 nuclear extracts were prepared as described above. Each sample contained electrophoretic mobility shift assay (EMSA) buffer [20 mM HEPES (pH 7.5), 4% Ficoll 400-DL (Sigma), 2.5 mM MgCl2, 40 mM KCl, 0.1 mM EGTA, 2 mM spermine, 0.5 mM DTT, 0.5 μg salmon sperm DNA, 0.01 U poly(dI-dC), 10 μg bovine serum albumin] and 5 μg of nuclear extract. For samples containing cold competitors, 40 ng of either wild-type or mutant unlabeled oligonucleotide containing an E2F consensus site was included. Following addition of 400 pg of the labeled oligonucleotide, each reaction mixture was incubated on ice for 10 min before loading on a gel. For antibody supershifts, the indicated antibodies were subsequently added and samples were incubated a final 25 min on ice prior to loading reactions. EMSA reaction mixtures were loaded onto a 4% polyacrylamide gel (containing 0.25× Tris-borate-EDTA and 2.5% glycerol) and electrophoresed at 4°C for 4 h at 180 V. All EMSA gels were prerun for 30 min at 100 V. Gels were dried and autoradiographed.
Viral infections.
IMR90 cells were grown until confluent, and cells were further incubated for two additional days to ensure maximal growth arrest. Infections were carried out using adenovirus type 5, strain dl309 according to standard procedures at a multiplicity of infection of 50. Quiescence and subsequent induction of cell cycle entry during these infections were confirmed by SDS-PAGE and Western blotting to identify hypo- and hyperphosphorylated forms of pRB, respectively (data not shown). Nuclear extracts were prepared at 0, 8, 24, and 48 h postinfection as described above, and pRB was immunoprecipitated as described above for one round of HEK293 nuclear extract IPs.
Ecotropic retroviruses were prepared by transfecting Bosc23 cells with control or E1A-encoding vectors. Two days later, medium was transferred from packaging cell cultures to proliferating MEFs supplemented with 4 μg/ml Polybrene and allowed to infect. Infected control and E1A-expressing cells were selected in 4 μg/ml puromycin for 2 days. Cells were used to generate extracts for IPs as described for transfections above. Cell viability was measured by plating cells in a 96-well dish, treating as described above, and analyzing metabolic conversion of Alamar blue to a fluorescent product according to the manufacturer's instructions (Biosource, Camarillo, CA).
Antibodies.
For Western blot assays, pRB was detected with C36 hybridoma supernatant, G3-245 monoclonal antibodies, or a rabbit antiserum raised against the C-terminal 15 amino acids of pRB (C15). Blots for HA-tagged proteins were probed with 12CA5 supernatant, and E1A was detected with M73. E2F1 was detected with KH95, E2F2 with monoclonal CC11 (Sigma), E2F3 with PG37 (Upstate), and E2F4 with WUF2. Immunoprecipitations were carried out using 12CA5 supernatant for HA-tagged proteins, C15 antiserum for pRB, and sc430AC (Santa Cruz) or M73 supernatant for E1A. In EMSA experiments, the following antibodies or culture supernatants were used: 12CA5 (anti-HA), 21C9 (anti-pRB; a gift from Sybille Mittnacht, London, United Kingdom), KH20 (anti-E2F1), sc633x (anti-E2F2; Santa Cruz), PG-37 (anti-E2F3), WUF10 (anti-E2F4), and WTH10 (anti-DP1).
RESULTS
GST-E1A is unable to disrupt pRB-E2F1 complexes.
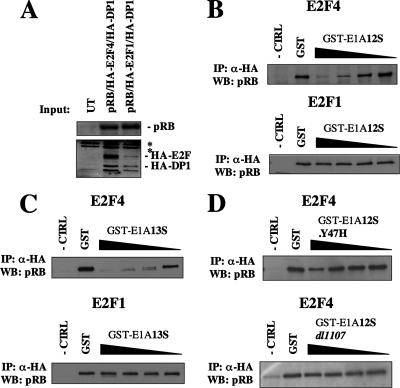
Based on evidence that pRB uses two distinct interaction mechanisms to contact E2F transcription factors (7, 10, 24), we sought to investigate how E1A affects these different interaction types. To this end we devised an in vitro competition assay to study pRB-E2F interactions. Briefly, RB-1-deficient C33A cells were transfected with pRB, HA-E2F1, and HA-DP1 or pRB, HA-E2F4, and HA-DP1. Extracts were analyzed by Western blotting, and input amounts were normalized to the expression level of pRB (Fig. 1A). Extracts were supplemented with a range of GST-E1A12S protein concentrations or GST only, as a control. E2F transcription factors were immunoprecipitated with anti-HA antibodies, and coprecipitated pRB was detected by Western blotting (Fig. 1B). Addition of GST-E1A12S resulted in a clear disruption of E2F4-pRB-containing complexes (Fig. 1B, top panel); however, E2F1-pRB interactions were insensitive to GST-E1A12S at all concentrations tested (Fig. 1B, bottom panel). This suggests that there is an inherent resistance of E2F1-pRB-containing complexes to GST-E1A12S-mediated disruption that is orders of magnitude stronger than E2F4. This is even more remarkable considering that the input level of E2F1-DP1 was lower than the E2F4-containing extract (Fig. 1A), indicating that our experiment was biased toward disruption of the E2F1-containing complexes. To ensure that regions contained in E1A12S were sufficient for disruption of pRB-E2F complexes, we also tested the ability of GST-E1A13S to disrupt pRB-E2F1- or pRB-E2F4-containing complexes. In a similar experiment we found that pRB is readily displaced from binding to E2F4 by the introduction of GST-E1A13S (Fig. 1C), while E2F1-pRB-containing complexes remain resistant to E1A-mediated disruption. This demonstrates that both the 12S and 13S forms of E1A can dissociate pRB-E2F4 complexes with as little as 100 ng of GST-E1A protein, while pRB-E2F1 interactions are resistant, even at GST-E1A protein levels that are 100 times higher.
