Abstract
Several families of endogenous retroviruses (ERVs) have been identified in the mouse genome, in several instances by in silico searches, but for many of them it remains to be determined whether there are elements that can still encode functional retroviral particles. Here, we identify, within the GLN family of highly reiterated ERVs, one, and only one, copy that encodes retroviral particles prone to infection of mouse cells. We show that its envelope protein confers an ecotropic host range and recognizes a receptor different from mCAT1 and mSMIT1, the two previously identified receptors for other ecotropic mouse retroviruses. Electron microscopy disclosed viral particle assembly and budding at the cell membrane, as well as release of mature particles into the extracellular space. These particles are closely related to murine leukemia virus (MLV) particles, with which they have most probably been confused in the past. This study, therefore, identifies a new class of infectious mouse ERVs belonging to the family Gammaretroviridae, with one family member still functional today. This family is in addition to the two MLV and mouse mammary tumor virus families of active mouse ERVs with an extracellular life cycle.
Complete sequencing of the mouse genome has led to the identification of multiple families of endogenous retroviruses (ERVs), each with a variable number of elements ranging from a few copies to several hundred (33; reviewed in references 6 and 20). These elements can now be classified according to their pol gene homology, and the resulting phylogenetic analysis recapitulates, in part, the diversity that can be found at the level of the present-day infectious retroviruses of animals. In the cases where functional copies have been identified and characterized, this classification can be further refined according to criteria unrelated to their sequences but involving both the site of assembly and the morphology of the associated virus-like particles. In fact, the latter classification corresponds to the historical one, essentially based on pioneering electron microscopic analyses of the A-, B-, C-, or epsilon-type virus-like particles that can be observed in mouse cells and tissues (5, 36; reviewed in reference 24). Along these lines, and as illustrated in Fig. 1B, the first significant fraction of the mouse ERVs belongs to the family Betaretroviridae and includes the endogenous mouse mammary tumor virus elements (∼10 copies in the C57BL/6 genome) and the highly reiterated Mus musculus type D/early transposon (MusD/ETn) ERVs (∼350 copies), whose particles have recently been demonstrated to assemble in a strictly intracellular location (28), as observed for type B/D retroviruses. A second large family of ERVs includes the intracisternal A-type particle (more than 1,000 copies) and intracisternal A-type particle-related, envelope-encoding elements (∼250 copies), which are also phylogenetically close to betaretroviruses but can be distinguished from the latter by the site of particle assembly, which is not within the cell cytoplasm but rather at the level of membranes, where budding occurs, as observed for type C retroviruses. Assembly takes place at the endoplasmic reticulum membrane in the case of intracisternal A-type particles (10, 15, 19) and at the cell plasma membrane in the case of intracisternal A-type particle-related, envelope-encoding elements (28a). A third large family of elements is composed of the rather atypical murine ERV-L (MuERV-L) elements, which are among the most ancient ERVs present in mammals, with ∼500 full-length copies dispersed throughout the mouse genome (3, 4). MuERV-L elements are unrelated to any known present-day infectious retroviruses at the levels of both their sequence (although their pol gene is distantly related to that of foamy viruses [9]) and the morphology of the virus-like particles they encode. We have recently demonstrated that they correspond to the epsilon particles previously reported to occur in two-cell mouse embryos (29), with a unique morphology among retroviral elements. Finally, the most diverse group of mouse ERVs belongs to the gammaretroviruses. Rather paradoxically, it includes both a very well-characterized family of elements, namely, the murine leukemia viruses (MLVs), and a series of little-studied elements, including murine retrovirus-related DNA sequences (30), murine retrovirus Y-associated element (13), MuERV-C (37), Mus musculus ERV (8), and murine retrovirus using tRNAGln (GLN) (18, 25). These are present at high copy numbers, with only the last two families showing copies with coding-competent env genes (reference 12 and data not shown). Whereas MmERV is closely related to Mus dunni endogenous virus, a well-characterized infectious ERV from Mus dunni (7, 34, 35), the GLN family has no known infectious close relative. This family was thus characterized to tentatively identify functional proviral copies and to investigate their structure and life cycle.
FIG. 1.
