Abstract
Splicing factors SF1 and U2AF associate cooperatively with pre-mRNA and play a crucial role in 3′ splice site recognition during early steps of spliceosome assembly. Formation of the active spliceosome subsequently displaces SF1 in a remodeling process that stabilizes the association of U2 snRNP with pre-mRNA. Fluorescence microscopy shows SF1 and U2AF distributed throughout the nucleoplasm, where transcription occurs, with additional concentration in nuclear speckles, where splicing factors accumulate when not engaged in splicing. Fluorescence recovery after photobleaching analysis in live cells shows that the mobilities of SF1 and the two subunits of U2AF (U2AF65 and U2AF35) are correlated with the abilities of these proteins to interact with each other. Direct binding of SF1 to U2AF65 was demonstrated by fluorescence resonance energy transfer in both the nucleoplasm and nuclear speckles. This interaction persisted after transcription inhibition, suggesting that SF1 associates with U2AF in a splicing-independent manner. We propose that SF1 and U2AF form extraspliceosomal complexes before and after taking part in the assembly of catalytic spliceosomes.
In eukaryotes, protein-coding regions (exons) within precursor messenger RNAs (pre-mRNAs) are separated by intervening sequences (introns) that must be removed to produce a functional mRNA. Pre-mRNA splicing requires accurate recognition of splice sites by the spliceosome, a large and dynamic machine composed of five major small nuclear ribonucleoprotein particles (the U1, U2, U4, U5, and U6 snRNPs) and more than 100 non-snRNP protein splicing factors (reviewed in references 21 and 31). In mammalian in vitro splicing systems, spliceosome assembly follows an ordered sequence of events that begins with formation of early complexes E′ and E. The E′ complex contains the U1 snRNP bound to the 5′ splice site and the splicing factor 1 protein (SF1, or mammalian branch point binding protein) at the branch point (23). Binding of U2AF (U2 small nuclear ribonucleoprotein auxiliary factor) to the polypyrimidine (Py) tract and 3′ splice site then forms complex E (23). In the presence of ATP, the E complex converts into the A complex, which is characterized by the stable association of U2 snRNP with the branch point. Joining of the U4/U6.U5 tri-snRNP forms the B complex, which undergoes an ATP-dependent rearrangement to become the catalytic C complex spliceosome (reviewed in reference 9).
Mammalian U2AF is a heterodimer composed of a 65-kDa subunit (U2AF65) that contacts the Py tract (38, 54, 55) and a 35-kDa subunit (U2AF35) that interacts with the AG dinucleotide at the 3′ splice site (30, 53, 56). Binding of U2AF65 to SF1 increases by 20-fold the affinity of SF1 to the pre-mRNA branch point sequence (6). Thus, the cooperative association of SF1 with U2AF65 plays an important role for initial spliceosome assembly. However, the U2AF65-SF1 interaction appears to be transient, as SF1 is absent from the A complex (39). During A complex formation, SF1 is thought to be displaced from U2AF65 and replaced by the U2 snRNP protein SF3b155/SAP155 (18).
The U2AF65 protein contains an arginine- and serine-rich (RS) domain and three RNA recognition motifs (RRMs). The two central motifs (RRM1 and RRM2) are canonical RRM domains responsible for recognition of the Py tract in the pre-mRNA, while the third RRM (called UHM, for U2AF homology motif) has unusual features and is specialized in protein-protein interaction (25). This motif interacts with the N-terminal domain of both SF1 and SF3b155. Recent data indicate that the SF1/U2AF65 complex is stabilized by 3.3 kcal mol−1 relative to the SF3b155/U2AF65 complex, consistent with the need for ATP hydrolysis to drive exchange of these partners during E-to-A spliceosome complex conversion (44). Interaction between the two subunits of the U2AF heterodimer involves amino acids 85 to 112 of U2AF65 and the central UHM domain of U2AF35 (reviewed in reference 25).
As spliceosomes form anew on nascent pre-mRNAs and disassemble after introns are excised and exons ligated, splicing factors in the nucleus are either actively engaged in splicing or waiting for the next turn to assemble a spliceosome. When they are not forming spliceosomes, splicing factors accumulate in so-called nuclear speckles, which are largely devoid of pre-mRNA (for reviews, see references 27 and 42). Although most (if not all) spliceosomal components colocalize in nuclear speckles, little is known about the intermolecular interactions that occur at this compartment. Do splicing factors assemble into extraspliceosomal complexes located at the nuclear speckles? Are there distinct types of such complexes? Can extraspliceosomal complexes contribute to regulate splicing? To start addressing these questions we performed fluorescence recovery after photobleaching (FRAP) and fluorescence (Förster) resonance energy transfer (FRET) analysis of U2AF65, U2AF35, and SF1 in HeLa cell nuclei. Our results reveal that SF1 interacts with U2AF in a splicing-independent manner and suggest that subsets of splicing proteins form distinct extraspliceosomal complexes localized in nuclear speckles.
MATERIALS AND METHODS
Cell culture, RNAi, and transfection procedures.
Human HeLa cells (ECACC 93021013) were grown as monolayers in minimum essential medium with Earle's salts supplemented with 10% (vol/vol) fetal calf serum and 1% (vol/vol) nonessential amino acids (Gibco, Invitrogen). For live imaging, cells were plated in glass-bottom chambers (MatTek, Ashland, MA), and the medium was changed to minimal essential medium with Earle's salts-F-12 without phenol red and supplemented with 15 mM HEPES buffer (Invitrogen). HeLa subconfluent cells were transiently transfected with either FuGENE6 reagent (Roche Biochemicals, Indianapolis, IN) or calcium chloride. Cells were analyzed at 16 to 24 h after transfection. 5,6-dichloro-1-b-d-ribofuranosylbenzimidazole (DRB; Sigma-Aldrich, St. Louis, MO) was used at 75 μM where indicated. RNA inhibition (RNAi) was performed as previously described (32). The sequences of the oligonucleotides used for targeting U2AF65 and U2AF35 were as follows: h65, 5′-GCA CGG UGG ACU GAU UCG U-dTdT-3′ (GenBank accession number NM_007279; nucleotides 1271 to 1289); h35a, 5′-CCA UUG CCC UCU UGA ACA U-dTdT-3′ (GenBank accession number NM_006758; nucleotides 218 to 238).
Immunofluorescence, immunoprecipitation, and Western blot analysis.
For immunofluorescence microscopy, cells grown on coverslips were rinsed briefly in phosphate-buffered saline (PBS), fixed in 3.7% formaldehyde-PBS for 10 min at room temperature, and washed with PBS. Cells were then permeabilized with 0.5% (wt/vol) Triton X-100-PBS for 10 min at room temperature and washed with PBS. For immunoprecipitation (IP), the cells were lysed in radioimmunoprecipitation buffer plus Complete protease inhibitors cocktail (Boehringer Mannheim), and lysates were precleaned with protein A-Sepharose beads (Sigma). Precleaned extracts were incubated for 1 h at 4°C with antibodies. Protein A-Sepharose beads were added, and immunoprecipitates were washed four times with extraction buffer. Antigen-antibody complexes were recovered by boiling in sodium dodecyl sulfate (SDS) sample buffer. Western blot analysis was performed as previously described (32).
Comparison between input and pulled down amounts of each protein in the IP was made by measuring the intensity ratio between the corresponding bands, after background correction and normalization for the amount of protein loaded.
The following primary antibodies were used in this study: mouse monoclonal antibodies directed against U2AF65 (MC3), β-actin (clone AC-15; Sigma), U2B" (4G3) (19), Sm proteins (Y12) (29), green fluorescent protein (GFP; anti-GFP clones 7.1 and 13.1; Boehringer Mannheim), human autoantiserum C45 specific for Sm proteins (1/75; kindly provided by W. van Venrooij, University of Nijmegen, The Netherlands), rabbit polyclonal serum directed against U2AF35 (kindly provided by Angus Lamond) (12), SF1 (kindly provided by Angela Kramer, University of Geneva, Switzerland), and hemagglutinin (HA) epitope (Y-11; Santa Cruz Biotechnology). For immunofluorescence, secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Immunoblots were developed using horseradish peroxidase-coupled secondary antibodies and detected by enhanced chemiluminescence (Amersham Biosciences).
Gel filtration analysis.
HeLa cell nuclear extracts were separated by chromatography through a Superose 6 HR 16/50 gel filtration column (Amersham Biosciences) that was previously equilibrated with two column volumes of eluent buffer (50 mM Tris-HCl, pH 7.9, 20% glycerol, 0.1 M KCl, 0.5 mM EDTA, 1 mM dithiothreitol). The eluted fractions were concentrated by trichloroacetic acid precipitation before analysis by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting.
Plasmids and constructs.