FIG. 1.
pRB/E2F1 complexes are resistant to GST-E1A disruption. (A) Whole-cell extracts from transfected cells were analyzed by Western blotting to show relative pRB and HA-E2F expression levels in extracts used in these experiments. Each lane represents 2.5% of the input used in the IP experiments shown in panels B to D. Asterisks mark irrelevant bands that cross-react with HA antibodies. A 10-fold dilution of GST-E1A12S (B) or GST-E1A13S (C) from 10 μg to 10 ng was added to extracts expressing pRB and either HA-E2F4 or pRB and HA-E2F1. The quantity of pRB captured with E2Fs by IP was analyzed by Western blotting. (D) Tenfold dilutions of GST-E1A12S.Y47H or GST-E1A12S dl1107 were used similarly in extracts containing HA-E2F4 and pRB. The negative control (-CTRL) lane was generated by anti-HA IPs of cell extracts expressing pRB but not HA-E2Fs.
To verify that E2F4-pRB complexes were being dissociated by functional GST-E1A, we utilized GST-tagged E1A proteins with mutations in the conserved regions to test if our competition assay relies on the same regions of E1A as previously described (16, 39). The LXCXE-containing region (CR2) normally mediates the initial contact between E1A and pRB, while conserved region 1 (CR1) contains an E1A-pRB interaction region that competes with E2Fs for binding to pRB. GST-E1A12S containing a Y47H amino acid substitution within CR1 has a diminished ability to compete with E2F4 for binding to pRB (Fig. 1D, top panel). GST-E1A12S dl1107 contains a deletion of residues 111 to 123 within CR2 that interrupts the LXCXE motif and causes a loss of pRB binding. This CR2 mutation abrogates E1A's ability to disrupt E2F4-pRB complexes (Fig. 1D, bottom panel). These observations validate the experiments outlined in Fig. 1A to C, which indicate that GST-E1A12S is a functional E1A protein capable of E2F4-pRB dissociation activity but lacking the ability to disrupt E2F1-pRB.
E1A uses a separate mechanism to regulate pRB-E2F1 interactions.
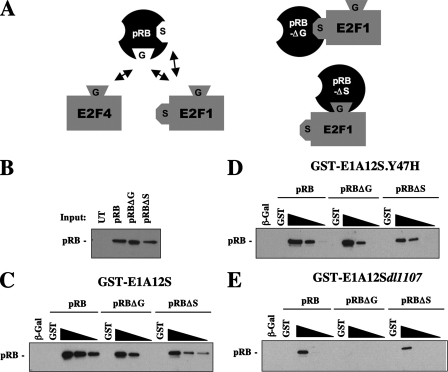
We recently discovered that pRB contains a second E2F interaction site that uniquely regulates E2F1 (10, 24). This interaction was mapped to distinct regions of both E2F1 and pRB, involving the C-terminal region of the large pocket of pRB and a small portion of the E2F1 marked box region (24, 41). Thus, E2Fs can bind to pRB by a common mechanism at a location that we have termed the “general” site (or G, for brevity). The interaction mechanism that is unique to E2F1 occurs in a region referred to as the “specific” site (or S), and these are depicted in a cartoon in Fig. 2A. Since E2F1 can interchangeably use these two interaction mechanisms, we have generated mutants defective for each type of interaction alone to study their functions in isolation (10, 24). These mutations allowed us to examine both interaction sites independently, because a combination allele carrying both types of mutations in the same molecule eliminates all E2F1 binding (24).
FIG. 2.
E1A CR1 contacts the general E2F binding site on pRB. (A) Schematic diagram illustrating the two different E2F-pRB interaction types. The general (G) E2F binding site on pRB interacts with E2Fs without preference. E2F1 can bind to pRB through either the G or the E2F1 S (specific) site. Mutant forms of pRB that lack either the general or specific interaction site allow the other E2F interaction type to be studied in isolation. (B) C33A whole-cell extracts from cells expressing the indicated RB constructs were analyzed for relative pRB expression. Each lane represents 2% of the input used in each pull-down experiment. (C) Recombinant GST-E1A12S was added to transfected cell extracts in a 10-fold titration range from 10 μg to 100 ng. Bound wild-type or mutant pRB was collected on glutathione-Sepharose beads and detected by Western blotting. Similar analyses of CR1 (Y47H) and CR2 (dl1107) mutants in GST-E1A12S were carried out (D and E).
To examine if one of the pRB-E2F1 interactions was responsible for the resistance to GST-E1A-mediated disruption, as depicted in Fig. 1, we sought to investigate how E1A binds to pRB and ultimately competes with E2Fs. C33A cells were transiently transfected with plasmids encoding pRB, pRBΔG, or pRBΔS, and expression levels of these proteins are shown in Fig. 2B. The ability of these expressed proteins to bind to E1A was measured by GST-pull-down assays. When GST-E1A12S was mixed with extracts expressing pRBΔG, the E1A-pRB interaction was lost in the 100-ng GST-E1A12S pull-down sample (Fig. 2C). This suggests that E1A binds in part through the general E2F binding site and the pRBΔG mutant reduces the affinity of this interaction. To investigate pRB binding further, we used CR1 and CR2 mutants to examine the contribution of each region of E1A in isolation. The use of GST-E1A12S.Y47H demonstrates that the affinity of E1A CR2 for each of the wild-type, pRBΔG, and pRBΔS proteins is the same and thus the LXCXE interaction with pRB is unaffected by either pRB mutation (Fig. 2D). In contrast, when GST-E1A12S dl1107 was mixed with extracts expressing pRB and pRB-E2F binding mutants, a complete loss of E1A-pRB association was detected for pRBΔG (Fig. 2E). This finding demonstrates that the CR1 region is essential for contacting the general E2F binding site. Thus, CR1 competition for E2F binding is through direct contact with the same region of pRB. This further suggests that E2F1, but not E2F4, may evade this disruption by using a separate binding site on pRB.
