Abstract
Human MRE11 is a key enzyme in DNA double-strand break repair and genome stability. Human MRE11 bears a glycine-arginine-rich (GAR) motif that is conserved among multicellular eukaryotic species. We investigated how this motif influences MRE11 function. Human MRE11 alone or a complex of MRE11, RAD50, and NBS1 (MRN) was methylated in insect cells, suggesting that this modification is conserved during evolution. We demonstrate that PRMT1 interacts with MRE11 but not with the MRN complex, suggesting that MRE11 arginine methylation occurs prior to the binding of NBS1 and RAD50. Moreover, the first six methylated arginines are essential for the regulation of MRE11 DNA binding and nuclease activity. The inhibition of arginine methylation leads to a reduction in MRE11 and RAD51 focus formation on a unique double-strand break in vivo. Furthermore, the MRE11-methylated GAR domain is sufficient for its targeting to DNA damage foci and colocalization with γ-H2AX. These studies highlight an important role for the GAR domain in regulating MRE11 function at the biochemical and cellular levels during DNA double-strand break repair.
Genome stability relies on the accurate repair of double-strand breaks (DSBs) that arise naturally during DNA replication or from treatment with exogenous DNA-damaging agents. DSBs in human cells may be repaired by nonhomologous end joining or homologous recombination. The MRE11-RAD50-NBS1 (MRN) complex has essential functions in numerous facets of genome stability. During the cell cycle, MRN is associated to chromatin, which is consistent with a role in the surveillance of genome integrity. Chromatin association is not increased following ionizing radiation (41). However, a redistribution of the MRN complex from the chromatin to sites of DNA damage occurs. When cells are challenged with ionizing radiation, ataxia-telangiectasia mutated protein (ATM) phosphorylates histone H2AX (30), a key event in this process. The phosphorylation of H2AX is thought to recruit MRN directly to DSBs, since NBS1 interacts directly with phosphorylated H2AX (17). MDC1 is also a key molecular linker responsible for bridging NBS1 with phosphoepitopes generated in DSB-flanking chromatin, as MRN components do not form foci in the absence of MDC1 (22). MRN is a central player during checkpoint signaling of DNA damage. MRN stimulates ATM kinase activity on p53, CHK2, and H2AX (19). ATM phosphorylates NBS1 on serine 343, which is necessary for the proper S-phase checkpoint following ionizing radiation (21). Consistent with key functions in DNA damage signaling and repair, ataxia-telangiectasia-like disorder (34) and Nijmegen breakage syndrome (40), which are caused by mutations in MRE11 and NBS1, respectively, are characterized by an increased sensitivity to ionizing radiation, checkpoint problems, and the accumulation of DSBs and aberrant chromosomes, leading to cancer.
DNA repair functions of the MRN complex involve the processing of DNA ends during homologous recombination, nonhomologous end joining, and maintenance of telomere length (9). By using yeast (Saccharomyces cerevisiae) and human proteins, biochemical studies have shown that Mre11 has a 3′-to-5′, Mn2+-dependent exonuclease activity on DNA substrates with blunt or 5′ protruding ends. MRE11 displays endonuclease activity on hairpin and single-stranded DNA (ssDNA) substrates (26, 37). The MRN complex can also mediate limited DNA duplex unwinding and hairpin opening in a reaction that is stimulated by ATP. The presence of ATP also allows the complex to endonucleolytically cut a 3′ overhang at a single-strand-double-strand transition (25). In view of the effects of deleting the Saccharomyces MRE11, RAD50, or XRS2 gene on the processing of DSBs in vivo, it was surprising to observe that MRE11 has in vitro 3′-to-5′ exonuclease activity on double-stranded DNA (dsDNA) (26). Indeed, DSBs induced in mitotic cells by the HO endonuclease are resected in a 5′ to 3′ fashion and this degradation of DSB ends is markedly retarded when any one gene among MRE11, RAD50, and XRS2 is deleted (15, 20). It is now clear that the nuclease activity of MRE11 is important for DSB signaling and repair. A nuclease-defective MRE11 protein containing two amino acid substitutions (H129L/D130V) that reduce its endonuclease activity (35) failed to fully restore damage-induced ATM activation in human ataxia-telangiectasia-like disorder cells (39). The nuclease activity of MRE11 is required for the processing of DNA DSBs to generate the replication-protein-A-coated ssDNA that is needed for ATR recruitment and the subsequent phosphorylation and activation of Chk1 (16). In this context, the processing of DSBs by MRE11 is modulated by the CtIP protein (32).
