Abstract
We report the identification and characterization of p33, the product of Kaposi's sarcoma-associated herpesvirus (KSHV) open reading frame 69 (ORF69), a positional homolog of the conserved herpesvirus protein UL31. p33 is expressed upon induction of viral lytic cycle with early kinetics. Immunofluorescence analysis revealed that in infected cell lines, the protein is localized in the nucleus, both in dotted spots and along the nuclear membrane. Nuclear fractionation experiments showed that p33 partitions with the nuclear matrix, and both immunoblotting of purified virions and immunoelectron microscopy indicated that the novel protein is not a component of the mature virus. Following ectopic expression in KSHV-negative cells, the protein was never associated with the nuclear membrane, suggesting that p33 needs to interact with additional viral proteins to reach the nuclear rim. In fact, after cotransfection with the ORF67 gene, the KSHV positional homolog of UL34, the p33 intranuclear signal changed and the two proteins colocalized on the nuclear membrane. A similar result was obtained when ORF69 was cotransfected with BFRF1, the Epstein-Barr virus (EBV) positional homolog of UL34 and ORF67. Finally, upon cotransfection, ORF69 significantly increased nuclear membrane reduplications induced by BFRF1. The above results indicate that KSHV p33 shares many similarities with its EBV homolog BFLF2 and suggest that functional cross-complementation is possible between members of the gammaherpesvirus subfamily.
A number of proteins are conserved among all herpesviruses and are designated “core proteins.” Among these, UL34 and UL31 protein family members have been studied extensively in the alphaherpesviruses herpes simplex virus type 1 (HSV-1) (54, 41), HSV-2 (53), pseudorabies virus (15), and equine herpesvirus 1 (EHV) (37) and in the betaherpesviruses human cytomegalovirus (CMV) (10) and mouse CMV (35, 47). For gammaherpesviruses, we initially identified and characterized the product of the Epstein-Barr virus (EBV) homolog of UL34, BFRF1 (12, 13), which has subsequently been shown to interact with the UL31 homolog BFLF2 (16, 26) and to play an essential role in viral envelopment at the nuclear membrane (14).
In all cases analyzed, UL34 and UL31 formed a complex at the inner nuclear membrane referred to as the nuclear egress complex (NEC), which is essential for viral egress (reviewed in reference 31), by interacting with viral (22, 44) and cellular kinases (35, 39) or with nuclear lamins (3, 16, 34, 43). These interactions contribute to the dismantling of the rigid nuclear lamina (35) and facilitate access of nucleocapids to the nuclear membrane for primary envelopment.
Very little is known of the intracellular viral maturation pathway of the second human gammaherpesvirus, Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), a lymphotropic herpesvirus detected in biopsies of all clinical and epidemiological forms of Kaposi's sarcoma and also linked to lymphoproliferative disorders, such as primary effusion lymphomas (PEL) and multicentric Castleman's disease (reviewed in reference 4). One recent report described the role of the KSHV envelope glycoprotein gB in viral egress (24), but KSHV members of the NEC have not been identified thus far.
Thus, to begin to assess the role of the NEC in KSHV intracellular maturation, we set up the present study to identify and characterize the product of open reading frame 69 (ORF69), the predicted UL31/BFLF2 homolog in KSHV. In addition, given the relatively high homology between EBV and KSHV, we attempted to investigate potential functional interactions between these two human gammaherpesviruses and we showed that ectopical expression of BFRF1, the EBV partner of BFLF2, modifies ORF69 intracellular localization and that ORF69, in a manner similar to that of its EBV homolog BFLF2 (16), significantly increases nuclear membrane reduplications induced by BFRF1.
MATERIALS AND METHODS
Cell culture and induction of viral lytic cycle.
BC1, BC3, BCBL-1, JSC-1, and TRExBCBL-1-Rta are human B-cell lines derived from PEL (6, 7, 36, 40) carrying latent KSHV. BC1 and JSC-1 are dually infected with KSHV and EBV. DG75 is an EBV-negative B-cell line derived from a Burkitt lymphoma (1). B95-8 is a marmoset B-cell line transformed with EBV (32). All cell lines were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (Invitrogen) in the presence of 5% CO2. The human embryonic kidney 293 cell line (17) was maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum.
In order to induce the KSHV lytic cycle, BC1, BC3, and BCBL-1 cells were treated with 20 ng/ml of 12-tetradecanoylphorbol-13-acetate (TPA), whereas TRExBCBL-1-Rta cells were cultured in the presence of doxycycline (final concentration, 1 μg/ml) (36). For virus production, BCBL-1 cells were synchronized in S phase and treated with 0.3 to 0.5 mM sodium butyrate (Sigma) for 72 h (30).
RNA analysis.
Total cellular RNA was prepared with TRIzol (Invitrogen). Twenty micrograms of total RNA per sample was separated by electrophoresis onto 1.2% agarose-6% formaldehyde gel in 20 mM morpholinepropanesulfonic acid (MOPS), pH 7.0, and transferred to a Nytran Plus membrane (Schleicher & Schuell) with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (3 M NaCl, 300 mM sodium citrate) by capillary transfer for 16 to 20 h (45). Hybridization for ORF69 was performed with the 32P-labeled RS3 oligonucleotide (5′-ACAGAGCGCACTGGAACCCCATGTTTGAGG-3′; genomic coordinates, 117103 to 117073). After stripping, the same blot was reprobed with a β-actin oligonucleotide (5′-TGTTGGCGTACAGGTCTTTGCGGATGTCCA-3′) for loading quantitation. Hybridization was carried out as described previously (16).
To characterize the ORF68/ORF69 polycistronic transcript, 5 μg of RNA extracted from TPA-induced BCBL-1 (48 h) was incubated with DNase I and amplified by reverse transcriptase PCR (RT-PCR). cDNA was synthesized with Moloney murine leukemia virus (Promega) by using an oligo(dT) primer (Invitrogen). PCR amplification was carried out with primer I (5′-ATGCATCTGCACTTTCCA-3′; genomic coordinates, 115607 to 115625), primer II (5′-TACCACCTGAACATGGTCAG-3′; genomic coordinates, 117687 to 117707), and the Expand polymerase (Roche) at 55°C. The PCR product was cloned in the pGEMT-easy vector and confirmed by sequencing. As a control, PCR was carried out without the reverse transcriptase reaction.
To identify the 5′ and 3′ ends of the ORF69 transcript, 5 μg of total RNA from TPA-induced BCBL-1 (48 h) was treated with DNase I (Promega) and amplified with a 5′/3′ rapid amplification of cDNA ends (RACE) second generation kit (Roche) according to the manufacturer's instructions. Briefly, to obtain the 5′ RACE fragment, first-strand cDNA was synthesized by using an ORF69-specific primer (RACE1, 5′-ACGGTAGACAGGTCTAGGG-3′; genomic coordinates, 117012 to 117031). After tailing, the purified cDNA was amplified by PCR at 56°C by using an oligo(dT)-anchor primer (supplied with the kit) and the RACE3 ORF69-specific primer (5′-GTGCATATCGGGGTCTTCCAT-3′; genomic coordinates, 116973 to 116994). The 3′ end of the ORF69 transcript was identified through PCR amplification of cDNA synthesized using an oligo(dT) anchor primer (supplied with the kit). PCR was performed with the UP4 primer (5′-GACATTCAAAAGGGAACCGCA-3′; genomic coordinates, 117612 to 117632) and the PCR anchor primer (supplied with the kit) at 56°C. Bands of 250 bp and 120 bp were observed on agarose gel from the 5′ and 3′ RACE, respectively. Both PCR products were cloned in pGEMT-easy vector (Promega) and sequenced.