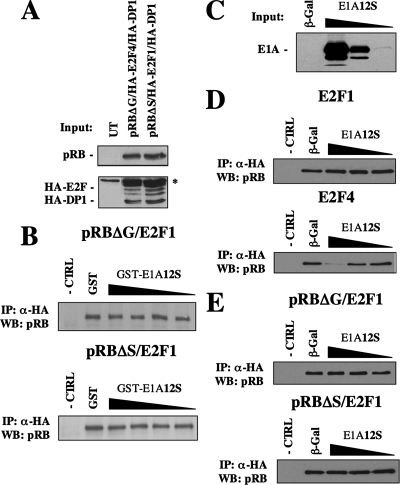
To test if E2F1 resistance to E1A displacement is dependent on the specific binding site, cells were transfected with plasmids encoding HA-E2F1 and HA-DP1 and either pRBΔG or pRBΔS. Input levels of pRB and HA-E2F1/HA-DP1 used in these assays are shown in Fig. 3A. Using the same competition assay approach as in Fig. 1, we analyzed the ability of GST-E1A12S to disrupt HA-E2F1/HA-DP1 binding to either pRBΔG or pRBΔS. As shown in Fig. 3B, neither interaction was disrupted. Surprisingly, these findings indicate that regardless of the configuration and regulatory role of pRB-E2F1 complexes, GST-E1A12S is insufficient to displace E2F1 from pRB under conditions where E2F4 can be readily removed. This was also found in competition assays using E1A13S (data not shown).
FIG. 3.
Both configurations of E2F1-pRB complexes show resistance to E1A12S disruption in vitro. (A) Input levels of pRB and HA-E2F1/HA-DP1 from transfected C33A extracts are shown. Each lane represents 2.5% of the input used in the IPs. The asterisk denotes a cross-reacting band detected by anti-HA antibodies. (B) Tenfold dilutions of GST-E1A12S from 10 μg to 10 ng were titrated into whole-cell extracts from cells expressing pRBΔG/HA-E2F1 or pRBΔS/HA-E2F1 complexes. RB protein bound to E2F1 was detected by Western blotting. (C) Nuclear extracts were prepared from C33A cells transfected with E1A or β-Gal expression plasmids. Western blots of E1A show the relative amounts used in competition assays. (D) E1A- (or β-Gal)-expressing nuclear extracts containing 1 μg, 100 ng, or 10 ng of the relevant protein were titrated against pRB/HA-E2F1 or pRB/HA-E2F4 complexes. HA-E2F-bound pRB was isolated on protein G-Sepharose and detected by Western blotting. (E) Complexes containing pRBΔG/HA-E2F1 or pRBΔS/HA-E2F1 interactions were analyzed similarly for sensitivity to E1A competition. The negative control (-CTRL) lane controls for specificity of anti-HA IPs by using whole-cell extracts containing pRB but lacking a transfected HA-E2F.
E1A associates with a number of kinases and is phosphorylated at several residues (15, 45). Previously, phosphorylation has been suggested to improve E1A's ability to disrupt pRB-E2F interactions (32); thus, it may explain differences in activity toward different E2F proteins in Fig. 1. E1A12S was expressed in C33A cells by transfection, and nuclear extracts were harvested to obtain E1A carrying the appropriate posttranslational modifications (Fig. 3C). E1A-containing nuclear extracts were added to E2F/pRB-containing extracts, and HA-E2F/HA-DP1 was immunoprecipitated. When E1A12S-expressing extracts were titrated against HA-E2F1-pRB-containing complexes, no dissociation of pRB from HA-E2F1 was detected (Fig. 3D). However, E1A12S expressed in mammalian cells was capable of competing complexes containing E2F4 and pRB, indicating that E1A used in these experiments is active. We also tested the ability of this endogenous form of E1A to compete for binding with E2F1 at each individual binding site on pRB. As shown in Fig. 3E, neither RBΔG-E2F1 nor RBΔS-E2F1 complexes could be disrupted by E1A12S expressed in mammalian cells. These findings indicate that there is a fundamental difference between E2F1 and E2F4 binding to pRB, and it is manifested in resistance to E1A-mediated disruption, even when these E2Fs bind to the same region of pRB.