Although the biochemical properties of MRE11 are well defined, little is known about the regulation of these activities by posttranslational modifications. Amino acid side chain methylation was first reported in the mid-sixties and proposed to regulate transcription (2, 8, 24). For many years, fewer than 20 proteins, including myelin basic proteins and histones, had been identified as containing dimethylated arginines (24). Arginine methylation is a posttranslational modification that can result in symmetrical or asymmetrical dimethylarginines (3). Nine protein arginine methyltransferases (PRMTs) catalyze this reaction by using S-adenosyl-l-methionine as a methyl donor (13). This modification occurs frequently on nucleic acid binding proteins containing a glycine-arginine-rich (GAR) motif, and arginine methylation has previously been shown to regulate the cellular localization of proteins as well as protein-protein interactions (3). A proteomic study conducted in 2003 identified more than 200 proteins that may be arginine methylated. Components required for pre-mRNA splicing, polyadenylation, transcription, and signal transduction as well as the DNA repair protein MRE11 were found to be arginine methylated, suggesting that this modification regulates many cellular processes (4). DNA repair proteins, such as MRE11 (5), 53BP1 (1, 7), and DNA polymerase β (11), are now emerging as key targets of PRMTs.
We have previously shown the role of arginine methylation in S-phase progression and nuclear compartmentalization (5, 6). These observations left open several important questions that we sought to address further in this study. What are the roles of the GAR motif and arginine methylation in MRE11 enzymatic activity? How many residues need to be methylated for optimal MRE11 biochemical activity? Does MRE11 relocalization to DNA damage sites depend on the GAR domain? We present in vitro and in vivo data to answer these questions. We have purified MRE11 from Sf9 cells and demonstrated that methylation by PRMT1 is conserved in insect cells. By using purified MRE11 mutant proteins, we show that the first six arginines modulate MRE11 biochemical activities. The inhibition of arginine methylation decreases MRE11 focus formation on a unique DSB in vivo. Moreover, the GAR domain is sufficient to localize MRE11 to nuclear focus after DNA damage and for DNA binding in vitro. The crucial role of the GAR domain for MRE11 protein function is supported by a high conservation in multicellular organisms.
MATERIALS AND METHODS
Cell culture.
DR95hyg-xt (29), SKN-SH neuroblastoma cells, and DR95 carrying an I-SceI cassette were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Sf9 cells were grown in Grace's medium supplemented with 10% fetal calf serum. When indicated, methylation inhibitor MTA (5-deoxy-5-methylthioadenosine) and/or ADOX (adenosine-2′,3′-dialdehyde) was added to the medium 24 h prior to the experiment.
Antibodies.
Asym25, anti-R587, and anti-MeMRE11 antibodies have previously been described (5, 6). Commercial antibodies used were anti-MRE11 rabbit (Oncogene), anti-MRE11 mouse (GeneTex), anti-c-Myc (Santa Cruz Biotechnology), anti-NBS1 and anti-RAD50 (Novus Biologicals), anti-Flag (Sigma), anti-γH2AX (Upstate), and anti-RAD51.
DNA constructs and MRE11 mutagenesis.