Plasmids for recombinant protein expression and antibody production.
ORF69 was cloned from TPA-induced (for 48 h) BCBL-1 total RNA by RT-PCR after treatment with DNase I. cDNA synthesis was performed with SuperScript II (Invitrogen) and ORF69-down primer (5′-TTATAGGGCGTTGACAAGTGC-3′; genomic coordinates, 117629 to 117650). Control reaction was carried out without reverse transcriptase. Two microliters of cDNA was amplified by PCR with ORF69-up-BamHI (5′-ATGCCGAAATCAGTGTCCAG-3′; genomic coordinates, 116742 to 116763) and ORF69-down-EcoRI primers. The RT-PCR product was digested with BamHI and EcoRI restriction enzymes and then cloned into pET30a(+) (Novagen), previously cut with the same enzymes, to give rise to pET30a-ORF69. This plasmid codes for the His6-ORF69 fusion protein. Furthermore, the BamHI-ORF69-EcoRI fragment was subcloned into the pGEX2T expression plasmid (Pharmacia) containing glutathione S-transferase (GST) as N-terminal tag. His-ORF69 and GST-ORF69 fusion proteins were expressed in Escherichia coli BL21(DE3)Lys by adding 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 30°C. Fusion proteins were successively purified through column chromatography according to the manufacturer's protocols. The purified His-ORF69 protein was used to generate monoclonal antibody in 4-week-old BALB/c mice as described previously (16). Among the positive hybridoma clones, we selected the one producing the H8 monoclonal antibody. Furthermore, to produce a rabbit anti-ORF69 antibody, two peptides were chosen from the ORF69 amino acid (aa) sequence (PP27); peptide selection, immunization, and antibody production were performed by Eurogentec (Belgium). Peptides sequences were NH2-VSSKHRNGLRKFISD (aa 43 to 58) and NH2-CPSTENPTVAQGSRPQ (aa 121 to 136). The anti-ORF69 polyclonal antibody was named PP27.
KSHV-negative cell transient transfection.
To construct the pCMV-ORF69 plasmid, the BamHI-ORF69-EcoRI fragment was subcloned from pET30a-ORF69 in the pHD1013 vector (11) previously digested with BamHI and EcoRI. Moreover, ORF69 was cloned in pEGFP-C3 vector (Clontech) (pEGFP-ORF69) to generate the green fluorescent protein (GFP)-ORF69 fusion protein. Both pCMV-ORF69 and pEGFP-ORF69 lead ORF69 expression in eukaryotic cells. pEGFP-ORF67 was produced by cloning the BamHI-ORF67cDNA-EcoRI in pEGFP-C1 vector digested previously. ORF67 cDNA was amplified by RT-PCR performed on mRNA extracted from TPA-stimulated BCBL-1 cells, using specific primers for the 3′ end and for the 5′ end of the ORF. The purified ORF67 cDNA was then cloned and sequenced. ORF68-ORF69 cDNA was cloned in pCDNA3 vector (Invitrogen). pCMV-BFRF1 was described previously (12). Slide-cultured 293 cells were grown in 24-well plates to reach 80% of confluence and transfected with an equal amount of plasmid DNA (0.5 to 1 μg) by using FuGENE 6 (Roche) according to the supplier's recommendations. Empty vectors were used as a negative control.
Immunoblot analysis.
Cells were harvested by centrifugation at 1,500 × g at 4°C and rinsed in ice-cold phosphate-buffered saline (PBS). Since p33 was difficult to solubilize, cellular pellets were resuspended in PBS-1% Triton-2 M urea and briefly sonicated. Thirty to 50 μg of protein from total cell lysates was boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer for 5 min, resolved on a 10 to 12% SDS-PAGE gel, and electrophoretically transferred onto Protran nitrocellulose membranes (Schleicher & Schuell). Membranes were blocked for 1 h at room temperature (RT) in 5% nonfat dry milk-PBS-0.1% Tween-20 (blocking solution) and probed with different primary antibodies either for 1 h at room temperature or overnight at 4°C. After three washes in blocking solution, the membranes were further incubated with horseradish peroxidase-conjugated secondary antibodies (Sigma) for 45 min at room temperature and washed as described above. Bands were revealed using an enhanced chemiluminescence kit (Pierce). The primary antibodies used included anti-ORF69 monoclonal antibody (H8), anti-ORF69 polyclonal antibody (PP27; dilution, 1:200), anti-K8.1A monoclonal antibody (1:100) (purchased from ABI), goat anti-lamin B (1:100) (purchased from Santa Cruz), and anti-p53 polyclonal antibody (1:50) (purchased from Santa Cruz).
Indirect immunofluorescence assay (IFA).
Untreated or TPA-induced HHV-8-positive cell lines were washed with PBS, applied onto multispot microscope slides (Roth), and air dried. According to the antibody used for the staining, cells were either fixed and permeabilized in acetone-methanol (1:1) for 5 min at −20°C or fixed with 4% paraformaldehyde in PBS for 30 min at room temperature, incubated with 0.1 M glycine for 20 min at room temperature, and permeabilized with 0.1% Triton X-100 for 5 min at room temperature. Slide-cultured 293 cells and slide-cultured transfected 293 cells were treated with the paraformaldehyde protocol 24 to 48 h after transfection. The fixed cells were incubated with primary antibodies against ORF69 (mouse monoclonal H8, undiluted; rabbit polyclonal PP27, diluted 1:100), K8.1A (mouse monoclonal, diluted 1:400), BFRF1 (mouse monoclonal E10, diluted 1:50; rabbit polyclonal R319, 1:500) (12, 13), ORF50-Rta (rabbit polyclonal, diluted 1:500; kind gift of D. Ganem) (29), nuclear pore complex (monoclonal antibody 414, diluted 1:50; Berkeley Antibody Co.), and calreticulin (rabbit polyclonal, diluted 1:50; Stressgen Biotechnologies) for 1 h at room temperature. Slides were washed three times with PBS and further incubated with secondary antibody conjugated with either fluorescein isothiocyanate (Jackson) or Texas Red (Jackson) for 30 min at room temperature. DNA was stained with 4′,6′-diamino-2-phenylindole (DAPI) for 1 min at room temperature. After three washes in PBS, the slides were mounted with Mowiol (Calbiochem) and observed under an epifluorescence microscope (Zeiss Axiophot). For optical section fluorescence microscopy, cells were scanned with an ApoTome system (Zeiss, Oberkochen, Germany) connected with an Axiovert 200 inverted microscope (Zeiss, Oberkochen, Germany). Images were generated by stacking multiple section scans acquired in series of 0.5 μm with a charge-coupled device camera (Zeiss, Oberkochen, Germany), obtaining at least 10 sections per cell. Image analysis of optical sections was performed by transferring the data sets to Axiovision software (Zeiss, Oberkochen, Germany). The colocalization of fluorescence signals was evaluated with a Zeiss confocal laser scan microscope. To prevent cross talk between the two signals, the multitrack function was used.
