Abstract
The adenovirus (Ad) early transcription unit 3 (E3) encodes multiple immunosubversive functions that are presumed to facilitate the establishment and persistence of infection. Indeed, the capacity of E3/19K to inhibit transport of HLA class I (HLA-I) to the cell surface, thereby preventing peptide presentation to CD8+ T cells, has long been recognized as a paradigm for viral immune evasion. However, HLA-I downregulation has the potential to render Ad-infected cells vulnerable to natural killer (NK) cell recognition. Furthermore, expression of the immediate-early Ad gene E1A is associated with efficient induction of ligands for the key NK cell-activating receptor NKG2D. Here we show that while infection with wild-type Ad enhances synthesis of the NKG2D ligands, major histocompatibility complex class I chain-related proteins A and B (MICA and MICB), their expression on the cell surface is actively suppressed. Both MICA and MICB are retained within the endoplasmic reticulum as immature endoglycosidase H-sensitive forms. By analyzing a range of cell lines and viruses carrying mutated versions of the E3 gene region, E3/19K was identified as the gene responsible for this activity. The structural requirements within E3/19K necessary to sequester MICA/B and HLA-I are similar. In functional assays, deletion of E3/19K rendered Ad-infected cells more sensitive to NK cell recognition. We report the first NK evasion function in the Adenoviridae and describe a novel function for E3/19K. Thus, E3/19K has a dual function: inhibition of T-cell recognition and NK cell activation.
The Adenoviridae family includes clinically important viruses associated with acute episodes of pharyngitis, conjunctivitis, diarrhea, and cystitis in immunocompetent individuals. In immunosuppressed patients, particularly pediatric transplant recipients, adenoviruses (Ads) are capable of systemic viremic spread that produces significant levels of morbidity and mortality. More than 50 human Ads have been described, of which the common species C serotypes 2 and 5 (HAdV-2 and HAdV-5) have been studied most intensively (52), and demonstrated to encode functions that act in vitro and in vivo to promote evasion of both innate and adaptive host immune responses (11). Immunomodulatory functions are concentrated in the early transcription unit E3 (33, 51). An E3 protein, E3/19K, became an immunological paradigm when it was demonstrated to impede cell surface antigen presentation by retaining the major histocompatibility class I complex (MHC-I) in the endoplasmic reticulum (ER) (1, 8, 10, 17). E3/10.4K and E3/14.5K (the RID complex) together remove Fas, tumor necrosis factor-related apoptosis-inducing ligand receptors 1 and 2, and the epidermal growth factor receptor from the cell surface and promote their subsequent degradation in lysosomes (5, 12, 18, 24, 44, 45, 51). Moreover, E3/14.7K suppresses inflammation and cytolysis by inhibiting tumor necrosis factor alpha-mediated signaling independently of the RID complex (11, 26).
We are interested in the role of NK cells in controlling Ad infections. NK cells are a heterogeneous population of cells expressing a wide range of activating and inhibitory receptors. NKG2D is a homodimeric activating receptor that is ubiquitously expressed on NK cells, γδ T cells, and NKT cells and also on certain αβ T-cell subsets (3). To date, seven human cellular NKG2D ligands (NKG2DLs) have been identified: the MHC-I chain-related A (MICA) and B (MICB) proteins, the UL16 binding proteins ULBP1 to -3, and the retinoic acid early inducible proteins E (ULBP4) and G (2, 3, 16). While MICA and MICB are closely related, sharing 84% amino acid (aa) identity and similar tertiary structures (25, 32), they are also highly polymorphic, with 61 MICA and 30 MICB alleles having been described (IMGT; http://imgt.cines.fr/). The induction of NKG2DLs has been reported to occur in response to stress, including infection, genotoxic stimuli, cell transformation, and heat shock (22, 23). Crucially, HAdV-5 E1A has been demonstrated to upregulate NKG2DLs in both transformed human and rodent cell lines (15, 39), rendering them susceptible to NK cell attack. More recently, we have shown that breakthrough expression from replication-deficient HAdV-5 E1-negative vectors also upregulated NKG2DLs and thereby sensitized vector-transduced cells to NK attack (48). Ad thus encodes E1-dependent and E1-independent functions that act separately to stimulate NKG2DL expression.
The efficient upregulation of NKG2DLs combined with the simultaneous intracellular retention of endogenous MHC-I theoretically should render Ad-infected cells extremely vulnerable to NK cell attack. We therefore sought to investigate the impact of NKG2DL activation during productive Ad infection. Unexpectedly, cell surface expression of MICA and MICB was efficiently suppressed during wild-type (wt) HAdV-5 infection. Ad was shown to encode a function that acts posttranslationally to sequester both MICA and MICB within the ER. This is the first description of an Ad NK cell evasion function and provides novel insights into the role of E3/19K as a potential virulence factor.
MATERIALS AND METHODS
Cells.
Human fetal foreskin fibroblasts (HFFF), HFFF immortalized with human telomerase reverse transcriptase (37) and transduced with a retrovirus expressing the human coxsackievirus Ad receptor (31) (HFFF-hTERT-hCAR), human embryonic retinoblast 911 cells, and human embryonic kidney 293 cells were all grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (Invitrogen). HFFF-hTERT-hCAR were used in all experiments using fibroblasts except those shown in Fig. 1A and are referred to as HF throughout. The 911 MICA-YFP cell line was generated by transfection with a MICA-yellow fluorescent protein (YFP) fusion construct (a kind gift from Daniel Davis, Imperial College, London) (36) and selection with G418 (0.75 mg/ml). The 293 cell lines, E3-45 and E22.7, were established following transfection with the EcoRV C and EcoRI D fragments of HAdV-2, containing all or a part of the E3 region, respectively (30). All E3/19K mutants have been generated in the EcoRI D fragment, which was then transfected into 293 cells together with the neophosphotransferase gene. After selection, 293 transfectants were routinely cultured in Dulbecco modified Eagle medium-10% fetal calf serum containing 200 μg/ml G418. Substituting alanine for the conserved tryptophan in position 96 of HAdV-2 E3/19K created mutant W96A. Transfectants were selected on the basis that they expressed a similar amount of E3/19K as detected by a rabbit antiserum against the cytoplasmic tail of E3/19K (40) or additional monoclonal antibodies (MAbs) (B. Menz and H.-G. Burgert, unpublished). EKE*1 cells express a modified E3/19K molecule with the transmembrane segment replaced by that of MHC-I and the lysines of the ER retrieval signal altered to serines (KKMP→SSMP). This E3/19K mutant is efficiently transported to the cell surface. The cysteine mutants have been described previously (40).