Full-length human MRE11, NBS1, and RAD50 cDNAs cloned in pFASTBAC1 were generously provided by Tanya Paull. Human PRMT1 cDNA fused to the c-Myc tag from the pcDNA3-PRMT1 construct was cloned in pFASTBAC by using BamHI and XhoI sites. Fusions to green fluorescent protein (GFP) (with or without nuclear localization signal [NLS]) were performed by using modified pcDNA3 vectors containing FLAG-GFP or FLAG-GFP-NLS. pCBASce is an I-SceI expression vector (29). Mutations were introduced into the GAR motif of MRE11 by using the QuikChange II site-directed mutagenesis kit (Stratagene). All constructs were confirmed by DNA sequencing.
Protein expression and purification from insect cells.
Recombinant wild-type (WT) MRE11 and MRE11 mutants, GAR-Sf9 (encompassing amino acids 498 to 615 of human MRE11) and MRE11-ΔGAR, were fused with a six-histidine tag and purified as described previously (5). Recombinant GAR-Bacto was purified from Escherichia coli BL21(DE3) RP (Stratagene) by using the same scheme of purification. NBS1, RAD50, and PRMT1-Myc were produced from baculovirus-infected Sf9 cells by using the Bac-to-Bac expression system (Invitrogen).
Gel filtration analysis.
The molecular mass of purified MRE11 proteins was determined by a comparison with gel filtration standards (250 μg; bovine thyroglobulin [670 kDa], bovine gamma globulin [158 kDa], chicken ovalbumin [44 kDa], horse myoglobin [17 kDa], and vitamin B-12 [1.35 kDa]). Proteins were analyzed on an Explorer 10 (fast-performance liquid chromatography) system fitted with a 24-ml Superdex 200 PC 3.2/30 column (Pharmacia) equilibrated in R150 buffer (25 mM Tris-Cl, pH 8.0, 50 mM NaCl, 5% glycerol, 1 mM dithiothreitol [DTT], 0.05% Tween 20). Fractions (500 μl) were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blotting with a monoclonal antihistidine antibody.
Coimmunoprecipitation.
Immunoprecipitations using DR95 or 293T cells (collected 24 h after transfection) were performed as described previously (29). Immunoprecipitations from Sf9 cells were performed as described above, with the following modifications. Sf9 cells (20 × 106) were infected with MRE11, NBS1, RAD50, or PRMT1 baculovirus (multiplicity of infection of ∼10) for 2 days at 27°C and stored in two aliquots at −80°C. Cells were lysed in P5 buffer (50 mM NaHPO4, pH 7.0, 500 mM NaCl, 5 mM imidazole, 10% glycerol, 0.05% Triton X-100), and soluble extracts were prepared to verify the level of expression of the proteins of interest in the first aliquot. When similar levels were achieved, the remaining cells were lysed in P5 buffer and used for immunoprecipitation and immunoblotting analysis.
Endonuclease, exonuclease assays, EMSA, and electron microscopy.
DNA substrates used in endonuclease and exonuclease assays were generated with purified oligonucleotides (18). Endonuclease and exonuclease reactions were performed as described previously (5). Electrophoretic mobility shift assay (EMSA) reactions were performed in 25 mM MOPS (morpholinepropanesulfonic acid), pH 7.0, 60 mM KCl, 0.2% Tween, 2 mM DTT, 1 mM Mg(CH3COO)2. Protein and DNA were incubated for 15 min at 37°C, followed by 10 min of fixation in 0.2% glutaraldehyde. Reaction mixtures were subjected to electrophoresis on 6% acrylamide/bis-acrylamide (29:1), 50 mM Tris-glycine gels.
Electron microscopy reactions contained 5 μM øX174 ssDNA in 25 mM MOPS, pH 7.0, 60 mM KCl, 0.2% Tween, 2 mM DTT, 1 mM Mg(CH3COO)2. After 5 min at 37°C, MRE11 proteins (0.2 μM) were added and incubation was continued for a further 10 min. When fixation was required, protein-DNA complexes were fixed by the addition of glutaraldehyde to 0.2%, followed by 15 min of incubation at 37°C. Samples were diluted and washed in 5 mM Mg(CH3COO)2 prior to uranyl acetate staining (33). Complexes were visualized at a magnification of ×22,000 by using a Philips CM12 electron microscope.