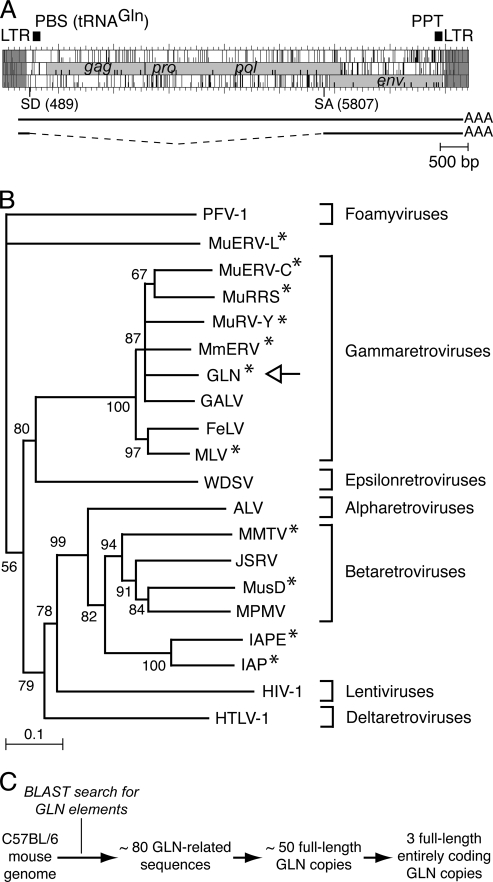
Structure and phylogeny of GLN elements. (A) Genomic organization of a full-length coding-competent GLN element, with the LTRs (in dark gray) flanking three complete ORFs homologous to the retroviral gag, pro-pol, and env genes (in light gray). A primer binding site (PBS) complementary to tRNAGln and a polypurine tract (PPT) can be identified, as indicated. The structures of the full-length and spliced viral transcripts are indicated (SD, splice donor site; SA, splice acceptor site). (B) Phylogeny of retroviruses, based on their RT domains. The tree was determined by the neighbor-joining method using the seven blocks of conserved residues found in the RT domains of all retroelements and was rooted using non-LTR retrotransposons. All sequences are readily accessible from GenBank and previous reports (e.g., reference 21). Percent bootstrap values, obtained from 1,000 replicates, are indicated. The retroviruses “endogenized” in the mouse genome are marked with asterisks. The seven retroviral families are indicated on the right. PFV, primate foamy virus; MuERV-L, murine ERV-L element; MuRRS, murine retrovirus-related DNA sequence; MuRV-Y, murine retrovirus Y-associated element; MmERV, Mus musculus ERV; GALV, gibbon ape leukemia virus; FeLV, feline leukemia virus; WDSV, walleye dermal sarcoma virus; ALV, avian leukosis virus; MMTV, mouse mammary tumor virus; JSRV, jaagsiekte sheep retrovirus; MusD, Mus musculus type D element; MPMV, Mason-Pfizer monkey virus; HIV-1, human immunodeficiency virus type 1; HTLV-1, human T-cell leukemia virus type-1. (C) In silico identification of the full-length, coding-competent GLN copies contained in the C57BL/6 genome. A Blast search was carried out with the NCBI m36 Mouse Assembly (April 2006 release).
MATERIALS AND METHODS
Plasmids.
Each coding-competent GLN env gene was PCR amplified from the corresponding bacterial artificial chromosome DNA (BACPAC Resources) by using the proofreading PfuTurbo Hotstart DNA polymerase (Stratagene) and appropriate primers (sequences are available on request). Each PCR product was then cloned in place of the vesicular stomatitis virus G gene into the phCMV-VSV-G vector (GenBank accession no. AJ318514), opened by XhoI.
The plasmid containing the GLN-2 copy was obtained by cloning into pBR322 the complete proviral DNA contained in a HindIII-SpeI fragment from bacterial artificial chromosome RP23-356G7 DNA (nucleotides [nt] 184912 to 193326). The defective neo-marked GLN element was obtained by inserting the blunt-ended HindIII-XhoI fragment from pSVneo* (14) into the GLN-2 plasmid, between the SnaBI and BlpI sites of the env gene (nt 6273 and 7795, respectively), and by further deleting an internal 4,568-bp gag-pro-pol fragment (from nt 1238 to 5805) upon digestion of the above-mentioned plasmid with AleI and religation.
Cell culture, pseudotyping, and infectivity assays.
Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (VWR International), 100 μg/ml streptomycin, and 100 U/ml penicillin.
HeLa cells expressing mSMIT1 or mCAT1 (or control HeLa cells) were obtained by infection, followed by G418 selection with neo-containing MLV-derived expression vectors for mCAT1 (SFEV-mCAT1-neo) or mSMIT1 (MPEV-mSMIT1-neo) or with a control vector (MPEV-neo) (17, 26).