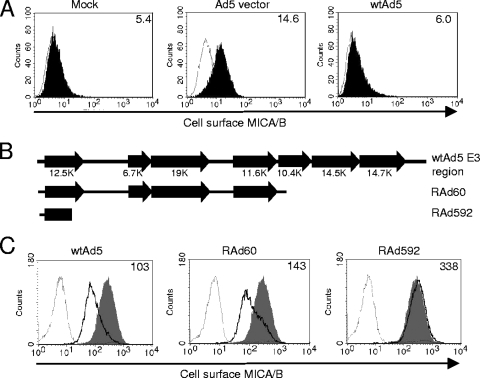
FIG. 1.
Ad carries an E3 gene that downregulates cell surface MICA/B. (A) HFFF infected at a multiplicity of infection of 50 with a replication-deficient (ΔE1,E3) HAdV-5 vector (RAd60) or wt HAdV-5 were stained at 48 h p.i. with an anti-MICA/B antibody or control IgG and analyzed by flow cytometry. The mouse IgG control is shown by the thin line, and the filled histograms represent staining with anti-MICA/B. The mean fluorescence intensity of MICA/B staining of mock-, RAd60-, or HAdV-5-infected cells is depicted in the top right corner of the histogram. (B) E3 genes present in each of the three viruses. (C) 911 cells infected at a multiplicity of infection of 5 with wt HAdV-5, RAd60, or RAd592 were stained at 24 h p.i. with anti-MICA/B antibody or control IgG before analysis by flow cytometry. The mouse IgG control is shown by the thin line, staining of mock-infected cells is in gray fill, and staining of infected cells is represented by the bold line. The mean fluorescence intensity of MICA/B staining of HAdV-5-, RAd60-, or RAd592-infected cells is depicted in the top right corner of the histogram.
MICA genotyping.
Genomic DNA was used to amplify a 2.2-kb fragment of the MICA gene encompassing exons 2, 3, 4, and 5. Sequencing-based typing was performed using primers to sequence exons 2, 3, and 4 in both orientations (29, 55). Sequence analysis and assignment of MICA allele type were performed using AssignSBT (Connexio Genomics, Australia). Ambiguous types were analyzed further if distinguishable by the number of GCT repeats in exon 5 using a reverse sequencing primer to sequence exon 5. The combination of GCT triplets was identified by the heterozygous sequence as described previously (54).
Viruses.
The generation of the Ad-Easy-1-based vector RAd592 (48) and the pJM17-based vector RAd60 (50) has been described previously. Both of these vectors are E1 deleted, and neither expresses a transgene. Ad-Easy-1-based vectors have a deletion in the E3 region spanning bp 28130 to 30820 of the Ad5 genome, eliminating all E3 open reading frames (ORFs) except 12.5K, which is present but incomplete and thus is likely to be nonfunctional. pJM17-based vectors are based on the HAdV-5 dl309 virus, which has a deletion running from bp 30005 to 30750 leading to partial or complete deletion of the 10.4K, 14.5K, and 14.7K ORFs; hence, the protein products of these genes are not expressed (6). wt HAdV-5 was provided by Vivien Mautner, University of Birmingham, United Kingdom. wt Ad5 and the E3/19K mutants were grown and titers were determined on A549 cells. The Ad5-based vectors RAd60 and RAd592 were grown and titers were determined on 293 cells. A549 and 293 exhibit comparable infectivities. Virus-infected cells were recovered by centrifugation and washed with phosphate-buffered saline (PBS), and virus was extracted using Arklone P as described previously (28).
To clone the HAdV-5 genome into a single-copy vector, Ad genomic DNA was isolated from purified virions using the QIAamp blood kit (Qiagen). Purified viral DNA was then cotransformed into induced SW102 bacteria (49) with a modified bacterial artificial chromosome (BAC)-based vector based on pBeloBAC11 (New England Biolabs) in which the left (bp 1 to 480) and right (bp 34951 to 35938) ends of the Ad5 genome had been cloned. The BAC construct was linearized with NheI, which cuts between the arms of homology, to promote homologous recombination. Chloramphenicol-resistant colonies were grown, and BAC DNA was extracted and digested with the appropriate enzymes to confirm the HAdV-5 genome had been successfully cloned. The generated construct, pAL908, contains the wt HAdV-5 genome. pAL908 was used as a parental construct to generate a number of E3/19K mutants using “recombineering.” Recombineering was performed essentially as described previously (49), using a selectable marker cassette that allows both positive and negative selection (sacB lacZ Ampr). To modify the parental genome in the BAC vector, the selectable marker was amplified using PCR primers (sequences are available on request) that contain homology to the appropriate regions of the HAdV-5 genome to target homologous recombination. Following successful recombination, the selectable marker was then removed using single-stranded or double-stranded DNA oligonucleotides containing the appropriate deletion, insertion, or substitution. Prior to transfection, BAC DNA was extracted from a 500-ml overnight culture using the BacMax 100 kit (Macherey-Nagel, Duren, Germany). BAC DNA was digested with PacI, whose recognition sites flank the HAdV-5 inverted terminal repeats in the BAC vector, and then transfected into 911 cells. Infectious virus was isolated, digested, and sequenced to confirm the appropriate genomic structure. The viruses generated contain the following mutations: ΔE3/19K (RAd918) has a 290-bp deletion beginning 56 bp into the E3/19K ORF, E3/19KSTOP (RAd930) has a premature stop codon and frameshift inserted 33 bp into the ORF, and E3/19KW96A (RAd936) has a mutation that converts a TGG(W) codon to a GCT(A) codon 339 bp into the ORF.
Flow cytometry and immunofluorescence.
For flow cytometry analysis, a FACSCalibur or FACscan with CELLQuest Pro software (Becton Dickinson) was used. Staining was performed as previously described (18, 40). Before immunofluorescence, cells were fixed with paraformaldehyde and were permeabilized with 0.5% NP-40. Fluorescence was detected and analyzed with a Leica DMIRBE microscope with Improvision Openlab software. The following mouse MAbs were used: Tw1.3 against E3/19K (17), W6/32 against HLA-I (ATCC HB95), HC10 against HLA-I (42), DX2 against Fas (Calbiochem), MAB3126 against calnexin (Chemicon International), 6D4 against MICA/B, AF1300 against MICA/B (R&D), MAB1300 against MICA (R&D), 1599 against MICB (R&D), 2211S against phospho-S6-ribosomal protein (Cell Signaling), and M58+M73 against Ad E1A (Abcam ab3648). Secondary antibodies included goat anti-mouse immunoglobulin G (IgG) conjugated to AlexaFluor 594 (A-11020), AlexaFluor 488 (A-11001), and AlexaFluor 647 (A-21237), all from Invitrogen.