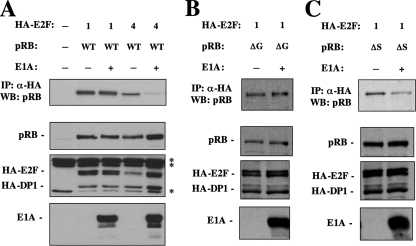
To investigate E1A regulation of E2F1-pRB complexes further, we tested competition in transfection assays to see if E1A could break this interaction in vivo. C33A cells were cotransfected with constructs for the expression of HA-E2F1 or HA-E2F4, pRB, and E1A12S where indicated. HA-tagged E2F-DP1 complexes were immunoprecipitated, and the persistence of pRB in these E2F complexes was detected by Western blotting. As shown in Fig. 4A, wild-type pRB associates with E2F1 in the presence of E1A to the same extent as when E1A is absent, indicating that E1A is unable to quantitatively disrupt E2F1-pRB complexes. However, when E1A is coexpressed with E2F4, disruption of E2F4-pRB complexes is observed. To explore the two types of E2F1 interactions in isolation, RBΔG- and RBΔS E2F-binding mutants were coexpressed in combination with HA-E2F1/HA-DP1 either in the absence or presence of E1A (Fig. 4B and C). HA-E2F1 complexes captured by IP coprecipitate with pRBΔG equally, indicating that interactions through the specific E2F1 regulatory site are not disrupted by E1A. However, when E1A is coexpressed with complexes consisting of pRBΔS and HA-E2F1, a clear competition with E2F1 binding to pRB is detected (Fig. 4C). This indicates that although resistant to E1A-mediated disruption, pRB-E2F1 complexes can be dissociated by E1A from the general, cell cycle regulatory binding site, but the specific E2F1 only configuration remains undisturbed. The fact that the pRB-E2F1 general interaction is only competed by E1A in an intact cell suggests that there are mechanistic distinctions between disruption of this interaction and disruption of E2F4-pRB, and these possibilities will be elaborated on in the Discussion section, below.
FIG. 4.
E1A12S disrupts pRB/E2F1 general interactions in vivo. (A) C33A whole-cell extracts expressing the indicated proteins were immunoprecipitated with anti-HA antibodies. Coprecipitated pRB was detected by Western blotting. Input levels of pRB, HA-E2F, HA-DP1, and E1A are shown in the lower three panels. Each lane represents 2.5% of the input used in the IPs. Asterisks mark irrelevant cross-reacting bands detected by HA antibodies. (B and C) C33A whole-cell extracts expressing the indicated proteins were also subjected to anti-HA IPs. The amount of pRB bound to E2F1 was determined by Western blotting. The relative expression level of input proteins is also shown.
Analysis of in vivo E2F-pRB interactions reveals the maintenance of endogenous pRB-E2F1 complexes in the presence of E1A.
Most analyses of E1A-mediated disruption of pRB-E2F interactions have used heterogeneous pocket protein-E2F complexes found in cell lysates (2, 8, 9, 16, 39). Because most of these reports predate the molecular cloning of individual E2F family members, we wondered if E2F1 is retained in endogenous complexes with pRB despite the presence of E1A.
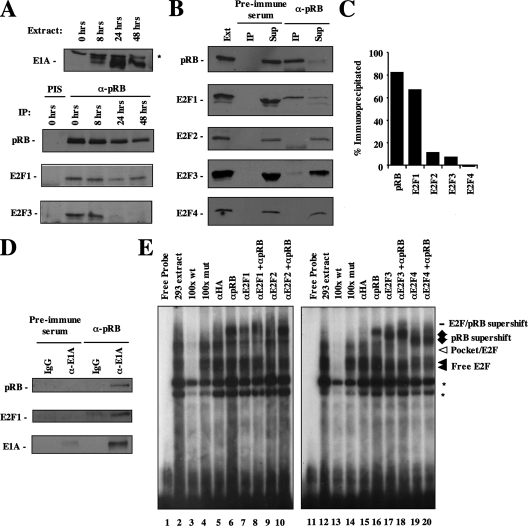
We investigated whether E2F1 is retained in complexes with pRB during a productive adenoviral infection. We used primary human IMR90 cells that were confluent, and therefore growth arrested, to determine if this interaction is preserved throughout viral replication. As shown in Fig. 5A, E1A expression is detectable across the time course of infection and peaks at 24 h, which is indicative of the infection, replication, and lysis that take place over a complete infectious cycle. Using nuclear extracts from these cells, we immunoprecipitated pRB and performed Western blotting to determine the relative abundance of E2F proteins that associate with pRB. These experiments revealed that E2F1 remains associated with pRB when E2F3's interaction is clearly disrupted. To examine the selective retention of E2F1-pRB complexes in more detail, we devised a quantitative immunoprecipitation assay to detect relative pRB-E2F abundance for all E2F complexes. HEK293 cells stably express E1A, making them an ideal source of material for these experiments. Nuclear extracts were subjected to three rounds of immunoprecipitation using antiserum directed against the C terminus of pRB. The pooled IP fractions contain nearly all pRB from the nuclear extract, and comparison with the unbound supernatant fraction allows the estimation of the percentage of the protein immunoprecipitated. Figure 5B illustrates that E2Fs 2 to 4 remained largely unassociated with pRB in the presence of E1A, but the majority of nuclear E2F1 was bound to pRB. Quantification of pRB binding to E2Fs in Fig. 5B was assessed by densitometry (Fig. 5C). Over 60% of the E2F1 nuclear population associates with pRB, whereas less than 15% of E2Fs 2 to 4 associate with pRB in the presence of E1A. Given the high-affinity interaction between CR2 of E1A and the pRB pocket domain, we further investigated whether this abundant pRB-E2F1 complex is stably associated with E1A. Figure 5D shows a sequential IP experiment where pRB immune complexes were eluted from antibodies using the antigenic peptide as a competitor. Eluted proteins were then reprecipitated using anti-E1A antibodies, and the resultant complex was analyzed by Western blotting for E2F1, pRB, and E1A. This experiment demonstrated that a ternary complex composed of E2F1-pRB-E1A is stably formed in HEK293 cells.
FIG. 5.
