Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) latency is central to the evasion of host immune surveillances and induction of KSHV-related malignancies. The mechanism of KSHV latency remains unclear. Here, we show that the KSHV latent gene vFLIP promotes viral latency by inhibiting viral lytic replication. vFLIP suppresses the AP-1 pathway, which is essential for KSHV lytic replication, by activating the NF-κB pathway. Thus, by manipulating two convergent cellular pathways, vFLIP regulates both cell survival and KSHV lytic replication to promote viral latency. These results also indicate that the effect of the NF-κB pathway on KSHV replication is determined by the status of the AP-1 pathway and hence provide a mechanistic explanation for the contradictory role of the NF-κB pathway in KSHV replication. Since the NF-κB pathway is commonly activated during infection of gammaherpesviruses, these findings might have general implications for the control of gammaherpesviral latency.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a human tumor virus associated with Kaposi's sarcoma (KS), the most dominant form of cancer in AIDS patients, and several other lymphoproliferative diseases, including primary effusion lymphoma (PEL) and multicentric Castleman's disease (8, 9, 40, 51). Like that of other herpesviruses, the life cycle of KSHV consists of latent and lytic phases (22). Following an acute viral infection, KSHV establishes a latent infection in the host. During this phase of the life cycle, KSHV replicates as episomes in the nucleus with a restricted viral transcriptional program, which confers to it the advantage of escaping the host immune surveillances. Upon stimulation by host or environmental factors, KSHV can be reactivated into lytic replication, during which it replicates as linear genomes, expresses most of the viral genes, and produces infectious virions (15). If the immune system loses control of viral replication, the infected host could be at high risk for developing KSHV-related malignancies (39). Uncontained KSHV lytic replication produces virus-encoded cytokines, spreads infectious virions, and induces inflammatory cytokines, all of which could contribute to the progression of malignancies (22). Unsurprisingly, these pathological conditions are frequently seen in human immunodeficiency virus-infected patients or patients undergoing immunosuppressive therapy, such as recipients of organ transplantation (22).
Similar to that of other oncogenic gammaherpesviruses, KSHV latent infection is essential for the development of KSHV-related malignancies, in part because of its dysregulation of cell growth and survival and induction of inflammatory cytokines, in addition to its essential role in sustaining a persistent viral infection (22, 38). Thus, the molecular mechanism mediating KSHV control of viral latency is a fundamental dilemma not only for the virus life cycle but also for KSHV-induced pathogenesis.
Only a few viral genes are expressed during KSHV latency (62). These include the latency-associated nuclear antigen (LANA or LNA) encoded by open reading frame 73 (ORF73), a viral cyclin D homolog (vCyclin) encoded by ORF72, and a viral Fas-associated protein with death domain-like interleukin-1β-converting enzyme/caspase-8-inhibitory protein (vFLIP) encoded by ORF71 (14, 20, 26, 46). These three latent genes are located in the latent locus of the KSHV genome and share the same set of transcripts, which consists of two polycistronic transcripts containing all three genes and a biscistronic transcript containing vCyclin and vFLIP (14, 26). Extensive studies in the last decade have defined the functions of these genes. LANA is essential for KSHV episome persistence and promotes cell growth by regulating the cellular tumor suppressors p53 and pRb and by targeting the β-catenin pathway (5, 17, 18, 45, 60). vCyclin also regulates cell growth by promoting cell cycle progression (10, 21, 32). vFLIP activates both the classic and the alternative NF-κB pathways, leading to enhanced cell survival and the secretion of inflammatory cytokines such as interleukin-6 (IL-6) and IL-8 (2, 11, 23, 36, 54). More-recent studies have shown that the KSHV latent locus also encodes a cluster of microRNAs, which are expressed during viral latency (7, 24, 44, 49); however, their functions remain largely unknown.
It is postulated that KSHV latent genes might directly regulate viral lytic replication and modulate viral latency. Indeed, LANA suppresses KSHV lytic replication by inhibiting transcription and the function of the central viral lytic replication activator RTA (ORF50) (29-31) and suppressing the expression of other viral lytic genes through epigenetic silencing of the viral genome (33, 34, 48). However, whether vCyclin and vFLIP, as well as KSHV microRNAs, also mediate viral latency is unclear. vFLIP is known to activate NF-κB (11, 36). While inhibition of NF-κB led to the reactivation and expression of KSHV lytic proteins (6), it was also shown that induction of NF-κB was required for the production of KSHV virions in PEL cells following induction with 12-O-tetradecanoyl-phorbol-13-acetate (TPA) (50). In the present study, we investigated the role of vFLIP in KSHV latency by using a reverse genetics approach. We showed that vFLIP indeed promotes KSHV latency by inhibiting the expression of RTA and viral lytic replication. Furthermore, we showed that vFLIP inhibits KSHV lytic replication by suppressing the AP-1 pathway. Our results demonstrate a novel function of vFLIP in mediating control of KSHV latency.
MATERIALS AND METHODS
Cell lines and culture conditions.
BCBL-1 cells carrying a recombinant KSHV cloned in a bacterial artificial chromosome (BAC), BAC36 (BCBL-1-BAC36) (63), were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml of gentamicin. BAC36 contains a green fluorescent protein (GFP) cassette, which facilitates the tracking of virus infection (19). BCBL-1 cells transduced with retrovirus murine stem cell virus-vFLIP (MSCV-vFLIP) or MSCV were cultured in RPMI 1640 supplemented with 10% FBS, 100 μg/ml of gentamicin, and 500 μg/ml of G418. Human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 100 μg/ml of gentamicin. To obtain stable recombinant KSHV 293T cell cultures, 200 μg/ml of hygromycin b was added to the media.
Plasmids and retroviruses.
vFLIP (ORF71) was cloned in frame into the plasmid pCMV-myc (ClonTech Laboratories, Inc., Mountain View, CA) to generate the expression construct pCMV-vFLIP. The luciferase reporter plasmid pGL3-Basic and the AP-1 reporter plasmid pAP-1-Luc were purchased from Stratagene (La Jolla, CA). The KSHV RTA promoter luciferase reporter plasmid R-914 and its deletion construct R-259 were previously described (42, 59). The reporter R-259 plasmid was used as a template to generate two reporter mutants with mutations in putative transcription factor binding sites, using a QuickChange II XL site-directed mutagenesis kit (Stratagene). The mutant reporter R-259mut-NF-κB had a mutation in the −148 putative NF-κB site, while the mutant reporter R-259mut-AP-1 had a mutation in the −81 AP-1 site, both of which were verified by DNA sequencing (see Fig. 7A). An NF-κB reporter plasmid containing tandem repeats of consensus NF-κB binding sites was obtained from Bill Sudgen of the University of Wisconsin—Madison through Kenneth Izumi of the University of Texas Health Science Center at San Antonio. The NF-κB dominant negative (DN) plasmid pIκB-αM was kindly provided by Paul J. Chiao of the University of Texas M.D. Anderson Cancer Center. The retrovirus constructs MSCV-vFLIP and MSCV were kindly provided by Preet Chaudhary of the University of Pittsburgh, and the production of infectious retroviruses with these constructs was carried out as previously described (11).
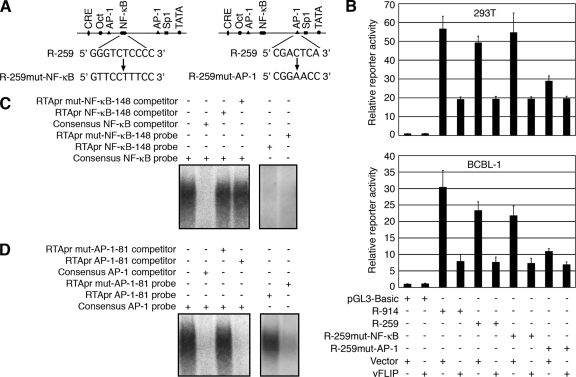
FIG. 7.
KSHV vFLIP inhibits RTA promoter activity by suppressing the AP-1 pathway. (A) Generation of mutant reporter constructs R-259mut-NF-κB and R-259mut-AP-1 with mutations in the −148 putative NF-κB site and the −81 AP-1 site, respectively, using the wild-type R-259 reporter construct as a template. (B) Relative reporter activities of the vector reporter pGL3-Basic, the RTA promoter reporters R-914 and R-259, and the mutant reporters R-259mut-NF-κB and R-259mut-AP-1 cotransfected with the control plasmid pCMV-myc (Vector) or pCMV-vFLIP (vFLIP) in 293T and BCBL-1 cells. (C) EMSA examination of the RTA promoter −148 putative NF-κB site. (Left panel) Neither the wild-type RTA promoter −148 putative NF-κB site probe (RTApr NF-κB-148) nor its mutant probe (RTApr mut-NF-κB-148) efficiently competed with the consensus NF-κB probe for the formation of a NF-κB DNA-protein complex. (Right panel) Neither RTApr NF-κB-148 nor its mutant RTApr mut-NF-κB-148 probe formed a DNA-protein complex. (D) EMSA examination of the RTA promoter −81 AP-1 site. (Left panel) Wild-type RTA promoter −81 AP-1 site probe (RTApr AP-1-81) but not its mutant probe (RTApr mut-AP-1-81) efficiently competed with the consensus AP-1 probe for the formation of an AP-1 DNA-protein complex. (Right panel) The RTApr AP-1-81 probe but not its mutant RTApr mut-AP-1-81 probe formed a DNA-protein complex. +, presence; −, absence.
Chemical induction and titration of infectious KSHV.