NK cell CD107a mobilization assays.
A total of 106 alpha interferon (IFN-α)-activated peripheral blood mononuclear cells (PBMC) were incubated for 6 h with 2 × 105 HF (48 h postinfection [p.i.]) and anti-CD107a-FITC MAb (3 μl for 106 PBMC; BD Biosciences), with 6 μl/ml BDGolgiStop (BD Biosciences) present for the last 5 h. PBMC were then harvested, washed in cold PBS, and stained for 30 min at 4°C with anti-CD3-PerCP (clone SK7; BD Biosciences) and anti-CD56-allophycocyanin (clone N901; Beckman Coulter) MAbs. Cells were washed twice in cold PBS, and a minimum of 4 × 103 CD3− CD56+ NK cells were acquired from each sample.
Protein analysis.
Protein extracts were separated on 10% bis-Tris gels (Invitrogen) before Western blotting and incubation with an anti-MICA/B polyclonal antibody (R&D, AF1300) and donkey anti-goat IgG-horseradish peroxidase (Santa Cruz, Sc-2056). Glycosylation was analyzed by digestion with endoglycosidase (EndoH) or peptide-N-glycosidase F (PNGase F) according to the manufacturer's instructions (New England Biolabs).
RESULTS
Identification of an NK cell evasion function in the HAdV-5 E3 region.
The effect of Ad infection on NKG2DL expression was investigated first in primary HFFF. Consistent with previous observations (48), delivery of a replication-deficient HAdV-5 ΔE1,E3 vector clearly induced MICA/B surface expression (Fig. 1A). However, HFFF infected in parallel with wt HAdV-5 did not exhibit an increase in surface MICA/B (Fig. 1A). This finding indicated that HAdV-5 may encode a function to control MICA/B during productive infection. Expression of viral genes from replication-deficient vectors in HFFF is restricted by the E1 deletion. To complement this genetic defect in replication-deficient vectors, the E1-positive 911 helper cell line was infected with two such vectors: RAd592 effectively lacks all E3 functions, whereas RAd60 has a partial deletion (Fig. 1B). 911 cells constitutively express high levels of surface MICA/B, which was efficiently downregulated following infection with both RAd60 and wt HAdV-5 but not with RAd592 (Fig. 1C). Since the function responsible for suppressing MICA/B was retained in RAd60 yet was absent from RAd592, it was provisionally mapped to E3 and more specifically to the 12.5K, 6.7K, 19K, and/or 11.6K ORF (Fig. 1B).
Downregulation of MICA/B is mediated by E3/19K.
A panel of 293 cell lines containing the complete HAdV-2 E3 region and targeted deletions therein was exploited to identify the function responsible for suppression of MICA/B surface expression (Fig. 2A). Since HLA-I and Fas are recognized targets of E3/19K and E3/10.4-14.5K (RID), respectively, their expression serves as a measure of E3 function and was determined alongside that of MICA/B. As expected, Fas surface expression was downregulated only in E3-45 cells, as only these cells express an intact RID (10.4K and 14.5K) complex (18). Cell surface MICA/B and HLA-I were efficiently downregulated in both the E3-45 and E22.7 cells; both cell clones express unmodified versions of E3/19K. HAdV-2 E3/19K is a glycoprotein comprising an N-terminal ∼105-aa ER-luminal domain, a ∼22-aa hydrophobic transmembrane sequence, and a 15-aa cytoplasmic tail (Fig. 2B). E3/19K has two functional entities: the luminal part containing two disulfide bonds essential for binding HLA (40) and a C-terminal ER retrieval signal mediating retrograde transport of E3/19K and associated HLA from the cis-Golgi apparatus to the ER (27). In the EKE*1 cell line, the C-terminal ER retrieval signal has been mutated and the substitutions made in the transmembrane domain so that the modified E3/19K escapes to the cell surface (51). Neither HLA-I nor MICA/B was efficiently downregulated in the EKE*1 cells. Thus, we conclude that E3/19K is responsible for suppressing cell surface expression of MICA/B.
FIG. 2.
The Ad E3/19K glycoprotein inhibits surface expression of MICA/B. (A) Composition of the E3 region in the E3-45 cells, E22.7 cells, and EKE*1, E2-C83S, and E2-W96A transfectants. (B) Schematic representation of the E3/19K structure, with the conserved cysteines numbered and positions of N-linked glycosylation sites indicated with filled circles. E3/19K expressed by EKE*1 has a mutated cytoplasmic tail and a substituted transmembrane region and reaches the cell surface efficiently (data not shown). E2-C83S and E2-W96A have the indicated amino acid substitution in E3/19K. (C) 293 cell lines stably transfected with HAdV-2 E3 region constructs were stained for surface expression of HLA-I (black fill), MICA/B (gray fill), and Fas (unfilled) and analyzed by flow cytometry. The bars denote the relative percentage of expression compared to untransfected 293 cells, whose mean fluorescence intensity was set to 100%. Data were compiled from >5 independent experiments. Differences in cell surface expression of Fas, MICA/B, and HLA in the various E3/19K mutant cell lines were analyzed using the analysis of variance test with Bonferroni's posttest. This showed that the actual level of reduction of MICA/B and HLA in EKE*1 was not significantly different (P > 0.05). However, in E2-W96A cells the reduction of MICA/B cell surface expression was significantly more pronounced (reduction by 58%) than the downregulation of HLA (reduction by 22.1%) (P < 0.001). (D) Expression of HLA (black fill) and MICA/B (gray fill) in stable 293 transfectants expressing cysteine mutants of E3/19K (indicated by the number on the x axis) relative to untransfected cells and cells expressing wt E3/19K. Two clones from each construct were analyzed in at least four independent experiments. The error bars represent standard errors.