Selective retention of endogenous E2F1/pRB complexes. IMR90 and HEK293 cells were used to examine the effect of E1A on endogenous pRB/E2F complexes. (A) Confluent IMR90 cells were infected with adenoviral strain dl309 and nuclear extracts were prepared at the indicated time points. The extract lanes contain 10% of the amount used for the subsequent IPs. Anti-pRB IPs and Western blot assays were carried out to detect the presence of coprecipitating E2F transcription factors. The preimmune serum control IP is indicated by the abbreviation PIS. (B) Immunoprecipitations were carried out to quantitatively deplete pRB from HEK293 nuclear extracts. Preimmune and anti-pRB antibodies were used, and bound (IP) along with supernatant (Sup) fractions were probed to detect pRB and E2Fs 1 to 4. The extract lane represents 1.5% of the input used in these IPs. (C) Densitometry was used to quantify E2F association with pRB and is represented as the percentage of protein that was immunoprecipitated. (D) HEK293 nuclear extracts were sequentially precipitated with anti-pRB antibodies, eluted with a competitor peptide, and precipitated with anti-E1A antibodies followed by Western blotting. (E) EMSAs were used to detect the ability of pRB/E2F complexes to bind to a consensus E2F recognition sequence. Addition of cold competitor probes, or antibodies directed against E2F and pRB proteins, were used to identify the different DNA binding complexes present in the gel. The asterisks denote nonspecific bands.
In previous work we discovered that when pRB-E2F1 complexes form through the “specific” interaction, they display a markedly reduced affinity for the consensus E2F DNA sequence element TTTCGCGC in an EMSA (10). As such, EMSA analysis can be used to characterize pRB-E2F interactions that occur via the “general” interaction, and we used this assay to investigate the nature of pRB-E2F interactions in E1A-expressing HEK293 cells. A radiolabeled oligonucleotide containing the E2F consensus site was combined with nuclear extracts from HEK293 cells and electrophoresed through native polyacrylamide gels (Fig. 5E). The combination of nuclear extract and radiolabeled probe yields a characteristic banding pattern containing a prominent E2F band and a fainter pocket protein-E2F species (lanes 2 and 12). Addition of an unlabeled wild-type oligonucleotide at 100 times the probe concentration competes away E2F-DNA interactions, and excess mutant E2F oligonucleotides confirm the identity of bona fide E2F complexes (lanes 3 and 13 versus lanes 4 and 14). Addition of an antibody specific to pRB produces a supershift above the pocket protein/E2F complexes, as indicated (lane 6). When antibodies specific for E2Fs 1, 2, 3, and 4 are added (lanes 7, 9, 17, and 19, respectively), again supershifts are detected, indicating that E2Fs 1 through 4 can be detected in these extracts. However, when E2F antibodies are combined with an antibody for pRB (lanes 8, 10, 18, and 20), only modest super-supershifts that appear as a smear are found higher on the gel. These EMSA results indicate that E2F1/pRB complexes identified in these experiments are no more abundant than any other pRB-E2F combination. This suggests that the abundant pRB-E2F1 complexes in HEK293 cells that we identified by immunoprecipitation are likely present in the specific configuration because they aren't readily detectable by EMSA.
These experiments indicate that pRB retains association with E2F1 in the presence of E1A as a means to regulate E2F1 or potentially facilitate a new function as a ternary E2F1-pRB-E1A complex. Based on our competition assays, immunoprecipitations, and EMSAs, there is a strong possibility that this endogenous pRB-E2F1 complex uses the “specific” interaction configuration to allow E2F1 to bind to pRB in the presence of E1A.
E2F1 functions to promote viability following E1A expression.
Our experiments reveal the existence of a pRB-E2F1 complex that is selectively retained during adenoviral infection. Furthermore, they indicate that E2F1, pRB, and E1A form a stable complex, with E2F1 most likely bound to pRB through its specific interaction. To investigate the physiological purpose for this complex, we devised assays to test the functions for pRB and E2F1 following ectopic expression of E1A, a situation that mimics early adenoviral infection. E1A expression alone is capable of inducing cell death in this primary cell culture system under conditions of serum deprivation or DNA damage (33, 42). Likewise, E2F1 expression is induced by DNA damage, suggesting that it offers the most physiologically relevant condition under which to investigate how activation of E2F1 function can modulate viability or induce apoptosis early in adenoviral infection (4, 28, 37, 44, 49). In these experiments we used RbΔL/ΔL MEFs, which express a mutant form of pRB that contains mutations in its LXCXE binding cleft to prevent binding to CR2 in E1A (12, 21), and E2f1−/− cells as well as wild-type controls. Expression of E1A in these cells was facilitated by retroviral transduction and drug selection (42).
The Western blots in Fig. 6A show the uniform expression of E1A and pRB in the three genotypes of MEFs. In addition, E1A can coprecipitate pRB in wild-type and E2f1−/− cells, but not RbΔL/ΔL MEFs. In this way our assay system allows us to determine the effect of blocking E1A access to all pRB-E2F complexes, as well as allowing it to interact with pRB (and disrupt E2F2, -3, and -4 interactions) in the absence of E2F1. We assayed the effect of expressing E1A on cell viability in the different genotypes of MEFs to determine if pRB-E2F regulation plays a role in maintaining viability. In Fig. 6C, E1A or vector control cells were treated with etoposide to induce double-stranded DNA breaks that would activate E2F1. This analysis revealed that preventing E1A from contacting pRB provides protection from cell death. Interestingly, allowing E1A to disrupt other pRB-E2F complexes in the absence of E2F1 renders cells more sensitive to DNA damage-induced death. In a similar experiment, cells were also subjected to serum deprivation (Fig. 6B). Again, the RbΔL mutation protected cells, while the lack of E2F1 conferred an increased sensitivity to cell death.