MLV and simian immunodeficiency virus (SIV) pseudotypes containing GLN, amphotropic-MLV, or ecotropic-MLV (Moloney MLV [MoMLV]) envelope (Env) proteins (or no Env protein for the negative control) were produced by cotransfecting 7.5 × 105 293T cells with 0.5 μg of the corresponding Env expression vector (or pcDNA3 for the negative control), 2.25 μg of vectors encoding the retroviral proteins (except the envelope) of MLV or SIV (23), and 2.25 μg of the corresponding defective retroviral vectors (pMFGsnlslacZ [16] or RqSA [22], both marked with a β-galactosidase reporter gene, by calcium phosphate precipitation (MBS transfection kit; Stratagene). M813-MLV recombinant pseudotypes were produced by cotransfecting 293T cells with 2.75 μg of an MLV retroviral vector containing the 5′ domain of the M813 env gene (encompassing the receptor binding domain, Mo-M813 [26]) and 2.25 μg of pMFGsnlslacZ. Target cells were seeded in 24-well plates the day prior to infection. Supernatants from transfected 293T cells were harvested 48 h posttransfection, filtered through 0.45-μm-pore-size polyvinylidene difluoride membranes, supplemented with Polybrene (4 μg/ml), and added to the target cells, followed by spinoculation at 1,200 × g for 2 h 30 min at 25°C. After an additional 60-h incubation period, viral titers were determined by in situ X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of target cells and quantification of LacZ-positive (LacZ+) CFU.
To assay for GLN infectivity, 293T cells were cotransfected with the defective neo-marked GLN reporter and the wild-type GLN-2 cloned provirus (or pBR322 as a control plasmid). Mouse 3T3 (WOP) cells were infected with the particles released into the transfected 293T cell supernatant. After removal of the supernatant and incubation in regular medium for 72 h at 37°C, 3T3 (WOP) cells were split into 100-mm dishes (5 × 105 cells per dish), allowed to settle for 24 h, and subjected to G418 selection. G418r foci were fixed, stained, and counted.
RT assay activity and detection of viral RNA in cell supernatants.
Supernatants of 293T cells were harvested 48 h posttransfection, centrifuged for 5 min at 1,500 rpm, filtered through a 0.45-μm-pore-size polyvinylidene difluoride membrane, and used for a product-enhanced reverse transcriptase (PERT) assay (27) or for detection of GLN viral RNA.
For the PERT assay, the RT contained in 2.5 μl of supernatant was used to reverse transcribe 0.3 μg of MS2 phage RNA previously annealed to 16 pmol of RT-1 primer (5′-CACAGGTCAAACCTCCTAGGAATG-3′). cDNA synthesis was then assayed using 1/50 of the RT reaction mixture as a template for a 25-cycle PCR conducted with RT-1 and RT-2 (5′-TCCTGCTCAACTTCCTGTCGAG-3′) primers, using Tth polymerase (Promega), leading to the amplification of a 112-bp fragment.
For detection of GLN viral RNA, RNAs contained in 30 μl of supernatant were purified using the RNeasy Microkit (Qiagen). One-fourth of an RT mixture from a reaction run with the GLN-RT primer (5′-CTGGTCCTTCCTGAAAAACA-3′) was then used as a template for a 30-cycle PCR conducted with GLN-F (5′-TGTGTAAGTCCAGACGCAGA-3′) and GLN-R (5′-CCAACCTACTCCAAAAACAG-3′) primers, leading to the amplification of a 239-bp fragment.
Electron microscopy.
For ultrastructural studies, transfected 293T cells were fixed in phosphate buffer, pH 7.2, 1.6% glutaraldehyde for 1 h and postfixed in 0.1 M cacodylate buffer, 1% osmium tetroxyde for 2 h. After being rinsed for 5 min in water and 15 min in 0.1 M cacodylate buffer, the cells were transferred to 0.2 M cacodylate buffer for 30 min. The cells were washed in 30% methanol for 10 min, stained in 2% uranyl acetate in 0.1 M cacodylate buffer-30% methanol for 1 h, and washed in 30% methanol. The cells were then dehydrated through a graded ethanol series and embedded in Epon 812. Ultrathin sections were stained with uranyl acetate and lead citrate and examined with a Zeiss 902 microscope at 80 kV.
RESULTS AND DISCUSSION
In silico analysis of the GLN family.