All GFP fusion proteins used in this study were previously described, except for GFP-SF1. The SF1 cDNA (Y08766) was obtained from pGEM/SF1 (a gift from Angela Krämer) and cloned in the BamHI site of pEGFP-C1 (Clontech). Yellow fluorescent protein (YFP)- and cyan fluorescent protein (CFP)-tagged splicing factors were obtained by subcloning the cDNAs from the pEGFP vector into the appropriate pECFP and pEYFP vectors (Clontech). All constructs were purified using a plasmid DNA Midi-prep kit (Qiagen, Hilden, Germany) and sequenced.
Dextran purification.
Fluorescein isothiocyanate (FITC)-labeled dextrans with average molecular masses of 40 and 70 kDa (Sigma-Aldrich, St. Louis, MO) were dissolved to 10 mg/ml in water. One milliliter of each sample was fractionated on a Superdex 200 column (Amersham Biosciences, Piscataway, NJ). For each dextran, the column fractions with higher absorbency were pooled and lyophilized. The 500-kDa FITC-labeled dextran was used without further purification. All samples were diluted to 200 μg/ml in water and microinjected into the nucleus of HeLa cells, as described previously (2).
Confocal microscopy.
Live cells were imaged at 37°C, which was maintained by a heating/cooling frame (LaCon, Staig, Germany) in conjunction with an objective heater (PeCon, Erbach, Germany). Images were acquired on a Zeiss LSM 510 confocal microscope (Carl Zeiss, Jena, Germany) using a PlanApochromat 63×, 1.4 numerical aperture objective. FITC and enhanced GFP (EGFP) fluorescence were detected using the 488-nm laser line of an Ar laser (25-mW nominal output) and an LP 505 filter. Cy3 fluorescence was detected using a 543-nm HeNe laser (1 mW) and an LP 560 filter. The pinhole aperture was set to 1 Airy unit.
Quantitative FRAP analysis and diffusion coefficients estimation.
FRAP experiments were performed essentially as described elsewhere (7). Each FRAP experiment of FITC-labeled dextrans started with three image scans followed by a bleach pulse of 242 ms on a spot with a diameter of 21 pixels (1.19-μm radius). A series of 97 single-section images (of size 256 by 30 and pixel width 114 nm) was then collected at intervals of 29.82 ms, with the first image acquired 2 ms after the end of bleaching. For EGFP-tagged splicing factors, bleaching was performed on a spot with a diameter of 25 pixels (0.59-μm radius) for 110 ms. A series of 97 single-section images (of size 512 by 50 and pixel width 48 nm) was then collected at intervals of 78.40 ms, again with the first image acquired 2 ms after the end of bleaching. For imaging, the laser power was attenuated to 1% of the bleach intensity.
For each FRAP time series, the background and nuclear regions were identified using an implementation of the ICM segmentation algorithm (10) in the Matlab software (Mathworks, Natick, MA). The average fluorescence in the nucleus, T(t), and the average fluorescence in the bleached region, I(t), were calculated for each background-subtracted image at time t after bleaching. FRAP recovery curves were normalized as described previously (35) with the equation Irel(t) = I(t)/I0 × T0/T(t), where T0 is the fluorescence in the nucleus before bleaching and I0 is the fluorescence in the bleached region before bleaching. This normalization corrected for the loss of fluorescence caused by imaging, which was typically <5%.
FRAP recovery curves were fitted to a recovery function that takes into account diffusion of highly mobile molecules during the bleach phase, essentially as described previously (7), with the following equation: Irel(t) = (1 − γ)F̂M(t) + γF̂im, where γ is the fraction of immobile molecules and F̂M and F̂im are the normalized fluorescence values for the mobile fraction and the immobile molecules, respectively. The fitting procedure yielded diffusion coefficient and immobile fraction values. Image processing routines also yielded the normalized fluorescence profile of the first postbleach image, from which the values of the parameters wM and KM (used in the determination of F̂M) were obtained (see reference 7 for more details). All fitting procedures were performed using the NonLinearRegress function of Mathematica 5.0 (Wolfram Research, Champaign, IL).
Estimation of diffusion coefficients based on the molecular weight of a protein compared to GFP were made using the Stokes-Einstein equation: D = KBT/ 6πηRH, where KB is the Boltzmann constant, T is the absolute temperature, η is the viscosity of the medium, and RH is the hydrodynamic radius (or Stokes' radius) of the molecule. RH is determined from the following equation: RH = (3M/4πρNA)⅓, where M is the molecular mass of the protein, ρ is the mean density of the molecule, and NA is Avogadro's number. Assuming that the molecular density ρ of a splicing protein is similar to that of GFP, the predicted diffusion coefficient for a freely diffusing splicing factor (SF) can be estimated by the following equation: DSF = DGFP × (MGFP/MSF)⅓, where MGFP and MSF are the molecular masses of GFP and a given splicing factor and DGFP and DSF are their corresponding diffusion coefficients. Using the same molecular mass parameters, the Stokes' radius of a given splicing factor (RSF) can be estimated by the following equation: RSF = RGFP × (MSF/MGFP)⅓, where RGFP represents the Stokes' radius of GFP, 2.35 nm (37). Stokes' radius values for FITC-dextrans were obtained from online supplier data, except for the 500-kDa dextran, which was measured by fluorescence correlation spectroscopy as reported in reference 28.
Estimation of a lower limit for the proportion of GFP-tagged splicing factors that are bound in an effective diffusion regimen was made using the generalized effective diffusion relation Deff = (1 − p)DF + pDC, where DF and DC are the diffusion coefficients of the unbound and bound fractions of splicing factors, respectively, Deff is the diffusion coefficient estimated by FRAP, and p is the proportion of bound molecules (8). Using the diffusion coefficients of GFP-U2AF65Δ35 and GFP-SF1R21D as the DF values for unbound GFP-U2AF65 and GFP-SF1, the diffusion coefficient of the complex DC is given by the following: DC = [DGFP-U2AF65 − (1 − pU2AF65) × DGFP-U2AF65Δ35]/pU2AF65 = [DGFP-Sp1 − (1 − pSF1) × DGFP-SF1R21D]/pSF1, where the D's and p's represent the FRAP-determined diffusion coefficients and the proportion of bound molecules of the subscripted splicing factors, respectively. In the case of an immobile complex, DC is 0. Solving for U2AF65 and SF1 separately yields the following: pU2AF65 = 1 − (DGFP-U2AF65/DGFP-U2AF65Δ35) ≈ 0.86 and pSF1 = 1 − (DGFP-SF1/DGFP-SF1R21D) ≈ 0.82. If DC is >0, solving the inequality separately for U2AF65 and SF1 yields the following: pU2AF65 > 1 − (DGFP-U2AF65/DGFP-U2AF65Δ35) ≈ 0.86 and pSF1 > 1 − (DGFP-SF1/DGFP-SF1R21D) ≈ 0.82.
Thus, a lower limit for the proportion of molecules transiently bound to the complex at any given time is obtained for a DC of 0. If the complex is also diffusing, then the proportion of bound molecules must necessarily increase to account for the difference between the different DF values and the FRAP-measured Deff.
Acceptor photobleaching FRET.
FRET between splicing factors tagged with the donor CFP and the acceptor YFP was measured using the acceptor photobleaching method (24) on a Zeiss LSM 510 confocal microscope (Carl Zeiss) operating a 25-mW argon laser. Cells were imaged using the PlanApochromat 63×, 1.4 numerical aperture oil immersion objective at zoom 5 (pixel width, 57 nm). CFP fluorescence was detected using the 458-nm laser line and a BP 470-500 nm filter, while YFP was excited with the 514-nm laser line and its fluorescence detected using an LP 530 nm filter. Detector gains were adjusted in order to eliminate cross talk and achieve a good dynamic range. In the acceptor photobleaching method, if FRET is occurring between the donor and the acceptor, then photobleaching of the acceptor (YFP) should yield a significant fluorescence increase of the donor (CFP). Bleaching of YFP was performed in a rectangular region of interest (ROI) in the cell, using the 514-nm argon laser line at 100% intensity and 40 bleach iterations (the time of bleach ranged from 8 to 12 s, depending on ROI size). A series of four images from the donor channel were taken before and after bleaching, with the laser intensity set to 30%. The pre- and postbleach image series were then background subtracted and processed with ImageJ (http://rsb.info.nih.gov/ij/) using a rigid body registration algorithm to correct for cell displacement during image acquisition. FRET energy transfer efficiency is given by the equation EFRET = 1 − (FDA/FD), where FDA is the donor fluorescence in the presence of the acceptor (before YFP bleaching) and FD is the donor fluorescence alone (after YFP bleaching). FRET efficiency maps were generated using Mathematica 5.0 (Wolfram Research). The pre- and postbleach image series were corrected for fluorescence loss due to scanning by multiplying each pixel intensity value by the ratio of the total average image intensities (excluding the bleached ROI) between each image and the first image of the series. The pre- and postbleach image series were then averaged to create single pre- and postbleach images, which were further convoluted with an 11- by 11-pixel filter to reduce image noise. FRET efficiency values were then calculated for each pixel in the image that had an intensity value above a certain threshold, thereby avoiding calculation of EFRET in the background and in areas of reduced intensity where the signal-to-noise ratio is lower. The calculated FRET efficiencies were then color coded for each pixel, ranging from 0 (blue) to 0.3 (red), and superimposed to the prebleach donor image, yielding the FRET efficiency maps.
