Abstract
The human immunodeficiency virus type 1 (HIV-1) protease (PR) has recently been shown to be inhibited by its propeptide p6* in vitro. As p6* itself is a PR substrate, the primary goal of this study was to determine the importance of p6* cleavage for HIV-1 maturation and infectivity. For that purpose, short peptide variants mimicking proposed cleavage sites within and flanking p6* were designed and analyzed for qualitative and quantitative hydrolysis in vitro. Proviral clones comprising the selected cleavage site mutations were established and analyzed for Gag and Pol processing, virus maturation, and infectivity in cultured cells. Amino-terminal cleavage site mutation caused aberrant processing of nucleocapsid proteins and delayed replication kinetics. Blocking the internal cleavage site resulted in the utilization of a flanking site at a significantly decreased hydrolysis rate in vitro, which however did not affect Gag-Pol processing and viral replication. Although mutations blocking cleavage at the p6* carboxyl terminus yielded noninfectious virions exhibiting severe Gag processing defects, mutations retarding hydrolysis of this cleavage site neither seemed to impact viral infectivity and propagation in cultured cells nor seemed to interfere with overall maturation of released viruses. Interestingly, these mutants were shown to be clearly disadvantaged when challenged with wild-type virus in a dual competition assay. In sum, we conclude that p6* cleavage is absolutely essential to allow complete activation of the PR and subsequent processing of the viral precursors.
The production of mature infectious human immunodeficiency virus type 1 (HIV-1) critically depends on complete processing of the precursor polyproteins by the virus-encoded aspartic protease. Unlike in Rous sarcoma virus, where the protease (PR) is an integral part of the Gag precursor, the HIV-1 PR is expressed from a separate pol reading frame. Due to a ribosomal −1 frameshift initiated by cis-active signals within the unspliced genomic 9-kb RNA, two polyprotein precursors, Gag and Gag-Pol are translated at a ratio of 20:1 (reviewed in reference 5). Whereas Gag comprises the structural components matrix protein (MA), capsid protein (CA), nucleocapsid protein (NC), the late domain p6, and two spacer peptides p2 and p1 (also referred to as SP1 and SP2, respectively), the Gag-Pol precursor provides the viral enzymatic activities PR, integrase (IN), and reverse transcriptase (RT) (reviewed in reference 20).
Despite comprehensive analyses of PR-mediated processing events giving rise to infectious virions (40-44, 57), the mechanism stringently controlling spatiotemporal PR activation during the late phase of viral replication is not yet fully understood. Correct folding of the enzyme within the dimerized Gag-Pol precursors is a prerequisite to allow formation of a functional active site (32, 58). As dimerization events become more frequent with Gag-Pol polyproteins accumulating beneath the cytoplasmic membrane during virus assembly, increasing PR activity results in stepwise autoprocessing and subsequent cleavage of the viral precursors while particles are being readied for release (24).
These sequential processing steps driving maturation of Gag and Gag-Pol precursors have largely been clarified (10, 30, 43, 44, 57), and kinetic analyses revealed significantly differing hydrolysis rates for individual PR cleavage events (15, 25, 42, 43). It is widely accepted that cleavage of the Gag precursor is initiated at the carboxyl terminus of the spacer peptide p2, separating the MA-CA-p2 polypeptide from the NC-p1-p6 moiety (39, 43). Subsequently, MA and p6 are cleaved off the corresponding intermediates at an approximately 10-fold-lower rate (43). Finally, the rate-limiting release of the spacer peptides p2 and p1 is catalyzed, allowing CA and NC domains to adopt their final conformation (16, 57). While most cleavages within Gag are mediated by the mature PR in trans, initial processing steps within Gag-Pol are accomplished by the precursor-associated immature enzyme, for which intrinsic activity has repeatedly been demonstrated (26, 30, 46, 61). However, sequential proteolysis of Gag-Pol via clearly defined intermediates remains difficult to determine under ex vivo conditions, as PR-containing precursors are characterized by very short half-lives (28, 52).
Apart from sterical and biochemical processes governing PR activation, we and others have ascribed a role for the transframe domain p6* (p6pol) in the regulation of PR activity (38, 51, 63). Nuclear magnetic resonance analysis of recombinant p6* protein revealed a widely flexible structure (3, 22), and its presence directly upstream of the PR is reminiscent of the propeptides flanking eukaryotic aspartic proteases and has been proven to prevent folding of the PR to its dimeric structure (6). Indeed, we could provide evidence that recombinant p6* protein is a strong competitive inhibitor of PR activity in vitro, and we proposed a model in which the carboxyl-terminal tetrapeptide of p6* following release from the Pol precursor, blocks the substrate binding cleft of the PR, thereby delaying overall processing (37).
Nevertheless, the in vivo function of p6* is still not clearly understood, and our recent data indicate that p6* functional domains are confined to the highly conserved amino- and carboxyl-terminal regions, whereas the sequence context of the entire central part does not appear essential for productive viral replication (8, 38). It is noteworthy though that p6* itself is a substrate of PR and is therefore likely involved in stepwise autoactivation of the enzyme. Whereas functional relevance of amino-terminal p6* cleavage separating the Pol moiety from the NC domain has not yet been demonstrated, there is accumulating evidence that carboxyl-terminal cleavage of p6* is a prerequisite for complete activation of the PR (8, 38, 53). Furthermore, p6* has been shown to contain a conserved internal PR cleavage site between Phe8 and Leu9 (see Fig. 1) hydrolyzed early during precursor maturation (1, 7, 29, 40, 41, 64). Interestingly, natural polymorphisms of the p6* cleavage sites have been reported to impact PR release with considerable effects on susceptibility toward PR inhibitors (56).
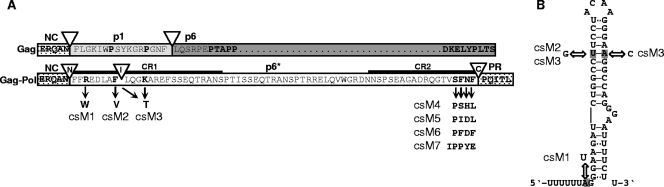
FIG. 1.
Mutagenesis of p6*-associated PR cleavage sites in the proviral context. (A) The entire HIV-1 BH10-derived p6* amino acid sequence (white box) is shown in the context of the Gag-Pol precursor with adjacent p7 and PR sequences (dotted boxes). The p6* region is overlapped by the gag open reading frame encoding the p1 (gray box) and p6 (dark gray box) proteins. PR cleavage sites are indicated by open triangles (N, amino-terminal; I, internal; C, carboxyl-terminal), and functional residues are depicted in boldface type. Amino acids replaced in p6* to alter PR cleavage sites are indicated by black arrows and are represented by provirus clones csM1 to csM7. CR1 and CR2 are conserved regions within p6*. (B) Proposed structure of the HIV-1 frameshift region according to Dulude et al. (13). Mutations introduced to modify amino-terminal (csM1) or internal (csM2, csM3) p6* cleavage are indicated by arrows. See text for more details.
On the basis of variant p6*-derived peptide sequences that were proven to exhibit strictly altered hydrolysis rates in an in vitro cleavage assay, proviral clones comprising mutations in the three proposed PR cleavage sites of p6* were generated and analyzed in different cell cultures to elucidate the importance of correct p6* cleavage for virus maturation and infectivity.
MATERIALS AND METHODS
Generation of provirus p6* mutants.
For mutation of the p6* sequence, the pUC18-derived vector plin8Pr55gag (55) comprising the Gag-encoding region of the HIV-1IIIB isolate BH10 (48; GenBank accession number M15654) was used as the template. Mutations csM1, csM2, csM3, and csM6 were introduced by PCR with primer pairs M1, M2, M3 and M6, respectively (Table 1) using a QuikChange site-directed mutagenesis kit (Stratagene, Heidelberg, Germany). Likewise, csM4 mutations were introduced using primer pairs M4a and M4b in successive PCRs. csM6 mutant served as the template for the generation of csM5 mutant using primer pair M5. The final provirus constructs csM1 to csM6 were obtained by replacing the SpeI-BclI fragment of the infectious provirus clone HX10 (48) with the corresponding SpeI-BclI fragments from the modified plin vectors. The entire SpeI-BclI region of the provirus clones csM1 to csM6 comprising Gag, p6*, and part of the PR was then verified by sequencing with primers S1f, S2f, S3f, S4r, and S5r. The provirus mutant csM7 has been described previously (38), where it was designated M8.
TABLE 1.