The sequences of previously described GLN elements (18, 25) were used to screen the mouse genome (BLAST-Like Alignment Tool at the University of California—Santa Cruz Genome Bioinformatics website; http://genome.ucsc.edu). About 80 GLN-related sequences were identified in the C57BL/6 mouse genome (Fig. 1C). Among them, about 50 were full-length elements, and only 3 were found to have complete open reading frames (ORFs) within the gag-pro-pol and env genes (GenBank accession numbers: GLN-1, AC136922, positions 167843 to 176257; GLN-2, AC153548, positions 184912 to 193326; and GLN-3, AL669853, positions 63151 to 54735). These three proviral copies show high amino acid sequence identity (i.e., >98.8%), and among the 50 full-length GLN copies, nucleotide sequence identity ranges from 95.5 to 100%, without any evidence for subclasses (deletion or truncation associated). The overall genomic organization of the prototypic GLN provirus is illustrated in Fig. 1A. These elements are 8.4 kbp long, with long terminal repeats (LTRs) of 430 bp, a clearly identified primer binding site complementary to tRNAGln, and a polypurine tract. Analysis of mouse expressed sequence tags allowed identification of spliced GLN transcripts, most probably corresponding to the subgenomic RNA for the env ORF (splice donor site at nt 489; splice acceptor site at nt 5807). The overall gene organization is clearly reminiscent of that of MLVs, with the gag-pol ORFs within the same frame and a stop codon between gag and pol. However, comparison of the GLN-2 and MoMLV genomes (NCBI GenBank accession number AF033811) disclosed only limited amino acid sequence similarities, with 60%, 71%, and 53% identity for the gag, pol, and env ORFs, respectively. As illustrated in Fig. 1B, phylogenetic analyses based on the pol genes of both exogenous retroviruses and ERVs revealed that GLN elements cluster with gammaretroviruses (a similar conclusion can be derived from an Env transmembrane [TM] subunit-based tree [12]). A search for a functional GLN copy among the three full-length elements with complete ORFs identified by in silico analysis was then performed using appropriate assays.
A GLN copy with a functional env gene.
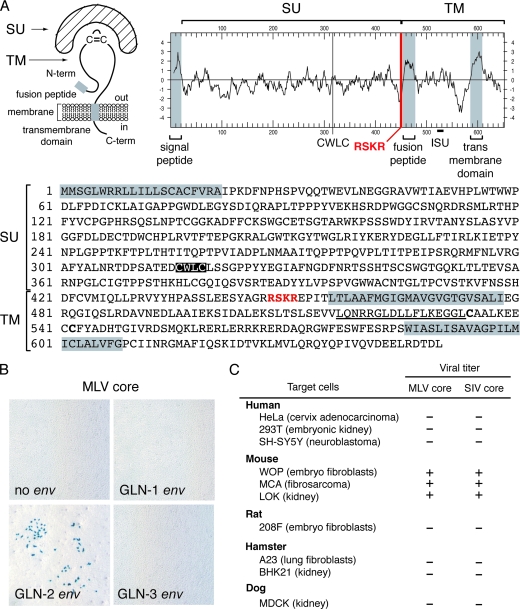
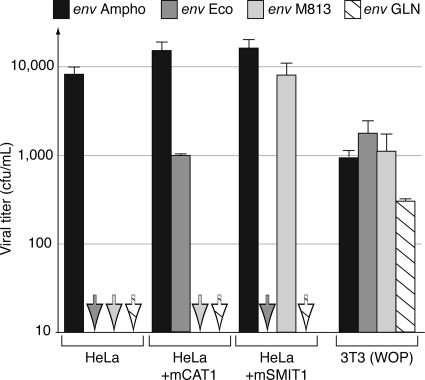
As illustrated in Fig. 2A, the structural organization of the GLN env is canonical, with a signal peptide at the N-terminal end and an R-X-(K/R)-R consensus cleavage site for the cellular furin protease that splits the Env protein into the surface (SU) and TM subunits, a hydrophobic fusion domain at the N-terminal end of the TM subunit, and a hydrophobic transmembrane anchor domain. To experimentally characterize the Env glycoproteins of the three coding-competent GLN copies, we first cloned the corresponding ORFs into a cytomegalovirus promoter-driven expression vector, starting from the env initiation codon to the end of the 3′ LTR. The functionality of the GLN env was then assayed by pseudotyping. To do so, human 293T cells were cotransfected with expression vectors for the retroviral proteins (except Env) from a gammaretrovirus (MLV) or a lentivirus (SIV), a corresponding lacZ gene-marked defective retroviral vector, and the above-mentioned expression vector for the GLN env gene to be tested (or an empty vector as a negative control). Then, the pseudotyped virions produced in the supernatant of the transfected cells were assayed for infectivity on test target cells. Among the three env genes tested, only one was found to be positive in the assay, namely, that from GLN-2. Interestingly, infection could be detected using either type of viral core, with viral titers in the range of 100 to 1,000 LacZ+ CFU per ml, but was found only with mouse cells as targets (Fig. 2C). No infection events were detected with