Induction of infectious virions from cells infected with recombinant KSHV was carried out with 20 ng/ml of TPA and 0.3 mM of sodium butyrate for BCBL-1 cells and 20 ng/ml of TPA and 1.5 mM of sodium butyrate for 293T cells, as previously described (63). For BCBL-1 cells, 5 × 106 cells per 10 ml of medium were induced with TPA and sodium butyrate (T/B) for 2 days. The medium was washed away by centrifugation and replaced with the same volume of fresh medium without T/B. For 293T cells, 2 × 106 cells in a 25 cm2 flask were induced with T/B for 2 days. The medium was washed away and replaced with the same volume of fresh medium without T/B. After 3 additional days, the culture supernatants were collected and centrifuged at 4,000 × g for 30 min to eliminate cells and cellular debris. The supernatants were then used to infect 293T cells for 48 h. Images of GFP-positive cells were then taken, and the cells were trypsinized and counted with a hemacytometer under a fluorescence microscope. The relative virus titer was determined by the number of GFP-positive cells in each milliliter of inoculum (GFU) (19). Each sample was examined in three to five wells of a 96-well plate. Six random microscopic fields at a magnification of ×20 were examined for each well. Results were calculated as averages with standard deviations.
Transient transfection and luciferase reporter assays.
Transient transfection was carried out in 293T cells, using F2 transfection reagent (Targeting Systems, El Cajon, CA). For luciferase reporter assays, cells transfected with luciferase reporter plasmids were lysed and collected at 48 h posttransfection, using a luciferase assay lysis buffer from Promega (Madison, WI). The luciferase activity was then measured with a Veritas microplate luminometer (Turner BioSystems, Sunnyvale, CA). Transfection efficiency was calibrated by cotransfection with a pSV-β-galactosidase construct (Promega). All reporter assays were carried out three times, each in triplicate. Results were calculated as averages with standard deviations from one representative experiment.
Generation of a vFLIP deletion mutant (BAC36ΔvF) and revertant (BAC36ΔvF-Rv).
The procedures for generating the vFLIP deletion mutant BAC36ΔvF and its revertant, BAC36ΔvF-Rv, are schematically illustrated in Fig. 2A. To generate BAC36ΔvF, a PCR product was first obtained with the primers 5′-GAGCACCCTGAAATCCAGGCTCTACAGGTAGGCCACATACGCTCGCCACTCTATATGGTGTAGGCTGGAGCTGCTTC-3′ (forward) and 5′-CCGCCCTAAACAAAATCACAAGCTTAATAGCTGTCCAGAATGCGCAGATCAAAGTCCCATATGAATATCCTCCTTAG-3′ (reverse), using plasmid pMS102-Zeocin as a template. This PCR fragment contained a Zeocin resistance cassette (ZeoR) flanked by LoxP sites and 50-bp KSHV sequences from the two ends (genomic positions 122711 and 122145) of vFLIP. The product was then electroporated into Escherichia coli strain DH10B containing BAC36, which had been pretransformed with the helper plasmid pGET-rec and induced with 0.3% arabinose to enhance homologous recombination. The selected Zeocin-resistant colonies contained the KSHV genome with vFLIP deleted and replaced by ZeoR through homologous recombination. To remove ZeoR, the Cre-expression plasmid pCTP-T (a kind gift from Yoshinaga Sacki of Harvard Medical School), which carries a tetracycline resistance gene (Tcr), a temperature-sensitive origin of replication, and the Cre recombinase gene under the control of the Tc-responsive promoter, was introduced into the Zeocin-resistant bacteria by transformation. The bacteria were then cultured in LB media in the presence of tetracycline (50 μg/ml) for 3 h at 30°C to allow the expression of Cre and site-specific recombination between the two LoxP sites and the excision of ZeoR from the KSHV genome. The resulting tetracycline-resistant colonies were selected, and the presence of the KSHV genome and the removal of ZeoR were verified. Finally, the Cre expression plasmid was eliminated by culturing the bacteria at 42°C.
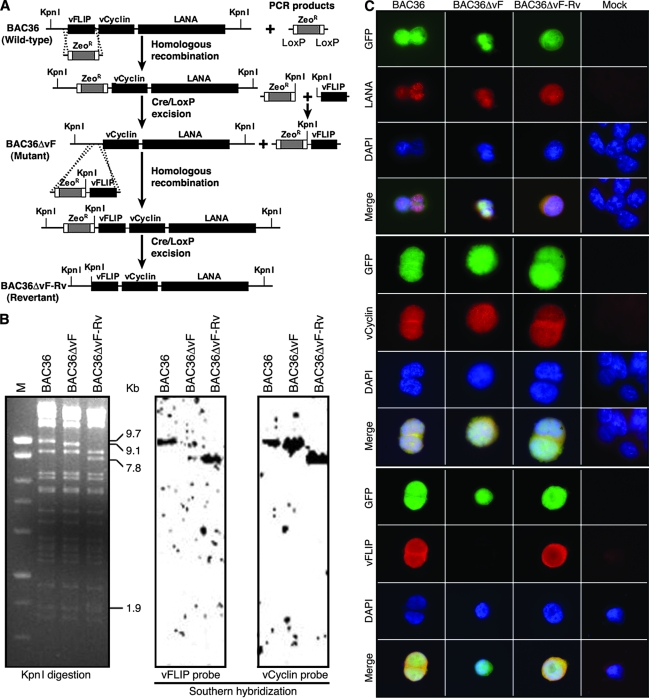
FIG. 2.
Generation of KSHV vFLIP gene deletion mutant and revertant. (A) Strategy for generating KSHV vFLIP gene deletion mutant BAC36ΔvF and revertant BAC36ΔvF-Rv using wild-type recombinant KSHV BAC36. The details are described in Materials and Methods. (B) Restriction analysis of BAC36, BAC36ΔvF, and BAC36ΔvF-Rv (left panel) and Southern blot hybridization with vFLIP probe (middle panel) and vCyclin probe (right panel). The 9.7-kb band in the BAC36 lane containing the vFLIP gene was shifted to a 9.1-kb band in the BAC36ΔvF lane because of the deletion of the vFLIP gene and to a 7.8-kb band and a 1.9-kb band in the BAC36ΔvF-Rv lane because of the insertion of a KpnI site downstream of the vFLIP gene. M, marker. (C) Immunofluorescence antibody staining for LANA, vCyclin, and vFLIP proteins in BAC36, BAC36ΔvF, and revertant BAC36ΔvF-Rv cell lines.
BAC36ΔvF-Rv was generated by using BAC36ΔvF as a template. First, two PCR products were generated. One was obtained by using the primers 5′-AAATCAGATACATACATTCTACGGACCAAAAATTAGCAACAGCTTGTTATTAATTAATGTTGACAATTAATCATCGGCATAGTA-3′(forward) and 5′-AAGGTACCTTAATTAACAGTCCTGCTCCTCGGCCACGAAGTGC-3′(reverse) and pMS102-Zeocin as a template. This fragment contained the entire ZeoR cassette flanked by LoxP sites with a 50-bp KSHV sequence (genomic position, 122096 to 122144) at one end and a KpnI restriction site at the other end. A second PCR product was obtained by using the primers 5′-AAGGTACCTATGGTGTATGGCGATAGTGTTGGG-3′(forward) and 5′-GCTTGTGATTTTGTTTAGGGCGGA-3′ (reverse) and BAC36 as a template. This product contained the entire vFLIP sequence, with a KpnI site at one end and a KSHV sequence in the intergenic region between ORF72 and ORF71 at the other end (genomic position, 122145 to 122793). Second, the two PCR products were ligated after KpnI digestion, resulting in a ZeoR-vFLIP cassette, which was then electroporated into E. coli DH10B containing BAC36ΔvF. The selection of colonies containing KSHV genomes with vFLIP repaired, followed by the excision of ZeoR, led to the generation of BAC36ΔvF-Rv.
Restriction analysis and Southern blot hybridization.
DNA preparations of recombinant viruses were digested with KpnI restriction enzyme, separated on 0.8% agarose gels by electrophoresis, and transferred to Zeta-Probe GT membranes. The membranes were hybridized with 32P-labeled vFLIP and vCyclin probes, and specific signals were detected with a Typhoon 9410 imaging system (GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
RT-qPCR.
The total amount of RNA (5 μg) was reverse transcribed into first-strand cDNAs by using a Superscript III first-strand synthesis system (Invitrogen, Carlsbad, CA). PCRs were also carried out in parallel without the reverse transcription step to ensure the absence of genomic DNA contamination in the RNA samples. All primers, the melting temperatures of the amplified products, and the lowest copy numbers that could be detected are listed in Table 1. Real-time quantitative PCR (RT-qPCR) was carried out in a DNA engine Opticon 2 continuous fluorescence detector (Bio-Rad, Hercules, CA) (61). Each sample was measured in triplicate.
TABLE 1.
PCR primers used, melting temperatures of the amplified products, and copy numbers detected
| Gene | Primer and sequence | Tma (7deg;C) | Nb |
|---|---|---|---|
| ORF50 | ORF50F: 5′-CACAAAAATGGCGCAAGATGA-3′ | 82 | 50 |
| ORF50R: 5′-TGGTAGAGTTGGGCCTTCAGTT-3′ | |||
| ORF57 | ORF57F: 5′-ACGAATCGAGGGACGACG-3′ | 82.5 | 50 |
| ORF57R: 5′-CGGGTTCGGACAATTGCT-3′ | |||
| ORF59 | ORF59F: 5′-CGAGTCTTCGCAAAAGGTTC-3′ | 80.2 | 50 |
| ORF59R: 5′-AAGGGACCAACTGGTGTGAG-3′ | |||
| ORF65 | ORF65F: 5′-ATATGTCGCAGGCCGAATAC-3′ | 77.4 | 5 |
| ORF65R: 5′-CCACCCATCCTCCTCAGATA-3′ | |||
| ORF71 | ORF71F: 5′-GGATGCCCTAATGTCAATGC-3′ | 82 | 5 |
| ORF71R: 5′-GGCGATAGTGTTGGGAGTGT-3′ | |||
| ORF72 | ORF72F: 5′-GCTGATAATAGAGGCGGGCAATGAG-3′ | 80 | 5 |
| ORF72R: 5′-GTTGGCGTGGCGAACAGAGGCAGTC-3′ | |||
| ORF73 | ORF73F: 5′-GCAGACACTGAAACGCTGAA-3′ | 81.5 | 50 |
| ORF73R: 5′-AGGTGAGCCACCAGGACTTA-3′ | |||
| K8.1 | K8.1F: 5′-AAAGCGTCCAGCCCACCACAGA-3′ | 80.2 | 5 |
| K8.1R: 5′-GGCAGAAAATGGCACACGGTTAC-3′ | |||
| K8 | K8F: 5′-CATGCTGATGCGAATGTGC-3′ | 82 | 50 |
| K8R: 5′-AGCTTCAACATGGTGGGAGTG-3′ | |||
| GAPDH | GAPDH-F: 5′-CCCCTGGCCAAGGTCATCCA-3′ | 83 | 5 |
| GAPDH-R: 5′-ACAGCCTTGGCAGCGCCAGT-3′ |
Melting temperature of the amplified product.