Nuclear matrix fractionation.
Nuclear matrix isolation protocol was carried out as described previously (2). Briefly, cells were washed twice with ice-cold PBS, harvested, and resuspended in three volumes of cytoskeleton buffer (CSK) {10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM mgCl2, 1 mM EGTA, 1 mM dithiothreitol, 0.5% [vol/vol] Triton X-100, 1× Roche protease inhibitor cocktail}. After 5 min at 4°C, the cytoskeletal framework was separated from soluble proteins by centrifugation at 5,000 × g for 3 min. Chromatin was digested with 1 mg/ml of RNase-free DNase I in 2 volumes of CSK buffer containing the protease inhibitor cocktail for 15 min at 37°C and precipitated by adding ammonium sulfate (1 M stock in CSK) to a 0.25-M final concentration. Samples were incubated for 5 min at 4°C and then centrifuged. To remove all the DNA and histones from the nucleus, the pellet was further extracted with 2 M NaCl in CSK buffer for 5 min at 4°C and spun down. Finally, the remaining pellet was solubilized in urea buffer (8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris, pH 8); this fraction contains the nuclear matrix.
Purification of KSHV virions.
BCBL-1 cells (5 × 105/ml) were synchronized at G0 by 24 h of incubation in serum-free medium. After 24 h, fetal calf serum was added at a 10% final concentration. Cells were grown for 16 h to reach S phase (30), and the HHV-8 lytic cycle was induced with 0.3 mM Na-butyrate. Seventy-two hours postinduction, cells were spun down. The supernatant was collected and further cleared by centrifugation at 8,000 × g for 30 min and by filtration through 0.45-mm-pore-size filters. Virions were precipitated with polyethylene glycol (PEG) as described previously (14, 48). After the addition of NaCl to a final concentration of 2%, one-fifth the volume of 50% PEG 8000 in 0.5 M NaCl was added. Virions were then collected by centrifugation at 9,000 × g for 20 min. Pellets containing purified virion lysates lysates were resuspended in SDS-PAGE loading buffer, run on SDS-PAGE, and immunoblotted. Total cellular lysates were included as a positive control, while TPA-treated DG75 protein extracts were loaded as a negative control.
Electron microscopy.
For ultrathin cryosections, cells were fixed with a mixture of 2% paraformaldehyde and 0.2% glutaraldehyde in PBS for 1 h at 4°C, washed, and embedded in 12% gelatin in 0.1 M phosphate buffer that was solidified on ice. Gelatin blocks were infused with 2.3 M sucrose for 3 h at 4°C, frozen in liquid nitrogen, and cryosectioned. Ultrathin cryosections were collected using sucrose and methyl cellulose and incubated with anti-BFRF1 monoclonal antibody diluted 1:20 in PBS-1% bovine serum albumin. Following several washes in PBS-0.1% bovine serum albumin, the sections were incubated with 18 nm colloidal gold (prepared by the citrate method) conjugated with protein A diluted 1:10 in PBS. Control experiments were performed by the omission of the primary antibody from the labeling procedure. Finally, ultrathin cryosections were stained with a solution of 2% methyl cellulose and 0.4% uranyl acetate before examination by electron microscopy.
For conventional transmission electron microscopy, cells were washed three times and fixed with 2% glutaraldehyde in the same buffer, pH 7.4, for 2 h at 4°C, stained with uranyl acetate (5 mg/ml), dehydrated in acetone, and embedded in Epon 812. Thin sections, unstained or stained with acetate and lead hydroxide, were examined under a Philips Morgagni electron microscope. Quantitative image analysis of the ultrastructural alterations of the nuclear membrane was performed on 30 ultrathin cell sections crossing the nucleus for each transfectant randomly recorded by a digital charge-coupled device camera and analyzed by analySIS software (Soft Imaging System, Münster, Germany). The increase in the thickness of the nuclear membranes due to reduplications (measured as μm2 membrane area) was evaluated by μm2 area/10 μm length.
RESULTS AND DISCUSSION
In a recent paper, Jenner and colleagues (20) showed by DNA microarray that ORF69 belongs to a class of KSHV ORFs transcribed in the first hours after lytic cycle induction. KSHV genome analysis using the NCBI Entrez Nucleotide database showed that ORF69 starts at nucleotide (nt) 116742, proceeds to nt 117650, and codes for a putative protein of 302 aa. ORF69 primary amino acid sequence computer analysis indicated an apparent molecular mass of 33,156 kDa and a potential nuclear localization. Moreover, the sequence contains 21 possible phosphorylation residues (14 serine, 5 threonine, and 1 tyrosine), 6 potential sites for protein kinase C, and 2 for casein kinase II phosphorylation. As mentioned in the introduction, ORF69 is the positional homolog of UL31, which has been studied extensively in numerous human and nonhuman herpesviruses (10, 15, 27, 35, 37, 41, 47, 53, 54). Figure 1, showing the UL31 family members of human herpesviruses, illustrates that KSHV ORF69 shares identities of 37% with EBV BFLF2, 21% with HSV-1 UL31, and 19% with CMV UL53, suggesting a potential related function.
FIG. 1.
Amino acid sequence comparison between the putative protein encoded by KSHV ORF69 (p33) and its homologs in human herpesviruses: HSV-1 UL31, CMV UL53, and EBV BFLF2. Sequences were aligned with the MacVector 7.0 program. Identical and similar amino acid residues are highlighted by dark and light solid boxes, respectively. Numbers indicate amino acid positions.
In this study, KSHV ORF69 cDNA was amplified according to nucleotide sequence by RT-PCR performed on mRNA extracted from TPA-stimulated BCBL-1 cells by using primers specific for the ORF69 stop codon at the 3′ end and for the predicted translational initiation site at the 5′ end of the ORF. The purified ORF69 cDNA was then cloned and sequenced as described in Materials and Methods.
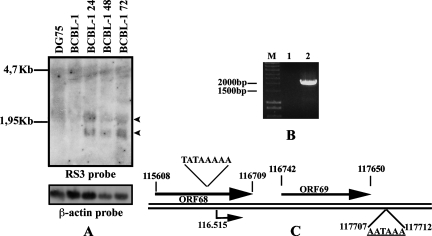
To identify the ORF69 transcript and to describe its expression kinetics, we performed a Northern blot analysis on total RNA isolated from TPA-treated BCBL-1 cells at different time points. It has recently been suggested that ORF69 belongs to KSHV ORFs insensitive to the DNA polymerase inhibitor cidofovir; so, it is an early gene (28). As shown in Fig. 2A, hybridization with an ORF69 oligonucleotide probe revealed two transcripts, 1.2 and 2.1 kb, after KSHV lytic activation. Both signals were still high at 72 h postinduction. No specific bands were observed in unstimulated BCBL-1 cells or the KSHV-negative DG75 cell line. The 1.2- and 2.1-kb transcripts were supposed to correspond to the ORF69 monocistronic and ORF68-ORF69 bicistronic transcripts, respectively. β-Actin expression was included as a loading control.