cells from other rodents, such as rats or hamsters, or with human cells, such as the 293T cells, which harbor the Pit1 receptor for GaLV and 10A1-MLV, as well as the Pit2 and Syg1 receptors for the ampho-, xeno-, and polytropic (MCF247) MLV (reference 32 and data not shown). Such a cell tropism, which classifies the GLN Env protein as ecotropic, has previously been found for several MLV strains (e.g., MoMLV) and for two murine ERVs, namely, HEMV in Mus spicilegus (32) and M813-MLV in Mus cervicolor (26), with the last two sharing the same receptor (17, 32). Despite severe sequence divergence between GLN and MLV Env proteins, even in the receptor binding domain (46% and 37% sequence homology between that of GLN and that of the ecotropic MoMLV and M813-MLV, respectively), we investigated whether GLN elements could recognize one of the receptors for these ecotropic retroviruses, i.e., mCAT1 (1) or mSMIT1 (17). To this end, we assayed whether expression of the mCAT1 or mSMIT1 receptor could confer on cells susceptibility to viral pseudotypes bearing the GLN Env glycoprotein. Nonpermissive human HeLa cells were first stably transduced by infection, followed by G418 selection, with neo-containing MLV-derived expression vectors for mCAT1 (SFEV-mCAT1-neo), mSMIT1 (MPEV-mSMIT1-neo), or a control vector (MPEV-neo) (17, 26). These receptor-expressing HeLa cells were then infected with various pseudotypes: MLV pseudotypes with the GLN Env glycoprotein, the amphotropic or ecotropic MLV Env glycoproteins as controls, and a M813-MLV pseudotype in which the N-terminal Env sequence is derived from M813 (26). As illustrated in Fig. 3, neither mCAT1 nor mSMIT1 confers susceptibility to GLN Env, although they do confer susceptibility to MoMLV Env and M813-MLV Env, respectively, indicating that GLN elements recognize a third kind of mouse ecotropic receptor.
FIG. 2.
Characterization of the GLN Env glycoprotein. (A) Schematic structure, hydrophobicity profile, and primary sequence of the GLN-2 Env protein. The SU and TM subunits are delineated, with a canonical furin cleavage site (RSKR) between the two subunits indicated in red. The hydrophobic signal peptide and fusion peptide and the transmembrane domain are shaded in gray. The CWLC motif (highlighted in black) and the putative immunosuppressive domain (ISU) (underlined) are indicated. (B) Infectivity assay of MLV particles pseudotyped with the GLN Env proteins. Supernatants of human 293T cells cotransfected with an expression vector for the MLV core proteins, a lacZ gene-marked defective retroviral vector, and an expression vector for the GLN-1, -2, or -3 Env protein, or a control plasmid (no env), was used to infect murine 3T3 (WOP) target cells. Infection events were detected by X-Gal staining of target cells. (C) Cell tropism of GLN-2 Env. The infectivities of MLV or SIV cores pseudotyped with the GLN-2 Env were determined using human, mouse, rat, hamster, or dog cell lines (the mouse MCA cells are from C3H mice, 3T3 [WOP] cells from NIH/Swiss mice, and LOK cells from 129/B6/DBA2 mice). Viral titers, corresponding to the number of LacZ+ CFU/ml of supernatant, are indicated as follows: −, <10; +, 100 to 1,000.
FIG. 3.
The GLN Env recognizes an unkown ecotropic receptor. Infectivities of MLV cores pseudotyped with either an MLV amphotropic (Ampho) Env, an MLV ecotropic (Eco) Env, an M813-derived Env (M813), or the GLN-2 Env (GLN) were assayed as in Fig. 2B. 3T3 (WOP) cells and human HeLa cells stably transduced with mSMIT1, mCAT1, or a control plasmid were used as target cells. Viral titers were determined by counting the LacZ+ CFU/ml of supernatant (the means of three independent experiments, with standard deviations, are shown). The arrows indicate the absence of LacZ+ foci.
A GLN provirus encoding infectious particles.