FLIM.
Fluorescence lifetime imaging microscopy (FLIM) images were recorded on an instrument described extensively elsewhere (46) and based on frequency domain lifetime detection (45). Basically, excitation light is modulated at 75.1 MHz, and fluorescence images are recorded by a charge-coupled-device camera through an image intensifier with the gain modulated also at 75.1 MHz. By recording fluorescent images at different phases between excitation light and intensifier, changes in phase and modulation depth of the emitted light relative to the excitation can be detected. From this, the two lifetimes are derived, τϕ and τΜ, based on the phase shift and modulation, respectively. The instrument is based on an inverted wide-field microscope (Axiovert 200 M; Carl Zeiss). CFP lifetime images were recorded using 442-nm excitation, a 63×, numerical aperture 1.3 oil objective (Carl Zeiss), a 455LP dichroic, and a 480/40-nm band-pass filter (all filters from Chroma). Per lifetime recording, eight phase images were recorded with an exposure time of 800 ms each. The order in which the images were recorded was chosen in such a way to limit the effects of photobleaching (47). Recorded images were analyzed for the occurrence of photobleaching and corrected for this if necessary. Lifetime images were generated by calculating τϕ and τΜ for each pixel in the cell, after threshold segmentation. After calculation of the lifetimes, the average phase and modulation lifetime of each cell was determined by averaging the pixels constituting the cell. FRET efficiencies can be calculated using either τϕ or τΜ. We used τϕ to calculate the energy transfer efficiency EFLIM = 1 − (τDA/τD), where τDA is the donor phase lifetime determined in the presence of the acceptor and τD is the donor phase lifetime determined in the absence of the acceptor.
RESULTS
A complex of SF1 and U2AF is detected in HeLa nuclear extracts.
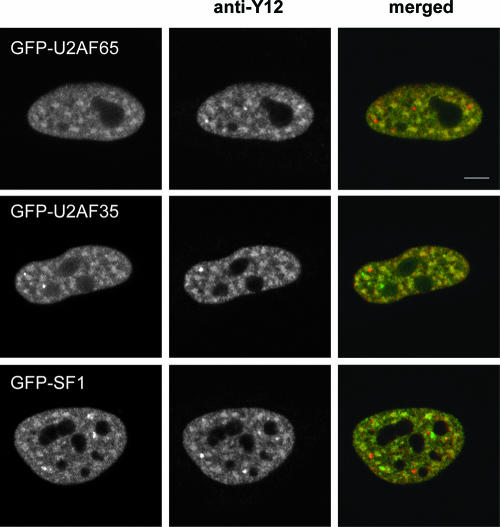
To study the dynamics and interactions of splicing factors U2AF65, U2AF35, and SF1 in vivo, we tagged these proteins with either GFP or its variants, YFP and CFP. Fluorescence microscopy revealed that all GFP-fusion proteins were diffusely distributed throughout the nucleoplasm with additional concentration in nuclear speckles (Fig. 1). Double labeling with monoclonal antibody Y12 directed against Sm proteins showed that the GFP-fusion proteins colocalized with splicing snRNPs in the nucleoplasm and in speckles but not in Cajal bodies, as previously reported (reviewed in reference 27). Using a splicing reporter minigene assay, we further confirmed that GFP-labeled U2AF proteins are functional in splicing (see Fig. S1 in the supplemental material), consistent with what was previously reported for enhanced YFP (EYFP)-labeled U2AF65 and U2AF35 (12).
FIG. 1.
GFP tagging does not alter the subnuclear distribution of U2AF65, U2AF35, and SF1. HeLa cells expressing GFP-tagged U2AF65, U2AF35, and SF1 were immunolabeled with Y12 monoclonal antibody. The merged images represent a superimposition of GFP (green) and Y12 (red) immunofluorescence. Note that the GFP-fusion proteins colocalize with endogenous Sm proteins throughout the nucleoplasm and speckles, but not in Cajal bodies (red foci). Bar, 5 μm.
To determine whether the GFP-tagged forms of U2AF65, U2AF35, and SF1 interact with endogenous partners, we prepared extracts from HeLa cells expressing either GFP alone or the GFP-fusion proteins. Proteins immunoprecipitated by monoclonal antibody anti-U2AF65 were separated by SDS-PAGE and probed in a Western blot assay with anti-GFP antibody (Fig. 2A). The results showed that the anti-U2AF65 antibody coimmunoprecipitated GFP-U2AF65 (Fig. 2A, lane 8), GFP-U2AF35 (Fig. 2A, lane 6), and GFP-SF1 (Fig. 2A, lane 7), but not GFP alone (Fig. 2A, lane 5). To further analyze how these proteins interact in HeLa cells, we fractionated nuclear extracts by size exclusion column chromatography. As shown in Fig. 2B, panel a, endogenous U2AF65 coeluted with U2AF35 and SF1 in fractions estimated to be in the 200- to 400-kDa range. By contrast, the U2 snRNP U2B" protein was eluted in fractions of a much higher molecular weight range. Fractionation of extracts prepared from cells expressing GFP-U2AF65 (Fig. 2B, panel b) or GFP alone (Fig. 2B, panel c) showed that the elution profile of GFP-U2AF65 partially overlapped that of endogenous U2AF65, while the majority of GFP was eluted in fractions estimated to be less than 100 kDa.
FIG. 2.
Association of U2AF65 with U2AF35 and SF1 in nuclear extracts. (A) HeLa cells expressing either GFP alone or the indicated GFP-fusion proteins were lysed and immunoprecipitated with anti-U2AF65 (MC3) antibody. Immunoprecipitates were subjected to SDS-PAGE followed by Western blotting with anti-GFP antibody (lanes 5 to 8). Ten percent of each immunoprecipitation input lysate was loaded in lanes 1 to 4. The arrowheads indicate the light and heavy chains of MC3 antibody. Molecular weight markers are shown on the left. (B) Nuclear extracts from nontransfected HeLa cells (a) or cells expressing either GFP-U2AF65 (b) or GFP alone (c) were separated by using Superose 6 gel filtration chromatography. Column fractions were subjected to SDS-PAGE followed by Western blotting with the indicated antibodies. The elution positions of marker proteins (calibration kit from Amersham Biosciences) are indicated by arrowheads, and SDS-PAGE molecular weight markers are shown on the left.
Taken together, these results suggest that the GFP tag does not interfere with the normal localization, binding properties, and splicing function of U2AF and SF1 proteins.
The mobilities of GFP-U2AF65, GFP-U2AF35, and GFP-SF1 correlate with the abilities of these proteins to interact with each other.
The finding that U2AF65 cofractionates and coimmunoprecipitates with U2AF35 and SF1 (Fig. 2) suggested that these proteins assemble into a complex. If such a complex existed in the cell nucleus, the mobility of U2AF65, U2AF35, and SF1 would be significantly slower than expected if these proteins were diffusing as isolated molecules. To determine the intranuclear dynamics of U2AF65, U2AF35, and SF1, we performed FRAP. The fluorescence of a small area in the nucleus was irreversibly photobleached using a high-intensity laser. Subsequent fluorescence recovery, due to movement of nonbleached molecules into the bleached area, was recorded by time-lapse imaging. By fitting an appropriate theoretical function to the recovery curve (7), we could determine both the effective diffusion coefficient (D) of the GFP-tagged protein and its immobile fraction (Fig. 3).
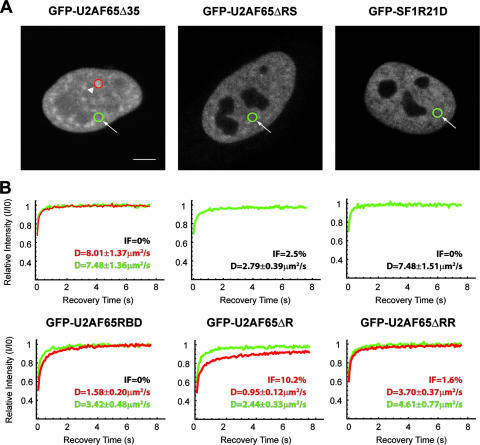
FIG. 3.
Splicing proteins are less mobile than dextrans. A. Fluorescence recovery was measured after photobleaching a circular region in a nuclear speckle (arrowhead; red circle) or in the nucleoplasm (arrow; green circle) of HeLa cells expressing GFP-tagged splicing factors. Fluorescence intensities of prebleach, postbleach, and 8-s recovery images are shown in pseudocolor on the right. Bar, 5 μm. B. FRAP recovery curves for the indicated splicing proteins in the speckles and in the nucleoplasm. The curves correspond to a pool of three independent experiments, with at least 10 different cells analyzed per experiment. C. Recovery curves for FITC-labeled dextrans with average molecular masses of 40, 70, and 500 kDa that were microinjected into the nucleus of HeLa cells. The curves correspond to a pool of four independent experiments with 10 to 20 different cells analyzed per experiment. Error bars represent standard deviations. D values represent means ± standard errors. IF, immobile fraction.