Primers used for generation of p6* mutants, reporter constructs, and sequencing
| Primer | Primer sequence (5′→3′) | Nucleotide positions within HX10a |
|---|---|---|
| M1 | AGACAGGCTAATTTTTTTGGGAAGATCTGGCCTTCC | 1284-1319 |
| GGAAGGCCAGATCTTCCCAAAAAAATTAGCCTGTCT | 1319-1284 | |
| M2 | GCTAATTTTTTAGGGAAGATCTGGCCGTCCTACAAG | 1290-1325 |
| CTTGTAGGACGGCCAGATCTTCCCTAAAAAATTAGC | 1325-1290 | |
| M3 | CAAGGGACGGCCAGGGAATTTTCTTCAGAGC | 1322-1352 |
| GCTCTGAAGAAAATTCCCTGGCCGTCCCTTG | 1352-1322 | |
| M4a | GGAACTGTACCCTTTAACCTCCCTCAGATCACTCTTTGG | 1478-1516 |
| CCAAAGAGTGATCTGAGGGAGGTTAAAGGGTACAGTTCC | 1516-1478 | |
| M4b | GGAACTGTACCCTCTCACCTCCCTCAGATCACTCTTTGG | 1478-1516 |
| CCAAAGAGTGATCTGAGGGAGGTGAGAGGGTACAGTTCC | 1516-1478 | |
| M5 | GGAACTGTACCCATTGACCTCCCTCAGATCACTCTTTGG | 1478-1516 |
| CCAAAGAGTGATCTGAGGGAGGTCAATGGGTACAGTTCC | 1516-1478 | |
| M6 | GGAACTGTACCCTTTGACTTCCCTCAGATCACTCTTTGG | 1478-1516 |
| CCAAAGAGTGATCTGAGGGAAGTCAAAGGGTACAGTTCC | 1516-1478 | |
| fs1 | AGACAGGCTAATTTTTTTGGGAAGATCTGGCCTTCC | ND |
| GGAAGGCCAGATCTTCCCAAAAAAATTAGCCTGTCT | ||
| fs2 | GCTAATTTTTTAGGGAAGATCTGGCCGTCCTACAAG | ND |
| CTTGTAGGACGCCAGATCTTCCCTAAAAAATTAGC | ||
| fs3 | CCTACAAGGGACGGCCAGGGAAATCC | ND |
| GGATCCTTCCCTGGCCGTCCCTTGTAGG | ||
| S1f | GCACCAGGCCAGATGAGAGAACC | 669-690 |
| S2f | GCATTGGGACCAGCGGCTACAC | 1005-1025 |
| S3f | CTACAAGGGAAGGCCAGGG | 1319-1337 |
| S4r | GGATACAGTTCCTTGTCTATCGGC | 1489-1466 |
| S5r | CCATTGTTTAACTTTTGGGCC | 1866-1847 |
| S6f | GGAAATGTGGAAAGGAAGGACACC | 1240-1262 |
| S7r | CTGTCTTACTTTGATAAAACCTCC | 1671-1649 |
Nucleotide positions within HX10 (48) with respect to Gag ATG. ND, not determined.
Generation of frameshift reporter constructs.
Extension of the 5′ end of the firefly luciferase gene from pGL2 (Promega) by the frameshift region of BH10 (pGLfs-wt) and generation of the control plasmids pGL-fsΔ and pGL-fsM1 has been described previously in detail (38). Frameshift modifications of the p6* mutants csM1, csM2, and csM3 were introduced into fs-wt sequence by site-directed mutagenesis using a QuikChange kit and primer pairs fs1, fs2, and fs3 (Table 1). All reporter constructs were verified by sequencing.
Synthetic oligopeptides.
The chromogenic substrate Lys-Ala-Arg-Val-Nle-Phe (p-NO2)-Glu-Ala-Nle-NH2 was obtained from Bachem Biochemica (Bubendorf, Switzerland), and all unmodified synthetic oligopeptides were purchased from G. J. Arnold (Gene Center, Munich, Germany). The chemical composition and purity of the synthetic oligopeptides were analyzed by electrospray ionization mass spectrometry (17). Concentrations of peptide dilutions were determined by Analytical Research and Services (University of Bern, Switzerland) by quantitative analysis of amino acid composition.
Peptide cleavage assay.
Affinity-purified HIV-1 PR for peptide cleavage was purchased from Bachem Biochemica. Oligopeptides were cleaved at concentrations of 140 μM at 25°C in 0.1 M sodium acetate (pH 5.0), 4 mM EDTA, 5 mM dithiothreitol, and 0.3% dimethyl sulfoxide in a PCR cycler with a heated lid to avoid condensation. Reactions were started by adding 450 nM PR dimer and were stopped by adding trifluoroacetic acid at a total concentration of 0.1%. To exclude unspecific cleavage, samples were incubated with 5 μM PR inhibitor RO 31-5989 (49) from Roche (Mannheim, Germany). Reaction products were separated via a linear acetonitrile gradient (0 to 50% in 0.1% trifluoroacetic acid) within 20 min at a flow rate of 0.2 ml/min using a μRPC C2/C18 SC 2.1/10 column (Amersham Pharmacia Biotech) and were detected at 215 nm. Cleavage products were collected and analyzed by electrospray ionization mass spectrometry (17) and gas phase sequencing. Total substrate turnover was calculated using external standards from the integrated peak area.
Cell lines, transfections, and infections.
H1299 human lung carcinoma and 293T human kidney epithelial cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin. The CD4-positive human cervix carcinoma cell line HeLa-CD4-LTR-β-gal (Magi cells) was maintained in Dulbecco's modified Eagle's medium containing 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin supplemented with 0.2 mg/ml G418 and 0.1 mg/ml hygromycin B. The HIV-1 permissive human T-cell lines CEM4 (P. R. Clapham, AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) and MT-4 (27) were grown in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Adherent H1299 and 293T cells were transfected with 30 μg of provirus plasmid DNA or with 3 μg of frameshift reporter constructs using the calcium phosphate precipitation technique, and cells and supernatants were harvested as described in Results. For analysis of replication kinetics, 5 × 106 logarithmically growing CEM4 or MT-4 suspension cells were transfected with 1 μg of provirus plasmid DNA using Fugene 6 from Roche according to the manufacturer's protocol. Samples were collected, and cultures were diluted with 1 volume of fresh medium every 48 h over a total period of 20 to 30 days.
Single-round infection of Magi cells was performed as previously described (38) with serial dilutions of virus-containing supernatants from transfected H1299 cells.
Quantification of HIV-1 capsid antigens.
To determine the amount of HIV-1 capsid protein in culture medium, supernatants were clarified of cell debris (300 × g, 10 min), filtered through a 0.45-μm-pore-size filter and used in a CA-specific capture enzyme-linked immunosorbent assay (ELISA). Alternatively, particle-associated antigens were enriched by centrifugation of clarified supernatants through a 20% sucrose cushion. For quantification of CA, 96-well Maxi Sorb microtiter plates (Nunc, Wiesbaden, Germany) were coated overnight (4°C) with a 1:300 dilution (0.1 M carbonate buffer [pH 9.5]) of the CA-specific antibody M01 (Polymun, Vienna, Austria). After three washes (with phosphate-buffered saline [PBS] containing 0.05% Tween), the wells were incubated with the serially diluted samples (PBS containing 1% bovine serum albumin [BSA]) for 1 h at 37°C. Following six washes, the wells were incubated with a 1:20,000 dilution (PBS containing 1% BSA) of the biotinylated CA-specific antibody 37G12 (Polymun) for 1 h at room temperature. After 10 further washes, a 1:10,000 dilution of horseradish peroxidase-conjugated streptavidin (Roche) was added for 30 min at room temperature, and after 10 final washes, the plates were developed with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Becton Dickinson, Heidelberg, Germany). The reaction was stopped with 1 N H2SO4/well, and plates were measured at 450 nm. CA content was quantified using a calibration curve based on serial dilutions of a CA standard (Polymun).
Western blot analysis.
To characterize cell-associated virus proteins, lysates of transfected H1299 cells were prepared and quantified as previously described (38), and 100 μg of total protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. Capsid protein species were visualized with the CA-specific monoclonal antibody (MAb) 16/4/2 (59) and an alkaline phosphatase-labeled anti-mouse antibody (Bio-Rad), and proteins were detected with Nitro Blue Tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (BCIP) solutions from Roche.
For analysis of virion-associated proteins, particles were purified from cell supernatants as previously described (38), and virus proteins were separated by SDS-PAGE and detected by immunoblotting with the CA-specific MAb 13/5 (59), the p17-specific MAb 3-H-7 (33), the polyclonal IN-specific rabbit antiserum 757 (D. P. Grandgenett, NIH AIDS Research and Reference Reagent Program), the polyclonal PR-specific sheep antiserum ARP413 (D. Bailey and M. Page, NIH AIDS Research and Reference Reagent Program), the polyclonal RT-specific antiserum 4F8 (P. Chandra, Gustav-Embden-Center of Biological Chemistry, University of Frankfurt, Germany), and polyclonal rabbit antisera raised against glutathione S-transferase (GST)-p6gag and GST-NC fusion proteins. Proteins were visualized by enhanced chemiluminescence using SuperSignal WestFemto (Pierce, Rockford, IL).