Lowest copy number that could be detected.
Western blotting and immunofluorescence antibody assays (IFA).
Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes as previously described (20). The blots were blocked with 5% skim milk and incubated with primary antibodies followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. Specific bands were revealed with chemiluminescence substrates and recorded with an IS2000MM imaging scanner (Eastman Kodak Company, Rochester, NY). To detect the RTA protein, a rabbit polyclonal antibody (a generous gift from Charles Wood of the University of Nebraska, Lincoln) diluted at 1:2,000 was used. Specific signals were revealed with a goat anti-rabbit HRP conjugate (Sigma Life Science, St. Louis, MO). To detect the vFLIP protein, a rat monoclonal antibody (a kind gift from Mary Collins of Windeyer Institute of Medical Sciences, London, United Kingdom) diluted at 1:100 was used. Specific signals were revealed with a rabbit anti-rat HRP conjugate (Sigma). A mouse antibody to β-tubulin (Sigma) diluted at 1:200 was used to monitor sample loading. Specific signals were revealed with a rabbit anti-mouse HRP conjugate (Sigma). All the secondary antibodies were used at 1:5,000 dilutions.
For IFA, cells were first fixed with methanol or 1% paraformaldehyde and blocked with 10% FBS. To detect the vFLIP protein, cells were incubated with the rat monoclonal antibody at a 1:100 dilution. LANA was also detected with a rat monoclonal antibody at a 1:1,000 dilution (ABI, New York, NY). Specific signals were revealed with a rabbit anti-rat immunoglobulin G (IgG) Alexa Fluor 568 conjugate (Invitrogen). A vCyclin rabbit peptide polyclonal antibody generated by immunizing rabbits with a peptide (TANNPPSGLLDPTLC) was used at a 1:1,000 dilution to detect the vCyclin protein. A goat anti-rabbit IgG rhodamine conjugate was used to reveal specific signals (Santa Cruz Biotechnology, Santa Cruz, CA). Mouse monoclonal antibodies were used to detect the ORF59 protein (a gift from Bala Chandran) and the ORF65 protein (19), and specific signals were revealed with a goat anti-mouse IgG Alexa Fluor 568 conjugate (Invitrogen). Antibodies to ORF59 and ORF65 were used at a 1:100 dilution. All the secondary antibodies were used at 1:200 dilutions. In all the IFA experiments, the cells were counter stained with DAPI (4′,6′-diamidino-2-phenylindole). Images were observed and recorded with an epifluorescence microscope (Carl Zeiss, Inc., Thornwood, NY).
Electrophoretic mobility shift assays (EMSAs).
Nuclear extracts were prepared as previously described (58). Annealed double-stranded oligonucleotides containing the consensus binding site for AP-1 (5′-GGGTTATGAGTCAGTTGC-3′), the consensus binding site for NF-κB (5′-AGTTGAGGGACTTTCCTT-3′), the RTA promoter −81 AP-1 site RTApr AP-1-81 (5′-CTACCGGCGACTCATTAAGC-3′) and its mutant site, RTApr mut-AP-1-81 (5′-CTACCGGCGGAACCTTAAGC-3′), and the RTA promoter −148 putative NF-κB site RTApr NF-κB-148 (5′-ACGCTAGGGTCTCCCCACCCAA3-′) and its mutant, RTApr mut-NF-κB-148 (5′-ACGCTAGTTCCTAACCACCCAA-3′), were labeled with [γ-32P]ATP. For gel shift assays, 4 μg of the nuclear extract was incubated for 20 min at room temperature with 5 × 105 cpm of labeled probe in 20 μl of binding buffer containing 10 mM Tris-HCl at pH 7.6, 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5% glycerol, 1 μg/μl bovine serum albumin, and 2 μg of poly(dI-dC). A competition assay was carried out in the same manner except that the above reaction mixture was preincubated with an excess cold probe or an irrelevant oligonucleotide for 10 min at 4°C before the addition of the labeled probe. For a supershift assay to detect the AP-1 complex, 1 μg of a specific antibody to either c-Fos or c-Jun (Santa Cruz Biotechnology) was incubated with the reaction mixture for another 15 min. IgG from a mouse antibody was used as a control. To detect the NF-κB complex, an antibody to p65, p50, or cRel was used (EMD Biosciences, San Diego, CA). Samples were separated in 6% polyacrylamide gels.
RESULTS
Overexpression of vFLIP inhibits RTA expression and viral reactivation in PEL cells.
To determine the function of vFLIP in KSHV latency, we overexpressed it in BCBL-1 cells carrying BAC36 (63). We infected the cells with a vFLIP retrovirus, MSCV-vFLIP, and a control retrovirus, MSCV (vector), and established their respective stable cultures. We used mixed cultures rather than selected individual clones for the experiments to ensure representation of the stable cell cultures. The growth of these cultures did not differ significantly from that of the parental cultures (data not shown). The expression level of vFLIP protein in stable MSCV cells was similar to that of the parental cells (data not shown). Treatment with TPA and sodium butyrate, two chemicals known to reactivate KSHV into lytic replication, increased the vFLIP protein level by 1.8-fold (Fig. 1A and B). These results were expected, because the promoter controlling vFLIP transcription is responsive to a similar chemical induction (43). Overexpression of vFLIP increased the vFLIP protein level by 11.6-fold (Fig. 1A and B). In agreement with these protein results, RT-qPCR detected 3.1-fold-higher expression levels of vFLIP transcripts in stable MSCV-vFLIP cells than in stable MSCV cells (Fig. 1D). Treatment with T/B increased the expression levels of vFLIP transcripts by 1.8- and 5.6-fold in MSCV and MSCV-vFLIP cells, respectively (Fig. 1D).
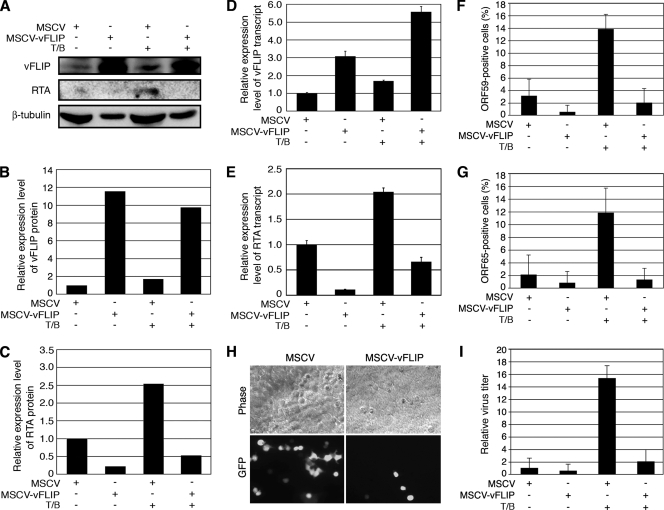
FIG. 1.
Overexpression of the vFLIP gene in PEL cells inhibited KSHV lytic replication. BCBL-1 cells stably transduced with vFLIP (MSCV-vFLIP) or a vector control (MSCV) with or without induction with T/B were analyzed for KSHV lytic replication. (A) Western blot analysis of vFLIP and RTA proteins. Western blotting for β-tubulin was also carried out to calibrate protein loading. (B and C) Quantification of expression levels of vFLIP (B) and RTA (C) proteins. The relative intensities of the bands were calibrated with their corresponding β-tubulin bands. Uninduced MCSV cells were set as 1. (D and E) Analysis by RT-qPCR of vFLIP (D) and RTA (E) transcripts. RT-qPCR for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcripts was also carried out for calibrating sample inputs. (F and G) Results of IFAs for ORF59 (F) and ORF65 (G) proteins. (H) Representative images showing that MSCV-vFLIP cells produced fewer KSHV virions than MSCV cells following induction with T/B. (I) Relative virus titers produced by MCSV and MSCV-vFLIP cells with or without induction with T/B. +, presence; −, absence.
We then examined the effect of vFLIP overexpression on the expression of RTA, a key viral transactivator that is necessary and sufficient for activating KSHV into lytic replication (35, 55). RTA transcripts were expressed at low levels because of a low rate of spontaneous viral lytic replication in PEL cells (12). Induction with T/B increased the expression of RTA transcripts by 2.1-fold in MSCV cells (Fig. 1E). Overexpression of vFLIP suppressed the expression of RTA transcripts before and after chemical induction (Fig. 1E). Without chemical induction, the level of RTA transcripts expressed in MSCV-vFLIP cells was only 11% of that expressed in MSCV cells. The level of RTA transcripts in MSCV-vFLIP cells was 32% of that in MSCV cells after chemical induction. The results of Western blotting carried out with an RTA-specific antibody were consistent with those for RTA transcripts (Fig. 1A and C). While treatment with T/B increased the RTA protein level in MSCV cells by 2.6-fold, overexpression of vFLIP almost abolished the expression of RTA protein with or without chemical induction (Fig. 1A and C).