We performed FRAP experiments in the nuclei of cells that were transfected with GFP-U2AF65 (predicted molecular mass, ∼93 kDa), GFP-U2AF35 (∼63 kDa), and GFP-SF1 (∼103 kDa). Only cells with minimal detectable levels of GFP fluorescence were selected for FRAP experiments, to avoid overexpression of the exogenous protein. The results show that GFP-tagged splicing factors are mostly mobile in the nucleoplasm (measured diffusion rates of 1.19 to 1.39 μm2 s−1 and no significant immobile fractions) (Fig. 3B). Compared to FITC-labeled dextrans (Fig. 3C), the three splicing proteins moved much slower than a 70-kDa particle (Stokes' radius, ∼6 nm; D, 5.9 μm2 s−1) and were even slower than a 500-kDa particle (Stokes' radius, ∼46.5 nm; D, 1.7 μm2 s−1). In contrast, GFP alone (∼28 kDa) is able to diffuse inside the nucleus, with an intranuclear diffusion coefficient of 33.3 μm2 s−1 (7). Taking into account the molecular mass of the fusion protein and assuming a molecular density similar to that of GFP (see Materials and Methods), GFP-U2AF65 (predicted Stokes' radius, ∼3.5 nm) should have a diffusion coefficient of ∼22 μm2 s−1, whereas GFP-SF1 (Stokes' radius, ∼3.6 nm) should diffuse at ∼21 μm2 s−1 and GFP-U2AF35 (Stokes' radius, ∼3.1 nm) at ∼25 μm2 s−1. Clearly, U2AF65, U2AF35, and SF1 move in the nucleus at a much slower rate than expected if these proteins diffuse as isolated noninteracting molecules.
Relative to the nucleoplasm, the mobility rate of the splicing factors in the nuclear speckles was reduced (D values of 0.49 μm2 s−1 for GFP-U2AF65, 1.16 μm2 s−1 for GFP-U2AF35, and 0.58 μm2 s−1 for GFP-SF1). GFP-U2AF65 showed an apparent immobile fraction of ∼18% that recovered completely when FRAP analysis was extended to longer periods of time (see Fig. S2 in the supplemental material).
To determine whether direct binding between U2AF65, U2AF35, and SF1 contributes to the reduced mobility of the individual proteins in the nucleus, we performed FRAP experiments on cells expressing GFP-tagged mutant variants of the splicing factors U2AF65 and SF1 (Fig. 4). U2AF65Δ35 has a deletion of amino acids 95 to 138, covering the region of interaction with U2AF35, and U2AF65ΔRS has a deletion of amino acids 23 to 65, covering the entire RS domain (16, 33). U2AF65RBD contains three point mutations in the RRM1 domain that compromise the ability to bind RNA (16). U2AF65ΔR has a deletion of amino acids 343 to 475, spanning the RRM3 domain, and U2AF65ΔRR combines the mutations of both U2AF65RBD and U2AF65ΔR. SF1R21D is a single-point mutant that is unable to bind to U2AF65 (40). Although all mutants recovered faster than wild-type proteins, the highest mobility was observed for GFP-U2AF65Δ35 and GFP-SF1R21D, with a D value of ∼8 μm2 s−1 and no significant difference between nucleoplasm and nuclear speckles. These two mutant proteins diffuse in the nucleus at a faster rate than a 70-kDa dextran (D, 5.9 μm2 s−1), and this is probably caused by the higher Stokes' radius of the dextran (6 nm) compared to the mutant proteins (3.5 nm). The higher mobility detected for U2AF65Δ35 could result from the smaller size of this deletion mutant. However, since the ratio of the diffusion coefficients of two proteins is inversely proportional to approximately the cubic root of the ratio of their molecular mass (36), the difference of less than 5 kDa between GFP-U2AF65Δ35 and GFP-U2AF65 would account for a 2% increase in the diffusion coefficient value, instead of the 500% increase measured. Moreover, FRAP experiments performed on other U2AF65 deletion mutants revealed distinct D values (Fig. 4). Taken together the results imply that interactions mediated by different binding domains of U2AF65, rather than the size of the mutant protein, contribute to its faster kinetics in the nucleus. Our data thus reveal that the mobility of GFP-U2AF65 is correlated with its ability to bind to U2AF35, and the mobility of GFP-SF1 correlates with its ability to bind to U2AF65. Since these correlations are detected both in the nucleoplasm and in nuclear speckles, it is most likely that U2AF65 binds U2AF35 and SF1 in both compartments.
FIG. 4.
Mutations that impair protein binding increase the mobility of U2AF65 and SF1. A. FRAP experiments were performed in cells expressing the indicated GFP fusion proteins. Fluorescence recovery was measured after photobleaching a circular region located either in a nuclear speckle (arrowhead; red circle) or in the nucleoplasm (arrow; green circle). Bar, 5 μm. B. FRAP recovery curves for the indicated proteins in the speckles (red recovery curves) and in the nucleoplasm (green recovery curves). The curves correspond to a pool of three independent experiments, with at least 10 different cells analyzed per experiment. D values in the speckles (red) and nucleoplasm (green) represent means ± standard errors. IF, immobile fraction.
We have further used the FRAP data to estimate the fraction of splicing proteins that are interacting in the nucleus. Since all FRAP recovery curves are well fitted by a single diffusion model, the calculated diffusion coefficients most likely correspond to effective diffusion regimens in which splicing proteins may bind to different partners over time, in a transient manner (8, 43). Assuming such an effective diffusive regimen, the diffusion coefficients determined by FRAP for each GFP-tagged protein are given by the equation Deff = (1 − p)DF + pDC, i.e., the weighted average of the unbound fraction diffusing at DF and the bound fraction diffusing at DC (8). Assuming that the mobility rates of GFP-U2AF65Δ35 and GFP-SF1R21D are similar to GFP-U2AF65 when it is not bound to U2AF35 and to GFP-SF1 when it is not bound to U2AF65 (i.e., DF), respectively, then we can calculate the proportion of bound molecules (p) for any given values for the complex diffusion coefficient (DC) and the experimentally determined FRAP diffusion coefficient (Deff). In the limit case of an immobile complex (i.e., DC = 0), we obtain P = 82% for GFP-SF1 and P = 86% for GFP-U2AF65 (see Materials and Methods). However, if the complex is also diffusing (DC > 0), then the values for the proportion of bound molecules increase for each protein (pSF1 > 82% and pU2AF65 > 86%), suggesting that at any given time, the vast majority of U2AF65 and SF1 molecules in the nucleus are making interactions.
FRET microscopy demonstrates interaction of U2AF65 with U2AF35 and SF1 in the nucleoplasm and nuclear speckles.
To directly visualize the predicted interactions between U2AF65, U2AF35, and SF1, we made use of FRET microscopy. FRET is a phenomenon that occurs when two different fluorophores (called donor and acceptor) with overlapping emission/absorption spectra are in close proximity (typically <1 to 10 nm) to each other and in a suitable orientation (24). FRET microscopy approaches include intensity-based methods, such as acceptor photobleaching FRET and fluorescence decay kinetics-based methods (52). The principle behind acceptor photobleaching FRET is that energy transfer is reduced or eliminated when the acceptor is irreversibly bleached, thereby causing an increase in donor fluorescence. However, acceptor photobleaching FRET has to be performed on fixed cells if the proteins of interest are mobile inside the nucleus, as is the case of splicing factors, as the redistribution of acceptor fluorescent molecules would interfere with the determination of unquenched donor emission. Kinetic-based approaches for FRET, such as FLIM (15, 49, 51) measure the decay kinetics of the donor fluorophore, instead of its intensity. In most FLIM implementations an average fluorescence lifetime value is determined for each position in an image. This average lifetime is a nonlinear function of the true lifetimes and the populations of bound and unbound donor molecules in that position (52). Compared to acceptor photobleaching FRET, FLIM has the advantage of allowing FRET detection to be performed in live cells, as no photobleaching is required. FLIM-calculated fluorescence lifetimes are also independent of the chromophore concentration, intensity variations due to absorption of light by the sample, microscope geometry, and moderate levels of donor photobleaching (45, 48).