Frameshift luciferase reporter assay.
For quantification of cell-associated luciferase activity, transfected cells were harvested and lysed as described previously (38), and 100 μg of total protein was used in a luciferase assay from Promega according to the manufacturer's protocol. Luciferase activity was measured by recording light emission for 10 s in a Lumat 9501 luminometer (Berthold, Bad Wildbach, Germany).
Growth competition assay.
Supernatants of 293T cells transfected with provirus plasmid DNA were harvested 48 h later, and the amounts of CA in the supernatants were quantified by ELISAs. Equal CA amounts (500 ng) of wild-type (wt) and mutant particles were mixed at ratios of 1:1, 4:1, and 1:4 and used to infect 2 × 106 CEM cells. After 6 h, infected cultures were transferred to flasks and cultivated for 3 weeks in a total volume of 4 ml. Every 2 days, samples were collected, and half of the cultures were replaced with fresh cells. Infected cells were pelleted (300 × g, 10 min), and genomic DNA was extracted using the QIAamp DNA Mini kit (Qiagen). DNA was PCR amplified with primers S6f and S7r (Table 1). Purified PCR products were then subjected to DNA sequencing with primer S7r. Chromatograms were analyzed using Vector NTI 10.3.0 software from Invitrogen (Karlsruhe, Germany).
RESULTS
Design of altered p6* PR cleavage sites.
The major goal of this study was to determine the critical contribution of PR cleavage sites within and flanking p6* to viral maturation and infectivity. For analysis of the p6* cleavage sites in the proviral context, we had to take into account maintenance of (i) the overlapping p1gag and p6gag reading frames limiting alterations to gag wobble positions (Fig. 1A), as well as (ii) the slippery sequence and (iii) a downstream stem-loop structure, the latter two regulating ribosomal frameshifting and the generation of Gag-Pol precursors (Fig. 1B).
To screen for cleavage site variants capable of (i) completely blocking cleavage or (ii) altering hydrolysis rates, an algorithm was used to predict the probability for any sequence of eight amino acids to be cleaved by the HIV-1 PR (9). Those residues predicted to either block cleavage or alter cleavage kinetics according to the algorithm were introduced into the infectious provirus HX10. To block cleavage of the amino-terminal scissile bond (PN) of p6*, Arg in the P3′ position was replaced by an aromatic Trp residue (54), yielding mutant csM1 (Fig. 1A). This mutation was associated with a nucleotide substitution turning the conserved slippery site UUUUUUA into UUUUUUU (Fig. 1B). However, this sequence has previously been shown to promote frameshifting in vitro (12). The amino-terminal p6* mutation also resulted in a Leu-to-Phe substitution at the P2′ position of the overlapping NC-p1 cleavage site (60), which according to the employed algorithm (9) was not expected to prevent cleavability of this modified site.
On the basis of the previous findings that the introduction of a β-branched amino acid at position P1 prevents cleavage by the PR (45, 54), Phe8 was replaced by Val to block internal cleavage (PI) of p6* (Fig. 1B). As the corresponding csM2 mutation reduced base pairing in the upper stem region, possibly affecting overall stem-loop stability (4, 35), a second mutant (csM3) was generated in which base pairing was restored (Fig. 1B). This resulted in a Lys-to-Thr substitution within a variable p6* portion without altering the gag open reading frame.
Since carboxyl-terminal cleavage of p6* from the Pol precursor is an essential prerequisite for PR autorelease and activation (37, 53), we further substituted the conserved carboxyl-terminal tetrapeptide of p6* by four different amino acid combinations predicted to be cleaved according to the criteria of Chou et al. (9) to analyze potential effects of altered hydrolysis rates on subsequent virus maturation. The modified tetrapeptides encoded by the resulting virus mutants contained either two (csM6), four (csM4 and csM5), or five (csM7) amino acid substitutions compared to wt p6* (Fig. 1A). Again the gag reading frame remained intact, as overlapping p6gag residues have been proposed to be involved in packaging of envelope proteins (34).
In vitro characterization of peptides with cleavage site variations.
In order to determine the influence of cleavage site modifications on hydrolysis rates in vitro, synthetic oligopeptides comprising wt cleavage site sequences (PN, PI, and PC) or mutated cleavage site sequences (P1 to P7, corresponding to virus mutants csM1 to csM7, respectively) were incubated with recombinant HIV-1 PR for at least 24 h to guarantee quantitative turnover of the peptide substrates. To confirm that cleavage products were originating from PR activity, parallel samples were incubated in the presence of an HIV-1 PR-specific inhibitor. Cleavage products were separated by reverse-phase high-performance liquid chromatography and analyzed by mass spectrometry and sequencing (data not shown). As expected, cleavage of the modified amino-terminal cleavage site (PN) was efficiently blocked in peptide P1. However, this substrate was cleaved two amino acids downstream of the wt site, instead (Table 2). Likewise, the original internal cleavage site (PI) was blocked by the mutations in P2 and P3 and replaced by a new scissile bond emerging two amino acids upstream. Consistently, altered internal cleavage was also observed when GST-p6* full-length proteins harboring the corresponding P2 and P3 mutations were processed in trans (data not shown), excluding the possibility that the observed cleavage site modifications were restricted to the short oligopeptide substrates. Deviating from predictions by the algorithm (Δ values in Table 2), only two of the four carboxyl-terminal cleavage site (PC) variations were processed by the PR in substrates P5 and P6, whereas cleavage of this site was completely blocked in P4 and P7.
TABLE 2.
In vitro cleavage of the p6* peptide substrates with recombinant HIV-1 PR
| Peptidea | Peptide sequenceb | Cleavage productsb,c | Cleavage site attributed | Calculated Δ valuee | Blockage of original sitef | Comparison of hydrolysis ratesg |
|---|---|---|---|---|---|---|
| PN | ERQANFFRED | ERQAN * FFRED | wt | ND | − | |
| P1 | ERQANFFWED | ERQANFF * WED | → | ND | + | 240-fold↑ |
| PI | EDLAFLQGKA | EDLAF * LQGKA | wt | ND | − | |
| P2 | EDLAVLQGKA | EDL * AVLQGKA | ← | ND | + | 5.3-fold↓ |
| P3 | EDLAVLQGTA | EDL * AVLQGTA | ← | ND | + | 10.5-fold↓ |
| PC | VSFNFPQITL | VSFNF * PQITL | wt | 1.38 | − | |
| P4 | VPSHLPQITL | VPSHLPQITL | − | 1.97 | + | |
| P5 | VPIDLPQITL | VPIDL * PQITL | wt | 1.14 | − | 15-fold↓ |
| P6 | VPFDFPQITL | VPFDF * PQITL | wt | 1.53 | − | 1.5-fold↓ |
| P7 | IPPYEPQITL | IPPYEPQITL | − | 1.16 | + |
Synthetic oligopeptides representing natural (PN, Pi, and PC) and modified (P1 to P7) p6* cleavage sites were incubated with mature HIV-1 PR for at least 24 h.
Amino acid substitutions relating to the wild-type sequence are underlined.
Cleavage products as determined by mass spectrometry and sequencing. The cleavage sites are indicated by asterisks.
Cleavage occurred at the wild-type site (wt) or 2 amino acids upstream (→) or downstream (←) of the wild-type site, or cleavage was blocked (−).
Δ values for the carboxyl-terminal p6* modifications were calculated by the method of Chou et al. (9). Values of >0 are predictive for cleavability by the HIV-1 PR. ND, not determined.
−, no blockage; +, blockage.
Relative hydrolysis rates of the modified peptide substrates compared to wild-type peptides (rate determined at 50% substrate turnover, respectively). Symbols: ↑, increase; ↓, decrease.