Immunofluorescence.
Immunofluorescence analyses of SKN-SH A1 cells transfected with pCBASce, or HeLa subjected to local laser damage, were performed as described previously (29). Images were collected with a Leica DMI3000B inverted microscope. Images were deconvoluted using Volocity software (Improvision).
Live-cell microscopy, FRAP analysis, and laser-induced DNA DSBs.
Live-cell imaging was performed on a PerkinElmer UltraVIEW system. A laser-scanning confocal microscope (Olympus FV1000) with a 25-mW, 405-nm laser diode was used to perform all photobleaching experiments by using a 100× objective (numerical aperture, 1.4). Transmitted light images were collected to monitor cell viability. We performed fluorescence recovery after photobleaching (FRAP) experiments by exposing defined regions of cells to 100% laser intensity for typically two iterations. The region bleached was 18 by 18 pixels, using the tornado function. Imaging was typically performed by using a 100-mW, 488-nm argon laser at 1% laser intensity. The interval between image scans was 127 ms for a total duration of 8.8 s. The interval between scans was set so that 70 image scans were required in total for the experiment. Recovery was considered complete when the intensity of the photobleached region stabilized (that is, when the curve flattened). For quantitative analysis, fluorescence intensity was measured at each time point for the photobleached region, a portion of the cell nucleus, and the extracellular background for normalization by using Olympus FluoView FV1000 (version 1.6) imaging software. Raw intensities were normalized following the double normalization as described previously (27). Briefly, the background is subtracted from the FRAP intensities before being normalized to prebleach intensities and corrected for the loss of total signal due to the bleach pulse. Half-recovery times were calculated by fitting an exponential model, I(t) = A(1 − e−ττ), on the normalized data by using R (www.r-project.org). In this equation, I(t) is the normalized data and t is the time elapsed since bleach in milliseconds. The two free parameters, A and τ, are fitted, and the half-recovery time is calculated with −ln(0.5)/τ. The significance of the difference between GAR RA and GAR WT half-recovery times was assessed using a t test assuming unequal variance and a two-sided hypothesis. Laser-induced DNA DSBs were created using a 25-mW, 405-nm laser diode by exposing defined regions of cells to 100% laser intensity for typically 10 iterations (19 ms per iteration). Immunofluorescence analysis was performed 1 h 30 min after the treatment.
RESULTS
Human MRE11 interacts with PRMT1.
We previously reported that MRE11 is methylated on nine arginines in human cells (5). Since our goal was to investigate the effect of arginine methylation on the biochemical properties of human MRE11, it was important to verify whether the GAR motif was methylated when human MRE11 was expressed in insect cells known to generate an active MRE11 nuclease (26). Indeed, MRE11 or MRN was methylated in insect cells (Fig. 1A). Methylation was abolished when insect cells expressing MRN or MRE11 were treated with methylation inhibitors MTA and ADOX (Fig. 1A, lanes 5 and 6, respectively). Since MRE11 is methylated in normal Sf9 insect cells, this result suggests that previous biochemical experiments with this protein were performed with methylated MRE11 (26).
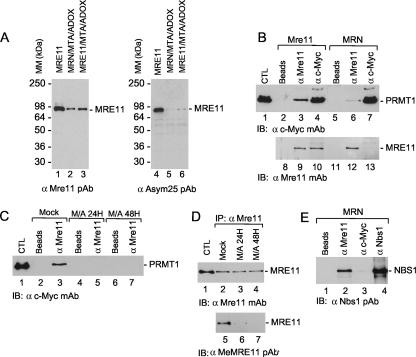
FIG. 1.