MICA and MICB are both cell surface glycoproteins with MHC-I-like α1, α2, and α3 domains, yet neither associates with β2-microglobulin or presents peptide (23). While MICA and MICB share 84% aa identity, they also exhibit 23 to 35% identity with domains in classical MHC-I heavy chains (32). Within the MHC-I heavy chain, the α1 and α2 domains appear to be critical for E3/19K binding (4, 9, 19, 20). To help identify sequence elements within E3/19K responsible for MICA/B suppression, a series of cysteine mutants of E3/19K were tested. The E3/19K luminal domain contains two intramolecular disulfide bonds between the conserved cysteines Cys11-Cys28 and Cys22-Cys83, which are essential for binding MHC-I, whereas the C-proximal cysteines 101, 109, and 122 are dispensable for this function (40) (Fig. 2B). A similar requirement was observed for control of MICA/B; downregulation was compromised when the four conserved N-proximal cysteines (aa 11, 22, 28, and 83) were replaced by serines, whereas mutation of the three C-proximal cysteines (aa 101, 109, and 122) had no significant effect (Fig. 2D). Some differences in the efficiency of downregulation of HLA and MICA/B were observed (e.g., with the Cys101 mutant). Interestingly, a point mutant with alanine substituted for tryptophan at position 96 (W96A) was unable to efficiently downregulate HLA-I surface expression in 293 cell lines yet substantially suppressed cell surface expression of MICA/B (Fig. 2C).
Characterization of HAdV-5 E3/19K mutants.
To evaluate E3/19K function in the context of productive infection, specific mutants were generated. The wt HAdV-5 genome was cloned into a BAC, and the following E3/19K mutants generated using DNA “recombineering”: (i) HAdV-5 ΔE3/19K, incorporating a deletion of aa 2 to 98; (ii) HAdV-5 E3/19KSTOP, with an insertion of a premature termination codon and frameshift after aa 11 in the signal sequence; and (iii) HAdV-5 E3/19KW96A, with an alanine substitution for tryptophan 96 (Fig. 3A). Ablation of E3/19K expression in cells infected with HAdV-5 ΔE3/19K and HAdV-5 E3/19KSTOP was confirmed by Western blotting (Fig. 3B). The point mutation in the E2-W96A cell line, associated with downregulation of MICA/B but not HLA-I (Fig. 2C), was essentially replicated in HAdV-5 E3/19KW96A; this modified protein was expressed in similar abundance by the mutant virus (Fig. 3B).
FIG. 3.
Deletion of E3/19K from Ad5 leads to an upregulation of cell surface MICA/B. (A) The wt HAdV-5 genome was cloned into a BAC, and three E3/19K mutants were generated by recombineering as depicted. (B) Extracts from infected HF were harvested at 24 h p.i. and were probed in a Western blot with anti-E3/19K MAb (Tw1.3) under nonreducing conditions and as a loading control with anti-phospho-S6-ribosomal protein antibody to confirm deletion of the E3/19K gene. (C) HF infected with the indicated viruses at a multiplicity of infection of 3 were stained at 48 h p.i. with anti-MICA/B, anti-MICA, or anti-MICB MAb before analysis by flow cytometry. The mouse IgG control is shown by the thin line, and the filled histograms represent staining with anti-MICA/B, anti-MICA, or anti-MICB. The mean fluorescence intensity of the MAb staining is depicted in the top right corners of the histograms. (D) HF infected at a multiplicity of infection of 3 with the indicated viruses at 48 h p.i. were incubated for 6 h with IFN-α-activated PBMC cultures before being stained, and the number of CD107+ NK (CD56+ CD3−) cells was enumerated by flow cytometry. Error bars indicate standard errors.
MICA/B surface expression was profoundly upregulated in HF (telomerase-immortalized human fibroblasts expressing hCAR) infected with the HAdV-5 mutants ΔE3/19K and E3/19KSTOP relative to the wt parental virus or mock-infected cells (Fig. 3C). Further analyses, using antibodies that could differentiate between MICA and MICB in flow cytometry, demonstrated comparable upregulation of MICA and MICB following infection with both HAdV-5 E3/19K-negative viruses that was not apparent with the parental wt virus. E3/19K-specific downregulation of both MICA and MICB was also observed in the continuous cell lines detailed in Fig. 2A when resolving antibodies were used to monitor surface expression (data not shown). The HAdV-5 E3/19KW96A mutant effectively reproduced the findings obtained with the E2-W96A cell line in that there was a substantial downregulation of MICA and MICB surface expression whereas the prevention of HLA-I maturation was less effective (see Fig. 4C). Most significantly, productive Ad infection was demonstrated to stimulate the efficient induction of NKG2DLs, with MICA and MICB surface expression being prevented only by the action of E3/19K.
FIG. 4.
E3/19K inhibits posttranslational maturation of MICA/B. (A) Lysates from the 293 cell lines indicated were either mock digested (M) or digested with EndoH (E) or PNGase F (P) before immunoblotting with a MICA/B-specific polyclonal antibody. Arrowheads denote the mature EndoH-resistant forms, and arrows denote the EndoH-sensitive forms. (B) Lysates generated from HF cells infected at a multiplicity of infection of 3 with the indicated viruses at 48 h p.i. were mock digested or digested with EndoH or PNGase F before immunoblotting with a MICA/B-specific polyclonal antibody. Arrowheads denote the EndoH-resistant forms. (C) Lysates as described for panel B were immunoblotted with anti-MHC-I monoclonal antibody (HC10). (D) Lysates as described for panel B were immunoblotted with an antiactin antibody as a loading control.
E3/19K promotes evasion of NK recognition.
The generation of HAdV-5 E3/19K mutants enabled the role of E3/19K in modulating NK cell recognition to be evaluated in a functional assay. The CD107a mobilization assay detects lysosome-associated membrane protein-1 released to the surface of NK cells following degranulation and therefore provides a direct readout of NK cell function. IFN-α-activated PBMC were incubated (6 h) with HF infected with HAdV-5 and the various E3/19K mutants (48 h p.i.), and NK cells (defined as CD3− CD56+) were assessed for CD107a expression by flow cytometry. A representative result from three independent experiments performed is depicted in Fig. 3D. Differences in CD107a expression on NK cells following incubation with mock- and Ad-infected cells were analyzed using the analysis of variance test with Bonferroni's posttest. This indicated that the ∼2-fold increase in CD107a expression following incubation with the HAdV-5 ΔE3/19K and E3/19KSTOP mutants compared to wt infection was statistically significant (P < 0.05 and P < 0.01, respectively). The difference in CD107a expression following incubation with cells infected with wt virus or the W96A E3/19K mutant was consistent but not statistically significant and thus should not be given weight. Since MHC-I is known to be a ligand for multiple NK cell inhibitory receptors, its downregulation by E3/19K may be expected to stimulate NK recognition. However, only a modest activation was observed with the wt HAdV-5 relative to mock-infected controls. In contrast, functional inactivation of E3/19K by deletion or mutation resulted in a marked stimulation of NK cell activation. Thus, protection against NK cell recognition in the context of a productive HAdV-5 infection is mediated by E3/19K expression.