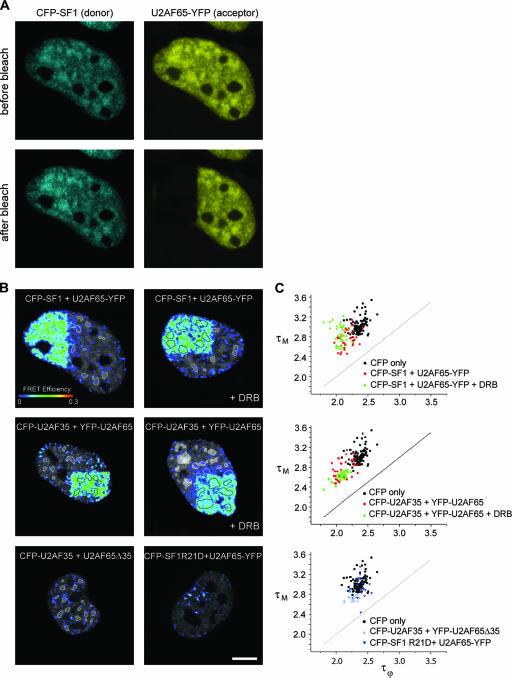
In our studies we first tagged the splicing proteins with CFP (donor) and YFP (acceptor) and performed confocal acceptor photobleaching experiments (22, 52). FRET between the donor (CFP-tagged splicing factor) and the acceptor (YFP-tagged splicing factor) is detected by irreversibly photobleaching the acceptor in a region of interest and comparing donor emission before and after bleaching. When FRET occurs, the donor fluorescence emission increases in the bleached region (Fig. 5A, left panels) and FRET efficiency maps can be generated, allowing for a spatial mapping of detected interactions (Fig. 5B; see also Materials and Methods). Acceptor photobleaching FRET performed on cells coexpressing CFP-U2AF35 and YFP-U2AF65 showed a clear FRET signal at both the speckles and nucleoplasm, indicating that these proteins are interacting directly in both compartments (Fig. 5B, middle left panel), with an average FRET efficiency of 9.30 ± 2.59% (Table 1). A similar result was obtained in cells that coexpressed CFP-SF1 and U2AF65-YFP (Fig. 5B, top left panel and Table 1). As negative controls, cells were cotransfected with either CFP-U2AF35 and the deletion mutant YFP-U2AF65Δ35 or U2AF65-YFP and the point mutant CFP-SF1R21D. As shown in Fig. 5B and Table 1, no significant FRET signal was detected.
FIG. 5.
FRET studies on fixed cells. A. HeLa cells expressing similar levels of SF1 tagged with CFP (blue) and U2AF65 tagged with YFP (yellow) were imaged before and after bleaching the YFP fluorescence in approximately one-third of the nucleus. Notice the increase in the donor fluorescence intensity at the bleached region. B. FRET efficiency maps were generated from acceptor photobleaching FRET experiments and superimposed to the corresponding prebleach donor images. Donor and acceptor pairs are indicated for each case. The upper left panel shows the FRET efficiency map obtained from the images depicted in panel A. The nuclear speckles are outlined (white contours). Cells were either mock treated or treated with the transcriptional inhibitor DRB (+DRB) for 30 min. Bar, 5 μm. C. FLIM was performed on HeLa cells expressing the indicated proteins in the absence or after 30 min of treatment with DRB (+DRB). The average phase (τϕ) and modulation (τΜ) lifetimes of the donor (CFP) were calculated for each cell (represented as a single dot in the graph).
TABLE 1.
FRET efficienciesa
| FRET pair | FRET efficiency
|
||||||
|---|---|---|---|---|---|---|---|
| EAP | Fixed
|
Live
|
|||||
| τφ | τM | EFLIM | τφ | τM | EFLIM | ||
| CFP-U2AF35 YFP-U2AF65 | 9.30 ± 2.59 (n = 8) | 2.11 ± 0.13 | 2.73 ± 0.19 | 7.46 ± 0.11 (n = 32) | 2.08 ± 0.08 | 2.62 ± 0.07 | 9.56 ± 0.07 (n = 6) |
| CFP-U2AF35 YFP-U2AF65 + 30 min DRB | 9.07 ± 2.01 (n = 7) | 2.08 ± 0.09 | 2.61 ± 0.09 | 8.77 ± 0.09 (n = 27) | 2.06 ± 0.08 | 2.84 ± 0.18 | 10.43 ± 0.06 (n = 6) |
| CFP-SF1 U2AF65-YFP | 10.19 ± 2.50 (n = 10) | 2.17 ± 0.10 | 2.76 ± 0.17 | 8.05 ± 0.07 (n = 38) | |||
| CFP-SF1 U2AF65-YFP + 30 min DRB | 9.81 ± 2.69 (n = 7) | 2.06 ± 0.08 | 2.84 ± 0.18 | 12.71 ± 0.06 (n = 38) | |||
| CFP-SF1 YFP-U2AF65 | 2.11 ± 0.07 | 2.58 ± 0.02 | 10.21 ± 0.06 (n = 7) | ||||
| CFP-U2AF35 YFP-U2AF65D35 | 2.69 ± 1.09 (n = 6) | 2.26 ± 0.07 | 2.73 ± 0.06 | 4.24 ± 0.08 (n = 21) | 2.15 ± 0.07 | 2.65 ± 0.07 | 6.52 ± 0.06 (n = 6) |
| CFP-SF1R21D U2AF65-YFP | 3.23 ± 2.20 (n = 6) | 2.39 ± 0.09 | 2.95 ± 0.20 | 0 ± 0.06 (n = 10) | |||
| CFP-SF1R21D YFP-U2AF65 | 2.24 ± 0.07 | 2.75 ± 0.06 | 2.61 ± 0.06 (n = 6) | ||||
| CFP-U2AF35 | 2.28 ± 0.12 | 3.01 ± 0.06 | 2.33 ± 0.03 | 2.84 ± 0.02 | |||
| CFP-SF1 | 2.36 ± 0.08 | 2.99 ± 0.15 | 2.48 ± 0.10 | 2.95 ± 0.07 | |||
| CFP-SF1R21D | 2.39 ± 0.07 | 3.17 ± 0.18 | 2.35 ± 0.05 | 2.85 ± 0.02 | |||
Average FRET efficiencies were calculated from acceptor photobleaching FRET (EAP) and FLIM (EFLIM) experiments performed on fixed and live cells in the absence of DRB or after 30 min of treatment with DRB. The phase (τφ) and modulation (τM) donor lifetimes are shown for each donor-acceptor pair. EFLIM was determined using the phase donor lifetime only (see Materials and Methods). For comparison, the values obtained for the donors (CFP-U2AF35, CFP-SF1, and CFP-SF1R21D) in the absence of acceptor are also indicated. Values represent means ± standard deviations.
The positive FRET signals observed between CFP-U2AF35 and YFP-U2AF65 as well as between CFP-SF1 and U2AF65-YFP persisted in cells treated with the polymerase II transcription inhibitor DRB (Fig. 5B, top and middle right panels; Table 1). DRB is a nucleoside analogue that inhibits the protein kinases which phosphorylate the C-terminal domain of the largest subunit of RNA polymerase II, thereby inhibiting elongation (11) and causing premature transcriptional termination in vitro (41) and in vivo (13). Studies performed on Balbiani ring genes in Chironomus tentans showed that after 30 to 35 min of DRB treatment, cells were completely devoid of nascent Balbiani ring RNA (14). During this period, the transcribing chromatin loops were gradually emptied of polymerases (3) and splicing factors (5) and condensed into more compact chromosome loci (4). Thus, the positive FRET signals observed between CFP-U2AF35 and YFP-U2AF65 as well as between CFP-SF1 and U2AF65-YFP in cells treated with DRB for 30 min indicate that the interaction between these proteins occurs even when they do not assemble with pre-mRNA.