MRE11 is methylated in insect cells and interacts with PRMT1. (A) SF9 cells were infected with MRE11 (lanes 3 and 6) or MRE11, RAD50, and NBS1 baculoviruses (lanes 2 and 5) and treated with methyltransferase inhibitors MTA and ADOX for 24 h (lanes 2 to 3 and 5 to 6). Lanes 1 and 4, purified MRE11-His6. MM, molecular mass; PAb, polyclonal antibody. (B) Interactions between MRE11 and PRMT1. SF9 cells were coinfected with MRE11 and c-Myc-PRMT1 baculoviruses. Immunoprecipitations were conducted using beads alone (lanes 2, 5, 8, and 11), anti-MRE11 (lanes 3, 6, 9, and 12), or anti-c-Myc (lanes 4, 7, 10, and 13) and visualized by Western blotting as indicated. Lane 1, whole-cell extract from Sf9 cells infected with c-Myc PRMT1 baculovirus. CTL, control; IB, immunoblot; mAb, monoclonal antibody. (C) Treatment of Sf9 cells coinfected with MRE11 and PRMT1 viruses with methyltransferase inhibitors MTA and ADOX disrupt MRE11-PRMT1 interaction. SF9 cells were coinfected with MRE11 and c-Myc-PRMT1 baculoviruses, and immunoprecipitations were conducted using beads alone (lanes 2, 4, and 6) or anti-MRE11 (lanes 3, 5, and 7) and visualized by Western blotting, as indicated. Lane 1, whole-cell extract from Sf9 cells infected with c-Myc PRMT1 baculovirus. (D) Control figure showing that MTA and ADOX treatment inhibit MRE11 methylation in the presence of PRMT1. SF9 cells coinfected with MRE11 and PRMT1 baculoviruses were mock treated (lanes 2 and 5) or treated with MTA and ADOX (M/A) for 24 h or 48 h (lanes 3 and 6 and 4 and 7, respectively). Lane 1, purified MRE11-His6. MRE11 was immunoprecipitated with anti-MRE11 polyclonal antibody and revealed with anti-MRE11 and anti-MeMRE11 polyclonal antibody, which detects methylated arginines in MRE11. IP, immunoprecipitate. (E) PRMT1 does not interact with the MRN complex. SF9 cells were coinfected with MRE11, RAD50, NBS1, and c-Myc-PRMT1 baculoviruses, and complexes were immunoprecipitated with beads alone (lane 1), anti-MRE11 (lane 2), anti-c-Myc (lane 3), and anti-NBS1 (lane 4) and revealed with an anti-Nbs1 polyclonal antibody.
We next examined the ability of MRE11 or the MRN complex to associate with the major arginine methyltransferase, PRMT1. Insect cells were coinfected with c-Myc-tagged PRMT1 and MRE11 or MRN baculoviruses. A complex between MRE11 and PRMT1 was observed (Fig. 1B, lanes 3 and 9). Conversely, the immunoprecipitation of PRMT1 allowed the detection of an MRE11-PRMT1 complex (Fig. 1B, lanes 4 and 10). The interaction was relatively strong, as washes containing up to 750 mM NaCl did not disrupt the interaction (data not shown). The addition of protein methyltransferase inhibitors MTA/ADOX to the culture medium abrogated the interaction between MRE11 and PRMT1 (Fig. 1C). Control experiments revealed that the same amount of MRE11 protein was immunoprecipitated, but methylation was not detected with a specific MRE11-GAR-methylated antibody (anti-MeMRE11) (Fig. 1D, lanes 6 and 7). We also verified whether PRMT1 interacted with MRE11-RAD50-NBS1. When MRE11, RAD50, NBS1, and PRMT1 were overexpressed together, the interaction between MRE11 and PRMT1 was greatly reduced (Fig. 1B, lanes 6 and 13). Under these conditions, the MRN complex is formed, but no PRMT1 is pulled down in NBS1 immunoprecipitates (Fig. 1E, lane 3). Taken together, these results suggest that arginine methylation of MRE11 occurs before its incorporation into the MRN complex.
The enzymatic properties of MRE11 are regulated by arginine methylation.