We further examined the protein expression of two other KSHV lytic genes, ORF59 and ORF65, by IFA (Fig. 1F and G). ORF59 and ORF65 proteins were expressed in 3.2% and 2.1% of MSCV cells, respectively, results that were consistent with the low level of spontaneous lytic replication observed in these cells (12). Induction with T/B increased the ORF59- and ORF65-positive cells to 13.8% and 11.8%, respectively. Overexpression of vFLIP reduced the number of ORF59- and ORF65-postive cells before and after chemical induction (Fig. 1F and G). MSCV-vFLIP cells had 0.7% and 0.9% ORF59- and ORF65-postive cells, respectively, before chemical induction, and 2% and 1.3% ORF59- and ORF65-postive cells, respectively, after chemical induction. Thus, overexpression of vFLIP also suppressed the expression of RTA downstream lytic genes in PEL cells.
Next, we examined whether vFLIP inhibition of viral lytic gene expression could be translated into virus production. Uninduced MSCV and MSCV-vFLIP cells produced low levels of infectious virions at the range of 1 × 104 to 2 × 104 GFU, which is similar to that of the parental cells (59). The level of infectious virions from MSCV-vFLIP cells was slightly lower than that from MSCV cells, but it was not statistically significant (Fig. 1I). Induction with T/B increased the level of infectious virions in MSCV cells by 15.3-fold; however, MCSV-vFLIP cells produced 7.5-fold-less infectious virions than MSCV cells (Fig. 1H-I). Thus, overexpression of vFLIP not only suppressed the expression of KSHV lytic genes but also reduced the production of infectious KSHV virions.
Deletion of vFLIP from the KSHV genome enhances the expression of viral lytic genes and lytic replication.
We used a loss-of-function approach to confirm the observed inhibitory effects of vFLIP on the expression of RTA and KSHV lytic replication. We generated a vFLIP deletion mutant and its revertant, using wild-type recombinant KSHV BAC36 as a template (Fig. 2A). In brief, we generated a PCR product containing ZeoR flanked by LoxP sites and two fragments of 50-bp KSHV sequences from the two ends of the vFLIP gene. Upon homologous recombination between this PCR product and BAC36 and Cre-mediated excision of ZeoR, we obtained a KSHV mutant with vFLIP deleted, BAC36ΔvF. A similar approach was used to generate a revertant of this mutant. We generated two PCR products, one containing ZeoR flanked by LoxP sites and a 50-bp KSHV sequence downstream of the vFLIP gene at one end and an added KpnI restriction site at the other end, and another containing the vFLIP gene sequence and an added KpnI site at one end and a 50-bp KSHV sequence outside the vFLIP gene at the other end. A replacement cassette was then generated by ligating the two PCR products after KpnI digestion. Upon recombination with BAC36ΔvF and Cre-mediated excision of ZeoR, we obtained a revertant, BAC36ΔvF-Rv, which was identical to the wild-type BAC36 with the exception of an added KpnI site immediately downstream of the vFLIP gene. The addition of the KpnI site should not affect the expression and function of the vFLIP gene but should allow the identification of the revertant through its distinct KpnI restriction pattern.
Restriction analysis by KpnI digestion showed that BAC36ΔvF and BAC36ΔvF-Rv had restriction patterns similar to that of BAC36, except for the 9.7-kb band containing the vFLIP gene in the BAC36 lane, which was shifted to a 9.1-kb band in the mutant lane because of the loss of the vFLIP gene and split into a 7.8-kb band and a 1.9-kb band in the revertant lane because of the added KpnI site (Fig. 2B). Southern blot hybridization with vFLIP and vCyclin probes confirmed that the vFLIP gene was indeed deleted in the mutant but restored in the revertant, while the vCyclin gene was not affected in the BAC36ΔvF and BAC36ΔvF-Rv genomes (Fig. 2B).
To examine the effect of the vFLIP deletion on the expression of KSHV genes, we reconstituted the recombinant viruses in 293T cells and generated stable cell lines of BAC36, BAC36ΔvF, and BAC36ΔvF-Rv, as previously described (63). PCR examination of the vFLIP locus showed its presence in BAC36 and BAC36ΔvF-Rv cells but not in BAC36ΔvF cells (data not shown). In contrast, both vCyclin and LANA loci were present in all cell lines (data not shown). IFA with specific antibodies further confirmed the presence of vFLIP protein in BAC36 and BAC36ΔvF-Rv cells but not in BAC36ΔvF cells (Fig. 2C). We also detected the expression of both LANA and vCyclin proteins in all cell lines (Fig. 2C). These results indicate that deletion of the vFLIP gene did not affect the expression of other adjacent viral genes.
We then examined the expression of KSHV genes in the three cell lines induced with T/B for different lengths of time by RT-qPCR (Fig. 3A). In uninduced cells, the expression level of RTA transcripts in BAC36ΔvF cells was 1.6-fold higher than in BAC36 cells. At 12 h postinduction (hpi), the level of RTA transcripts increased to 6.3-fold in BAC36ΔvF cells but to only 1.9-fold in BAC36 cells. The level of RTA transcripts remained elevated in BAC36ΔvF cells but low in BAC36 cells at 48 and 72 hpi (Fig. 3A). The expression pattern of RTA transcripts in BAC36ΔvF-Rv cells was similar to that in BAC36 cells.
FIG. 3.
Deletion of the KSHV vFLIP gene enhanced KSHV lytic replication. (A) Analysis by RT-qPCR of transcripts of KSHV genes in 293T cells carrying BAC36, BAC36ΔvF, and BAC36ΔvF-Rv after induction with T/B for indicated lengths of time. (B and C) Relative intracellular KSHV DNA levels (B) and production of KSHV virions (C) in BAC36 and BAC36ΔvF 293T cells following induction with T/B. (D) Analysis by RT-qPCR of RTA transcripts in BAC36 and BAC36ΔvF cells transfected with a vFLIP expression construct (pCMV-vFLIP) or a vector (pCMV-myc) followed with induction with T/B. +, presence; −, absence.
The lytic gene ORF57 is a direct transcriptional target of RTA. In uninduced cells, the expression of ORF57 transcripts was 4.7-fold higher in BAC36ΔvF cells than in BAC36 cells (Fig. 3A). The expression of ORF57 was highly induced in all cell lines, reaching to 6.7-, 17.0-, and 5.4-fold in BAC36, BAC36ΔvF, and BAC36ΔvF-Rv cells, respectively, at 12 hpi. Interestingly, the elevated levels of ORF57 transcripts were sustained for up to 72 hpi. Importantly, BAC36ΔvF cells had higher expression levels of ORF57 transcripts than BAC36 and BAC36ΔvF-Rv cells at all time points following induction with T/B. The expression patterns of the early lytic gene K8 and late lytic gene K8.1 were similar to that of ORF57, with BAC36ΔvF cells having higher expression levels than both BAC36 and BAC36ΔvF-Rv cells at all time points examined, albeit the K8 and K8.1 transcripts were induced at a lower level than that of ORF57 in all cell lines (Fig. 3A). At 48 hpi, K8 transcripts were induced to 2.4-, 3.6-, and 2.7-fold, while K8.1 transcripts were induced to 1.4-, 2.1-, and 1.5-fold in BAC36, BAC36ΔvF, and BAC36ΔvF-Rv cells, respectively.
The early lytic gene ORF59 displayed a distinct expression pattern, with the highest expression level observed at 48 hpi, reaching to 4.5-, 17.6-, and 8.7-fold in BAC36, BAC36ΔvF, and BAC36ΔvF-Rv cells, respectively (Fig. 3A). Again, the expression levels of ORF59 transcripts were higher in BAC36ΔvF cells than in BAC36 and BAC36ΔvF-Rv cells at all time points examined. The expression pattern of the late lytic gene ORF65 was similar to that of ORF59, but the ORF65 transcripts were induced at lower levels than those of ORF59 in all cell lines, reaching to 1.9-, 2.7-, and 1.4-fold in BAC36, BAC36ΔvF, and BAC36ΔvF-Rv cells, respectively, at 48 hpi.
In contrast to that of the viral lytic genes, the expression of the viral latent genes LANA and vCyclin were at similar levels in all cell lines (Fig. 3A). Both LANA and vCyclin genes were moderately induced at 12 hpi. At 72 hpi, their expression levels were similar to or slightly lower than those of uninduced cells. The expression pattern of vFLIP transcripts was similar to those of LANA and vCyclin in BAC36 and BAC36ΔvF-Rv cells. As expected, we did not detect any vFLIP transcripts in BAC36ΔvF cells (Fig. 3A).
If deletion of the vFLIP gene enhances the expression of RTA and other downstream viral lytic genes, it should also increase the production of KSHV virions. We measured both KSHV intracellular viral DNA copy numbers and virions in the culture supernatants. Without chemical induction, there were only marginal differences in the intracellular viral DNA copy numbers and the production of virions between BAC36 and BAC36ΔvF cells (Fig. 3B and C). Following induction with T/B, the intracellular viral DNA copy numbers in BAC36ΔvF cells were increased 2.3-fold, while those of BAC36 cells were increased only 1.3-fold, indicating a more-active viral lytic replication program in BAC36ΔvF cells. Indeed, the production of KSHV virions was increased 11.8-fold in BAC36ΔvF cells, while that in BAC36 cells was increased only 5.2-fold (Fig. 3C). In these experiments, BAC36 cells typically generated 104 GFU following induction with T/B. Consistent with these results, IFA staining showed that BAC36ΔvF cells had more ORF59- and ORF65-positive cells than BAC36 cells following chemical induction (data not shown).
To further confirm that the observed, more-active KSHV lytic replication program in BAC36ΔvF cells was indeed due to the deletion of the vFLIP gene from the KSHV genome rather than alterations in any other region of the viral genome, we carried out a complementation experiment by overexpressing the vFLIP gene in the BAC36ΔvF cells. We tracked the expression level of RTA transcripts, since it is a good indicator of viral lytic replication. As shown in Fig. 3D, while deletion of the vFLIP gene increased the expression of RTA transcripts by 2.4-fold in BAC36ΔvF cells, overexpression of the vFLIP gene reduced the expression of RTA transcripts to 1.6-fold. Induction with T/B increased the expression of RTA transcripts by 4.9-fold in BAC36ΔvF cells. Overexpression of the vFLIP gene reduced the expression level of RTA transcripts to 1.8-fold, which was close to the 1.5-fold level observed in the T/B-induced BAC36 cells (Fig. 3D). Thus, overexpression of the vFLIP gene rescued its suppression function in KSHV lytic replication in BAC36ΔvF cells.