FIG. 6.
E2F1 is necessary for maintenance of viability following E1A expression. MEF cells were infected with retroviruses expressing E1A and selected in puromycin-containing medium. (A) Expression levels of E1A and pRB were detected by Western blotting. Extracts were subjected to anti-E1A IPs and Western blotted to detect pRB. These extracts represent 8% of the input used in these IPs. The asterisk indicates a cross-reacting, unreduced antibody band. (B) E1A or control transduced cells were subjected to growth factor deprivation for 24 h in medium containing 0.1% serum. Cell viability was measured by the conversion of Alamar blue reagent into a fluorescent product and compared between serum-depleted and control cultures for each experimental group. (C) Control or E1A-expressing cells were subjected to etoposide treatment. Cell viability was measured as for panel B. Each error bar represents 1 standard deviation from the mean (n = 3).
Previous work by Samuelson et al. shows that E1A CR1 and CR2 are necessary for DNA damage-associated cytotoxicity (42); however, this activity of E1A only disrupts E2F2, -3, and -4 binding to pRB, suggesting that cell death is caused by a conflict in proliferative signals from E1A and arrest signals from DNA damage in a manner that is similar to what has been suggested for serum starvation combined with E1A expression (33). Our work indicates that E2F1, likely in complex with E1A and pRB, contributes to the maintenance of cell viability in response to DNA damage and E1A. This effect on cell viability suggests why it is advantageous for E1A to regulate E2F1 differently than other E2Fs immediately after viral infection.
DISCUSSION
This report describes a mechanism to explain how E1A disrupts the binding of E2Fs to one site of pRB while avoiding disruption of the E2F1 interaction at a second contact site on pRB. The selective activity of E1A is illustrated in Fig. 7, in which we show that an interaction between pRB and E2F4 is disrupted by E1A through the binding of CR1 to pRB to block E2F binding at the “general” interaction site (Fig. 7A). In contrast, when E2F1 is bound to pRB through the E2F1 “specific” site, E1A is unable to disrupt this protein complex (Fig. 7B). Furthermore, we demonstrate for the first time that E2F1 functions to maintain cell viability in response to external signals in the presence of E1A.
FIG. 7.
Model for E1A disruption of E2F/pRB interactions. (A) The general E2F binding site on pRB controls cell proliferation. E1A can bind to pRB through its CR2 domain and compete with E2Fs bound to the general site using CR1. (B) The RB protein can also interact with E2F1 through the specific binding interaction. Because of the distinct nature of this interaction, E1A is unable to disrupt this complex and E2F1 remains bound. This trimolecular complex may function in maintaining viability.
The in vitro competition assays used in this report reveal a striking difference in sensitivity between E2F4 and E2F1 in their dissociation from pRB by recombinant E1A. Since pRB preferentially regulates activator E2Fs over repressors like E2F4 (46), one possible interpretation is that this assay merely reflects differences in binding affinity for pRB. We do not think differences in affinity can explain this result. As shown in Fig. 4A, E2F1 and E2F4 immunoprecipitations of pRB in the absence of E1A result in only slightly less pRB in E2F4 IPs compared to E2F1. Thus, the dramatically more resistant behavior of pRB-E2F1 complexes to E1A is unlikely to be explained by a small difference in pRB-E2F affinity.
Our studies manipulated the region of pRB where E2F1 could bind and then tested the sensitivity to E1A-mediated disruption. These assays showed an inability of E1A to disrupt E2F1 from binding to either the specific or general regulatory sites in vitro but an ability to disrupt the general interaction in vivo (Fig. 3 and 4). It is possible that this difference is due to the fact that recombinant E1A12S doesn't have sufficient opportunity to disrupt E2F1-pRBΔS, whereas expression of E1A concurrently with pRB and E2F1 in vivo may prevent these complexes from forming. It is also possible that heat shock proteins coexpressed with E1A in vivo participate in liberating E2F1 from the general site in an intact cell but are functionless in an in vitro assay (35, 48). Regardless of the explanation for this discrepancy, the experimental differences between E1A disruption of E2F4 and E2F1 at the same site on pRB emphasize that E1A recognizes E2F1 as unique among E2Fs.
The analysis of pRB-E2F complexes during adenoviral infection and in HEK293 cells reveals that endogenous pRB readily interacts with E2F1 in the presence of E1A, but not with other E2Fs (Fig. 5). It seems surprising that this specificity has been overlooked until now. However, previous experiments to investigate the mechanism used by E1A to disrupt pRB-E2F complexes examined disruption of a mixed population of E2Fs and couldn't have made this distinction (2, 8, 9, 16, 39). The most obvious implication of the preferential maintenance of pRB-E2F1 interactions in the presence of E1A is that E2F1 functions differently than other E2F family members. Since E2F1 has previously been shown to preserve viability through the induction of DNA repair (4, 49), perhaps this unique interaction is not surprising. Our experiments reveal that E2F1 functions to maintain cell viability in response to growth factor deprivation, or following DNA damage, when E1A is present. We think this is highly provocative, because these experiments mimic the susceptibility of newly infected cells to exogenous signals that could kill the host cell and prevent viral replication. The mechanism by which E2F1 has this viability-promoting effect is presently unclear; however, we envision two general possibilities. One is that the stable E2F1-pRB-E1A complex actively provides negative regulation of apoptosis through further protein-protein interactions that modify cell viability signaling pathways, or regulation of apoptotic target gene expression. For example, this complex could occupy promoters that accommodate the pRB-E2F1 “specific” configuration and along with E1A regulate transcription of genes that affect cell viability. Alternatively, it is possible that the functional connection between E1A and loss of E2F1 in cell death is a relatively indirect effect of separate regulatory mechanisms. In this interpretation, the E1A and E2F1 functional connection could be mediated by something as simple as competition between these two transcription factors for a common cofactor like p300.