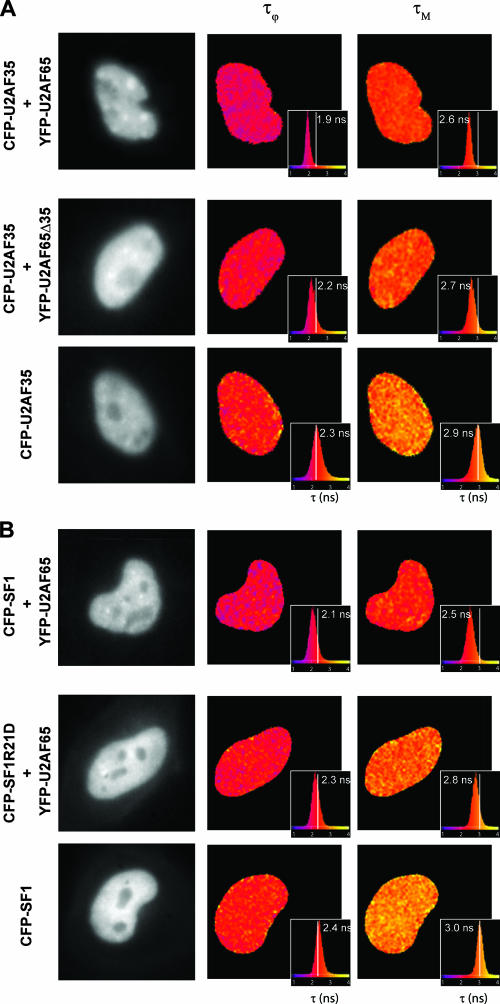
In order to validate the results observed by acceptor photobleaching FRET, we next performed wide-field frequency domain FLIM microscopy on live cells. We measured the mean donor fluorescence lifetime from the phase shift yielding the phase-determined lifetime (τϕ) and the modulation depth yielding the modulation lifetime (τΜ), both of them being typically on the order of a few nanoseconds. If FRET were occurring, then both the phase and modulation lifetimes of the donor would decrease in the presence of the acceptor, compared to the values of the phase and modulation lifetimes with the donor alone. FRET efficiency values could then be calculated using either the phase or modulation decrease (45). FLIM experiments performed on fixed cells expressing donor CFP-fusion proteins alone yielded an average fluorescence phase lifetime of 2.36 ± 0.09 ns and a modulation lifetime of 3.02 ± 0.15 ns (n = 101). When cells were coexpressing CFP-U2AF35 and YFP-U2AF65, FLIM showed a clear reduction of both phase and modulation donor lifetimes, corresponding to an average FRET efficiency of 7.46 ± 0.11% (Table 1). A similar effect was observed in cells treated with the transcriptional inhibitor DRB (E, 8.77 ± 0.09%) (Table 1 and Fig. 5C, middle graph). Similar FRET efficiencies (E) were detected for CFP-SF1 and U2AF65-YFP: 8.05 ± 0.07% and 12.71 ± 0.06% in cells treated with DRB (Fig. 5C, top graph, and Table 1). As expected, no significant FRET efficiencies were detected when cells coexpressed CFP-U2AF35 and YFP-U2AF65Δ35 (E, 4.24 ± 0.08%) (Table 1) or CFP-SF1R21D and U2AF65-YFP (E, 0 ± 0.06%) (Table 1 and Fig. 5C, bottom graph). Lifetime maps for live cells further showed that in the presence of the donor CFP-U2AF35 alone (Fig. 6A, bottom panels), the average fluorescence phase lifetime was 2.33 ± 0.03 ns and the modulation lifetime was 2.84 ± 0.02 ns. Inclusion of the acceptor YFP-U2AF65 (Fig. 6A, top panels) reduced the lifetime values to τϕ = 2.08 ± 0.08 ns and τΜ = 2.62 ± 0.07 ns (E, 9.56 ± 0.07%) (Table 1), whereas in the presence of YFP-U2AF65Δ35 (Fig. 6A, middle panels) the lifetime values were not significantly affected (τϕ = 2.15 ± 0.07 ns and τΜ = 2.65 ± 0.07 ns) (Table 1). For cells expressing only the donor CFP-SF1 (Fig. 6B, lower panels), the average phase and modulation lifetimes were τϕ = 2.36 ± 0.08 ns and τΜ = 2.99 ± 0.15 ns. In the presence of the acceptor YFP-U2AF65 the lifetimes were reduced to τϕ = 2.11 ± 0.07 ns and τΜ = 2.58 ± 0.02 ns (E, 10.21 ± 0.06%) (Table 1), whereas no significant reduction was observed in cells expressing CFP-SF1R21D and YFP-U2AF65 (Ε, 2.61 ± 0.06%) (Table 1 and Fig. 6B, middle panels). The lifetime values were uniformly reduced throughout the nucleus (Fig. 6B, top panels), indicating that U2AF65, U2AF35, and SF1 establish similar interactions in the speckles and in the nucleoplasm.
FIG. 6.
FLIM-FRET studies on living cells. FLIM was performed on HeLa cells expressing the indicated proteins. The phase (τϕ) and modulation (τΜ) lifetimes of the donor (CFP) were calculated for each pixel in the cell and color coded into lifetime maps. The histogram for each lifetime map is also shown, together with the corresponding average lifetime value for each cell (inlay) and the reference mean lifetime value obtained for the donor alone (vertical white line). FRET occurs when both τϕ and τΜ are reduced relative to the values of the donor alone.
Self-interaction of U2AF65 in vivo.
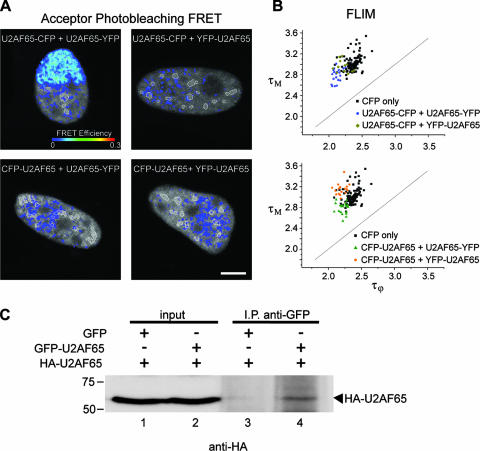
During our FRET studies, we tagged U2AF65 with either CFP or YFP and we fused both fluorophores at either the amino or carboxyl terminus of the protein. We then performed FRET analysis on cells expressing the four different donor-acceptor combinations, namely, U2AF65-CFP plus U2AF65-YFP, U2AF65-CFP plus YFP-U2AF65, CFP-U2AF65 plus U2AF65-YFP, and CFP-U2AF65 plus YFP-U2AF65 (Fig. 7A and B). Using either acceptor photobleaching (Fig. 7A, lower right panel) or FLIM (Fig. 7B, lower graph), no FRET signal was detected in cells coexpressing CFP-U2AF65 and YFP-U2AF65 (as reported by others [12]), U2AF65-CFP and YFP-U2AF65, or CFP-U2AF65 and U2AF65-YFP. However, a clear FRET signal (EAP, 7.82 ± 1.52%; EFLIM, 9.01 ± 0.05%) was measured in the nucleoplasm and speckles of cells coexpressing U2AF65-CFP and U2AF65-YFP (Fig. 7A, upper left panel). To further demonstrate that U2AF65 forms homodimers, we cotransfected HeLa cells with HA- and GFP-tagged versions of U2AF65. As shown in Fig. 7C, HA-U2AF65 was immunoprecipitated with an anti-GFP antibody. Taken together, these results strongly suggest that U2AF65 can self-interact in vivo.
FIG. 7.
Self-interaction of U2AF65. A. Acceptor photobleaching FRET efficiency maps of HeLa cells expressing the indicated combinations of CFP- and YFP-tagged U2AF65. FRET is only detected when both the donor CFP and the acceptor YFP are placed in the U2AF65 C terminus. The nuclear speckles are outlined (white contours). Bar, 5 μm. B. FLIM measurements performed on HeLa cells expressing the indicated proteins. The average phase (τϕ) and modulation (τΜ) lifetimes of the donor were calculated for each cell (represented as a single dot in the graph). Consistent with the results shown in panel A, FLIM-FRET is only detected if both the donor CFP and the acceptor YFP are placed in the C terminus of U2AF65. C. HeLa cells expressing the indicated proteins were lysed and immunoprecipitated with anti-GFP antibody. Immunoprecipitates were subjected to SDS-PAGE followed by Western blotting with anti-HA antibody (lanes 3 and 4). Ten percent of each immunoprecipitation input lysate was loaded in lanes 1 and 2. Molecular weight markers are shown on the left.
Formation of the U2AF65-U2AF35-SF1 complex is required for retention in nuclear speckles.
Our FRAP measurements showed that both SF1 and U2AF are less mobile in the speckles than in the nucleoplasm. However, the mutants U2AF65Δ35, which fails to bind U2AF35, and SF1R21D, which fails to bind U2AF65, diffuse sixfold faster throughout the nucleus and are no longer retarded in the speckles (Fig. 4). This suggests that formation of a complex between U2AF65, U2AF35, and SF1 contributes to retention in nuclear speckles. According to this view, we predicted that depletion of one member of the complex would prevent association of the remaining proteins with the speckles. We therefore used RNAi to knock down expression of either U2AF35 or U2AF65, as previously described (32). Western blot analysis showed that U2AF65-targeting small interfering RNA (siRNA) caused a significant knockdown of both U2AF65 and U2AF35 protein levels, whereas treatment with siRNAs directed against U2AF35 decreased predominantly the level of the U2AF35 protein (32, 34). As shown in Fig. 8, treatment with the GL2 control siRNA duplex did not affect the normal distribution of U2AF65 and SF1 proteins that were detected throughout the nucleoplasm, with a higher concentration in nuclear speckles. In contrast, U2AF65 no longer concentrated in speckles when the level of U2AF35 protein was down-regulated (Fig. 8A). Similarly, SF1 (Fig. 8B) and U2AF35 (data not shown) failed to accumulate in speckles following U2AF65 depletion. Double labeling with Y12 antibody showed that Sm proteins persist normally associated with the speckles in U2AF35-depleted cells (Fig. 8A), suggesting that the association of spliceosomal snRNPs with nuclear speckles is independent of the U2AF65-U2AF35-SF1 complex. Based on these findings we propose that after each round of splicing, the disassembled components of the spliceosome reassemble into distinct types of extraspliceosomal complexes that colocalize in nuclear speckles.