Taken together, the above results indicate that deletion of the vFLIP gene results in enhanced expression of viral lytic genes and lytic replication during KSHV latent infection and reactivation.
vFLIP inhibits RTA promoter activity.
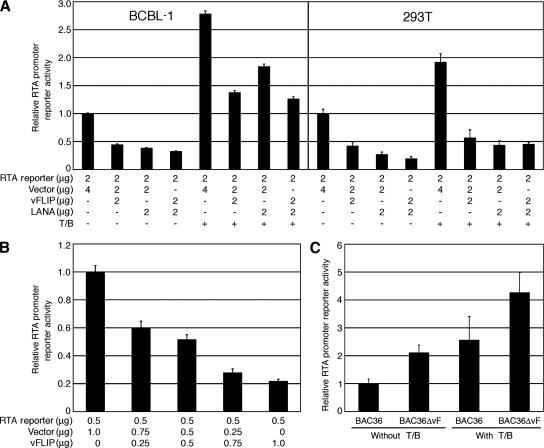
Since both overexpression and deletion of the vFLIP gene in the context of KSHV infection altered the expression of RTA transcripts, we postulated that vFLIP might regulate RTA promoter activity. As shown in Fig. 4A, cotransfection of the RTA promoter reporter R-914 with a vFLIP expression construct, pCMV-vFLIP, reduced the reporter activity by 55% and 58% in BCBL-1 and 293T cells, respectively. In addition, the inhibitory effect of vFLIP on the RTA promoter was dose dependent (Fig. 4B). Cotransfection of R-914 with a LANA expression construct, pcDNA3-His-LANA, reduced the reporter activity by 61% and 73% in BCBL-1 and 293T cells, respectively, which confirmed the previous finding that LANA suppresses the RTA promoter (29). Cotransfection of R-914 with both vFLIP and LANA constructs reduced the reporter activity by 67% and 80% in BCBL-1 and 293T cells, respectively, suggesting an additive effect of vFLIP and LANA on the suppression of the RTA promoter.
FIG. 4.
Inhibition of KSHV RTA promoter activity by vFLIP. (A) Relative activities of the RTA promoter reporter R-914 in BCBL-1 and 293T cells cotransfected with a vFLIP expression construct (pCMV-vFLIP) alone or together with a LANA expression construct, pcDNA3-His-LANA, with or without induction with T/B. Cotransfection with the vector was used as a control. +, presence; −, absence. (B) Dose-dependent inhibition of the RTA promoter activity by vFLIP in 293T cells. The relative activities of the RTA promoter reporter R-914 were examined following cotransfection with increasing doses of pCMV-vFLIP. (C) Relative activities of the RTA promoter reporter R-914 in 293T cells carrying BAC36 and BAC36ΔvF with or without induction with T/B.
We further examined whether vFLIP could inhibit the activation of the RTA promoter during KSHV reactivation from latency. As shown in Fig. 4A, treatment with T/B increased the RTA reporter activity by 2.8- and 1.9-fold in BCBL-1 and 293T cells, respectively. However, cotransfection with the vFLIP construct reduced 50% and 70% of the reporter activity in BCBL-1 and 293T cells, respectively. Similar inhibitory effects were also observed with the LANA construct. These results indicate that similar to LANA, vFLIP potently suppresses the RTA promoter activity with and without chemical induction. It was also noted that neither LANA nor vFLIP could completely inhibit the RTA promoter activity.
To examine the effect of vFLIP on RTA promoter activity in the context of a virus infection, we carried out the reporter assay in BAC36 and BAC36ΔvF cells. As shown in Fig. 4C, the RTA promoter activity in BAC36ΔvF cells was 2.1-fold higher than that in BAC36 cells. Upon induction with T/B, the RTA promoter activity was increased 4.3-fold in BAC36ΔvF cells but only 2.6-fold in BAC36 cells.
vFLIP inhibits RTA promoter activity by suppressing the AP-1 pathway.
Considering that the vFLIP protein does not have any molecular motif for direct regulation of gene expression, we reasoned that it might control RTA expression and viral replication by regulating specific cellular pathways. vFLIP activates the NF-κB pathway (11, 36), and this pathway has an inhibitory effect on KSHV reactivation (6). Nevertheless, the mechanism of NF-κB pathway-mediated inhibition of KSHV reactivation remains unclear, and contradictory observations have been reported (50). Previous studies have shown that activation of the AP-1 pathway is essential for KSHV lytic replication during both de novo primary infection and reactivation, in part by regulating the expression of RTA (42, 57, 59).
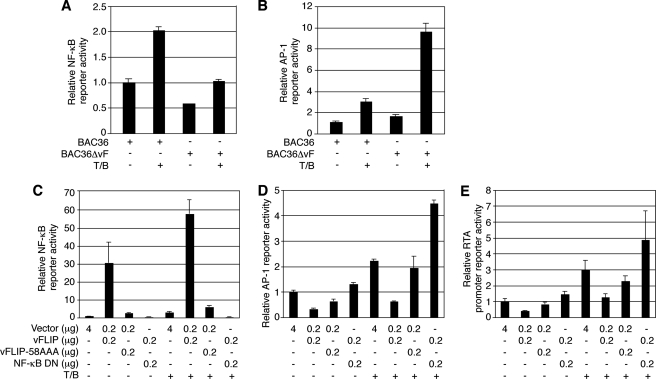
We first examined vFLIP regulation of the NF-κB and AP-1 pathways in the context of a viral infection. As shown in Fig. 5A, NF-κB reporter activity in BAC36ΔvF cells was only 58% of that in BAC36 cells. Treatment with T/B increased the NF-κB activity by 2.1-fold in BAC36 cells; however, it restored NF-κB activity only to the level of that in uninduced BAC36 cells in BAC36ΔvF cells (Fig. 5A). By contrast, AP-1 activity was 1.5-fold higher in BAC36ΔvF cells than in BAC36 cells (Fig. 5B). Treatment with T/B increased AP-1 reporter activity by 6.4-fold in BAC36ΔvF cells but only 3.0-fold in BAC36 cells (Fig. 5B). Thus, the RTA promoter reporter activity positively correlated with that of the AP-1 reporter and negatively correlated with that of the NF-κB reporter in both uninduced and induced cells (Fig. 4C and 5B).
FIG. 5.
KSHV vFLIP suppressed the AP-1 pathway and RTA promoter activity by activating the NF-κB pathway. (A and B) Relative activities of the NF-κB (A) and AP-1 (B) reporters in BAC36 and BAC36ΔvF cells with and without induction with T/B. (C to E) Relative reporter activities of the NF-κB (C), AP-1 (D), and RTA promoter (E) reporters in 293T cells cotransfected with the control plasmid pCMV-myc (Vector), pCMV-vFLIP (vFLIP), and pCMV-vFLIP-58AAA (vFLIP-58AAA) alone or together with the NF-κB DN construct, pIκB-αM. +, presence; −, absence.
To further determine the role of the NF-κB pathway in the regulation of the AP-1 pathway and RTA promoter activity, we carried out transient reporter assays. As shown in Fig. 5C, cotransfection of a NF-κB reporter with the vFLIP expression construct activated the reporter by 31-fold, which was abolished by a DN construct of NF-κB, pIκB-αM. In fact, cells cotransfected with the NF-κB DN had an even lower level of luciferase activity than cells cotransfected with the control vector pCMV-myc, suggesting that the DN construct might have knocked down some endogenous NF-κB activity. As previously reported, cotransfection of a NF-κB reporter with a vFLIP mutant expression construct, vFLIP-58AAA, activated the reporter by only threefold (Fig. 5C). Treatment with T/B increased the NF-κB reporter activity by 3.1-, 1.9-, and 2.3-fold in cells cotransfected with vector, vFLIP, and vFLIP-58AAA, respectively, while no induction was observed in cells cotransfected with vFLIP and NF-κB DN (Fig. 5C).
In contrast to results for the NF-κB reporter, cotransfection of an AP-1 reporter with the vFLIP expression construct resulted in a 67% reduction of the reporter activity. Cotransfection with the NF-κB DN construct not only fully restored but also slightly increased the AP-1 activity by 1.3-fold (Fig. 5D). Treatment with T/B increased the AP-1 reporter activity by 2.2- and 1.9-fold in cells cotransfected with vector and vFLIP, respectively, while cotransfection of vFLIP with NF-κB DN further increased the reporter activity by 3.4-fold. vFLIP-58AAA slightly inhibited the AP-1 reporter activity, which reflected its weak activation of the NF-κB reporter. These results suggest that vFLIP-mediated NF-κB activation could negatively regulate the AP-1 pathway. In agreement with this notion, the increased AP-1 activity in cells cotransfected with the NF-κB DN construct could be due to its inhibitory effects on both vFLIP-induced and endogenous NF-κB activities. As expected, the RTA promoter reporter activity measured in parallel in the same set of experiments displayed a pattern similar to that of the AP-1 reporter (Fig. 5E). Specifically, cotransfection with the vFLIP expression construct significantly reduced the RTA promoter activity with or without induction with T/B, while cotransfection with the NF-κB DN construct completely abolished the vFLIP inhibitory effect, resulting in an even higher level of reporter activity than that in cells transfected with the control plasmid. As expected, vFLIP-58AAA slightly inhibited RTA promoter activity (Fig. 5E).