Since human MRE11 is methylated in insect cells, which suggests an evolutionary conservation, we checked whether the GAR motif was conserved in various organisms. Pileup analysis revealed that the GAR motif is conserved in Xenopus, chicken, mouse, rat, and monkey and possesses at least nine arginines within 60 amino acids (Fig. 2A). The first region of the GAR motif was better conserved than the second. Within the C terminus, the GAR motif is the most conserved MRE11 region for these organisms. Specific arginine-glycine stretches were not found in budding and fission yeast protein sequences. We envisioned two roles for the GAR domain in the regulation of MRE11: (i) methyl-dependent protein-protein interactions on this domain could modulate MRE11 activity, and (ii) the GAR domain could directly control MRE11 biochemical activities.
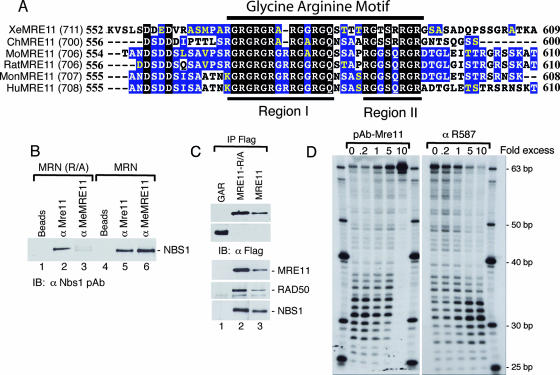
FIG. 2.
(A) Comparison of amino acid sequences encompassing the MRE11 GAR motif. The sequences include XeMRE11 from Xenopus laevis, ChMRE11 from Gallus gallus, MoMRE11 from Mus musculus, RatMRE11 from Rattus norvegicus, MonMRE11-from Macaca fascicularis, and HuMRE11 from Homo sapiens. Residues conserved in all the sequences are highlighted in black, residues conserved in more than 50% of the sequences are highlighted in blue, and residues of the same group are shown in yellow. (B) The GAR motif of MRE11 within the MRN complex is accessible. SF9 cells were infected with MRE11(R/A), RAD50, and NBS1 baculoviruses or WT MRE11, RAD50, and NBS1 baculoviruses, as indicated. Immunoprecipitations were conducted using beads alone (lanes 1 and 4), anti-MRE11 (lanes 2 and 5), anti-R587 (lanes 3 and 7), and anti-MeMRE11 (lanes 3 and 6). Proteins were detected with anti-Nbs1 antibodies. (C) The GAR motif of MRE11 does not interact with MRE11, RAD50, or NBS1. 293T cells were transfected with GFP-GAR-NLS (lane 1), MRE11-R/A (lane 2), or WT MRE11 (lane 3). Whole-cell extracts were prepared, and complexes were immunoprecipitated using anti-Flag. Complexes were revealed using anti-Flag, anti-MRE11, anti-RAD50, and anti-NBS1, as indicated. (D) MRE11 exonuclease activity is inhibited by an MRE11 polyclonal antibody and stimulated by a specific methylated GAR motif polyclonal antibody. The excess of MRE11 or R587 polyclonal antibodies is indicated. A total of 10 nM of purified MRE11 was used in this assay.