FIG. 2.
ORF69 transcription analysis in BCBL-1 cells. (A) Kinetics of ORF69 expression upon chemical induction in BCBL-1 cell line. Cells were untreated or induced with TPA for 24, 48, or 72 h; total RNA was prepared at the times indicated, and 20 mg was analyzed by Northern blotting. As a negative control, RNA from DG75 treated with TPA for 72 h was loaded. The filter was hybridized with an oligonucleotide specific for ORF69 (RS3 probe) and subsequently with a β-actin probe. The two specific ORF69 transcripts are shown with arrowheads. (B) ORF68-ORF69 bicistronic transcript. Reverse transcription was performed on total RNA from BCBL-1 cells treated with TPA for 48 h by using oligo(dT), and PCR was carried out with primers I and II. Nucleotide sequences of primers I and II are described in Materials and Methods. Lanes: M, marker; 1, PCR without reverse transcription; 2, RT-PCR. (C) 5′-3′ RACE analysis of ORF69 transcript. Schematic drawing of the location of ORF69 in the KSHV genome. Numbers correspond to nucleotide positions, and arrows indicate the direction ORF68 and ORF69. The ORF69 transcription start is at position 116515. TATAAAAA is the putative TATA box. AATAAA is the polyadenylation recognition sequence.
To demonstrate that the 2.1-kb band corresponds to the ORF68-ORF69 bicistronic transcript, total RNA from BCBL-1 treated with TPA for 48 h was reversed with oligo(dT). PCR was then performed with primer I (ORF68 specific), together with primer II, starting 37 nt downstream of the ORF69 stop codon (Fig. 2B, lane 2), and confirmed by sequencing. No bands were observed when PCR was carried out on sample not treated with reverse transcriptase (Fig. 2B, lane 1). We next determined the sequence of ORF69 monocistronic mRNA by performing 5′-3′ RACE on RNA extracted from TPA-induced BCBL-1 cells. Figure 2C shows a diagram representing the ORF68 and ORF69 locations in the KSHV genome and the results obtained after ORF69 5′ and 3′ end mapping. Sequence analysis of 5′-3′ RACE products indicated that the ORF69 transcription start site is located at nt 116515. Transcription proceeds 79 nt after the TAA, giving rise to a 1,214-nt product. Scanning of the regions upstream of the ORF69 transcription start and downstream of the stop codon revealed the presence of a putative TATA box and a polyadenylation recognition sequence. Since ORF69 monocistronic transcript sequence is identical to ORF69 genomic sequence, these data showed that the ORF69 transcript is not spliced. The bicistronic transcript we described is probably necessary to allow ORF68 expression, since there is a single shared polyadenylation signal downstream of ORF69 at nt 117707. It is known that the expression of several KSHV ORFs requires a pA site far downstream and that the resulting transcripts are usually bicistronic or tricistronic (55).
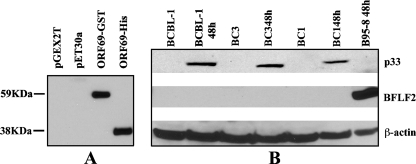
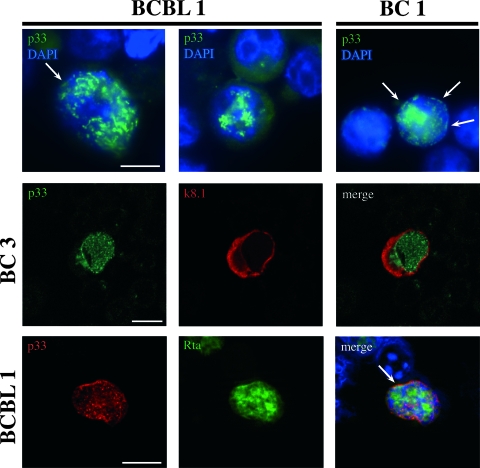
Next, to characterize the ORF69 product, a mouse monoclonal antibody was raised in mice immunized with a purified His-ORF69 fusion protein expressed in E. coli. His-ORF69 was generated as described in Materials and Methods. We selected the H8 monoclonal antibody based on its specific immunoreactivity in Western blotting against both His-ORF69 and GST-ORF69 fusion proteins (Fig. 3A). No cross-reactivity was detected with extracts from E. coli transformed with either pET30a or pGEX-2T empty vectors. To identify the ORF69 product, SDS lysates from different KSHV-positive cell lines and from B95-8 cells (KSHV negative and EBV positive) were run and immunoblotted with the H8 anti-ORF69 monoclonal antibody. As shown in Fig. 3B, H8 recognized a specific band of approximately 33 kDa (indicated as p33) in the BCBL-1, BC3, and BC1 cell lines after treatment with TPA. The observed molecular mass is very close to the predicted size of ORF69 product. No signal was present in uninduced BCBL-1, BC3, and BC1 cell lines. Furthermore, no cross-reaction with EBV BFLF2 was observed in B95-8 cells incubated for 48 h with TPA and sodium butyrate. p33 expression appeared to be maximal between 36 and 48 h postinfection. The protein was also expressed following KSHV lytic cycle induction by ORF50, as observed in TRExBCBL-1-Rta cells after treatment with doxycycline (data not shown). p33 intracellular localization was analyzed by IFA. TPA-induced and uninduced KSHV-positive cell lines were stained with a rabbit polyclonal antibody raised against p33. Figure 4 shows a selection of optical sections (0.5 μm thick) of TPA-treated BCBL-1 and BC1 cells immunolabeled for p33 and acquired by deconvolution microscopy. The staining pattern consisted of a dotted distribution of p33 inside the nucleus and at the nuclear rim, and the two localization patterns were equally frequent in the two cell types. In contrast, the protein was not detected in the cytoplasm. Nuclei were stained with DAPI. No signal was revealed on untreated cells (data not shown). To further confirm ORF69 protein localization, double staining with rabbit anti-p33 antibody and anti-K8.1A monoclonal antibody, which recognizes a KSHV membrane glycoprotein (56), was carried out on TPA-stimulated BC3 cells and analyzed by confocal microscopy. As shown in Fig. 4 (middle panels), K8.1A was expressed both in the cytoplasm and on the plasma membrane and, consistent with the observed p33 nuclear localization, the two proteins did not colocalize. Finally, since it was suggested that HSV-1 UL31 could assist the synthesis and packaging of viral DNA (9), we asked whether p33 could colocalize with viral replication compartments. For this purpose, we carried out a double immunofluorescence by using anti-KSHV-Rta polyclonal antibody, together with anti-p33 monoclonal antibody, on TRExBCBL-1-Rta cells after treatment with doxycycline. Rta was chosen since its association with KSHV replication compartments has recently been demonstrated (51). The results confirm that both p33 and Rta are localized in the nucleus, with sparse areas of colocalization (Fig. 4, bottom panel).