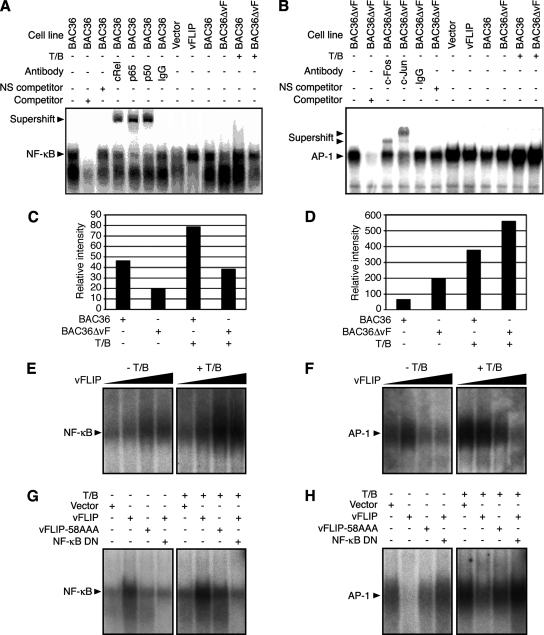
Taken together, these results indicate that vFLIP negatively regulates the AP-1 pathway by activating the NF-κB pathway, which correlates with the inhibition of RTA promoter activity. To further confirm these observations, we carried out EMSAs, using specific probes for the NF-κB and AP-1 pathways. As shown in Fig. 6A, the NF-κB probe formed a strong DNA-protein complex with nuclear extracts from BAC36 cells, which was abolished by a cold probe (specific competitor) but not by nonspecific oligonucleotides. This band was also supershifted to higher molecular weights by antibodies to p65, p50, and cRel, components of the NF-κB complex, but not by a control antibody, indicating that this complex was indeed the p65/p50/cRel NF-κB complex. As expected, BAC36ΔvF cells had a weaker NF-κB band than BAC36 cells (Fig. 6A and C). Upon induction with T/B, the intensity of the NF-κB band was increased in both cell lines, but it remained weaker in BAC36ΔvF cells than in BAC36 cells (Fig. 6A and C). Thus, in the context of the whole KSHV genome, vFLIP contributed significantly to the activation of the NF-κB pathway.
FIG. 6.
Examination of KSHV vFLIP modulation of NF-κB and AP-1 DNA-protein complexes by EMSA. (A) A nuclear extract from BAC36 cells had a strong NF-κB DNA-protein complex band, which was abolished by competition with the cold probe (competitor) but not with an irrelevant oligonucleotide (NS competitor) and was supershifted by an antibody to cRel, p65, or p50 but not a control antibody. BAC36 cells had a stronger NF-κB band than BAC36ΔvF cells with and without induction with T/B. Cells transiently transfected with the vFLIP expression construct pCMV-vFLIP also had a stronger NF-κB band than cells transfected with the vector pCMV-myc. (B) A nuclear extract from the BAC36ΔvF cells had a strong AP-1 DNA-protein complex band, which was abolished by competition with the cold probe but not with an irrelevant oligonucleotide and was supershifted by an antibody to c-Fos or c-Jun but not a control antibody. The BAC36ΔvF cells had a stronger AP-1 band than the BAC36 cells with and without induction with T/B. Cells transiently transfected with pCMV-vFLIP also had a weaker AP-1 band than cells transfected with the vector control, pCMV-myc. (C and D) Quantification of NF-κB (C) and AP-1 (D) band intensities in BAC36 and BAC36ΔvF cells with and without T/B induction. (E and F) Positive dose-response of NF-κB band (E) and negative dose-response of AP-1 band (F) in 293T cells transfected with pCMV-vFLIP with and without T/B induction. (G and H) NF-κB (G) and AP-1 (H) DNA-protein complexes in 293T cells transfected with the control plasmid pCMV-myc (Vector), vFLIP mutant (vFLIP-58AAA), and pCMV-vFLIP (vFLIP) alone or together with the NF-κB DN with and without T/B induction. NS competitor, an irrelevant oligonucleotide; +, presence; −, absence.
Next, we carried out EMSAs to examine the regulation of the DNA-AP-1 complex by vFLIP. While a DNA-protein complex was detected in BAC36 cells by using a specific AP-1 probe, the intensity of this complex was threefold stronger in BAC36ΔvF cells (Fig. 6B and D). This complex was abolished by a cold probe (specific competitor) but not by nonspecific oligonucleotides and was supershifted to higher molecular weights by antibodies to c-Fos and c-Jun, components of the AP-1 complex, but not by a control antibody, indicating that this complex was indeed the c-Fos/c-Jun AP-1 complex. As expected, treatment with T/B increased the intensity of the AP-1 complex in both BAC36 and BAC36ΔvF cells, with the latter having a stronger AP-1 band (Fig. 6B and D). Furthermore, overexpression of vFLIP reduced the intensity of the AP-1 band while increasing the intensity of the NF-κB band in a dose-dependent fashion (Fig. 6E and F), thus supporting a role for the NF-κB pathway in vFLIP regulation of AP-1 activity.
To further determine the role of the NF-κB pathway in vFLIP-mediated suppression of the AP-1 pathway, we employed the vFLIP-58AAA mutant and NF-κB DN in the EMSAs. While overexpression of the NF-κB DN inhibited vFLIP activation of the NF-κB complex as expected, it also restored the AP-1 band abolished by vFLIP (Fig. 6G and H). vFLIP-58AAA only slightly reduced the intensity of the AP-1 band, which was consistent with its weak activation of the NF-κB complex (Fig. 6G and H). Treatment with T/B increased the intensity of the NF-κB band in vFLIP-, vFLIP-58AAA-, and vector-transfected cells but not in cells cotransfected with both vFLIP and NF-κB DN, suggesting that vFLIP and T/B could synergize with each other to activate the NF-κB complex and the NF-κB DN could inhibit the NF-κB complex activated by both vFLIP- and T/B (Fig. 6G and H). Interestingly, treatment with T/B also increased the intensity of the AP-1 band in vFLIP-, vFLIP-58AAA-, and vector-transfected cells, as well as in cells cotransfected with both vFLIP and NF-κB DN, suggesting that vFLIP could not totally inhibit the AP-1 complex activated by T/B (Fig. 6G and H). Together, these results indicate that vFLIP suppression of the AP-1 pathway is mediated by the NF-κB pathway.
To further define the mechanism by which vFLIP might regulate KSHV replication, we examined the roles of the NF-κB and AP-1 pathways in vFLIP-mediated suppression of RTA expression. Our previous studies have shown that the upstream −259 promoter sequence contributes the most to RTA promoter activity and identified the −81 AP-1-binding site as the essential site for the activation of RTA promoter activity (42, 59). In addition to the AP-1 site, a putative NF-κB binding site is present in this region (Fig. 7A). Using the RTA promoter reporter R-259 as a template, we mutated the putative NK-κB site by in vitro mutagenesis and generated a mutant reporter, R-259mut-NF-κB (Fig. 7A). A mutant reporter with the key −81 AP-1-binding site mutated, R-25mut-AP-1, was previously described (59). As expected, mutation of the −81 AP-1 site within the RTA promoter drastically reduced the reporter activity (Fig. 7B). Consistent with these results, cotransfection with DN of c-Fos or c-Jun alone or in combination significantly reduced R-259 reporter activity in both 293T and BCBL1 cells (data not shown). In contrast to that for the −81 AP-1 site, mutation of the −148 putative NF-κB binding site had little effect on reporter activity, indicating that the NF-κB pathway had no direct effect on RTA promoter activity. In agreement with these results, cotransfection of either the wild-type R-259 or the mutant R-259mut-NF-κB reporter with the vFLIP expression construct reduced 70% and 70.5%, respectively, of the promoter activity (Fig. 7B). In contrast, cotransfection of the mutant R-259mut-AP-1 reporter with the vFLIP expression construct had little effect on promoter activity. Consistent with the reporter assays, the −148 putative NF-κB binding site failed to compete with the consensus NF-κB probe and did not form any NF-κB DNA-protein complex in EMSAs (Fig. 7C). In contrast, the −81 AP-1 site efficiently competed with the consensus AP-1 probe. The −81 AP-1 site probe, but not its mutant, also formed a strong AP-1 DNA-protein complex (Fig. 7D). These results indicate that vFLIP inhibition of the RTA promoter activity is mediated through the AP-1 site but not the putative NF-κB site. Taken together, these results indicate that vFLIP inhibits RTA expression and KSHV lytic replication by suppressing the AP-1 pathway.
DISCUSSION
Like other herpesviruses, KSHV establishes latent infections in immunocompetent hosts following primary infections. Latent infection allows KSHV to evade the host immune system as a consequence of the restricted expression of viral genes. Thus, latent infection is also a default phase of the KSHV life cycle that ensures persistent viral infection. Nevertheless, the mechanism by which KSHV controls the expression of viral lytic genes during latency is still not fully understood. We postulated that the three viral latent genes and the KSHV-encoded microRNAs in the viral latent locus could regulate the expression of viral lytic genes and lytic replication and therefore contribute to the control of latency. Indeed, genetic disruption of LANA abolished KSHV latent infection (60). In addition to regulating KSHV episome persistence by participating in episome replication and controlling the proper segregation of the episome into daughter cells during cell division (5), LANA suppresses the expression of KSHV lytic genes and lytic replication by inhibiting the expression and function of RTA and mediating epigenetic modifications of KSHV genomes (29-31, 33, 34, 48). Consequently, genetic disruption of LANA also led to the enhanced expression of KSHV lytic genes and the lytic replication program (our unpublished data). Nevertheless, the roles of other viral latent products in KSHV latency remain unknown. In this study, we showed that overexpression of the vFLIP gene in PEL cells inhibited the expression of RTA and other viral lytic genes, as well as the production of virions. Consistent with these results, genetic deletion of the vFLIP gene enhanced the expression of RTA and viral lytic replication. Furthermore, we demonstrated that vFLIP reduced the expression of RTA by suppressing its promoter activity. To our knowledge, this is the first time that vFLIP has been shown to promote viral latency by regulating the expression of KSHV lytic genes and lytic replication. Several gammaherpesviruses also possess the vFLIP gene. It would be interesting to further investigate whether this function of the vFLIP gene is conserved among other gammaherpesviruses. Furthermore, our results indicate that KSHV uses multiple mechanisms to control lytic replication and latency. It remains to be determined whether other latent viral products, including vCyclin and microRNAs, could regulate KSHV latency.
vFLIP protein has been found to be involved mainly in cell survival. The knockdown of vFLIP protein in PEL cells by small interfering RNA induced apoptosis, indicating an essential role of the vFLIP gene in lymphomagenesis (25, 53). Nevertheless, whether the vFLIP gene also plays an important role in KS tumor development should be examined in appropriate models, such as the recently described KSHV-induced KS tumor model (41). Indeed, recent studies have demonstrated the ability of vFLIP to induce inflammatory cytokines such as IL-6 and IL-8 (2, 23, 54) and the spindle phenotype in endothelial cells (23), which are features of KS tumors. Results from this study indicate that vFLIP might contribute to tumor development by promoting persistent latent infection with KSHV, which is generally necessary for the induction of tumorigenesis by gammaherpesviruses (1).