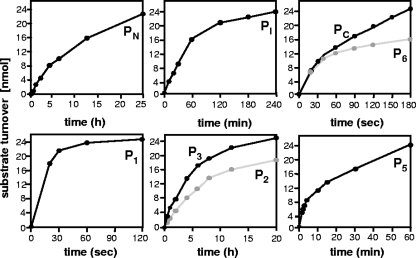
In order to determine cleavage rates of the modified sites, turnover of the oligopeptide substrates was monitored over time. As illustrated in Fig. 2 and summarized in Table 2, all mutated cleavage sites exhibited significantly altered hydrolysis rates compared with the original wt sites PN, PI, and PC. Whereas the novel site in P1 was processed much faster than the wt site, the internal p6* substitutions led to retarded cleavage of the substrates P2 and P3 which was consistent with relative cleavage rates determined for the corresponding GST-p6* full-length proteins (data not shown). Lower hydrolysis rates were also calculated for the carboxyl-terminal cleavage site mutations in P5 and P6. It is worth mentioning though that P6 was initially cleaved at the wt rate with hydrolysis decelerating after approximately 30% turnover, suggesting product inhibition of the PR. In sum, we have established a series of p6* cleavage site mutations exhibiting cleavage attributes completely differing from the respective wt sites.
FIG. 2.
Cleavage of wild-type and modified peptide substrates with recombinant PR. Synthetic oligopeptides containing wt (PN, PI, and PC) and modified cleavage sites (P1, P2, P3, P5, and P6) of p6* were cleaved with recombinant HIV-1 PR in a total volume of 200 μl. To monitor cleavage kinetics, 15-μl samples were collected at given time points and analyzed by reverse-phase high-performance liquid chromatography. The data shown are representative of three independent experiments.
Influence of p6* mutations on frameshifting in mammalian cells.
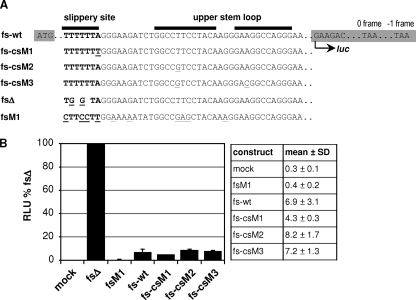
In order to exclude the possibility that p6* mutations interfered with frameshift function on the RNA level, amino-terminal and internal mutations were tested in a previously described frameshift reporter assay (38). To render expression of a luciferase reporter frameshift dependent, the gene encoding firefly luciferase was fused to the frameshift region of the HIV-1 isolate BH10, comprising the slippery site and stem-loop region. The resulting reporter sequence fs-wt was then further modified to introduce csM1-, csM2-, and csM3-specific point mutations (Fig. 3A). For quantification of frameshift rates, construct fsΔ was generated allowing for constitutive full-length luciferase expression from the −1 frame (corresponding to a frameshift rate of 100%). In addition, we used the formerly described frameshift mutant fsM1 as a negative control to detect luciferase background activity (38). This mutant harbors a destroyed slippery site driving expression of luciferase exclusively from the 0 frame (corresponding to a 0% frameshift rate), which results in premature translation termination. To analyze frameshift modifications, H1299 cells were transfected with the corresponding reporter plasmids, and luciferase activity was quantified after 48 h and related to values determined for fsΔ. As shown in Fig. 3B, frameshift-dependent luciferase expression from the reporter constructs fs-wt, fs-csM1, fs-csM2, and fs-csM3 ranged between ∼4 and 8% and was therefore clearly distinguishable from fsM1-transfected cells, which gave luciferase activities around the mock-transfected level. Basically, all tested mutants were capable of driving frameshift-dependent luciferase expression. However, taking into account the limited sensitivity of this assay system, a slight increase in frameshifting could be observed for the stem-loop variants fs-csM2 (8.2%) and fs-csM3 (7.2%) in H1299 cells, whereas luciferase activity derived from expression of fs-csM1 (4.3%) was clearly diminished compared to fs-wt (6.9%). Similar frameshift ratios were obtained in multiple transfection experiments performed with different DNA preparations, basically excluding the possibility that differential luciferase expression resulted from variations in transfection efficiencies. Furthermore, comparable results were obtained in 293T cells (data not shown), indicating that the altered heptamer sequence in fs-csM1 was less efficient in promoting ribosomal slippage.
FIG. 3.
Luciferase frameshift reporter assay. (A) The sequence encoding firefly luciferase (highlighted in gray) was extended at its 5′ end by the frameshift region of BH10 to render luciferase expression frameshift dependent (fs-wt). Reporter constructs fs-csM1, fs-csM2, and fs-csM3 carrying the frameshift regions of the corresponding cleavage site mutants are indicated. For a control, the slippery site has been destroyed in fsΔ, allowing for constitutive expression of luciferase from the −1 frame. In contrast, luciferase is exclusively expressed from the 0 frame in mutant fsM1, resulting in a premature translational stop soon after the luciferase ATG. Point mutations introduced in the slippery site (boldface) or the upper stem-loop region are underlined, and deletions are expressed by spaces. The positions of start and stop signals within the 0 frame and the −1 frame are indicated. (B) H1299 cells were transiently transfected with the indicated frameshift reporter plasmids or pcDNA3 (mock), and cell-associated luciferase activity was quantified after 48 h. Luciferase activity (relative light units [RLU]) determined for fsΔ was set at 100%, and all other values were related to the fsΔ value accordingly. The results from one representative blot comprising mean values plus standard deviations (SDs) (error bars) from four independent transfection experiments are shown.
Influence of the mutated PR cleavage sites on expression of viral proteins.
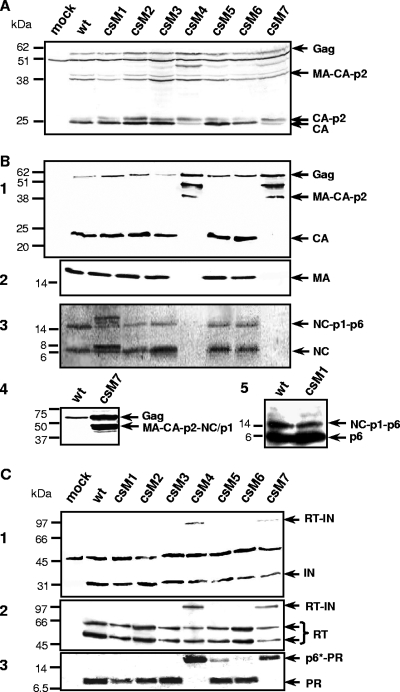
To further examine the modified p6* cleavage sites in the viral context, H1299 cells were transiently transfected with the infectious provirus clone HX10 (wt) or HX10 derivatives carrying the respective p6* mutations (csM1 to csM7). Forty-eight hours posttransfection, cell lysates were analyzed by immunoblotting with a CA-specific antibody to reveal potential effects of the cleavage site modifications on expression and cell-associated processing of viral Gag proteins. As shown in Fig. 4A, all cells yielded protein patterns comparable to wt virus transfections with trace amounts of uncleaved 55-kDa Gag precursors and MA-CA-p2 intermediates. This demonstrates that the p6* mutations do not affect expression or stability of viral Gag proteins. However, significant levels of an approximately 48-kDa protein and increased amounts of the CA-p2 intermediate were detected in cells transfected with csM4 and csM7 proviruses, whereas the mature CA species predominated in all other lysates. These findings suggest that both carboxyl-terminal mutations shown to block cleavage in the peptide cleavage assay (Table 2) are associated with Gag-specific processing defects, which might result from either decreased PR activity or altered substrate specificity. Comparable protein patterns were observed in transfected 293T cells (data not shown), confirming that the above findings are not cell specific. An additional 50-kDa product also present in mock-transfected controls likely results from cross-reactivity of the anti-CA antibody with an unknown cellular protein.
FIG. 4.
Influence of p6* mutations on expression and maturation of Gag-Pol proteins. H1299 cells were transfected with the indicated provirus plasmids or pcDNA3.1 (mock). (A) For analysis of Gag expression, cells were harvested at 48 h posttransfection, and equal amounts of cell lysates were subjected to immunoblot analysis using a CA-specific antibody. (B and C) To characterize virion-associated Gag and Pol proteins, particles released into cell supernatants were harvested 72 h posttransfection and sedimented through a 20% sucrose cushion. Viral proteins were separated by SDS-PAGE and analyzed by immunoblotting. For detection of Gag-specific products, CA (B1)-specific or MA (B2)-specific monoclonal antibodies or polyclonal sera directed to NC (B3 and B4) or p6gag (B5) were used. Pol-derived products were analyzed using IN (C1)-, RT (C2)-, and PR (C3)-specific polyclonal antisera. The positions of molecular mass markers (in kilodaltons) are shown to the left of the gels, and the arrows to the right of the gels indicate specific protein bands.
Influence of p6* cleavage site mutations on composition and maturation of virus particles.