FIG. 3.
Identification of p33. (A) Reactivity of H8 monoclonal antibody. Protein extracts from BL21(DE3)Lys transformed with pGEX2T, pET30a, pGEX2T-ORF69 (ORF69-GST), or pET30a-ORF69(ORF69-His) were analyzed by Western blotting after IPTG induction. (B) Expression of ORF69-encoded protein in different PEL cell lines analyzed by Western blotting. The H8 monoclonal antibody revealed a 33-kDa-specific band (p33) after KSHV lytic cycle induction with TPA in BCBL-1, BC3, and BC1. B95-8 cells were treated with TPA and butyrate. Anti-BFLF2 antibody showed EBV activation. Anti-β-actin was used to calibrate the protein loading.
FIG. 4.
p33 intracellular localization. (Top panels) Indirect immunofluorescence on BCBL-1 and BC1 cell lines stimulated with 20 ng/ml of TPA for 48 h and incubated with PP27 rabbit polyclonal antibody against p33. At least 10 optical sections (0.5 μm) per cell were obtained, and a selection is shown as a gallery. Nuclei are stained with DAPI. p33 is distributed in dots within the nucleus as well as on portions of the nuclear membrane (arrows). (Middle panels) Confocal microscopy performed for the BC3 cell line after 48 h of treatment with TPA; staining was with rabbit anti-p33 antibody and anti-K8.1A monoclonal antibody. K8.1A is present both in the cytoplasm and on the plasma membrane. Merged images do not show any p33 colocalization with K8.1A. (Bottom panels) Confocal microscopy of TRExBCBL-1-Rta cells after treatment with doxycycline and incubation with anti-p33 monoclonal antibody, together with anti-Rta polyclonal antibody. Both p33 and Rta are localized within the nucleus, as expected. Merge reveals colocalization of the two signals in a few intranuclear spots. Bars = 10 μm.
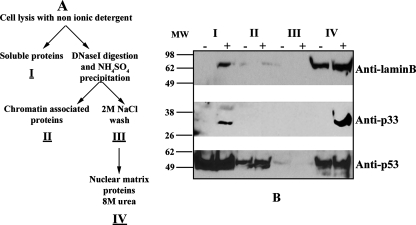
Additional experiments were then undertaken to evaluate whether the ORF69 product is associated with the nuclear matrix. We examined this possibility also, since the pattern of intranuclear distribution of p33 and its difficult solubility are characteristics of other cellular and viral proteins, such as the HSV-1 UL31 homolog (8), known to partition with the aforementioned nuclear component. The nuclear matrix is a nuclear substructure that remains after the majority of DNA and soluble and chromatin-bound proteins have been removed from the nucleus. It is involved in chromatin organization and in different aspects of nucleic acid metabolism and consists of the nuclear pore complexes embedded in the nuclear lamina. Further, the main components of the lamina network and of nuclear architecture are lamins which are grouped as A and B types: A-type lamins are expressed in a tissue-specific manner, whereas B-type lamins are essential for cell viability (reviewed in reference 18). Figure 5A shows a scheme of the high-salt nuclear matrix extraction procedure adapted from the method of Ben-Yehoyada et al. (2) and used to separate the various nuclear fractions from untreated and TPA-induced BCBL-1 cells. Briefly, in the first step, nonionic detergent was employed to extract soluble proteins (fraction I). In the next step, chromatin was solubilized by RNase-free DNase I digestion and the addition of ammonium sulfate allowed the precipitation of the chromatin-associated proteins (fraction II). After we washed the pellet with 2 M NaCl (fraction III), the remaining pellet, which contains the structural nuclear matrix and the nuclear matrix-associated proteins, was solubilized in 8 M urea (fraction IV). Fractions were then analyzed by SDS-PAGE and immunoblotting. As shown in Fig. 5B, the antibody anti-lamin B, one of the major components of nuclear matrix (25), was used to verify the effectiveness of nuclear matrix fractionation (top panel). Lamin B was detected mainly in the nuclear matrix fraction in both uninduced and TPA-treated BCBL-1 cells. The reaction with the specific polyclonal antibody anti-p33 demonstrated a strong association of the ORF69 product with the nuclear matrix fraction in the TPA-induced BCBL-1 cells (Fig. 5B, middle panel). A small amount of p33 was also present in the soluble fraction, whereas no signal was detectable in fractions obtained from untreated cells, as expected. Finally, an anti-p53 antibody was used as a control for the fractions collected (Fig. 5B, bottom panel) (21). These experiments demonstrate that p33 cofractionates with the nuclear matrix.
FIG. 5.
p33 is a nuclear matrix-associated protein. (A) Scheme of high-salt nuclear matrix fractionation protocol from Ben-Yehoyada et al. (2). (B) Distribution of p33 in the different fractions. Nuclear matrix fractionation was performed on the BCBL-1 cells that were untreated (−) or were induced with TPA for 48 h (+). Proteins were analyzed by SDS-PAGE and immunoblotting. The membrane was incubated with goat anti-lamin B (top panel) and mouse anti-p53 (bottom panel) as a control for the fractions obtained. H8 monoclonal antibody detects p33 predominantly in the nuclear matrix fraction (middle panel). I, II, III, and IV: fractions after high-salt purification. MW, molecular mass in kilodaltons.
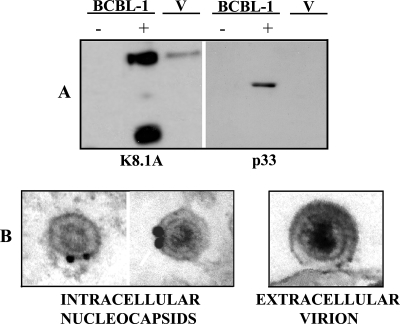
In addition, to examine whether p33 is a virion component, extracellular KSHV particles were precipitated from the supernatant of induced BCBL-1 cells (30) by using PEG (43). Untreated and induced BCBL-1 cell extracts (Fig. 6A, lanes − and +, respectively) as well as purified virion lysates (Fig. 6A lane V) were resolved on SDS-PAGE gels and immunoblotted. As shown in Fig. 6, p33 was detected in induced BCBL-1 cells but not in untreated BCBL-1 cells or in virion lysates. K8.1A monoclonal antibody was used as a positive control; in this case, the signal was also revealed in virion extracts, as expected (55). To further analyze the presence of p33 in virions, we performed immunoelectron microscopy of induced BCBL-1 cells. Immunogold labeling in cryosections of the induced cells was seen on intracellular nucleocapsids, whereas extracellular virions were consistently unlabeled (Fig. 6B). This finding is in accordance with the results reported for p33 homologs in other herpesviruses (15, 16, 42).