We found that vFLIP inhibited KSHV RTA expression by suppressing the AP-1 pathway via activation of the NF-κB pathway. vFLIP activation of the NF-κB pathway has been well documented (11, 36), and our results support the previously published data. In fact, results from our vFLIP genetic knockout experiments indicate that vFLIP is the dominant viral factor that contributes to KSHV activation of the NF-κB pathway (Fig. 6A). Our conclusion that vFLIP suppressed the AP-1 pathway was derived from the results of reporter assays and EMSAs with vFLIP expressed either alone or in the context of a virus infection (Fig. 5 and 6). Intriguingly, our results for vFLIP suppression of the AP-1 pathway contradicted those of a previous study showing that a vFLIP-GFP fusion protein actually activated the JNK/AP-1 pathway (2). We speculate that these discrepancies could be attributed to the use of different cell lines in the experiments. Alternatively, the GFP-vFLIP fusion protein used in the previous study might behave differently from the vFLIP expression construct used in this study. Fusion of a protein with GFP has been shown to have an adverse effect on the function of the protein (4). Nevertheless, our results are supported by two recently published works (37, 54). In one study, it was demonstrated that vFLIP activated the NF-κB pathway but failed to activate the JNK/AP-1 pathway in two different cell lines, K562 and 293T (54). In another recent study, it was demonstrated that vFLIP bypassed the TNF receptor-associated factor family of proteins and directly interacted with the IκB kinase complex to selectively activate the NF-κB pathway without affecting the JNK pathway (37). In order to resolve these controversies, further studies with different cell lines are warranted. In addition, it would be advantageous to examine the function of vFLIP in its native form during KSHV infection. In this context, the three genetically modified KSHV genomes described in this study would be valuable.
Our results confirm the findings of a previous study that demonstrated the suppression of KSHV lytic replication by the NF-κB pathway (6). Similar results have also been observed with murine gammaherpesvirus type 68 and Epstein-Barr virus (6, 28). Thus, the NF-κB pathway appears to have a general function in regulating the viral latency of gammaherpesviruses. Distinct from previous reports, our results indicate that the negative regulation of viral lytic replication by the NF-κB pathway is indirect, through its modulation of the AP-1 pathway. This is true at least for the RTA promoter, since mutations in the NF-κB binding site did not change its reporter activity, while mutations in the AP-1 site significantly reduced its activity (Fig. 7). Thus, we concluded that the NF-κB pathway did not directly suppress the RTA promoter. Rather, it did so by suppressing the AP-1 pathway. Since the expression of several other KSHV lytic genes such as ORF57, ORF-K8, and the origin of lytic replication (oriLyt) also depend on AP-1 activation (3, 57), it can be postulated that the NF-κB pathway might also negatively regulate the expression of other viral lytic genes to affect viral lytic replication. Consequently, we inferred that vFLIP might inhibit the expression of other viral lytic genes by regulating either the AP-1 pathway or the expression of RTA. Indeed, results from a reporter assay indicated that vFLIP inhibited the reporter activity of a KSHV oriLyt reporter construct (data not shown) that is known to depend on AP-1 activation (3). Similarly, while deletion of vFLIP enhanced the expression of RTA and other viral lytic genes, the expression patterns of some viral lytic genes were different from that of RTA (Fig. 3A). Although these distinct patterns could be due to the different expression kinetics of individual viral genes, they could also be attributed to their differential activation by RTA and AP-1. As a result, vFLIP is likely to target multiple KSHV lytic and oriLyt genes to inhibit viral lytic replication.
The finding that vFLIP activation of the NF-κB pathway led to the suppression of the AP-1 pathway is intriguing. Both NF-κB and AP-1 are complex pathways that are involved in many different biological processes. Convergence of the NF-κB pathway with the AP-1 upstream mitogen-activated protein kinase pathways has been observed in other biological systems, with both mutual promotion and inhibition existing (47, 56). The NF-κB pathway is constitutively activated in cells latently infected with KSHV because of the expression of vFLIP protein in these cells. We have previously shown that mitogen-activated protein kinase pathways are also constitutively activated in cells latently infected with KSHV, and the extracellular signal-regulated kinase and p38 pathways can be transiently enhanced further following induction with TPA (59). It would be of interest to further dissect the molecular mechanism by which vFLIP suppresses the AP-1 pathway, which could certainly provide some insights into the regulation of KSHV latency and lytic replication.
Whether KSHV goes into latency or lytic replication is a complex issue involving multiple intracellular and extracellular factors. Our finding that the NF-κB pathway regulates KSHV lytic replication by inhibiting the AP-1 pathway could provide a mechanistic explanation for some of the contradictory observations regarding the role of the NF-κB pathway in KSHV lytic replication (6, 50). If the NF-κB pathway controls the activation of the AP-1 pathway, it could be speculated that the pathway would tilt KSHV into latency. However, the AP-1 pathway is often activated by many different viral and cellular genes or extracellular inflammatory cytokines (58). It is likely that in some cases, the NF-κB pathway fails to control the activation of the AP-1 pathway, which would result in the expression of RTA and activation of KSHV lytic replication. This could occur even when the NF-κB pathway was strongly activated, such as in the case of induction with T/B. Thus, the role of the NF-κB pathway in KSHV replication might be strictly context dependent; i.e., its role may depend on its ability to control the AP-1 pathway. Such interplay of the two pathways in KSHV replication is perhaps best illustrated during primary infection with KSHV. While some studies have shown that the default KSHV replication pathway following primary infection in human foreskin fibroblasts and human dermal microvascular endothelial cells is latency (27), other studies have demonstrated active viral lytic replication at the early stage of KSHV primary infection (13, 16, 19), which correlates with the activation of the AP-1 pathway (42, 58). It would be interesting to examine the relationship of KSHV replication programs with the status of the AP-1 and NF-κB pathways during KSHV primary infection in different cell types.
A number of cellular factors, including RBP-JK, EBPα, Oct-1, CRE, and Sp1, have been shown to regulate RTA expression and KSHV lytic replication (52). Most of these factors have been examined alone without taking other factors into consideration. Analysis of the RTA promoter has identified AP-1 as a key factor that is necessary and sufficient for activating the expression of RTA and KSHV lytic replication (42, 57, 59). It would be interesting to determine how other factors coordinate with AP-1 in this process.
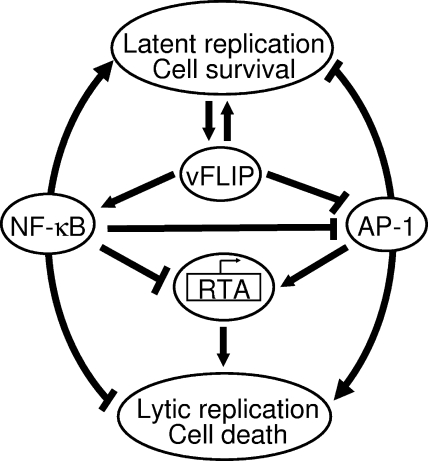
Taken together, the results from this study demonstrate that NF-κB and AP-1 are two central pathways involved in the regulation of the KSHV life cycle and that these two pathways have opposing effects on viral latency and lytic replication (Fig. 8). By regulating both the NF-κB and AP-1 pathways, vFLIP contributes to the control of KSHV latency.
FIG. 8.
vFLIP regulation of KSHV latent and lytic replication mediated by the NF-κB pathway. Activation of the NF-κB pathway promotes the survival of latent cells but suppresses RTA expression and viral lytic replication by inhibiting the AP-1 pathway.
Acknowledgments
This work was supported by an American Cancer Society research scholar grant (RSG-04-195) and grants from the National Institutes of Health (CA096512, CA124332, and DE017333) to S.-J.G.
We thank Charles Wood of the University of Nebraska, Lincoln, for providing the RTA antibody, Bala Chandran of Rosalind Franklin University of Medicine and Science for providing the ORF59 antibody, and Mary Collins of Windeyer Institute of Medical Sciences, London, for the vFLIP antibody. We are also grateful to Preet Chaudhary of the University of Pittsburgh for the vFLIP retrovirus construct, to Bill Sudgen of the University of Wisconsin—Madison and Kenneth Izumi of the University of Texas Health Science Center at San Antonio for the NF-κB reporter construct, and to Paul J. Chiao of the University of Texas M.D. Anderson Cancer Center for the NF-κB DN plasmid pIκB-αM. We thank Kenneth Izumi and Anthony Griffiths for their valuable suggestions for this work and appreciate the technical assistance of members of the Gao laboratory.
Footnotes
Published ahead of print on 27 February 2008.