FIG. 8.
Depletion of U2AF prevents accumulation of SF1 in nuclear speckles. HeLa cells were treated with either control siRNA (GL2) or siRNAs against U2AF35 (A) or U2AF65 (B) and analyzed 48 to 72 h after transfection. A. Cells were double labeled with monoclonal antibody directed against U2AF65 (green staining) and human autoimmune antibodies against Sm proteins (red staining). The merged panels correspond to superimposition of green and red images. B. Cells were labeled with either anti-U2AF65 or anti-SF1 antibodies, as indicated.
DISCUSSION
Here, we provide the first evidence that extraspliceosomal complexes composed of SF1 and U2AF are present in the nuclei of living human cells. Although a stable, RNA-independent complex of SF1 and both U2AF subunits was detected in nuclear extracts from the fission yeast Schizosaccharomyces pombe (20), whether such a complex forms in mammalian cells was unclear. Interactions between mammalian SF1 and U2AF65 were observed using recombinant proteins in yeast two-hybrid assays and detected by coimmunoprecipitation in HeLa splicing extracts (1), but when and where these interactions occur were unknown.
In this work we show that the majority of U2AF65, U2AF35, and SF1 cofractionate when HeLa nuclear extracts are analyzed by size exclusion column chromatography (Fig. 2B). U2AF65, U2AF35, and SF1 coeluted between ∼200 and ∼400 kDa, suggesting either a stoichiometry higher than 1:1:1 (the predicted mass for a 1:1:1 complex is <200 kDa) or the presence of additional proteins in the complex. Immunoprecipitation experiments further showed that most of the GFP-U2AF35 in the nuclear extract (Fig. 2A, input) was precipitated by anti-U2AF65 antibody (Fig. 2A, IP), whereas only a minor fraction of GFP-SF1 was immunoprecipitated (Fig. 2A). This is consistent with results from previous studies indicating that the affinity of SF1 for U2AF65 (KD, 50 to 100 nM) is significantly lower than the affinity of U2AF65 for U2AF35 (KD, 1.7 nM) (26) and that the interface of the U2AF65/SF1 complex compared to that of the U2AF35/U2AF65 complex is 25% smaller (40).
Our FRAP experiments revealed that U2AF65, U2AF35, and SF1 are drastically less mobile in the nucleus than expected for noninteracting freely diffusing individual particles, consistent with the view that these proteins establish interactions with each other and probably also with other nuclear components. These interactions can be either stable or transient. In the case of transient interactions, each splicing protein may bind to different partners over time. Thus, although our measured FRAP recovery curves are well fitted by a single diffusion model, the diffusion coefficients that are calculated can correspond to effective or pseudo-effective diffusion regimens (8, 43) in which transient binding to immobile and/or diffusing partners also occurs.
In the speckles, U2AF35 was significantly more mobile than U2AF65 and SF1 (Fig. 3B). This was surprising, taking into account that most of the U2AF35 and U2AF65 in nuclear extracts are found as a heterodimer (J. Valcárcel, personal communication). To explain this difference, we speculate that although at steady state most U2AF65 and U2AF35 molecules form heterodimers, the partners in each complex are constantly exchanging. Components of nuclear speckles may specifically bind U2AF65 but not U2AF35. Individual U2AF65 molecules will therefore move with slower kinetics, while U2AF35 proteins establish sequential transient interactions with different U2AF65 partners.
Introducing mutations in either U2AF65 or SF1 increased the mobility of these proteins, in agreement with the view that they are slowed down by establishing interactions with other nuclear components. As shown in Fig. 4, deletion of different U2AF65 domains caused distinct effects on mobility, as expected since each protein domain is involved in specific interactions. The most prominent increase in mobility was observed for GFP-U2AF65Δ35, a deletion mutant of U2AF65 that lacks the U2AF35 interaction domain (17). Notably, an SF1 variant with a single point mutation that prevents binding to U2AF65 (GFP-SF1R21D [40]) shows a similar mobility rate. For both proteins, the mobility became indistinguishable in the nucleoplasm and nuclear speckles. From this we conclude that binding of U2AF65 to U2AF35 and SF1 is a major determinant to the kinetic behavior of the complex in the nucleus. Using data from our FRAP experiments we were able to estimate a lower limit for the fraction of interacting splicing proteins in the nucleus. Assuming an effective diffusion regimen in which splicing factors bind transiently to the complex, we estimate that the vast majorities of U2AF65 (at least 86%) and SF1 (at least 82%) are interacting at any given time in the nucleus.
The view that SF1 binds to U2AF65 both in the nucleoplasm and in nuclear speckles, as suggested by the previous results, was further demonstrated by FRET methods. Using both acceptor photobleaching FRET in fixed cells (Fig. 5B) and wide-field frequency domain FLIM in fixed (Fig. 5C) and live (Fig. 6) cells, we have shown that SF1 interacts with U2AF65 both in the nucleoplasm and in nuclear speckles and that the interaction persists after treatment with the transcription inhibitor DRB. We also confirmed that U2AF65 interacts with U2AF35 both in the nucleoplasm and in nuclear speckles, as previously reported (12). The possibility that FRET was the result of nonspecific interactions is unlikely, because no FRET was detected when either CFP-U2AF35 and the deletion mutant YFP-U2AF65Δ35 or U2AF65-YFP and the point mutant CFP-SF1R21D were coexpressed (Table 1).
We further detected that U2AF65 can self-interact in vivo, based on both FRET analysis and immunoprecipitation experiments (Fig. 7). Detection of this self-interaction by FRET has been attempted before (12), although with negative results. We attribute this discrepancy to the positioning of the donor and acceptor fluorophores in the tagged splicing factor. In fact, when both CFP and YFP are fused to the amino terminal of U2AF65, as in the study report in reference 12, no FRET signal is detected. Only when both fluorophores are fused to the carboxy terminal of U2AF65 do we obtain a clear FRET signal (Fig. 7). Because FRET is highly dependent on the distance between donor and acceptor fluorophores, having both CFP and YFP at the C terminal of U2AF65 is probably the only configuration in which the two fluorophores are close enough for FRET to occur. This in turn suggests that a putative U2AF65 self-interaction domain could be located close to the C-terminal part of the protein. However, we cannot rule out the possibility that the FRET signal is caused by a close proximity between fluorescently tagged domains of two different U2AF65 molecules that are part of a complex but not directly interacting with each other. The self-interaction of U2AF65 (this work) and U2AF35 (12) could contribute to the high molecular masses of the complexes containing U2AF65, U2AF35, and SF1 that were observed by size exclusion column chromatography (Fig. 2B). Alternatively (or additionally), these complexes may contain other still-unidentified proteins.
Finally, we have shown that depletion of U2AF35 and U2AF65 by RNAi altered the normal distribution of U2AF65 and SF1, which no longer concentrated in nuclear speckles (Fig. 8). By contrast, the distribution of Sm proteins was unaffected, suggesting that the association of spliceosomal snRNPs with nuclear speckles is independent from the U2AF65-U2AF35-SF1 complex.
In conclusion, our results show that SF1 associates with both subunits of U2AF in human cells, forming a complex that assembles in the absence of pre-mRNA. The measured mobilities of U2AF65, U2AF35, and SF1 are consistent with the model that these proteins diffuse in the nucleus as part of a complex. However, individual proteins may constantly exchange between different complexes, making it impossible to conclude that U2AF65, U2AF35, and SF1 are recruited to pre-mRNA as a preformed complex. Our results further strengthen the importance of understanding in more detail the mechanism responsible for displacement of SF1 with replacement by SF3b155/SAP155 during formation of the catalytic spliceosome. This replacement is presumably regulated by phosphorylation of Ser20 in SF1 by PKG-I (50). Based on the observation that U2AF associates with SF1 outside the spliceosome, we speculate that displacement of SF1 is controlled by a transient modification reverted after splicing.
Supplementary Material
Acknowledgments
We are grateful to Angus Lamond, Juan Valcárcel, and Angela Kramer for kindly providing reagents and to Eduardo Pinho e Melo for the use of the gel filtration facilities at Instituto Superior Técnico, Portugal. We thank Erik B. van Munster for his assistance with the FLIM experiments. We are also thankful to our colleagues José Braga and Margarida Gama-Carvalho for help and stimulating discussions.
This work was supported by grants from Fundação para a Ciência e Tecnologia, Portugal (POCI/SAU-MMO/57700/2004) and the European Commission (LSHG-CT-2003-503259 and LSHG-CT-2005-518238).