FIG. 6.
p33 is not detected in HHV-8 mature virions. (A) Western blot analysis was performed to evaluate the presence of p33 in the extracellular virions. BCBL-1 cells were synchronized in S phase and treated with Na-butyrate for 72 h. Total cellular lysates from uninduced (−) BCBL-1 cells, from Na-butyrate-treated BCBL-1 cells (+), and from PEG-purified virion lysates (V) were run on SDS-PAGE gels and immunoblotted. Incubation with H8 shows that p33 is not present in extracellular virions. Anti-K8.1A monoclonal antibody was used as a control for extracellular KSHV. (B) Immunoelectron microscopy with rabbit anti-p33 of KSHV particles in induced BCBL-1. Only intracellular KSHV nucleocapsids in the perinuclear area (+), not extracellular virions (V), were immunogold labeled.
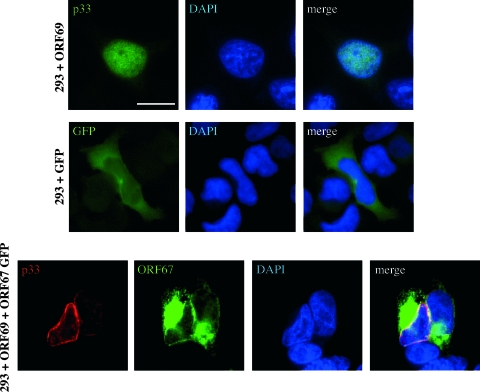
We next examined the ectopic expression of ORF69 in order to define whether the localization of p33 is affected by additional viral proteins, as is the case for its homologs in other herpesviruses (31). For this purpose, KSHV-negative 293 cells were transfected with ORF69 and subjected to IFA staining with rabbit anti-p33. As shown in Fig. 7 (top panels), p33 is localized in the nucleus. It is noteworthy that while we could also observe a nuclear dotted distribution in some 293-ORF69 transfected cells, no apparent association with the nuclear membrane was detected, suggesting that p33 needs to interact with additional KSHV proteins in order to reach the nuclear rim. Finally, the transfection of ORF68-ORF69 cDNA confirms that p33 is not produced from the bicistronic transcript, since we did not observe any p33 staining (data not shown).
FIG. 7.
Intracellular localization of ectopically expressed p33. Human embryonic kidney 293 cells were transfected with ORF69 and, after 24 h, were stained with H8 monoclonal antibody. IFA analysis shows that p33 is confined to the nuclei of transfected cells (top panels). ORF69 cotransfection with EGFP-ORF67 in 293 cells (bottom panels). Cells were immunostained with H8 monoclonal antibody detected with sheep anti-mouse Cy3 24 h after transfection. p33 is localized at the nuclear rim in the cotransfected cells, and merged images show colocalization of the two signals. As a control, 293 cells were transfected with pEGFP empty vector (middle panels). Nuclei were stained with DAPI. Bars = 10 μm.
Since a conserved feature in all herpesvirus subgroups studied so far is the interaction between UL34 and UL31 homologs, a similar interaction most likely occurs also in the case of KSHV. A systematic yeast two-hybrid interactome map has recently been reported for KSHV (50). By that analysis, no interaction between ORF69 and ORF67, the predicted homolog of UL34, has been revealed, but this could be due to technical limitations of yeast two-hybrid screening. To evaluate this hypothesis, ORF69 was transiently transfected, together with GFP-ORF67 plasmid, into 293 cells. As shown in Fig. 7 (bottom panel), a different picture emerged from this experiment, as IFA with H8 monoclonal antibody against p33 indicated that this protein localized at the nuclear rim in the presence of GFP-ORF67. Conversely, GFP-ORF67 localization appeared to be unaffected by ORF69 expression, since it was detected consistently both at the nuclear rim (although with a minor staining intensity) and within cytoplasmic aggregates, in accordance with a recent report (46). Furthermore, the two proteins appeared to colocalize in some areas of the nuclear membrane. pEGFP empty vector was used as a control (Fig. 7, middle panel). Altogether, these results suggest that p33 could interact with the ORF67 product, which seems to be required for p33 targeting at the nuclear rim, as described for the other herpesvirus homologs. Experiments are currently ongoing in the laboratory to identify and characterize the product of ORF67, which will allow us to validate this hypothesis for infected cells.
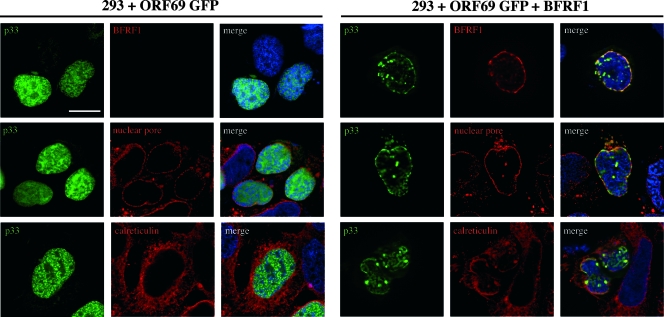
Finally, since it has recently been demonstrated that cross-complementation is possible between UL34/UL31 proteins of the same herpesviral subfamily (47) and KSHV and EBV are both human gammaherpesviruses, we asked whether p33 could interact with the EBV protein BFRF1, which is known to colocalize on the nuclear rim with BFLF2, the ORF69 homolog (16, 26). A proteomic analysis of EBV has recently confirmed the reported interaction between BFRF1 and BFLF2 and has highlighted BFLF2 as a possible “hub,” with several potential interactions with numerous cellular proteins, some of which are considered of great interest for membrane fusion events (5). We first attempted to analyze the potential ORF69-BFRF1 interactions in dually EBV- and KSHV-infected JSC-1 PEL cells, following induction of the viral replicative cycle. However, despite several attempts with different inducers, we have not been able to activate both the EBV and the KSHV lytic cycles in the same cell population (data not shown). This inability was not unexpected, since the majority of PEL cells are coinfected in a latent state and no previous attempts to contemporaneously activate the lytic cycle of both viruses have been successful, as reported by Miller et al. (33) for BC-1 cells and by Cannon et al. (6) for JSC-1 cells. We next examined the possible association of BFRF1 with p33 following ectopic expression of the two proteins. For this purpose, 293 cells were transfected with enhanced green fluorescent protein (EGFP)-ORF69 or cotransfected with EGFP-ORF69 and BFRF1, and p33 and BFRF1 localization was analyzed by IFA. As shown in Fig. 8 (right panels), coexpression of the two proteins resulted in their colocalization on the nuclear membrane. p33 also colocalized with nuclear pores, used as a marker of the nuclear membrane as well as with calreticulin, an endoplasmic reticulum marker, but was limited to the cisternae composing the nuclear membrane. The left panels of Fig. 8 show that in the absence of BFRF1, p33 maintains its diffused or dotted nuclear distribution, with no colocalization with either nuclear pore complex or endoplasmic reticulum. Thus, BFRF1 alters ORF69 intracellular localization and the two proteins colocalize on the nuclear membrane. In conclusion, p33 maintains its nuclear localization in both infected (Fig. 4) and transfected cells (Fig. 7 and 8). In infected cells, the nuclear localization is in dots within the nucleus as well as along the nuclear rim, observable in an equal proportion of cells, independently of the cell type or of the presence of EBV. Upon transfection, p33 is diffused in the nucleus, whereas it associates with the nuclear membrane if cotransfected with ORF67 or BFRF1.