If MRE11 was regulated by methyl-dependent protein-protein interactions, the GAR domain must be accessible. We verified the accessibility of the GAR domain by immunoprecipitation of MRN with the methyl-specific anti-MeMRE11 antibody (Fig. 2B). Anti-MeMRE11 could immunoprecipitate MRN at the same level as that of a polyclonal antibody raised against MRE11, suggesting that this region is accessible (Fig. 2B, lanes 5 and 6). Control experiments revealed that anti-MeMRE11 did not immunoprecipitate MRN(R/A), suggesting that the antibody was specific for the GAR-methylated region (lane 3). Second, we expressed GAR-GFP-NLS in cells and verified whether it could bind RAD50 and NBS1 (Fig. 2C). GAR-GFP-NLS was not associated with NBS1 or RAD50, whereas full-length MRE11 or MRE11(R/A) bound to NBS1 or RAD50. These results suggested that the GAR domain is not implicated in the formation of the MRN complex and is available for interactions with other proteins. As stated above, these proteins might modulate MRE11 activities in vivo by protein-protein contact. This effect can be reproduced using antibody contact. Different antibodies targeting the MRE11 protein were incubated with purified MRE11, and the exonuclease activity was assayed (Fig. 2D). As predicted, anti-MRE11 polyclonal antibodies had an inhibitory effect on MRE11 exonuclease activity. A 10-fold excess of antibody could effectively inhibit MRE11 exonuclease activity, probably due to a general coating of the MRE11 protein. On the other hand, polyclonal antibodies raised against the methylated region of the MRE11 GAR domain had an activating effect on exonuclease activity. As controls, two other monoclonal antibodies against MRE11 had no effect on MRE11 exonuclease activity (data not shown). Thus, our results show that protein-protein interactions involving the GAR domain may modulate MRE11 activity during DNA repair.
One of the key functions of MRE11 is its nuclease activity. Hence, we investigated whether arginine methylation can directly regulate MRE11 exonuclease and endonuclease activities. Thus, we separated the GAR motif into two regions and different mutants were generated (Fig. 3A) based on a sequence comparison (Fig. 2A). Mutations in all nine arginines were named “full” mutants, whereas mutants in region I (RI) comprised mutations in the first six arginines, and mutants in region II (RII) were mutated in the last three arginines. The first mutant group, designated R/A, represents mutants in which the arginines were changed to alanines, therefore allowing us to study the effects of absence of positive charges and methyl group on each residue. Members of the R/K group of mutants have arginines changed to lysines in order to conserve a long side chain with positive charges. Altogether, six different mutant proteins, along with WT MRE11, were purified to homogeneity (Fig. 3B). The methylation status of the different mutants was verified using antibodies that recognize methylated arginines in RI (anti-MeMRE11) or RII (anti-R587). Mutants in RI were not recognized by anti-MeMRE11, whereas mutants in RII were not detected by anti-R587. Therefore, all purified MRE11 mutants are region specific and mutations in one region do not affect the methylation of the other.
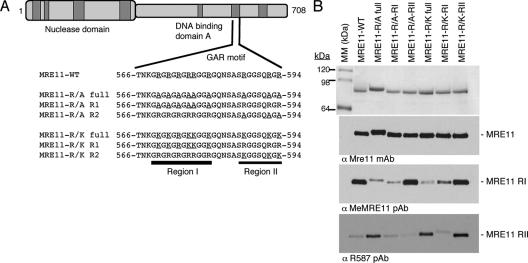
FIG. 3.
(A) Schematic representation of the various MRE11 mutants. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western analysis of the purified WT and mutant MRE11 proteins (top panels) and their methylation state were assessed by Western blotting with methylation-specific antibodies (middle and bottom panels). Anti-MeMRE11 recognizes arginines methylated in RI, whereas anti-R587 recognizes arginines methylated in RII. MM, molecular mass; mAb, monoclonal antibody; pAb, polyclonal antibody.