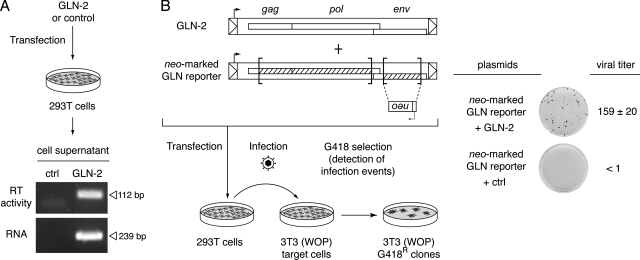
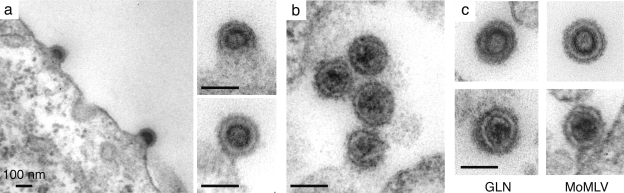
Preliminary experiments revealed that GLN LTRs are active promoters—at least in the series of cells in culture commonly used for ex vivo assays (e.g., human HeLa and 293T cells, mouse 3T3 [WOP] cells, and feline G355-5 cells) (data not shown). Assay of the identified GLN-2 copy for retroviral particle formation was therefore carried out by transfecting 293T cells with a plasmid containing the proviral copy expressed under its own LTR. As illustrated in Fig. 4A, RT activity was detected in the supernatant of the transfected cells using a PCR in vitro assay for reverse transcription (27), with no activity detected in the supernatant of mock-transfected cells. Consistently, GLN viral RNA could be detected by RT-PCR in the supernatant of the GLN-transfected cells. To determine whether GLN-2 encodes a functional retrovirus, we first constructed a neo-marked GLN reporter in which the neo gene was inserted into the env gene of the GLN-2 element and the gag-pro-pol genes were further inactivated by a large internal deletion (Fig. 4B). This defective neo-marked GLN-2 reporter was then complemented in trans by the full-length wild-type GLN-2 provirus (or a control plasmid), upon cotransfection of 293T cells with the two plasmids. Two days posttransfection, the supernatant was harvested and transferred to target 3T3 (WOP) cells. The cells were then subjected to G418 selection for 10 days. As illustrated in Fig. 4B, G418r clones could be recovered with the GLN-2 provirus, but not with the control plasmid, clearly indicating that the cloned GLN-2 ERV element is a bona fide retrovirus that generates functional “ecotropic” viral particles prone to stable infection. The structure and site of assembly of the GLN-2 particles was then investigated by electron microscopic analysis of GLN-2-transfected 293T cells (Fig. 5). As illustrated in Fig. 5, 293 cells transfected with the GLN-2 plasmid revealed viral particles budding at the cell membrane, as classically observed for type C retroviruses (no particles were observed with cells transfected by a control plasmid). Free particles could also be observed in the extracellular space, with two distinct morphologies, corresponding most probably to immature and mature particles. This was further assessed by introducing an in-frame deletion within the protease gene of the cloned GLN-2 copy (from nt 2605 to 2898, to preserve translation of the downstream genes), which then yielded only particles with the immature morphology (data not shown). Interestingly, 293T cells transfected with an expression vector for MoMLV and analyzed under the same experimental conditions (Fig. 5C) revealed particles that could not be distinguished, at the levels of their structure, site of assembly, and maturation, from the GLN particles.
FIG. 4.
GLN encodes infectious retroviral particles. (A) Analysis of GLN particles in cell supernatants. The supernatant of 293T cells transfected with the GLN-2 copy or a control plasmid was harvested 48 h posttransfection and assayed for RT activity (using the sensitive PERT assay described in reference 27, resulting in the amplification of a 112-bp DNA product) and for GLN viral RNA (using an RT-PCR assay on RNA extracted from the cell supernatant, resulting in the amplification of a 239-bp DNA product corresponding to part of the gag gene). (B) Infectivity assay of the GLN viral particles. Supernatant of 293T cells cotransfected with the neo-marked defective GLN-2 reporter (in which a backward neo resistance gene is introduced into a partly deleted env gene, with the gag-pro-pol genes further inactivated by a large internal deletion) and the full-length GLN-2 provirus or a control plasmid was harvested 48 h posttransfection and used to infect 3T3 (WOP) target cells. Infection events were detected upon G418 selection of the target cells, and viral titers were quantified by counting the G418r clones per milliliter of supernatant (the means of three independent experiments, with standard deviations, are shown).
FIG. 5.
Morphology of the GLN viral particles. Shown is electron microscopy of 293T cells transfected with GLN-2 or MoMLV. (a) GLN particles assembling and budding at the plasma membrane. (b) High magnification of mature extracellular GLN particles. (c) Morphological similarities between immature (top) and mature (bottom) GLN and MoMLV viral particles. Scale bars, 100 nm.