Footnotes
Published ahead of print on 19 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89403-412. [DOI] [PubMed] [Google Scholar]
- 2.Almeida, F., R. Saffrich, W. Ansorge, and M. Carmo-Fonseca. 1998. Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 142899-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, K., B. Bjorkroth, and B. Daneholt. 1984. Packing of a specific gene into higher order structures following repression of RNA synthesis. J. Cell Biol. 981296-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson, K., R. Mahr, B. Bjorkroth, and B. Daneholt. 1982. Rapid reformation of the thick chromosome fiber upon completion of RNA synthesis at the Balbiani ring genes in Chironomus tentans. Chromosoma 8733-48. [DOI] [PubMed] [Google Scholar]
- 5.Bauren, G., W. Q. Jiang, K. Bernholm, F. Gu, and L. Wieslander. 1996. Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J. Cell Biol. 133929-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berglund, J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braga, J., J. M. Desterro, and M. Carmo-Fonseca. 2004. Intracellular macromolecular mobility measured by fluorescence recovery after photobleaching with confocal laser scanning microscopes. Mol. Biol. Cell 154749-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braga, J., J. G. McNally, and M. Carmo-Fonseca. 2007. A reaction-diffusion model to study RNA motion by quantitative fluorescence recovery after photobleaching. Biophys. J. 922694-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36333-360. [DOI] [PubMed] [Google Scholar]
- 10.Calapez, A., H. M. Pereira, A. Calado, J. Braga, J. Rino, C. Carvalho, J. P. Tavanez, E. Wahle, A. C. Rosa, and M. Carmo-Fonseca. 2002. The intranuclear mobility of messenger RNA binding proteins is ATP dependent and temperature sensitive. J. Cell Biol. 159795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chodosh, L. A., A. Fire, M. Samuels, and P. A. Sharp. 1989. 5,6-Dichloro-1-beta-d-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 2642250-2257. [PubMed] [Google Scholar]
- 12.Chusainow, J., P. M. Ajuh, L. Trinkle-Mulcahy, J. E. Sleeman, J. Ellenberg, and A. I. Lamond. 2005. FRET analyses of the U2AF complex localize the U2AF35/U2AF65 interaction in vivo and reveal a novel self-interaction of U2AF35. RNA 111201-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois, M. F., V. T. Nguyen, S. Bellier, and O. Bensaude. 1994. Inhibitors of transcription such as 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole and isoquinoline sulfonamide derivatives (H-8 and H-7) promote dephosphorylation of the carboxyl-terminal domain of RNA polymerase II largest subunit. J. Biol. Chem. 26913331-13336. [PubMed] [Google Scholar]
- 14.Egyhazi, E. 1975. Inhibition of Balbiani ring RNA synthesis at the initiation level. Proc. Natl. Acad. Sci. USA 72947-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadella, T. W. J., and T. M. Jovin. 1995. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J. Cell Biol. 1291543-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gama-Carvalho, M., M. P. Carvalho, A. Kehlenbach, J. Valcarcel, and M. Carmo-Fonseca. 2001. Nucleocytoplasmic shuttling of heterodimeric splicing factor U2AF. J. Biol. Chem. 27613104-13112. [DOI] [PubMed] [Google Scholar]
- 17.Gama-Carvalho, M., R. D. Krauss, L. Chiang, J. Valcarcel, M. R. Green, and M. Carmo-Fonseca. 1997. Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137975-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gozani, O., J. Potashkin, and R. Reed. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 184752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Habets, W. J., M. H. Hoet, B. A. De Jong, A. Van der Kemp, and W. J. Van Venrooij. 1989. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J. Immunol. 1432560-2566. [PubMed] [Google Scholar]
- 20.Huang, T., J. Vilardell, and C. C. Query. 2002. Pre-spliceosome formation in S. pombe requires a stable complex of SF1-U2AF(59)-U2AF(23). EMBO J. 215516-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 125-14. [DOI] [PubMed] [Google Scholar]
- 22.Karpova, T. S., C. T. Baumann, L. He, X. Wu, A. Grammer, P. Lipsky, G. L. Hager, and J. G. McNally. 2003. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 20956-70. [DOI] [PubMed] [Google Scholar]
- 23.Kent, O. A., D. B. Ritchie, and A. M. Macmillan. 2005. Characterization of a U2AF-independent commitment complex (E′) in the mammalian spliceosome assembly pathway. Mol. Cell. Biol. 25233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenworthy, A. K. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods 24289-296. [DOI] [PubMed] [Google Scholar]
- 25.Kielkopf, C. L., S. Lucke, and M. R. Green. 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 181513-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kielkopf, C. L., N. A. Rodionova, M. R. Green, and S. K. Burley. 2001. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106595-605. [DOI] [PubMed] [Google Scholar]
- 27.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4605-612. [DOI] [PubMed] [Google Scholar]
- 28.Lenart, P., G. Rabut, N. Daigle, A. R. Hand, M. Terasaki, and J. Ellenberg. 2003. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J. Cell Biol. 1601055-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lerner, E. A., M. R. Lerner, C. A. Janeway, Jr., and J. A. Steitz. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA 782737-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcarcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402838-841. [DOI] [PubMed] [Google Scholar]
- 31.Nilsen, T. W. 2003. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 251147-1149. [DOI] [PubMed] [Google Scholar]
- 32.Pacheco, T. R., M. B. Coelho, J. M. Desterro, I. Mollet, and M. Carmo-Fonseca. 2006. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol. Cell. Biol. 268183-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pacheco, T. R., A. Q. Gomes, N. L. Barbosa-Morais, V. Benes, W. Ansorge, M. Wollerton, C. W. Smith, J. Valcarcel, and M. Carmo-Fonseca. 2004. Diversity of vertebrate splicing factor U2AF35: identification of alternatively spliced U2AF1 mRNAs. J. Biol. Chem. 27927039-27049. [DOI] [PubMed] [Google Scholar]
- 34.Pacheco, T. R., L. F. Moita, A. Q. Gomes, N. Hacohen, and M. Carmo-Fonseca. 2006. RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol. Biol. Cell 174187-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404604-609. [DOI] [PubMed] [Google Scholar]
- 36.Reits, E. A., and J. J. Neefjes. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3E145-E147. [DOI] [PubMed] [Google Scholar]
- 37.Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 201320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruskin, B., P. D. Zamore, and M. R. Green. 1988. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 52207-219. [DOI] [PubMed] [Google Scholar]
- 39.Rutz, B., and B. Seraphin. 1999. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA 5819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selenko, P., G. Gregorovic, R. Sprangers, G. Stier, Z. Rhani, A. Kramer, and M. Sattler. 2003. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell 11965-976. [DOI] [PubMed] [Google Scholar]
- 41.Serizawa, H., J. W. Conaway, and R. C. Conaway. 1993. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature 363371-374. [DOI] [PubMed] [Google Scholar]
- 42.Spector, D. L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9265-315. [DOI] [PubMed] [Google Scholar]
- 43.Sprague, B. L., R. L. Pego, D. A. Stavreva, and J. G. McNally. 2004. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 863473-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thickman, K. R., M. C. Swenson, J. M. Kabogo, Z. Gryczynski, and C. L. Kielkopf. 2006. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J. Mol. Biol. 356664-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Munster, E. B., and T. W. Gadella. 2005. Fluorescence lifetime imaging microscopy (FLIM). Adv. Biochem. Eng. Biotechnol. 95143-175. [DOI] [PubMed] [Google Scholar]
- 46.van Munster, E. B., and T. W. Gadella, Jr. 2004. phiFLIM: a new method to avoid aliasing in frequency-domain fluorescence lifetime imaging microscopy. J. Microsc. 21329-38. [DOI] [PubMed] [Google Scholar]
- 47.van Munster, E. B., and T. W. Gadella, Jr. 2004. Suppression of photobleaching-induced artifacts in frequency-domain FLIM by permutation of the recording order. Cytometry A 58185-194. [DOI] [PubMed] [Google Scholar]
- 48.Vermeer, J. E., E. B. Van Munster, N. O. Vischer, and T. W. Gadella, Jr. 2004. Probing plasma membrane microdomains in cowpea protoplasts using lipidated GFP-fusion proteins and multimode FRET microscopy. J. Microsc. 214190-200. [DOI] [PubMed] [Google Scholar]
- 49.Wallrabe, H., and A. Periasamy. 2005. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 1619-27. [DOI] [PubMed] [Google Scholar]
- 50.Wang, X., S. Bruderer, Z. Rafi, J. Xue, P. J. Milburn, A. Kramer, and P. J. Robinson. 1999. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J. 184549-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wouters, F. S., and P. I. Bastiaens. 1999. Fluorescence lifetime imaging of receptor tyrosine kinase activity in cells. Curr. Biol. 91127-1130. [DOI] [PubMed] [Google Scholar]
- 52.Wouters, F. S., P. J. Verveer, and P. I. Bastiaens. 2001. Imaging biochemistry inside cells. Trends Cell Biol. 11203-211. [DOI] [PubMed] [Google Scholar]
- 53.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402832-835. [DOI] [PubMed] [Google Scholar]
- 54.Zamore, P. D., and M. R. Green. 1989. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl. Acad. Sci. USA 869243-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zamore, P. D., J. G. Patton, and M. R. Green. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355609-614. [DOI] [PubMed] [Google Scholar]
- 56.Zorio, D. A., and T. Blumenthal. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402835-838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.