To analyze effects of the p6* mutations on virus maturation, particles released from H1299 cells 72 h posttransfection were subjected to immunoblot analysis with Gag- and Pol-specific antibodies. Like wt particles, most of the virus mutants contained largely the mature CA and MA species (Fig. 4B, panels 1 and 2). In contrast, only full-length Gag precursors and two smaller CA intermediates of approximately 41 and 48 kDa were found in csM4 and csM7 virions. Whereas the 41-kDa product exactly correlates with the molecular mass calculated for the MA-CA-p2 polyprotein, the 48-kDa product does not appear to be a natural product of the Gag processing cascade. These observations strongly argue for Gag-specific processing defects induced by the corresponding mutations, thus resulting in a lack of mature CA and MA proteins. These findings were confirmed and extended by analysis of virus-associated NC proteins. As shown in Fig. 4B, panel 3, the carboxyl-terminal p6* mutants csM4 and csM7 were lacking mature NC proteins and NC-p1-p6 precursor, both of which were found in all other virus mutants. Additional analysis of wt and csM7 particles for larger NC-associated products revealed a csM7-specific band likely corresponding to the ∼ 48-kDa product detected with the CA-specific antibody in csM4 and csM7 particles and which might thus represent a MA-CA-p2-NC-p1 intermediate (Fig. 4B, panel 4). The presence of a faint double band further suggests that the spacer peptide p1 has been partially released from this aberrant precursor.
Whereas csM2, csM3, csM5, and csM6 mutants showed wt-like processing of NC proteins, csM1 virions contained additional NC-specific bands that were not detected in any other particle preparation. Since the csM1 mutation is associated with an amino acid substitution in the p1gag domain, the prominent 8-kDa band found in csM1 particles might represent a NC-p1 intermediate, suggesting retarded hydrolysis of the corresponding cleavage site. However, the identities of two additional NC-specific products of 16 to 20 kDa remain unknown, as these proteins were not recognized by any other antibody used in this study. Immunoblot analysis of csM1 particles with a p6gag-specific antibody (Fig. 4B, panel 5) did not reveal any differences compared to wt virions. Hence, the csM1-associated processing defect appears to be limited specifically to the NC protein.
To further examine packaging and maturation of Pol proteins, particle preparations were analyzed with IN-, RT-, and PR-specific antibodies. As shown in Fig. 4C, none of the p6* mutations interfered with packaging of Pol proteins into virus particles. However, processing patterns revealed differing levels of maturation. Along with the mature IN and RT species, we could clearly detect traces of the 98-kDa RT-IN precursor in csM4 and csM7 particles (Fig. 4C, panels 1 and 2), whereas all other virions contained exclusively the mature enzymes. Finally, a PR-specific antibody failed to identify the mature 11-kDa PR in csM4 and csM7 particles (Fig. 4C, panel 3), where a protein of approximately 17 kDa was detected instead. This product likely represents a p6*-PR precursor as verified with a p6*-specific antiserum (data not shown). These results strongly indicate that the carboxyl-terminal csM4 and csM7 mutations completely block cleavage between p6* and PR. Although this processing defect clearly affected maturation of the Gag precursor, quantitative cleavage of the Pol components was influenced to a minor extent. Interestingly, in addition to the mature PR, trace amounts of this p6*-PR intermediate were also observed in csM5 and csM6 particles. These data suggest that amino-terminal autorelease of the PR was not completely impaired but rather delayed in csM5 and csM6 viruses, with csM5-specific mutations causing the most striking effect. This observation is in accordance with the results obtained by in vitro cleavage of the corresponding peptides (Fig. 2).
In contrast, the amino-terminal (csM1) and internal (csM2 and csM3) p6* mutations did not visibly influence the release of the viral PR or otherwise affect the maturation of virus-associated Pol proteins yielding only the mature IN, RT, and PR species. Since we could not detect any aberrant p6*-specific cleavage intermediates in those particles probably owing to the narrow time frame in which p6* cleavages occur, these results make it tempting to speculate that the modified cleavage sites of corresponding peptides P1, P2, and P3 were either not used in the viral precursors or did not impact on Pol-specific maturation.
Influence of p6* cleavage site mutations on viral replication and infectivity.
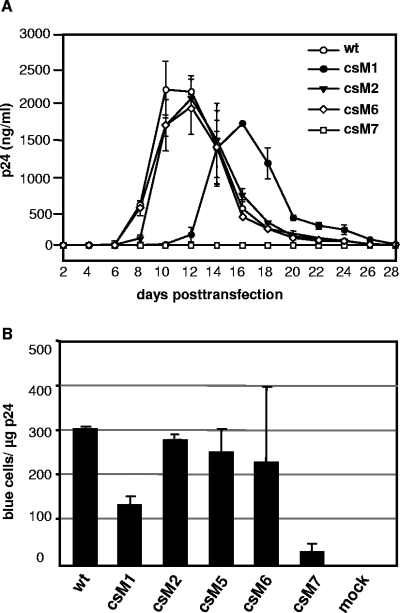
Next, the influence of p6* mutations on virus growth in permissive cells was studied following transfection of CD4-positive MT-4 lymphocytes with the various provirus clones. Virus production was followed over a period of 3 to 4 weeks by measuring the amount of total CA protein in culture supernatants. As depicted in Fig. 5A, wt viruses replicated to high titers in MT-4 cells peaking at days 10 to 12 posttransfection and followed by a rapid decline as cell populations died. p6* mutants csM2, csM3, csM5, and csM6 showed similar replication kinetics (csM3- and csM5-specific replication curves have been omitted for clarity), indicating that none of the underlying internal or carboxyl-terminal mutations had a major impact on viral growth in cultured cells. In contrast, replication of the csM1 mutant was significantly delayed and reached lower peak titers compared to wt viruses. This retarded phenotype was also observed in a second T-cell line (data not shown), confirming the above results. A reversion of the amino acid substitution to the wt or the emergence of compensatory mutations within the monitored period was widely excluded by PCR sequencing of the entire Gag-p6* region and part of the PR, suggesting that the delayed replication profile of these viruses was rather due to compromised frameshifting (Fig. 3B) and/or retarded NC-p1 cleavage (Fig. 4B, panel 4). As expected from deficient Gag processing, no productive replication of the csM4 (data not shown) and csM7 mutants in the analyzed lymphocytes was detectable within the monitored period.
FIG. 5.
Influence of p6* mutations on viral in vitro replication and infectivity. (A) MT-4 lymphocytes were transiently transfected with the indicated provirus clones. Cells were subcultivated at a 1:2 ratio every 48 h, and the total amount of virus-associated CA antigen in culture supernatants was determined by an ELISA. The data shown are mean values from two independent transfections performed in duplicate experiments with different DNA preparations, and the error bars indicate the standard deviations. (B) Magi indicator cells were infected with different dilutions of wt and mutant viruses obtained from transient transfections of H1299 cells and normalized to the amount of CA amount as described in Materials and Methods. Forty-eight hours postinfection, Magi cells were fixed and stained, and blue nuclei in each well were counted. The data shown are the mean values from three independent infections, and the standard deviations (error bars) are indicated.
To further reveal minor differences in viral infectivity, we assessed the capability of selected virus mutants to infect Magi indicator cells in a single-round replication assay. Thus, virus preparations of mutants csM1, csM2 (representative of csM3), csM5, csM6 and csM7 (representative of csM4) produced in H1299 cells were titrated on this cell line and compared to wt infectivity. Compatible with the results obtained from replication analyses in permissive lymphocytes, infectivity of csM1 particles was reduced about twofold on Magi cells (Fig. 5B), and csM7 viruses proved to be hardly infectious on this cell line, which confirms our former results (38). In contrast, the number of infectious units determined for csM2, csM5, and csM6 mutants ranged within 80 to 90% of the wt level, suggesting that corresponding mutations affected viral infectivity only to a moderate extent. Together, these data imply that internal and carboxyl-terminal p6* cleavage site mutations had only minor effects on viral in vitro replication and infectivity as long as PR was fully released.
Modifications in csM5 and csM6 mutants result in a loss of viral fitness when competing with wt virus.