FIG. 8.
Indirect immunofluorescence of 293 cells transfected with EGFP-ORF69 or cotransfected with EGFP-ORF69 and BFRF1. p33 is localized diffusely within the nucleus in cells transfected with EGFP-ORF69 alone and does not colocalize with nuclear pore complexes or the endoplasmic reticulum (calreticulin) (left panels). On the contrary, the presence of BFRF1 modifies the localization of p33, which colocalizes on the nuclear membrane with BFRF1, nuclear pores, and the endoplasmic reticulum (right panels). Bars = 10 μm.
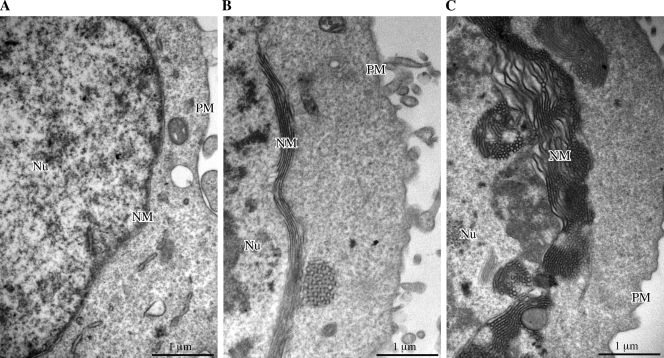
A second functional modification known to be induced by UL34 and UL31 coexpression consists of ultrastructural modifications of the nuclear membrane. We have recently reported (16) that EBV BFRF1, transfected in 293 cells, caused membrane reduplications corresponding to multilayered portions of the nuclear membrane and that this effect was markedly increased following cotransfection with BFRF1 and BFLF2. We previously reported (49) that these membrane duplications are a characteristic feature of EBV-producing cells, and they also have previously been described as occurring during KSHV replication in PEL cells (33, 38). Thus, by conventional transmission electron microscopy, we analyzed the possible alterations of the nuclear membrane following transfection of 293 cells with either ORF69 or BFRF1 or cotransfection with ORF69 and BFRF1, and the results are shown in Fig. 9. As expected (16), BFRF1 caused membrane duplications (Fig. 9B). When cotransfected with BFRF1, ORF69 accentuated this effect (Fig. 9C) in a manner similar to that of its EBV homolog BFLF2, probably rendering these areas of the nuclear membrane more suitable for viral envelopment. Quantitation of this effect is reported in Table 1. Neither in cells transfected with ORF69 (Fig. 9A) nor in cells untransfected or transfected with vector alone were similar nuclear membrane alterations observed (data not shown). Similarly, coexpression of HSV-1 UL34 and UL31, in the absence of any other viral factor, has very recently been shown to induce modifications of the nuclear membrane, resulting in vesicle formation (23).
FIG. 9.
Electron microscopic analysis of the nuclear membrane structure in transfected 293 cells. (A) Cells transfected with ORF69. (B) Cells transfected with BFRF1. (C) Cells cotransfected with ORF69 and BFRF1. In cells expressing BFRF1 alone (B), large areas of nuclear membrane reduplications are evident, which cannot be observed in ORF69-expressing cells (A). In cells expressing both BFRF1 and ORF69 (C), the overall ultrastructure of the nuclear membrane appears drastically modified, showing interrupted areas of thick and irregular multilayering.
TABLE 1.
Quantitation of nuclear membrane reduplications induced in 293 cells
| Cell type and transfection | Total nuclear membrane length (μm) | Total nuclear membrane thickness (μm2) | Nuclear membrane thickness (μm2)/10 μm of lengtha |
|---|---|---|---|
| 293+ORF69 | 163.23 | 7.1 | 0.40 ± 0.02 |
| 293+BFRF1 | 270.93 | 70.5 | 2.64 ± 0.18b |
| 293+BFRF1+ORF69 | 176.97 | 93.1 | 5.23 ± 0.37c |
Results are presented as means ± standard errors.
P value (by the Student t test) was < 0.0001 by a comparison with 293 plus ORF69 cells.
P value (by the Student t test) was 0.001 by a comparison with 293 plus BFRF1 cells.
In conclusion, our results indicate that p33 shares many similarities with its EBV homolog BFLF2 and suggest that in human gammaherpesviruses, cross-complementation is possible between KSHV and EBV proteins. EBV and KSHV coinfect the majority of PEL cell lines, and molecular interactions between the two viruses have previously been reported (52). Whether these interactions might be relevant for the pathogenesis of EBV- and KSHV-associated diseases deserves further study.
Future studies will focus on the generation of a recombinant KSHV, by the use of bacterial artificial chromosomal technology, that is deleted for expression of ORF69 to assign a more precise functional role to this protein in viral replication.
Finally, given the importance of viral lytic replication for viral transmission and pathogenesis (19), a better knowledge of KSHV intracellular maturation might ultimately lead to the development of novel therapeutical approaches in KSHV-associated diseases.
Acknowledgments
This work was partially supported by grants from the MIUR, Ministero della Sanità, Programma AIDS, and from the Associazione Italiana per la Ricerca sul Cancro.
We thank D. Ganem and J. Jung for the kind gift of the anti-ORF50 polyclonal antibody and the TRExBCBL-1-Rta cells, respectively.
Footnotes
Published ahead of print on 27 February 2008.