E3/19K promotes MICA/B retention in the ER.
To investigate the effect of E3/19K on posttranslational processing of MICA/B, extracts were prepared from the panel of E3-expressing cell lines (detailed in Fig. 2A) and subjected to EndoH or PNGase F treatment (Fig. 4A). EndoH will cleave only molecules containing high-mannose sugars, whereas those having acquired complex-type carbohydrates during the transport through the medium/trans-Golgi apparatus become resistant. Thus, EndoH resistance is indicative of MICA/B transport through the medium/trans-Golgi apparatus. As a control, extracts were also treated with PNGase F, which cleaves all N-linked carbohydrates. In the parental 293 cell line, MICA/B was detected as a diffuse EndoH-resistant, ∼60-kDa protein, with two species (40 and 33 kDa) observed following PNGase F digestion. 293 cells are homozygous for the truncated MICA*008 allele that lacks a cytosolic domain. The apparent molecular mass of the faster-migrating species (∼33 kDa) detected in Fig. 4A is consistent with it being the deglycosylated form of MICA*008, predicted to be 35 kDa. In contrast, in cells expressing wt E3/19K (E3-45 and E22.7) a lower-molecular-mass EndoH-sensitive species of ∼48 kDa was apparent, indicating incomplete posttranslational processing of MICA/B. MICA/B was resistant to EndoH digestion in cells expressing the E3/19K mutants EKE*1 and C83S; both forms of E3/19K were also impaired in suppressing MICA/B cell surface expression (Fig. 2C). In the W96A cell line, both the EndoH-sensitive and EndoH-resistant forms were present, with the EndoH-sensitive form predominating. This is consistent with this point mutant possessing an intermediate phenotype.
To corroborate these results during virus infection, Western blot experiments were also performed on extracts prepared from virus-infected cells. HAdV-5 infection resulted in enhanced expression of an EndoH-sensitive form of MICA/B (Fig. 4B) and HLA-I (Fig. 4C); HF consistently exhibit extremely low levels of constitutive MICA/B expression, which is clearly upregulated upon Ad infection. Thus, mature EndoH-resistant MICA/B forms of ∼70 kDa were more readily detected following infection with the HAdV-5 E3/19K mutants (ΔE3/19K and E3/19KSTOP) than in the uninfected controls or in wt infection. The E3/19KW96A mutant was again associated with an intermediate effect on MICA/B (Fig. 4B). Samples were immunoblotted for actin as a loading control (Fig. 4D). When expressed in continuous 293 cell lines or in the context of productive Ad infection, E3/19K of species C Ads (HAdV-2 and HAdV-5) acts to inhibit posttranslational maturation of MICA and MICB.
The differential migration of deglycosylated MICA/B in the two cell types can be attributed to allelic variation. While 293 cells are homozygous for the MICA*008 allele, HF carry full-length alleles (MICA*016,*027). Further studies with the HAdV-5 E3/19K mutants in additional genotyped cell lines (positive for MICA*004, MICA*018, and MICA*00901/*049) have demonstrated that E3/19K was capable of acting on a broad range of MICA alleles (data not shown).
Colocalization of E3/19K and MICA.
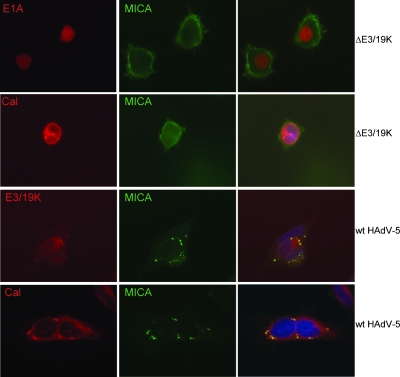
To monitor the intracellular localization of MICA during the course of an HAdV-5 infection, a plasmid encoding a MICA-YFP fusion protein was transfected into 911 cells, where cell surface expression was readily detected and confirmed by flow cytometry (not shown). Following infection with HAdV-5 ΔE3/19K, MICA-YFP remained predominantly associated with the cell surface (Fig. 5, top two rows). In contrast, in cells infected with wt HAdV-5 virus, the MICA-YFP fusion protein was found in distinct intracellular foci that overlapped significantly with E3/19K staining (Fig. 5, bottom two rows). These foci were found within calnexin-labeled areas of the cell, consistent with the E3/19K:MICA complex being retained within the ER. Indeed, calnexin appears to be enriched in these foci. These foci do not colocalize with any other intracellular compartment tested, including endosomes, the Golgi apparatus, and caveolae (not shown). The intracellular distribution of MICA-YFP following HAdV-5 infection is consistent with E3/19K sequestration of MICA/B in the ER.
FIG. 5.
MICA and E3/19K colocalize in the ER. 911 cells stably expressing a MICA-YFP fusion protein were infected at a multiplicity of infection of 5 with wt HAdV-5 or ΔE3/19K for 24 h. ΔE3/19K-infected cells were stained with an antibody cocktail against E1A or calnexin, and wt HAdV-5-infected cells were stained with anti-E3/19K or anticalnexin. The nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole). Antibody staining is in red, and the MICA-YFP fusion protein fluorescence is depicted in green.
DISCUSSION
E3/19K was shown to inhibit terminal glycosylation and to downregulate cell surface expression of MHC-I more than 20 years ago (8). In addition to conferring protection against MHC-I restricted cytotoxic T cells, we now show that the same Ad gene product protects against NK cell attack by intracellular sequestration of the NK cell activating ligands MICA and MICB. MICA/B downregulation from the cell surface was demonstrated in continuous cell lines expressing HAdV-2 E3/19K protein and with HAdV-5 E3/19K in the context of lytic virus infection. All characterized human Ad serotypes in species B, C, D, and E encode an E3/19K homolog (7). Suppression of both HLA-I and MICA/B requires both an ER retention/retrieval signal and formation of critical intramolecular disulfide bonds; these features are conserved in E3/19K across the different human Ad subtypes (11). Additional studies have shown this NK cell evasion function is intact in HAdV-4 (species E) (data not shown).