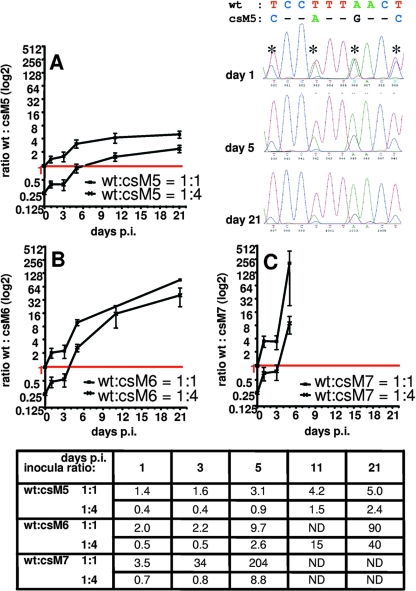
Although carboxyl-terminal csM5 and csM6 mutations showed significantly altered hydrolysis rates of the corresponding cleavage site in vitro (Fig. 2), resulting in a retarded autorelease of the PR (Fig. 4), we could not observe any severe effects of these mutations on viral replication or infectivity in the cell lines examined. This prompted us to further assess viral fitness of these candidates when challenged with wt virus. Therefore, a dual competition assay was designed, where CEM4 cells were infected by wt viruses mixed at different ratios with mutant csM5, csM6, or csM7. Spread of virus infection was monitored via quantification of integrated virus cDNA. Overlapping peaks at the modified p6* positions of the resulting chromatograms were then used to determine the relative amounts of wt versus mutant sequences in a sample. Taking into account the fact that peak heights were dependent on the base due to different kinetics in base incorporation during the sequencing reaction, the wt to mutant inoculation ratios of 1:1 or 1:4 were nicely reflected by the corresponding peak heights obtained at day 1 postinfection (see for example csM5 versus wt infection in Fig. 6). Surprisingly, csM7-specific peaks were found 1 day following infection, suggesting that csM7-derived cDNA was at least partially capable of integrating into target cell genomes. This observation is in accordance with the faint infection of Magi cells by csM7 viruses (Fig. 5B). However, no csM7-specific peaks could be detected beyond 5 days postinfection confirming the above experiments in lymphocytes, where no productive replication of this mutant was visible (Fig. 5A). To more precisely quantify the loss of viral fitness, ratios of wt versus mutant peak heights were calculated for each individual time point (Fig. 6). Accordingly, slopes of the resulting curves are indicative of the rate at which mutants are overgrown by wt viruses with the fastest shift determined for mutant csM7 (Fig. 6C). It is interesting, however, that at day 5 postinfection with 1:1 mixtures, there was a also a significant reduction of csM5- and csM6-specific peaks compared to wt peaks, indicating that wt viruses were more successful in the following infection rounds (Fig. 6A and B). Twenty-one days postinfection, wt virus was the dominant species in all samples even when mutant viruses were inoculated at a fourfold excess. These data clearly indicate that carboxyl-terminal p6* modifications in mutants csM5 and csM6, albeit allowing for amino-terminal release of the PR, result in a loss of viral fitness that becomes apparent when competing with wt virus.
FIG. 6.
Dual competition of wild-type and mutant viruses in permissive T lymphocytes. CEM4 cells were infected with CA-normalized amounts of wt and mutant viruses csM5, csM6, and csM7, mixed at 1:1 or 1:4 ratios prior to infection. At the indicated days postinfection (p.i.), genomic DNA from infected cells was prepared, and the p6* region of integrated virus genomes was sequenced. As an example, one chromatogram showing a section of mixed wt and csM5 sequences at day 1, 5, and 21 postinfection is shown. Ratios of wt versus mutant peak heights were determined for all mixed nucleotide positions (asterisks) and averaged to one mean ratio per analyzed time point (shown at the bottom of figure). Peak ratios were calculated for cells infected with wt and mutant viruses mixed at a ratio of 1:1 or 1:4, and resulting data were plotted as binary logarithms (A to C). n.d., not determined.
DISCUSSION
On the basis of a functional similarity with the propeptides of cellular aspartic proteases, the transframe protein p6* was hypothesized to play a role in the regulation of PR activation (36, 37, 51, 63). However, conflicting results have been reported regarding the rate and order of PR-mediated cleavages in Gag-Pol (references 1, 7, 29, 41, 42, and 64 and references therein) partially owing to different experimental conditions influencing PR activity, stability, or substrate specificity (23, 29, 31, 47). Moreover, truncated or in vitro-translated Gag-Pol precursors do not necessarily reflect the physiological situation where tight condensation of full-length precursors within the virion might affect accessibility of distinct PR cleavage sites (18, 53). Finally, viral cofactors, such as the accessory Vif protein, have been shown to modulate cleavage of individual sites in vivo (2). Owing to the limited informative value of reduced assay formats, our various in vitro approaches to characterize the biological function of p6* cleavage in this study have been extended to include an in-depth analysis of the p6* modifications in the context of viral replication.
Mutagenesis of p6* in the context of an infectious provirus appears challenging, as parts of the pol reading frame are superimposed by cis-active RNA frameshift signals and functional Gag protein domains. Thus, giving priority to frameshift function, only a single mutation predicted to affect amino-terminal cleavage of p6* appeared reasonable without completely destroying the slippery site. Albeit we could prove functionality of this modified slippery site in a cell culture-based frameshift reporter assay, frameshift rates were clearly reduced compared to the wt level, confirming recent observations by others (14). Further analysis of this modification in the csM1 virus mutant revealed a clearly delayed replication profile in cultured lymphocytes associated with a ∼50% loss of infectivity in single-round replication. Furthermore, we observed NC-associated processing defects in csM1 particles likely associated with a substitution in the NC-p1 cleavage site introduced by amino-terminal p6* mutation. Albeit hydrolysis of the NC-p1 site has been reported to be rate limiting in virus maturation (16, 62), recent analyses by Coren et al. (10) have provided evidence that NC-p1 cleavage is not required for viral replication and infectivity. Together, these findings suggest that the aberrant csM1 phenotype is mainly due to compromised frameshifting at the altered slippery site.
Surprisingly, the modified amino-terminal p6* cleavage site, although resulting in a 240-fold-increased hydrolysis rate in vitro, did not influence the order and rate of PR-mediated precursor maturation. In contrast to our in vitro analyses, Chen et al. (7) did not observe amino-terminal cleavage of p6* when deploying a similar peptide cleavage assay and hypothesized that NC released from the Gag-Pol precursor might be extended by eight residues of p6*. However, our in vitro cleavage experiments have shown that substrate turnover was not completed until 24 h of incubation, suggesting that an observation period of 60 min selected by Chen et al. might not be sufficient to draw this conclusion. Furthermore, peptides representing the amino-terminal octapeptide of p6* were formerly isolated from HIV-1 virions (19), and mutations in the NC-p6* cleavage site have been implicated in compensating for the loss of viral fitness in PR inhibitor-resistant mutants (11), which clearly points to a biological function of amino-terminal p6* cleavage.
As internal cleavage of p6* between Phe8 and Leu9 is an early event in the Gag-Pol processing cascade (1, 40, 53), modification of this site in the viral context appeared particularly informative. However, the mutation introduced to block this cleavage site resulted in a shift of the scissile bond associated with a 5- to 10-fold-decreased hydrolysis rate. Surprisingly, this mutation did not influence correct processing of the viral precursor proteins or replication of csM2 and csM3 viruses in permissive T lymphocytes. Further confirming our observations, a comparable mutation did not impact viral infectivity, when analyzed in the context of stem-loop stability studies (21). These results indicate that the original cleavage site might be functionally replaced by the novel proximal site which has also been used in full-length GST-p6* proteins carrying the respective mutation. In a current model, the internal cleavage site is processed intramolecularly by the precursor-embedded PR which exhibits a substrate specificity different than that of the mature free enzyme (30, 40, 41, 61). Taking into account the fact that in vitro peptide cleavage was performed by a mature PR dimer in trans, we cannot completely exclude the possibility that the internal mutation might have different effects in the context of the Gag-Pol precursor.
Although carboxyl-terminal mutagenesis of p6* was carefully designed, cleavability predicted by the algorithm used (9) was achieved only for the amino acid combinations in P5 and P6. In contrast, cleavage was completely blocked in peptides P4 and P7 and in the respective virus mutants csM4 and csM7. Corroborating previous work by others and us, completely blocking carboxyl-terminal p6* cleavage resulted in immature viruses with impaired infectivity and severe Gag processing defects (38, 53). Interestingly, the amino-terminally extended PR species found in the corresponding virions was not capable of processing the MA-CA scissile bond, indicating that this cleavage can be carried out only by the mature enzyme. Even though the amino-terminal PR extension is likely to restrict accessibility of the Gag substrates within the condensed viral shell (53), similar observations have been made with uncondensed, in vitro-translated Gag-Pol and Gag precursors, rather suggesting that the precursor-embedded PR exhibits a cleavage site affinity or substrate specificity different from that of the mature enzyme (40).
In contrast, those carboxyl-terminal p6* modifications allowing peptide cleavage in vitro (P5 and P6) at 15- and 1.5-fold-lower hydrolysis rates than the wt site, which was reflected perfectly by the retarded release of the PR in csM5 and csM6 virions, did not result in Gag-specific processing defects. As traces of a PR-specific 17-kDa precursor were detected in those particles along with the mature 11-kDa species, the 17-kDa protein likely represents the very last PR intermediate. This implies that internal hydrolysis of p6* precedes carboxyl-terminal cleavage in virus particles, albeit the carboxyl-terminal site was processed at a 60-fold-higher hydrolysis rate in the in vitro cleavage assay. Hence, our data are concordant with a recently proposed model where p6* cleavage is initiated in cis at an internal site (Pi) that appears to be best accessible to the active site of the precursor-embedded PR (40, 41). However, as mature RT and IN species together with an RT-IN intermediate were present in csM4 and csM7 virions where amino-terminal cleavage of the PR was blocked, our results also indicate that amino-terminal release of the PR is not a prerequisite for removal of the carboxyl-terminal RT-IN moiety.