REFERENCES
- 1.Ben-Bassat, H., N. Goldblum, S. Mitrani, T. Goldblum, J. M. Yoffey, M. M. Cohen, Z. Bentwich, B. Ramot, E. Klein, and G. Klein. 1977. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75). Int. J. Cancer 1927-33. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Yehoyada, M., I. Ben-Dor, and Y. Shaul. 2003. c-Abl tyrosine kinase selectively regulates p73 nuclear matrix association. J. Biol. Chem. 27834475-34482. [DOI] [PubMed] [Google Scholar]
- 3.Bjerke, S. L., and R. J. Roller. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boshoff, C., and R. Weiss. 2002. AIDS-related malignancies. Nat. Rev. Cancer 2373-382. [DOI] [PubMed] [Google Scholar]
- 5.Calderwood, M. A., K. Venkatesan, L. Xing, M. R. Chase, A. Vazquez, A. M. Holthaus, A. E. Ewence, N. Li, T. Hirozane-Kishikawa, D. E. Hill, M. Vida, E. Kieff, and E. Johannsen. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. USA 1047606-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, J. S., D. Ciufo, A. J. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma derived cell line yields a high infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 7410187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 1862708-2714. [PubMed] [Google Scholar]
- 8.Chang, Y. E., and B. Roizman. 1993. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein which partitions with the nuclear matrix. J. Virol. 676348-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 718307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dal Monte, P., S. Pignatelli, N. Zini, M. Maraldi, E. Perret, M. C. Prevost, and M. P. Landini. 2002. Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J. Gen. Virol. 831005-1012. [DOI] [PubMed] [Google Scholar]
- 11.Davis, M. G., and E. S. Huang. 1988. Transfer and expression of plasmids containing human cytomegalovirus immediate-early gene 1 promoter-enhancer sequences in eukaryotic and prokaryotic cells. Biotechnol. Appl. Biochem. 16-12. [PubMed] [Google Scholar]
- 12.Farina, A., R. Santarelli, R. Gonnella, R. Bei, R. Muraro, G. Cardinali, S. Uccini, G. Ragona, L. Frati, A. Faggioni, and A. Angeloni. 2000. The BFRF1 gene of Epstein-Barr virus encodes a novel protein. J. Virol. 743235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farina, A., G. Cardinali, R. Santarelli, R. Gonnella, J. Webster-Cyriaque, R. Bei, R. Muraro, L. Frati, A. Angeloni, M. R. Torrisi, and A. Faggioni. 2004. Intracellular localization of the Epstein-Barr virus BFRF1 gene product in lymphoid cell lines and oral hairy leukoplakia lesions. J. Med. Virol. 72102-111. [DOI] [PubMed] [Google Scholar]
- 14.Farina, A., R. Feederle, S. Raffa, R. Gonnella, R. Santarelli, L. Frati, A. Angeloni, M. R. Torrisi, A. Faggioni, and H. J. Delecluse. 2005. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 793703-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonnella, R., A. Farina, R. Santarelli, S. Raffa, R. Feederle, R. Bei, M. Granato, A. Modesti, L. Frati, H. J. Delecluse, M. R. Torrisi, A. Angeloni, and A. Faggioni. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 793713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 3659-74. [DOI] [PubMed] [Google Scholar]
- 18.Gruenbaum, Y., A. Margalit, R. D. Goldman, D. K. Shumaker, and K. L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 621-31. [DOI] [PubMed] [Google Scholar]
- 19.Herndiern, B., and D. Ganem. 2001. The biology of Kaposi's sarcoma. Cancer Treat. Res. 10489-126. [DOI] [PubMed] [Google Scholar]
- 20.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, M., T. Axe, R. Holgate, C. P. Rubbi, A. L. Okorokov, T. Mee, and J. Milner. 2001. p53 binds the nuclear matrix in normal cells: binding involves the proline-rich domain of p53 and increases following genotoxic stress. Oncogene 205449-5458. [DOI] [PubMed] [Google Scholar]
- 22.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 822363-2371. [DOI] [PubMed] [Google Scholar]
- 23.Klupp, B. G., H. Granzow, W. Fuchs, G. M. Keil, S. Finke, and T. C. Mettenleiter. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. USA 1047241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnan, H. H., N. Sharma-Walia, L. Zeng, S. J. Gao, and B. Chandran. 2005. Envelope glycoprotein gB of Kaposi's sarcoma-associated herpesvirus is essential for egress from infected cells. J. Virol. 7910952-10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzmina, S. N., T. V. Buldyaeva, S. B. Akopov, and I. B. Zbarsky. 1984. Protein patterns of the nuclear matrix in differently proliferating and malignant cells. Mol. Cell. Biochem. 58183-186. [DOI] [PubMed] [Google Scholar]
- 26.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 32099-106. [DOI] [PubMed] [Google Scholar]
- 27.Lötzerich, M., Z. Ruzsics, and U. H. Koszinowski. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 8073-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, M., J. Suen, C. Frias, R. Pfeiffer, M. H. Tsai, E. Chuang, and S. L. Zeichner. 2004. Dissection of the Kaposi's sarcoma-associated herpesvirus gene expression program by using the viral DNA replication inhibitor cidofovir. J. Virol. 7813637-13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252304-312. [DOI] [PubMed] [Google Scholar]
- 30.McAllister, S. C., S. G. Hansen, I. Messaoudi, J. Nikolich-Zugich, and A. V. Moses. 2005. Increased efficiency of phorbol ester-induced lytic reactivation of Kaposi's sarcoma-associated herpesvirus during S phase. J. Virol. 792626-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mettenleiter, T. C., B. G. Klupp, and H. Granzow. 2006. Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9423-429. [DOI] [PubMed] [Google Scholar]
- 32.Miller, G., and M. Lipman. 1973. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. USA 70190-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, G., L. Heston, E. Grogan, L. E. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chan. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mou, F., T. Forest, and J. D. Baines. 2007. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 816459-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297854-857. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 774205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neubauer, A., J. Rudolph, C. Brandmuller, F. T. Just, and N. Osterrieder. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300189-204. [DOI] [PubMed] [Google Scholar]
- 38.Orenstein, J. M., D. M. Ciufo, J. P. Zoeteweji, A. Blauvelt, and G. S. Hayward. 2000. Morphogenesis of HHV8 in primary human dermal microvascular endothelium and primary effusion lymphomas. Ultrastruct. Pathol. 24291-300. [DOI] [PubMed] [Google Scholar]
- 39.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2342-346. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 758803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 768939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 785564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Sander, G., A. Konrad, M. Thurau, E. Wies, R. Leubert, E. Kremmer, H. Dinkel, T. Schulz, F. Neipel, and M. Sturzl. 2008. Intracellular localization map of human herpesvirus 8 proteins. J. Virol. 821908-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnee, M., Z. Ruzsics, A. Bubeck, and U. H. Koszinowski. 2006. Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 8011658-11666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serio, T. R., A. Angeloni, J. L. Kolman, L. Gradoville, R. Sun, D. A. Katz, W. Van Grunsven, J. Middeldorp, and G. Miller. 1996. Two 21-kilodalton components of the Epstein-Barr virus capsid antigen complex and their relationship to ZEBRA-associated protein p21 (ZAP21). J. Virol. 708047-8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Torrisi, M. R., M. Cirone, A. Pavan, C. Zompetta, G. Barile, L. Frati, and A. Faggioni. 1989. Localization of Epstein-Barr virus envelope glycoproteins on the inner nuclear membrane of virus-producing cells. J. Virol. 63828-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uetz, P., Y. A. Dong, C. Zeretzke, C. Atzler, A. Baiker, B. Berger, S. V. Rajagopala, M. Roupelieva, D. Rose, E. Fossum, and J. Haas. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311239-242. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Y., Q. Tang, G. G. Maul, and Y. Yuan. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J. Virol. 8012171-12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, D., T. Coleman, J. Zhang, A. Fagot, C. Kotalik, L. Zhao, P. Trivedi, C. Jones, and L. Zhang. 2007. Epstein-Barr virus inhibits Kaposi's sarcoma-associated herpesvirus lytic replication in primary effusion lymphomas. J. Virol. 816068-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamauchi, Y., C. Shiba, F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 821423-1428. [DOI] [PubMed] [Google Scholar]
- 54.Ye, G. J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 9711002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng, Z. M. 2003. Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev. Med. Virol. 13173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, L., V. Puri, and B. Chandran. 1999. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology 262237-249. [DOI] [PubMed] [Google Scholar]