One implication of our work is that the differential effects that E1A exerts on the general and specific pRB-E2F complexes is a very precise means of selectively inducing cell cycle advancement in infection and transformation while manipulating E2F1 to maintain cell viability. This extends our understanding of the mechanism by which viral proteins such as E1A deregulate cellular growth control and raises the possibility that there is a class of E2F target genes, controlled by E2F1, which retain normal regulation despite the effects of E1A on pRB.
Acknowledgments
We are indebted to Sybille Mittnacht (Chester Beatty Labs, London, United Kingdom) for providing the 21C9 antibodies against pRB. We also thank numerous colleagues in the London Regional Cancer Program for helpful discussions in the course of this work; in particular, we are indebted to Jai Ablack for providing dl309 adenoviruses.
The Strategic Training Program in Cancer Research supported L.M.J. and L.A.S. L.M.J. also thanks the Women's Charity Cashspiel, and L.A.S. received additional support from the Ontario Graduate Scholarships in Science and Technology. M.C. acknowledges support from an M.D./Ph.D. studentship award from the Canadian Institutes of Health Research. S.T. is partially supported by an ERA award from the Ontario government. F.A.D. is a Research Scientist of the National Cancer Institute of Canada and Canadian Cancer Society. Funding from the National Cancer Institute of Canada (016191) supported this work.
Footnotes
Published ahead of print on 27 February 2008.
REFERENCES
- 1.Avvakumov, N., A. E. Kajon, R. C. Hoeben, and J. S. Mymryk. 2004. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 329477-492. [DOI] [PubMed] [Google Scholar]
- 2.Bagchi, S., P. Raychaudhuri, and J. E. Nevins. 1990. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A trans-activation. Cell 62659-669. [DOI] [PubMed] [Google Scholar]
- 3.Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 247673-7685. [DOI] [PubMed] [Google Scholar]
- 4.Berton, T. R., D. L. Mitchell, R. Guo, and D. G. Johnson. 2005. Regulation of epidermal apoptosis and DNA repair by E2F1 in response to ultraviolet B radiation. Oncogene 242449-2460. [DOI] [PubMed] [Google Scholar]
- 5.Binne, U. K., M. K. Classon, F. A. Dick, W. Wei, M. Rape, W. G. Kaelin, Jr., A. M. Naar, and N. J. Dyson. 2007. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell Biol. 9225-232. [DOI] [PubMed] [Google Scholar]
- 6.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391597-601. [DOI] [PubMed] [Google Scholar]
- 7.Chau, B. N., C. W. Pan, and J. Y. Wang. 2006. Separation of anti-proliferation and anti-apoptotic functions of retinoblastoma protein through targeted mutations of its A/B domain. PLoS ONE 1e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chellappan, S., S. Hiebert, M. Mudryj, J. Horowitz, and J. Nevins. 1991. The E2F transcription factor is a cellular target for the RB protein. Cell 651053-1061. [DOI] [PubMed] [Google Scholar]
- 9.Chellappan, S., K. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and the human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 894549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dick, F. A., and N. Dyson. 2003. pRB contains an E2F1 specific binding domain that allows E2F1 induced apoptosis to be regulated separately from other E2F activities. Mol. Cell 12639-649. [DOI] [PubMed] [Google Scholar]
- 11.Dick, F. A., and N. Dyson. 2002. Three regions of the pRB pocket domain affect its inactivation by human papillomavirus E7 proteins. J. Virol. 766224-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dick, F. A., E. Sailhamer, and N. J. Dyson. 2000. Mutagenesis of the pRB pocket domain reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 203715-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 122245-2262. [DOI] [PubMed] [Google Scholar]
- 14.Dyson, N., P. Guida, C. McCall, and E. Harlow. 1992. Adenovirus E1A makes two distinct contacts with the retinoblastoma protein. J. Virol. 664606-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faha, B. F., E. Harlow, and E. M. Lees. 1993. The adenovirus E1A-associated kinase consists of cyclin E/p33cdk2 and cyclin A/p33cdk2. J. Virol. 672456-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fattaey, A. R., E. Harlow, and K. Helin. 1993. Independent regions of adenovirus E1A are required for binding to and dissociation of E2F-protein complexes. Mol. Cell. Biol. 137267-7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Field, S. J., F.-Y. Tsai, F. Kuo, A. M. Zubiaga, W. G. Kaelin, Jr., D. M. Livingston, S. H. Orkin, and M. E. Greenberg. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85549-561. [DOI] [PubMed] [Google Scholar]
- 18.Figge, J., T. Webster, T. F. Smith, and E. Paucha. 1988. Prediction of similar transforming regions in simian virus 40 large T, adenovirus E1A, and myc oncoproteins. J. Virol. 621814-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flinterman, M. B., J. S. Mymryk, P. Klanrit, A. F. Yousef, S. W. Lowe, C. Caldas, J. Gaken, F. Farzaneh, and M. Tavassoli. 2007. p400 function is required for the adenovirus E1A-mediated suppression of EGFR and tumour cell killing. Oncogene 266863-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hurford, R., D. Cobrinik, M.-H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 111447-1463. [DOI] [PubMed] [Google Scholar]