Whereas blockage of PR release severely affected maturation and infectivity of csM4 and csM7 particles with a potential impact on the natural processing cascade, the retarded PR release in mutants csM5 and csM6 did not visibly interfere with virus infectivity or replication on cultured cells, suggesting that altered cleavage rates at the PR amino terminus are well tolerated. These findings are concordant with former reports that viral infectivity is not significantly impaired unless PR activity is reduced by more than fourfold (50). Nevertheless, these mutants were rapidly overgrown by wt viruses in a more sensitive dual competition assay. It is noteworthy that the csM5 mutant, albeit showing a significantly delayed amino-terminal release of the PR, performed better in this assay than csM6, for which PR release was scarcely affected. Interestingly, csM6-specific p6* residues seemed to cause product inhibition in the in vitro cleavage assay, suggesting that compromised growth of the csM6 mutant in the competition assay might be due at least partially to secondary effects exerted by the released carboxyl-terminal p6* tetrapeptide (37). Additional support for this assumption has been given by analysis of purified p6* proteins an in vitro PR inhibition assay, where a p6* variant carrying the csM6-specific carboxyl terminus showed a slightly stronger inhibitory capacity than the wt protein did (unpublished data).
In sum, this study has provided further evidence that correct processing of p6* is essential to drive temporal and stepwise activation of the PR. However, our results have also shown that individual cleavage site variations of p6* did not interfere with viral replication in cultured cells as long as overall processing of the viral Gag and Pol precursors was guaranteed.
Acknowledgments
We thank Hermann Katinger for providing reagents and antibodies for the capsid ELISA and Heribert Stoiber for the ELISA protocol. The RT-specific antiserum was kindly provided by Prakash Chandra. Moreover, we thank the contributors to the NIH AIDS Research and Reference Reagent Program for antisera and cell lines.
Part of the work was financed by DFG grant Wa873/1-4.
Footnotes
Published ahead of print on 5 March 2008.
REFERENCES
- 1.Almog, N., R. Roller, G. Arad, L. Passi-Even, M. A. Wainberg, and M. Kotler. 1996. A p6Pol-protease fusion protein is present in mature particles of human immunodeficiency virus type 1. J. Virol. 707228-7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bardy, M., B. Gay, S. Pebernard, N. Chazal, M. Courcoul, R. Vigne, E. Decroly, and P. Boulanger. 2001. Interaction of human immunodeficiency virus type 1 Vif with Gag and Gag-Pol precursors: co-encapsidation and interference with viral protease-mediated Gag processing. J. Gen. Virol. 822719-2733. [DOI] [PubMed] [Google Scholar]
- 3.Beissinger, M., C. Paulus, P. Bayer, H. Wolf, P. Rösch, and R. Wagner. 1996. Sequence-specific resonance assignments of the 1H-NMR spectra and structural characterization in solution of the HIV-1 transframe protein p6*. Eur. J. Biochem. 237383-392. [DOI] [PubMed] [Google Scholar]
- 4.Bidou, L., G. Stahl, B. Grima, H. Liu, M. Cassan, and J. P. Rousset. 1997. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA 31153-1158. [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley, I. 1995. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 761885-1892. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee, A., P. Mridula, R. K. Mishra, R. Mittal, and R. V. Hosur. 2005. Folding regulates autoprocessing of HIV-1 protease precursor. J. Biol. Chem. 28011369-11378. [DOI] [PubMed] [Google Scholar]
- 7.Chen, N., A. Morag, N. Almog, I. Blumenzweig, O. Dreazin, and M. Kotler. 2001. Extended nucleocapsid protein is cleaved from the Gag-Pol precursor of human immunodeficiency virus type 1. J. Gen. Virol. 82581-590. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, H. C., F. D. Wang, Y. M. Chen, and C. T. Wang. 2006. Effects of human immunodeficiency virus type 1 transframe protein p6* mutations on viral protease-mediated Gag processing. J. Gen. Virol. 872041-2046. [DOI] [PubMed] [Google Scholar]
- 9.Chou, K. C., A. G. Tomasselli, I. M. Reardon, and R. L. Heinrikson. 1996. Predicting human immunodeficiency virus protease cleavage sites in proteins by a discriminant function method. Proteins 2451-72. [DOI] [PubMed] [Google Scholar]
- 10.Coren, L. V., J. A. Thomas, E. Chertova, R. C. Sowder II, T. D. Gagliardi, R. J. Gorelick, and D. E. Ott. 2007. Mutational analysis of the C-terminal Gag cleavage sites in human immunodeficiency virus type 1. J. Virol. 8110047-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Côté, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyon, L., C. Payant, L. Brakier-Gingras, and D. Lamarre. 1998. Novel Gag-Pol frameshift site in human immunodeficiency virus type 1 variants resistant to protease inhibitors. J. Virol. 726146-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulude, D., M. Baril, and L. Brakier-Gingras. 2002. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 305094-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulude, D., Y. A. Berchiche, K. Gendron, L. Brakier-Gingras, and N. Heveker. 2006. Decreasing the frameshift efficiency translates into an equivalent reduction of the replication of the human immunodeficiency virus type 1. Virology 345127-136. [DOI] [PubMed] [Google Scholar]
- 15.Erickson-Viitanen, S., L. Manfredi, P. Viitanen, D. E. Tribe, R. Tritch, C. A. Hutchison III, D. D. Loeb, and R. Swanstrom. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retrovir. 5577-591. [DOI] [PubMed] [Google Scholar]
- 16.Fehér, A., I. T. Weber, P. Bagossi, P. Boross, B. Mahalingam, J. M. Louis, T. D. Copeland, I. Y. Torshin, R. W. Harrison, and J. Tozser. 2002. Effect of sequence polymorphism and drug resistance on two HIV-1 Gag processing sites. Eur. J. Biochem. 2694114-4120. [DOI] [PubMed] [Google Scholar]
- 17.Fenn, J. B., M. Mann, C. K. Meng, S. F. Wong, and C. M. Whitehouse. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science 24664-71. [DOI] [PubMed] [Google Scholar]
- 18.Hazebrouck, S., V. Machtelinckx-Delmas, J. J. Kupiec, and P. Sonigo. 2001. Local and spatial factors determining HIV-1 protease substrate recognition. Biochem. J. 358505-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson, L. E., M. A. Bowers, R. C. Sowder, S. A. Serabyn, D. G. Johnson, J. W. Bess, L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 661856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, M., G. Tachedjian, and J. Mak. 2005. The packaging and maturation of the HIV-1 Pol proteins. Curr. HIV Res. 373-85. [DOI] [PubMed] [Google Scholar]
- 21.Hill, M. K., M. Shehu-Xhilaga, S. M. Crowe, and J. Mak. 2002. Proline residues within spacer peptide p1 are important for human immunodeficiency virus type 1 infectivity, protein processing, and genomic RNA dimer stability. J. Virol. 7611245-11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishima, R., D. A. Torchia, S. M. Lynch, A. M. Gronenborn, and J. M. Louis. 2003. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J. Biol. Chem. 27843311-43319. [DOI] [PubMed] [Google Scholar]
- 23.Jordan, S. P., J. Zugay, P. L. Darke, and L. C. Kuo. 1992. Activity and dimerization of human immunodeficiency virus protease as a function of solvent composition and enzyme concentration. J. Biol. Chem. 26720028-20032. [PubMed] [Google Scholar]
- 24.Kaplan, A. H., M. Manchester, and R. Swanstrom. 1994. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 686782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konvalinka, J., A.-M. Heuser, O. Hruskova-Heidingsfeldova, V. M. Vogt, J. Sedlacek, P. Strop, and H.-G. Kräusslich. 1995. Proteolytic processing of particle-associated retroviral polyproteins by homologous and heterologous viral proteinases. Eur. J. Biochem. 228191-198. [DOI] [PubMed] [Google Scholar]
- 26.Kotler, M., G. Arad, and S. H. Hughes. 1992. Human immunodeficiency virus type 1 gag-protease fusion proteins are enzymatically active. J. Virol. 666781-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koyanagi, Y., Y. Hinuma, J. Schneider, T. Chosa, G. Hunsmann, N. Kobayashi, M. Hatanaka, and N. Yamamoto. 1984. Expression of HTLV-specific polypeptides in various human T-cell lines. Med. Microbiol. Immunol. 173127-140. [DOI] [PubMed] [Google Scholar]