Conclusion.
The present investigation identified, among the 80 copies of GLN ERVs found in the mouse genome, 1 copy that is fully functional and generates type C retroviral particles that can mediate infection with a tropism restricted to mouse cells. The GLN elements are therefore bona fide mouse ecotropic retroviruses. Interestingly, their LTRs are active promoters in various cell lines, and Northern blot analyses have revealed GLN transcripts in several mouse tissues, including the thymus, spleen, and liver (25). It is therefore possible that a significant fraction of the particles previously detected in mouse cells and tissues by electron microscopy are actually GLN elements and not MLV particles, from which they appear to be indistinguishable at the structural level (Fig. 5C).
GLN-related sequences have also been detected by Southern blotting experiments performed on the genomic DNAs from a large set of Mus species (in those cases, at a high copy number), as well as from other rodents, including woodchuck, ground squirrel, hamster, gerbil, and rat (18, 25). Consistently, an in silico search that we performed in rat genome databases identified about 15 GLN-related copies, clearly belonging to the GLN family but much more degenerate, and less numerous, than in the mouse, with no full-length coding-competent copy. Taking the data together, it is thus likely that the GLN progenitor entered the rodent germ line prior to the Mus/Rattus split (i.e., >15 million years ago) (31)—and possibly even much earlier, before the radiation of Muridae—with amplification bursts then occurring, especially in the mouse genome. Being infectious, the GLN elements have most probably amplified by reinfection, and not by intracellular retrotransposition, of the germ line, as was the case in humans for the human ERV HERV-K(HML2) (2, 11).
Acknowledgments
We thank E. Pichard for technical assistance, C. Stocking for providing the plasmids encoding the ecotropic receptors, and C. Lavialle for critical reading of the manuscript.
This work was supported by the CNRS, by a grant from the Ligue Nationale contre le Cancer (Equipe Labellisée), and by a fellowship from the Association pour la Recherche sur le Cancer to D.R.
Footnotes
Published ahead of print on 20 February 2008.
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57659-666. [DOI] [PubMed] [Google Scholar]
- 2.Belshaw, R., V. Pereira, A. Katzourakis, G. Talbot, J. Paces, A. Burt, and M. Tristem. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. USA 2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benit, L., N. de Parseval, J.-F. Casella, I. Callebaut, A. Cordonnier, and T. Heidmann. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 715652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benit, L., J. B. Lallemand, J. F. Casella, H. Philippe, and T. Heidmann. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J. Virol. 733301-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard, W. 1958. Electron microscopy of tumor cells and tumor viruses; a review. Cancer Res. 18491-509. [PubMed] [Google Scholar]
- 6.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p. 343-435. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 7.Bonham, L., G. Wolgamot, and A. D. Miller. 1997. Molecular cloning of Mus dunni endogenous virus: an unusual retrovirus in a new murine viral interference group with a wide host range. J. Virol. 714663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromham, L., F. Clark, and J. J. McKee. 2001. Discovery of a novel murine type C retrovirus by data mining. J. Virol. 753053-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordonnier, A., J.-F. Casella, and T. Heidmann. 1995. Isolation of novel human endogenous retrovirus-like elements with foamy virus-related pol sequence. J. Virol. 695890-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dewannieux, M., A. Dupressoir, F. Harper, G. Pierron, and T. Heidmann. 2004. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 36534-539. [DOI] [PubMed] [Google Scholar]
- 11.Dewannieux, M., F. Harper, A. Richaud, C. Letzelter, D. Ribet, G. Pierron, and T. Heidmann. 2006. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 161548-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupressoir, A., G. Marceau, C. Vernochet, L. Bénit, C. Kanellopoulos, V. Sapin, and T. Heidmann. 2005. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. USA 102725-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eicher, E. M., K. W. Hutchison, S. J. Phillips, P. K. Tucker, and B. K. Lee. 1989. A repeated segment on the mouse Y chromosome is composed of retroviral-related, Y-enriched and Y-specific sequences. Genetics 122181-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esnault, C., J. F. Casella, and T. Heidmann. 2002. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 30e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fehrmann, F., M. Jung, R. Zimmermann, and H. G. Krausslich. 