Although E3/19K and MICA/B colocalize, it has not yet been demonstrated that the proteins interact directly. Hence, the possibility that the interaction between E3/19K and MIC is mediated by a cellular intermediate cannot formally be ruled out. However, though quantitative phenotypic differences were observed, virtually all cysteine E3/19K mutants exhibited a simultaneous modulation of HLA-I, MICA, and MICB. The E3/19K point mutant (W96A) provided some discrimination by inducing suppression of MICA/B yet minimal downregulation of HLA-I in continuous cell lines. Potentially, the tryptophan at position 96 may preferentially promote recognition of HLA-I. This disparity was also observed, although it was less pronounced, when the equivalent substitution was introduced into the HAdV-5 genome. Further E3/19K mutagenesis will show whether other amino acids are preferentially required for HLA and MICA/B downregulation. In view of the limited amino acid identity in the extracellular domains between MIC and HLA alleles (∼25%), it seems likely that E3/19K is recognizing a conserved structure in these molecules. In this context, it is remarkable that E3/19K can accommodate the extreme polymorphism associated with HLA-I, MICA, and MICB.
MICA and MICB are cell surface glycoproteins with MHC-I-like α1, α2, and α3 domains, yet neither associates with β2-microglobulin or presents peptide (23). We have previously shown that E3/19K can bind MHC-I heavy chains in the absence of β2-microglobulin and prior to folding of the α1 and α2 domains (34, 41). E3/19K recognition of HLA-I also proceeds independently of peptide binding in vitro (34). Thus, E3/19K appears to bind at an early stage during the assembly of MHC-I-like proteins, presumably at the level of calnexin interaction. In this study, we used calnexin as an ER marker together with a MICA-YFP fusion protein to help identify the intracellular site of MICA retention. The MICA-YFP and E3/19K were observed to colocalize with calnexin in bright foci within the cells. In most cell types E3/19K exhibits a reticular ER-like staining rather than a focal distribution, and the concentration of complexes into foci may be a consequence of MICA-YFP overexpression. Nevertheless, the association of calnexin with these foci is consistent with ER retention.
Human cytomegalovirus (HCMV) encodes a number of functions that modulate NK cell recognition (16, 46, 47), including at least three genes that specifically target NKG2DLs. UL142 downregulates MICA alleles but not the truncated MICA*008 allele, and it does not affect MICB (13). UL16 encodes an ER-localized glycoprotein that acts to promote intracellular retention of MICB, ULBP1, and ULBP2 (16). In addition, a recent report indicates that an HCMV-encoded micro-RNA specifically targets the 3′ untranslated region of the MICB transcript (43). Although E3/19K and HCMV gpUL16 both promote ER retention and accumulation of NKG2DLs, their functions are quite distinct. E3/19K targets both MICA and MICB with similar efficiency and does not affect ULBP1 to -3 (data not shown), whereas UL16 does not target MICA or HLA-I. E3/19K thus has a novel function in being able to target a broad range of MICA and MICB alleles; in particular, it can target the MICA*008 allele, which is the most commonly expressed allele in populations studied to date (35, 38). It has been speculated that the MICA*008 allele may have emerged as a result of the host response to viral evasion functions acting on MICA (53). However, E3/19K can clearly retain both full-length and truncated MICA alleles and thus represents the first description of a viral protein that can downregulate surface expression of the commonly expressed MICA*008 allele.
The identification of this novel function for E3/19K has implications for the design and application of Ad vector systems (replication deficient and replication competent) and therapeutic oncolytic viruses. Murine models are used extensively to evaluate Ad vectors and oncolytic viruses in vivo. While a comparable murine NKG2D receptor exists, the NKG2DLs have diverged so there is no direct equivalent of MICA and MICB. It should therefore be borne in mind that the E3/19K-mediated NK cell evasion function described here would not be expected to operate in mice. NKG2D also acts as a costimulatory molecule for αβ CD8+ T cells (3), and E3/19K expression has been shown to protect infected cells from cytotoxic T-lymphocyte attack (1, 10, 21). Previously this was attributed solely to its ability to downregulate surface MHC-I, but the intracellular retention of potential costimulatory ligands by E3/19K can be expected to contribute to this effect. NKG2D is expressed on additional important immune effector cells, including γδ T cells and IFN-producing killer dendritic cells (3, 14). Thus, the inhibition of NKG2DL surface expression by E3/19K is expected to affect the recognition of Ad-infected cells by multiple effector cells of both the innate and adaptive immune systems.
Acknowledgments
We are grateful to Siân Llewellyn-Lacey and Carole Rickards for providing research support and to Andrew J. Davison and Kathleen Gallagher for helpful discussions.
This work was supported by grant funding from the Wellcome Trust, Biotechnology and Biological Sciences Research Council, and the Medical Research Council (United Kingdom).
Footnotes
Published ahead of print on 20 February 2008.