- 28.Lindhofer, H., K. von der Helm, and H. Nitschko. 1995. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology 214624-627. [DOI] [PubMed] [Google Scholar]
- 29.Louis, J. M., G. M. Clore, and A. M. Gronenborn. 1999. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 6868-875. [DOI] [PubMed] [Google Scholar]
- 30.Louis, J. M., N. T. Nashed, K. D. Parris, A. R. Kimmel, and D. M. Jerina. 1994. Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease from an analog of the Gag-Pol polyprotein. Proc. Natl. Acad. Sci. USA 917970-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Louis, J. M., E. M. Wondrak, A. R. Kimmel, P. T. Wingfield, and N. T. Nashed. 1999. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 27423437-23442. [DOI] [PubMed] [Google Scholar]
- 32.Miller, M., M. Jaskolski, J. K. Rao, J. Leis, and A. Wlodawer. 1989. Crystal structure of a retroviral protease proves relationship to aspartic protease family. Nature 337576-579. [DOI] [PubMed] [Google Scholar]
- 33.Niedrig, M., J. Hinkula, W. Weigelt, J. L'Age-Stehr, G. Pauli, J. Rosen, and B. Wahren. 1989. Epitope mapping of monoclonal antibodies against human immunodeficiency virus type 1 structural proteins by using peptides. J. Virol. 633525-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott, D. E., E. N. Chertova, L. K. Busch, L. V. Coren, T. D. Gagliardi, and D. G. Johnson. 1999. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6Gag protein produces a mutant that fails to package its envelope protein. J. Virol. 7319-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkin, N. T., M. Chamorro, and H. E. Varmus. 1992. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 665147-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Partin, K., G. Zybarth, L. Ehrlich, M. DeCrombrugghe, E. Wimmer, and C. Carter. 1991. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 884776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulus, C., S. Hellebrand, U. Tessmer, H. Wolf, H.-G. Kräusslich, and R. Wagner. 1999. Competitive inhibition of immunodeficiency virus type-1 protease by the Gag-Pol transframe protein*. J. Biol. Chem. 27421539-21543. [DOI] [PubMed] [Google Scholar]
- 38.Paulus, C., C. Ludwig, and R. Wagner. 2004. Contribution of the Gag-Pol transframe domain p6* and its coding sequence to morphogenesis and replication of human immunodeficiency virus type 1. Virology 330271-283. [DOI] [PubMed] [Google Scholar]
- 39.Pettit, S. C., J. C. Clemente, J. A. Jeung, B. M. Dunn, and A. H. Kaplan. 2005. Ordered processing of the human immunodeficiency virus type 1 GagPol precursor is influenced by the context of the embedded viral protease. J. Virol. 7910601-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettit, S. C., L. E. Everitt, S. Choudhury, B. M. Dunn, and A. H. Kaplan. 2004. Initial cleavage of the human immunodeficiency virus type 1 GagPol precursor by its activated protease occurs by an intramolecular mechanism. J. Virol. 788477-8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettit, S. C., S. Gulnik, L. Everitt, and A. H. Kaplan. 2003. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J. Virol. 77366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettit, S. C., J. N. Lindquist, A. H. Kaplan, and R. Swanstrom. 2005. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. Kaplan, P. V. Nantermet, C. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 688017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettit, S. C., N. Sheng, R. Tritch, S. Erickson-Viitanen, and R. Swanstrom. 1998. The regulation of sequential processing of HIV-1 Gag by the viral protease. Adv. Exp. Med. Biol. 43615-25. [DOI] [PubMed] [Google Scholar]
- 45.Pettit, S. C., J. Simsic, D. D. Loeb, L. Everitt, C. A. Hutchison III, and R. Swanstrom. 1991. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 26614539-14547. [PubMed] [Google Scholar]
- 46.Phylip, L. H., J. S. Mills, B. F. Parten, B. M. Dunn, and J. Kay. 1992. Intrinsic activity of precursor forms of HIV-1 proteinase. FEBS Lett. 314449-454. [DOI] [PubMed] [Google Scholar]
- 47.Porter, D. J. T., M. H. Hanlon, L. H. Carter III, D. P. Danger, and E. S. Furfine. 2001. Effectors of protease peptidolytic activity. Biochemistry 4011131-11139. [DOI] [PubMed] [Google Scholar]
- 48.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R. S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 357-69. [DOI] [PubMed] [Google Scholar]
- 49.Roberts, N., J. Martin, D. Kinchington, A. Broadharst, J. Craig, I. Duncan, S. Galpin, B. Handa, J. Kay, A. Krohn, R. Lambert, J. Merrett, J. Mills, K. Parks, S. Redshaw, D. Taylor, G. Thomas, and P. Machin. 1990. Rational design of peptide-based HIV proteinase inhibitors. Science 248358-361. [DOI] [PubMed] [Google Scholar]
- 50.Rosé, J. R., L. M. Babe, and C. S. Craik. 1995. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J. Virol. 692751-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schramm, H. J., A. Billich, E. Jaeger, K.-P. Rucknagel, G. Arnold, and W. Schramm. 1993. The inhibition of HIV-1 protease by interface peptides. Biochem. Biophys. Res. Commun. 194595-600. [DOI] [PubMed] [Google Scholar]
- 52.Speck, R. R., C. Flexner, C.-J. Tian, and X.-F. Yu. 2000. Comparison of human immunodeficiency virus type 1 Pr55Gag and Pr160Gag-Pol processing intermediates that accumulate in primary and transformed cells treated with peptidic and nonpeptidic protease inhibitors. Antimicrob. Agents Chemother. 441397-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tessmer, U., and H.-G. Kräusslich. 1998. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J. Virol. 723459-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tözsér, J., I. T. Weber, A. Gustchina, I. Blaha, T. D. Copeland, J. M. Louis, and S. Oroszlan. 1992. Kinetic and modeling studies of S3-S3′ subsites of HIV proteinases. Biochemistry 314793-4800. [DOI] [PubMed] [Google Scholar]
- 55.Wagner, R., H. Fliessbach, G. Wanner, M. Motz, M. Niedrig, G. Deby, A. von Brunn, and H. Wolf. 1992. Studies on processing, particle formation, and immunogenicity of the HIV-1 gag gene product: a possible component of a HIV vaccine. Arch. Virol. 127117-137. [DOI] [PubMed] [Google Scholar]
- 56.Whitehurst, N., C. Chappey, C. Petropoulos, N. Parkin, and A. Gamarnik. 2003. Polymorphisms in p1-p6/p6* of HIV type 1 can delay protease autoprocessing and increase drug susceptibility. AIDS Res. Hum. Retrovir. 19779-784. [DOI] [PubMed] [Google Scholar]
- 57.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H.-G. Kräusslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 722846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wlodawer, A., M. Miller, M. Jaskolski, B. K. Sathyanarayana, E. Baldwin, I. T. Weber, L. M. Selk, L. Clawson, J. Schneider, and S. B. Kent. 1989. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science 245616-621. [DOI] [PubMed] [Google Scholar]
- 59.Wolf, H., S. Modrow, E. Soutschek, M. Motz, R. Grunow, H. Döbl, and R. von Baehr. 1990. Production, mapping and biological characterization of monoclonal antibodies against core protein (p24) of the human immunodeficiency virus. AIDS-Forsch. 116-18. [Google Scholar]
- 60.Wondrak, E. M., J. M. Louis, H. de Rocquigny, J. C. Chermann, and B. P. Roques. 1993. The Gag precursor contains a specific HIV-1 protease cleavage site, between the NC (p7) and p1 proteins. FEBS Lett. 33321-24. [DOI] [PubMed] [Google Scholar]
- 61.Wondrak, E. M., N. T. Nashed, M. T. Haber, D. M. Jerina, and J. M. Louis. 1996. A transient precursor of the HIV-1 protease. Isolation, characterization, and kinetics of maturation. J. Biol. Chem. 2714477-4481. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., H. Imamichi, T. Imamichi, H. Lane, J. Falloon, M. Vasudevachari, and N. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 716662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zybarth, G., and C. Carter. 1995. Domains upstream of the protease (PR) in human immunodeficiency virus type 1 Gag-Pol influence PR autoprocessing. J. Virol. 693878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zybarth, G., H.-G. Kräusslich, K. Partin, and C. Carter. 1994. Proteolytic activity of novel human immunodeficiency virus type 1 proteinase proteins from a precursor with a blocking mutation at the N terminus of the PR domain. J. Virol. 68240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]