2003. Transport of the intracisternal A-type particle Gag polyprotein to the endoplasmic reticulum is mediated by the signal recognition particle. J. Virol. 776293-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferry, N., O. Duplessis, D. Houssin, O. Danos, and J. M. Heard. 1991. Retroviral-mediated gene transfer into hepatocytes in vivo. Proc. Natl. Acad. Sci. USA 888377-8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hein, S., V. Prassolov, Y. Zhang, D. Ivanov, J. Lohler, S. R. Ross, and C. Stocking. 2003. Sodium-dependent myo-inositol transporter 1 is a cellular receptor for Mus cervicolor M813 murine leukemia virus. J. Virol. 775926-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itin, A., and E. Keshet. 1986. A novel retroviruslike family in mouse DNA. J. Virol. 59301-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuff, E. L., and K. K. Lueders. 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51183-276. [DOI] [PubMed] [Google Scholar]
- 20.Maksakova, I. A., M. T. Romanish, L. Gagnier, C. A. Dunn, L. N. van de Lagemaat, and D. L. Mager. 2006. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik, H. S., and T. H. Eickbush. 2001. Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res. 111187-1197. [DOI] [PubMed] [Google Scholar]
- 22.Negre, D., and F. L. Cosset. 2002. Vectors derived from simian immunodeficiency virus (SIV). Biochimie 841161-1171. [DOI] [PubMed] [Google Scholar]
- 23.Negre, D., P. E. Mangeot, G. Duisit, S. Blanchard, P. O. Vidalain, P. Leissner, A. J. Winter, C. Rabourdin-Combe, M. Mehtali, P. Moullier, J. L. Darlix, and F. L. Cosset. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 71613-1623. [DOI] [PubMed] [Google Scholar]
- 24.Nermut, M. V., and D. J. Hockley. 1996. Comparative morphology and structural classification of retroviruses. Curr. Top. Microbiol. Immunol. 2141-24. [DOI] [PubMed] [Google Scholar]
- 25.Obata, M. M., and A. S. Khan. 1988. Structure, distribution, and expression of an ancient murine endogenous retroviruslike DNA family. J. Virol. 624381-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prassolov, V., S. Hein, M. Ziegler, D. Ivanov, C. Munk, J. Lohler, and C. Stocking. 2001. Mus cervicolor murine leukemia virus isolate M813 belongs to a unique receptor interference group. J. Virol. 754490-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyra, H., J. Boni, and J. Schupbach. 1994. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc. Natl. Acad. Sci. USA 911544-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribet, D., F. Harper, M. Dewannieux, G. Pierron, and T. Heidmann. 2007. Murine MusD retrotransposon: structure and molecular evolution of an “intracellularized” retrovirus. J. Virol. 811888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Ribet, D., F. Harper, A. Dupressoir, M. Dewannieux, G. Pierron, and T. Heidmann. 6 February 2008, posting date. An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res. doi: 10.1101/gr.073486.107. [DOI] [PMC free article] [PubMed]
- 29.Ribet, D., S. Louvet-Vallee, F. Harper, N. de Parseval, M. Dewannieux, O. Heidmann, G. Pierron, B. Maro, and T. Heidmann. 2008. Murine endogenous retrovirus MuERV-L is the progenitor of the “orphan” epsilon viruslike particles of the early mouse embryo. J. Virol. 821622-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt, M., T. Wirth, B. Kroger, and I. Horak. 1984. Structure and genomic organization of a new family of murine retrovirus-related DNA sequences (MuRRS). Nucleic Acids Res. 133461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Springer, M. S., W. J. Murphy, E. Eizirik, and S. J. O'Brien. 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. USA 1001056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tipper, C. H., C. E. Bencsics, and J. M. Coffin. 2005. Characterization of hortulanus endogenous murine leukemia virus, an endogenous provirus that encodes an infectious murine leukemia virus of a novel subgroup. J. Virol. 798316-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420520-562. [DOI] [PubMed] [Google Scholar]
- 34.Wolgamot, G., L. Bonham, and A. D. Miller. 1998. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J. Virol. 727459-7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolgamot, G., and A. D. Miller. 1999. Replication of Mus dunni endogenous retrovirus depends on promoter activation followed by enhancer multimerization. J. Virol. 739803-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yotsuyanagi, Y., and D. Szöllösi. 1981. Early mouse embryo intracisternal particles: fourth type of retrovirus-like particles associated with the mouse. J. Natl. Cancer Inst. 67677-683. [PubMed] [Google Scholar]
- 37.Zhao, Y., C. P. Jacobs, L. Wang, and S. C. Hardies. 1999. MuERVC: a new family of murine retrovirus-related repetitive sequences and its relationship to previously known families. Mamm. Genome 10477-481. [DOI] [PubMed] [Google Scholar]