REFERENCES
- 1.Andersson, M., A. McMichael, and P. A. Peterson. 1987. Reduced allorecognition of adenovirus-2 infected cells. J. Immunol. 1383960-3966. [PubMed] [Google Scholar]
- 2.Bacon, L., R. A. Eagle, M. Meyer, N. Easom, N. T. Young, and J. Trowsdale. 2004. Two human ULBP/RAET1 molecules with transmembrane regions are ligands for NKG2D. J. Immunol. 1731078-1084. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, S., V. Groh, J. Wu, A. Steinle, J. H. Phillips, L. L. Lanier, and T. Spies. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285727-729. [DOI] [PubMed] [Google Scholar]
- 4.Beier, D. C., J. H. Cox, D. R. Vining, P. Cresswell, and V. H. Engelhard. 1994. Association of human class I MHC alleles with the adenovirus E3/19K protein. J. Immunol. 1523862-3872. [PubMed] [Google Scholar]
- 5.Benedict, C. A., P. S. Norris, T. I. Prigozy, J. L. Bodmer, J. A. Mahr, C. T. Garnett, F. Martinon, J. Tschopp, L. R. Gooding, and C. F. Ware. 2001. Three adenovirus E3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and -2. J. Biol. Chem. 2763270-3278. [DOI] [PubMed] [Google Scholar]
- 6.Bett, A. J., V. Krougliak, and F. L. Graham. 1995. DNA sequence of the deletion/insertion in early region 3 of Ad5 dl309. Virus Res. 3975-82. [PubMed] [Google Scholar]
- 7.Burgert, H.-G., and J. H. Blusch. 2000. Immunomodulatory functions encoded by the E3 transcription unit of adenoviruses. Virus Genes. 2113-25. [PubMed] [Google Scholar]
- 8.Burgert, H.-G., and S. Kvist. 1985. An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell 41987-997. [DOI] [PubMed] [Google Scholar]
- 9.Burgert, H.-G., and S. Kvist. 1987. The E3/19K protein of adenovirus type 2 binds to the domains of histocompatibility antigens required for CTL recognition. EMBO J. 62019-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgert, H.-G., J. L. Maryanski, and S. Kvist. 1987. “E3/19K” protein of adenovirus type 2 inhibits lysis of cytolytic T lymphocytes by blocking cell-surface expression of histocompatibility class I antigens. Proc. Natl. Acad. Sci. USA 841356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgert, H.-G., Z. Ruzsics, S. Obermeier, A. Hilgendorf, M. Windheim, and A. Elsing. 2002. Subversion of host defense mechanisms by adenoviruses. Curr. Top. Microbiol. Immunol. 269273-318. [DOI] [PubMed] [Google Scholar]
- 12.Carlin, C. R., A. E. Tollefson, H. A. Brady, B. L. Hoffman, and W. S. Wold. 1989. Epidermal growth factor receptor is down-regulated by a 10,400 MW protein encoded by the E3 region of adenovirus. Cell 57135-144. [DOI] [PubMed] [Google Scholar]
- 13.Chalupny, N. J., A. Rein-Weston, S. Dosch, and D. Cosman. 2006. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem. Biophys. Res. Commun. 346175-181. [DOI] [PubMed] [Google Scholar]
- 14.Chan, C. W., E. Crafton, H. N. Fan, J. Flook, K. Yoshimura, M. Skarica, D. Brockstedt, T. W. Dubensky, M. F. Stins, L. L. Lanier, D. M. Pardoll, and F. Housseau. 2006. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nat. Med. 12207-213. [DOI] [PubMed] [Google Scholar]
- 15.Cook, J. L., C. K. Krantz, and B. A. Routes. 1996. Role of p300-family proteins in E1A oncogene induction of cytolytic susceptibility and tumor cell rejection. Proc. Natl. Acad. Sci. USA 9313985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosman, D., J. Mullberg, C. L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N. J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14123-133. [DOI] [PubMed] [Google Scholar]
- 17.Cox, J. H., J. R. Bennink, and J. W. Yewdell. 1991. Retention of adenovirus E19 glycoprotein in the endoplasmic reticulum is essential to its ability to block antigen presentation. J. Exp. Med. 1741629-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsing, A., and H.-G. Burgert. 1998. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. USA 9510072-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feuerbach, D., S. Etteldorf, C. Ebenau-Jehle, J. P. Abastado, D. Madden, and H.-G. Burgert. 1994. Identification of amino acids within the MHC molecule important for the interaction with the adenovirus protein E3/19K. J. Immunol. 1531626-1636. [PubMed] [Google Scholar]
- 20.Flomenberg, P., E. Gutierrez, and K. T. Hogan. 1994. Identification of class I MHC regions which bind to the adenovirus E3-19k protein. Mol. Immunol. 311277-1284. [DOI] [PubMed] [Google Scholar]
- 21.Flomenberg, P., V. Piaskowski, R. L. Truitt, and J. T. Casper. 1996. Human adenovirus-specific CD8+ T-cell responses are not inhibited by E3-19K in the presence of gamma interferon. J. Virol. 706314-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasser, S., S. Orsulic, E. J. Brown, and D. H. Raulet. 2005. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 4361186-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groh, V., S. Bahram, S. Bauer, A. Herman, M. Beauchamp, and T. Spies. 1996. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl. Acad. Sci. USA 9312445-12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilgendorf, A., J. Lindberg, Z. Ruzsics, S. Höning, A. Elsing, M. Löfqvist, H. Engelmann, and H.-G. Burgert. 2003. Two distinct transport motifs in the adenovirus E3/10.4-14.5 proteins act in concert to down-modulate apoptosis receptors and the epidermal growth factor receptor. J. Biol. Chem. 27851872-51884. [DOI] [PubMed] [Google Scholar]
- 25.Holmes, M. A., P. Li, E. W. Petersdorf, and R. K. Strong. 2002. Structural studies of allelic diversity of the MHC class I homolog MIC-B, a stress-inducible ligand for the activating immunoreceptor NKG2D. J. Immunol. 1691395-1400. [DOI] [PubMed] [Google Scholar]
- 26.Horton, T. M., T. S. Ranheim, L. Aquino, D. I. Kusher, S. K. Saha, C. F. Ware, W. S. Wold, and L. R. Gooding. 1991. Adenovirus E3 14.7K protein functions in the absence of other adenovirus proteins to protect transfected cells from tumor necrosis factor cytolysis. J. Virol. 652629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson, M. R., T. Nilsson, and P. A. Peterson. 1993. Retrieval of transmembrane proteins to the endoplasmic reticulum. J. Cell Biol. 121317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs, S. C., A. J. Davison, S. Carr, A. M. Bennett, R. Phillpotts, and G. W. Wilkinson. 2004. Characterization and manipulation of the human adenovirus 4 genome. J. Gen. Virol. 853361-3366. [DOI] [PubMed] [Google Scholar]