- 21.Isaac, C. E., S. M. Francis, A. L. Martens, L. M. Julian, L. A. Seifried, N. Erdmann, U. K. Binne, L. Harrington, P. Sicinski, N. J. Dyson, and F. A. Dick. 2006. The retinoblastoma protein regulates pericentric heterochromatin. Mol. Cell. Biol. 263659-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji, P., H. Jiang, K. Rekhtman, J. Bloom, M. Ichetovkin, M. Pagano, and L. Zhu. 2004. An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol. Cell 1647-58. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, D. G., and J. Degregori. 2006. Putting the oncogenic and tumor suppressive activities of E2F into context. Curr. Mol. Med. 6731-738. [DOI] [PubMed] [Google Scholar]
- 24.Julian, L. M., O. Palander, L. A. Seifried, J. E. G. Foster, and F. A. Dick. 24 September 2007, posting date. Characterization of an E2F1-specific binding domain in pRB and its implications for apoptotic regulation. Oncogene. doi: 10.1038/sj.onc.1210803. [DOI] [PubMed]
- 25.Kaczmarek, L., B. Ferguson, M. Rosenberg, and R. Baserga. 1986. Induction of cellular DNA synthesis by purified adenovirus E1A proteins. Virology 1521-10. [DOI] [PubMed] [Google Scholar]
- 26.Kovesdi, I., R. Reichel, and J. R. Nevins. 1986. Identification of a cellular transcription factor involved in E1A trans-activation. Cell 45219-228. [DOI] [PubMed] [Google Scholar]
- 27.Lai, A., J. M. Lee, W. M. Yang, J. A. DeCaprio, W. G. J. Kaelin, E. Seto, and P. E. Branton. 1999. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol. Cell. Biol. 196632-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, W. C., F. T. Lin, and J. R. Nevins. 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 151833-1844. [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, X., and R. Marmorstein. 2007. Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation or a tumor suppressor. Genes Dev. 212711-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92463-473. [DOI] [PubMed] [Google Scholar]
- 31.Magnaghi, L., R. Groisman, I. Naguibneva, P. Robin, S. Lorain, J. P. Le Villain, F. Troalen, D. Trouche, and A. Harel-Bellan. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391601-604. [DOI] [PubMed] [Google Scholar]
- 32.Mal, A., A. Piotrkowski, and M. L. Harter. 1996. Cyclin-dependent kinases phosphorylate the adenovirus E1A protein, enhancing its ability to bind pRb and disrupt pRb-E2F complexes. J. Virol. 702911-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mymryk, J. S., K. Shire, and S. T. Bayley. 1994. Induction of apoptosis by adenovirus type 5 E1A in rat cells requires a proliferation block. Oncogene 91187-1193. [PubMed] [Google Scholar]
- 34.Nevins, J. R. 1992. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science 258424-429. [DOI] [PubMed] [Google Scholar]
- 35.Nevins, J. R. 1982. Induction of the synthesis of a 70,000 dalton mammalian heat shock protein by the adenovirus E1A gene product. Cell 29913-919. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412561-565. [DOI] [PubMed] [Google Scholar]
- 37.Pediconi, N., A. Ianari, A. Costanzo, L. Belloni, R. Gallo, L. Cimino, A. Porcellini, A. Screpanti, I. Screpanti, C. Balsano, E. Alesse, A. Gulino, and M. Levrero. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 5552-558. [DOI] [PubMed] [Google Scholar]
- 38.Phelps, W. C., C. L. Yee, K. Munger, and P. M. Howley. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53539-547. [DOI] [PubMed] [Google Scholar]
- 39.Raychaudhuri, P., S. Bagchi, S. H. Devoto, V. B. Kraus, E. Moran, and J. R. Nevins. 1991. Domains of the adenovirus E1A protein that are required for oncogenic activity are also required for dissociation of E2F transcription factor complexes. Genes Dev. 51200-1211. [DOI] [PubMed] [Google Scholar]
- 40.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubin, S. M., A. L. Gall, N. Zheng, and N. P. Pavletich. 2005. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell 1231093-1106. [DOI] [PubMed] [Google Scholar]
- 42.Samuelson, A. V., and S. W. Lowe. 1997. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc. Natl. Acad. Sci. USA 9412094-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stabel, S., P. Argos, and K. Philipson. 1985. The release of growth arrest by microinjection of adenovirus E1A DNA. EMBO J. 42329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens, C., L. Smith, and N. B. La Thangue. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5401-409. [DOI] [PubMed] [Google Scholar]
- 45.Tremblay, M. L., C. J. McGlade, G. E. Gerber, and P. E. Branton. 1988. Identification of the phosphorylation sites in early region 1A proteins of adenovirus type 5 by amino acid sequencing of peptide fragments. J. Biol. Chem. 2636375-6383. [PubMed] [Google Scholar]
- 46.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 311-20. [DOI] [PubMed] [Google Scholar]
- 47.Wells, J., K. Boyd, C. Fry, S. Bartley, and P. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 205797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White, E., D. Spector, and W. Welch. 1988. Differential distribution of the adenovirus E1A proteins and colocalization of E1A with the 70-kilodalton cellular heat shock protein in infected cells. J. Virol. 624153-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wikonkal, N. M., E. Remenyik, D. Knezevic, W. Zhang, M. Liu, H. Zhao, T. R. Berton, D. G. Johnson, and D. E. Brash. 2003. Inactivating E2f1 reverts apoptosis resistance and cancer sensitivity in Trp53-deficient mice. Nat. Cell Biol. 5655-660. [DOI] [PubMed] [Google Scholar]
- 50.Yamasaki, L., T. Jacks, R. Bronson, E. Goillot, E. Harlow, and N. Dyson. 1996. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85537-548. [DOI] [PubMed] [Google Scholar]