REFERENCES
- 1.Ackermann, M. 2006. Pathogenesis of gammaherpesvirus infections. Vet. Microbiol. 113211-222. [DOI] [PubMed] [Google Scholar]
- 2.An, J., Y. Sun, R. Sun, and M. B. Rettig. 2003. Kaposi's sarcoma-associated herpesvirus encoded vFLIP induces cellular IL-6 expression: the role of the NF-κB and JNK/AP1 pathways. Oncogene 223371-3385. [DOI] [PubMed] [Google Scholar]
- 3.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318542-555. [DOI] [PubMed] [Google Scholar]
- 4.Baens, M., H. Noels, V. Broeckx, S. Hagens, S. Fevery, A. D. Billiau, H. Vankelecom, and P. Marynen. 2006. The dark side of EGFP: defective polyubiquitination. PLoS One 1e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284641-644. [DOI] [PubMed] [Google Scholar]
- 6.Brown, H. J., M. J. Song, H. Deng, T. T. Wu, G. Cheng, and R. Sun. 2003. NF-κB inhibits gammaherpesvirus lytic replication. J. Virol. 778532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 1025570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 3321186-1191. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 2661865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Chang, Y., P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, K. Godden, H. Paterson, R. A. Weiss, and S. Mittnacht. 1996. Cyclin encoded by KS herpesvirus. Nature 382410. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary, P. M., A. Jasmin, M. T. Eby, and L. Hood. 1999. Modulation of the NF-κ B pathway by virally encoded death effector domains-containing proteins. Oncogene 185738-5746. [DOI] [PubMed] [Google Scholar]
- 12.Deng, J. H., Y. J. Zhang, X. P. Wang, and S. J. Gao. 2004. Lytic replication-defective Kaposi's sarcoma-associated herpesvirus: potential role in infection and malignant transformation. J. Virol. 7811108-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dezube, B. J., M. Zambela, D. R. Sage, J. F. Wang, and J. D. Fingeroth. 2002. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 infection of human vascular endothelial cells: early events. Blood 100888-896. [DOI] [PubMed] [Google Scholar]
- 14.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 728309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foglieni, C., S. Scabini, D. Belloni, F. Broccolo, P. Lusso, M. S. Malnati, and E. Ferrero. 2005. Productive infection of HUVEC by HHV-8 is associated with changes compatible with angiogenic transformations. Eur. J. Histochem. 49273-284. [DOI] [PubMed] [Google Scholar]
- 17.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402889-894. [DOI] [PubMed] [Google Scholar]
- 18.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of β-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9300-306. [DOI] [PubMed] [Google Scholar]
- 19.Gao, S. J., J. H. Deng, and F. C. Zhou. 2003. Productive lytic replication of a recombinant Kaposi's sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J. Virol. 779738-9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, S. J., L. Kingsley, D. R. Hoover, T. J. Spira, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, P. Parry, Y. Chang, and P. S. Moore. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 335233-241. [DOI] [PubMed] [Google Scholar]
- 21.Godden-Kent, D., S. J. Talbot, C. Boshoff, Y. Chang, P. Moore, R. A. Weiss, and S. Mittnacht. 1997. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J. Virol. 714193-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greene, W., K. Kuhne, F. Ye, J. Chen, F. Zhou, X. Lei, and S. J. Gao. 2007. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat. Res. 13369-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossmann, C., S. Podgrabinska, M. Skobe, and D. Ganem. 2006. Activation of NF-κB by the latent vFLIP gene of Kaposi's sarcoma-associated herpesvirus is required for the spindle shape of virus-infected endothelial cells and contributes to their proinflammatory phenotype. J. Virol. 807179-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guasparri, I., S. A. Keller, and E. Cesarman. 2004. KSHV vFLIP is essential for the survival of infected lymphoma cells. J. Exp. Med. 199993-1003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 119-29. [PubMed] [Google Scholar]
- 27.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 783601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krug, L. T., J. M. Moser, S. M. Dickerson, and S. H. Speck. 2007. Inhibition of NF-κB activation in vivo impairs establishment of gammaherpesvirus latency. PLoS Pathog. 3e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lan, K., D. A. Kuppers, and E. S. Robertson. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jκ, the major downstream effector of the Notch signaling pathway. J. Virol. 793468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 786585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lan, K., D. A. Kuppers, S. C. Verma, N. Sharma, M. Murakami, and E. S. Robertson. 2005. Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J. Virol. 797453-7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, M., H. Lee, D. W. Yoon, J. C. Albrecht, B. Fleckenstein, F. Neipel, and J. U. Jung. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J. Virol. 711984-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, F., L. Day, S. J. Gao, and P. M. Lieberman. 2006. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi's sarcoma-associated herpesvirus lytic transcription. J. Virol. 805273-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, F., J. Zhou, A. Wiedmer, K. Madden, Y. Yuan, and P. M. Lieberman. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 7711425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252304-312. [DOI] [PubMed] [Google Scholar]
- 36.Matta, H., and P. M. Chaudhary. 2004. Activation of alternative NF-κ B pathway by human herpes virus 8-encoded Fas-associated death domain-like IL-1β-converting enzyme inhibitory protein (vFLIP). Proc. Natl. Acad. Sci. USA 1019399-9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matta, H., L. Mazzacurati, S. Schamus, T. Yang, Q. Sun, and P. M. Chaudhary. 2007. Kaposi's sarcoma-associated herpesvirus (KSHV) oncoprotein K13 bypasses TRAFs and directly interacts with the IκB kinase complex to selectively activate NF-κB without JNK activation. J. Biol. Chem. 28224858-24865. [DOI] [PubMed] [Google Scholar]
- 38.McAllister, S. C., and A. V. Moses. 2007. Endothelial cell- and lymphocyte-based in vitro systems for understanding KSHV biology. Curr. Top. Microbiol. Immunol. 312211-244. [DOI] [PubMed] [Google Scholar]
- 39.Means, R. E., J. K. Choi, H. Nakamura, Y. H. Chung, S. Ishido, and J. U. Jung. 2002. Immune evasion strategies of Kaposi's sarcoma-associated herpesvirus. Curr. Top. Microbiol. Immunol. 269187-201. [DOI] [PubMed] [Google Scholar]
- 40.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 3321181-1185. [DOI] [PubMed] [Google Scholar]
- 41.Mutlu, A. D., L. E. Cavallin, L. Vincent, C. Chiozzini, P. Eroles, E. M. Duran, Z. Asgari, A. T. Hooper, K. M. La Perle, C. Hilsher, S. J. Gao, D. P. Dittmer, S. Rafii, and E. A. Mesri. 2007. In vivo-restricted and reversible malignancy induced by human herpesvirus-8 KSHV: a cell and animal model of virally induced Kaposi's sarcoma. Cancer Cell 11245-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan, H., J. Xie, F. Ye, and S. J. Gao. 2006. Modulation of Kaposi's sarcoma-associated herpesvirus infection and replication by MEK/ERK, JNK, and p38 multiple mitogen-activated protein kinase pathways during primary infection. J. Virol. 805371-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearce, M., S. Matsumura, and A. C. Wilson. 2005. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter. J. Virol. 7914457-14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2269-276. [DOI] [PubMed] [Google Scholar]
- 45.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 61121-1127. [DOI] [PubMed] [Google Scholar]
- 46.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 715915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saha, R. N., M. Jana, and K. Pahan. 2007. MAPK p38 regulates transcriptional activity of NF-κB in primary human astrocytes via acetylation of p65. J. Immunol. 1797101-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakakibara, S., K. Ueda, K. Nishimura, E. Do, E. Ohsaki, T. Okuno, and K. Yamanishi. 2004. Accumulation of heterochromatin components on the terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus mediated by the latency-associated nuclear antigen. J. Virol. 787299-7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 799301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sgarbanti, M., M. Arguello, B. R. tenOever, A. Battistini, R. Lin, and J. Hiscott. 2004. A requirement for NF-κB induction in the production of replication-competent HHV-8 virions. Oncogene 235770-5780. [DOI] [PubMed] [Google Scholar]
- 51.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J.-P. Clauvel, M. Raphael, L. Degos, and F. Sigaux. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 861276-1280. [PubMed] [Google Scholar]
- 52.Staudt, M. R., and D. P. Dittmer. 2007. The Rta/Orf50 transactivator proteins of the gamma-herpesviridae. Curr. Top. Microbiol. Immunol. 31271-100. [DOI] [PubMed] [Google Scholar]
- 53.Sun, Q., H. Matta, and P. M. Chaudhary. 2003. The human herpes virus 8-encoded viral FLICE inhibitory protein protects against growth factor withdrawal-induced apoptosis via NF-κ B activation. Blood 1011956-1961. [DOI] [PubMed] [Google Scholar]
- 54.Sun, Q., H. Matta, G. Lu, and P. M. Chaudhary. 2006. Induction of IL-8 expression by human herpesvirus 8 encoded vFLIP K13 via NF-κB activation. Oncogene 252717-2726. [DOI] [PubMed] [Google Scholar]
- 55.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 9510866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang, G., Y. Minemoto, B. Dibling, N. H. Purcell, Z. Li, M. Karin, and A. Lin. 2001. Inhibition of JNK activation through NF-κB target genes. Nature 414313-317. [DOI] [PubMed] [Google Scholar]
- 57.Wang, S. E., F. Y. Wu, H. Chen, M. Shamay, Q. Zheng, and G. S. Hayward. 2004. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 784248-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie, J., H. Pan, S. Yoo, and S. J. Gao. 2005. Kaposi's sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways. J. Virol. 7915027-15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie, J. P., A. O. Ajibade, F. C. Ye, K. Kuhne, and S. J. Gao. 2008. Reactivation of Kaposi's sarcoma-associated herpesvirus from latency requires MEK/ERK, JNK and p38 multiple mitogen-activated protein kinase pathways. Virology 371139-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye, F. C., F. C. Zhou, S. M. Yoo, J. P. Xie, P. J. Browning, and S. J. Gao. 2004. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 7811121-11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoo, S. M., F. C. Zhou, F. C. Ye, H. Y. Pan, and S. J. Gao. 2005. Early and sustained expression of latent and host modulating genes in coordinated transcriptional program of KSHV productive primary infection of human primary endothelial cells. Virology 34347-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 936641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, F. C., Y. J. Zhang, J. H. Deng, X. P. Wang, H. Y. Pan, E. Hettler, and S. J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 766185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]