- 29.Katsuyama, Y., M. Ota, H. Ando, S. Saito, N. Mizuki, J. Kera, S. Bahram, Y. Nose, and H. Inoko. 1999. Sequencing based typing for genetic polymorphisms in exons, 2, 3 and 4 of the MICA gene. Tissue Antigens 54178-184. [DOI] [PubMed] [Google Scholar]
- 30.Körner, H., U. Fritzsche, and H.-G. Burgert. 1992. Tumor necrosis factor alpha stimulates expression of adenovirus early region 3 proteins: implications for viral persistence. Proc. Natl. Acad. Sci. USA 8911857-11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leon, R. P., T. Hedlund, S. J. Meech, S. Li, J. Schaack, S. P. Hunger, R. C. Duke, and J. DeGregori. 1998. Adenoviral-mediated gene transfer in lymphocytes. Proc. Natl. Acad. Sci. USA 9513159-13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, P., S. T. Willie, S. Bauer, D. L. Morris, T. Spies, and R. K. Strong. 1999. Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand. Immunity 10577-584. [DOI] [PubMed] [Google Scholar]
- 33.Lichtenstein, D. L., K. Toth, K. Doronin, A. E. Tollefson, and W. S. Wold. 2004. Functions and mechanisms of action of the adenovirus E3 proteins. Int. Rev. Immunol. 2375-111. [DOI] [PubMed] [Google Scholar]
- 34.Liu, H., W. F. Stafford, and M. Bouvier. 2005. The endoplasmic reticulum lumenal domain of the adenovirus type 2 E3-19K protein binds to peptide-filled and peptide-deficient HLA-A*1101 molecules. J. Virol. 7913317-13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marin, M. L., C. R. Savioli, J. H. Yamamoto, J. Kalil, and A. C. Goldberg. 2004. MICA polymorphism in a sample of the Sao Paulo population, Brazil. Eur. J. Immunogenet. 3163-71. [DOI] [PubMed] [Google Scholar]
- 36.McCann, F. E., P. Eissmann, B. Onfelt, R. Leung, and D. M. Davis. 2007. The activating NKG2D ligand MHC class I-related chain A transfers from target cells to NK cells in a manner that allows functional consequences. J. Immunol. 1783418-3426. [DOI] [PubMed] [Google Scholar]
- 37.McSharry, B. P., C. J. Jones, J. W. Skinner, D. Kipling, and G. W. Wilkinson. 2001. Human telomerase reverse transcriptase-immortalized MRC-5 and HCA2 human fibroblasts are fully permissive for human cytomegalovirus. J. Gen. Virol. 82855-863. [DOI] [PubMed] [Google Scholar]
- 38.Munoz-Saa, I., A. Cambra, L. Pallares, G. Espinosa, A. Juan, F. Pujalte, N. Matamoros, J. Mila, and M. R. Julia. 2006. Allelic diversity and affinity variants of MICA are imbalanced in Spanish patients with Behcet's disease. Scand. J. Immunol. 6477-82. [DOI] [PubMed] [Google Scholar]
- 39.Routes, J. M., S. Ryan, K. Morris, R. Takaki, A. Cerwenka, and L. L. Lanier. 2005. Adenovirus serotype 5 E1A sensitizes tumor cells to NKG2D-dependent NK cell lysis and tumor rejection. J. Exp. Med. 2021477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sester, M., and H.-G. Burgert. 1994. Conserved cysteine residues within the E3/19K protein of adenovirus type 2 are essential for binding to major histocompatibility complex antigens. J. Virol. 685423-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sester, M., D. Feuerbach, R. Frank, T. Preckel, A. Gutermann, and H.-G. Burgert. 2000. The amyloid precursor-like protein 2 associates with the major histocompatibility complex class I molecule K (d). J. Biol. Chem. 2753645-3654. [DOI] [PubMed] [Google Scholar]
- 42.Stam, N. J., T. M. Vroom, P. J. Peters, E. B. Pastoors, and H. L. Ploegh. 1990. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int. Immunol. 2113-125. [DOI] [PubMed] [Google Scholar]
- 43.Stern-Ginossar, N., N. Elefant, A. Zimmermann, D. G. Wolf, N. Saleh, M. Biton, E. Horwitz, Z. Prokocimer, M. Prichard, G. Hahn, D. Goldman-Wohl, C. Greenfield, S. Yagel, H. Hengel, Y. Altuvia, H. Margalit, and O. Mandelboim. 2007. Host immune system gene targeting by a viral miRNA. Science 317376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tollefson, A. E., T. W. Hermiston, D. L. Lichtenstein, C. F. Colle, R. A. Tripp, T. Dimitrov, K. Toth, C. E. Wells, P. C. Doherty, and W. S. Wold. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392726-730. [DOI] [PubMed] [Google Scholar]
- 45.Tollefson, A. E., A. R. Stewart, S. P. Yei, S. K. Saha, and W. S. Wold. 1991. The 10,400- and 14,500-dalton proteins encoded by region E3 of adenovirus form a complex and function together to down-regulate the epidermal growth factor receptor. J. Virol. 653095-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomasec, P., V. M. Braud, C. Rickards, M. B. Powell, B. P. McSharry, S. Gadola, V. Cerundolo, L. K. Borysiewicz, A. J. McMichael, and G. W. Wilkinson. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2871031. [DOI] [PubMed] [Google Scholar]
- 47.Tomasec, P., E. C. Wang, A. J. Davison, B. Vojtesek, M. Armstrong, C. Griffin, B. P. McSharry, R. J. Morris, S. Llewellyn-Lacey, C. Rickards, A. Nomoto, C. Sinzger, and G. W. Wilkinson. 2005. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 6181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomasec, P., E. C. Wang, V. Groh, T. Spies, B. P. McSharry, R. J. Aicheler, R. J. Stanton, and G. W. Wilkinson. 2007. Adenovirus vector delivery stimulates natural killer cell recognition. J. Gen. Virol. 881103-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warming, S., N. Costantino, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson, G. W., and A. Akrigg. 1992. Constitutive and enhanced expression from the CMV major IE promoter in a defective adenovirus vector. Nucleic Acids Res. 202233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Windheim, M., A. Hilgendorf, and H.-G. Burgert. 2004. Immune evasion by adenovirus E3 proteins: exploitation of intracellular trafficking pathways. Curr. Top. Microbiol. Immunol. 27329-85. [DOI] [PubMed] [Google Scholar]
- 52.Wold, W. S., and M. S. Horwitz. 2007. Adenoviruses, p. 2395-2436. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 53.Zou, Y., W. Bresnahan, R. T. Taylor, and P. Stastny. 2005. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J. Immunol. 1743098-3104. [DOI] [PubMed] [Google Scholar]
- 54.Zou, Y., M. Han, Z. Wang, and P. Stastny. 2006. MICA allele-level typing by sequence-based typing with computerized assignment of polymorphic sites and short tandem repeats within the transmembrane region. Hum. Immunol. 67145-151. [DOI] [PubMed] [Google Scholar]
- 55.Zwirner, N. W., C. Y. Marcos, F. Mirbaha, Y. Zou, and P. Stastny. 2000. Identification of MICA as a new polymorphic alloantigen recognized by antibodies in sera of organ transplant recipients. Hum. Immunol. 61917-924. [DOI] [PubMed] [Google Scholar]