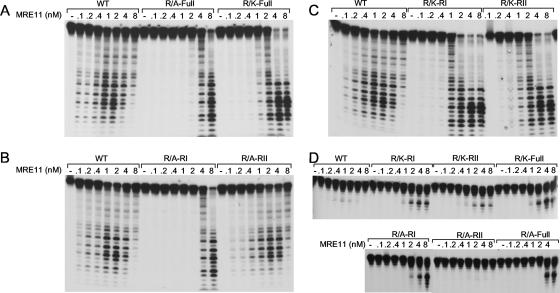
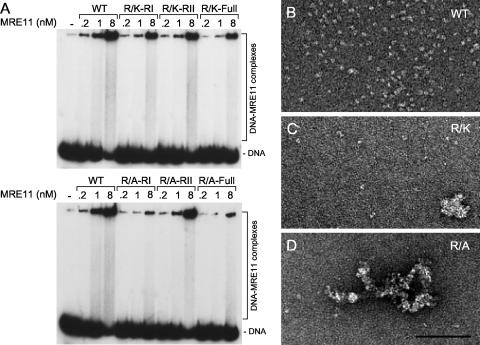
We next investigated whether the exonuclease activity of MRE11 was affected in the mutants (Fig. 4A, B, and C). MRE11-WT displayed strong Mn+2-dependent exonuclease activity at a concentration of 0.1 to 0.4 nM protein (which corresponds to an MRE11-to-DNA ratio of 1/1,000 to 1/250), whereas R/A-RI, R/K-RI, R/A-full, and R/K-full were impaired in DNA resection at these concentrations. At a 1-nM concentration of MRE11, the R/A-RI and R/A-full mutants displayed no activity, whereas the R/K-RI and R/K-full mutants showed weak activity. At an 8-nM protein concentration, DNA resection was reduced for WT MRE11, whereas strong exonuclease activity was observed for R/A-RI, R/K-RI, and R/A-full. The possibility of the presence of a contaminating protein was excluded, since MRE11 H217Y mutated in the phosphodiesterase motif shows no activity (data not shown). Our results could be explained by an increase of the local concentration of the MRE11 mutants at the extremity of DNA. To further confirm this hypothesis, we performed endonuclease assays (Fig. 4D). We expected to see a difference in endonuclease activity, since this reaction should not involve extensive movement of the protein on DNA. The endonuclease activity of MRE11 was monitored on 3′-tailed DNA substrates in which DNA can be cleaved at a dsDNA/ssDNA junction. As expected, WT MRE11 showed weak endonuclease activity. Surprisingly, the MRE11 mutants with the weakest exonuclease activity showed better endonuclease activity (Fig. 4D). Taken together, these results show that the GAR motif influences the balance between the exonuclease and the endonuclease activities of MRE11.
FIG. 4.
(A to C) Exonuclease activity of WT MRE11 and mutants on dsDNA. The indicated amounts of WT or mutant MRE11 proteins were incubated with 100 nM of blunt-ended dsDNA labeled at a single 5′ extremity, followed by deproteinization and analysis of the samples on a denaturing gel. (D) Endonuclease activity of WT MRE11 and mutants on dsDNA. The indicated amounts of WT or mutant MRE11 proteins were incubated with 100 nM of 3′-tailed DNA labeled at the 5′ extremity of the longest oligonucleotide, followed by deproteinization and analysis of the samples on a denaturing gel.
How could this difference between exonuclease and endonuclease activities of the mutants be explained? We used DNA binding analysis of MRE11 mutants in order to get clues to solve this question. The WT and mutant proteins were all proficient in DNA binding, albeit at different levels (Fig. 5A). For instance, the most affected mutants in exonuclease assays (R/A-full and R/A-RI) were also the most compromised for DNA binding. The R/K-full mutant showed intermediate exonuclease activity and DNA binding. However, the R/A-full, R/A-RI, and R/K-full mutants were the most effective in endonuclease assays. It is conceivable that the DNA binding of these mutants might be similar in gel assays but different when observed by electron microscopy. We did not observe aggregation of the WT, R/K-full, or R/A-full mutants in the absence of DNA (data not shown). However, we observed increased aggregation of the R/A-full mutant on ssDNA (Fig. 5D) compared to the level for the WT or the R/K-full mutant (Fig. 5C). Consistent with DNA binding assays, the number of bound DNA molecules by R/A-full and R/K-full was decreased compared to that for the WT protein. Collectively, these results suggest that the aggregation observed might be beneficial for endonuclease activity but deleterious for exonuclease activity.
FIG. 5.
DNA binding of MRE11 is influenced by the GAR motif. (A) EMSA assay of WT MRE11 and mutants on dsDNA. The indicated amounts of WT or mutant MRE11 proteins were incubated with 100 nM of blunt-ended DNA and analyzed on a Tris-glycine native gel. Electron microscopy of WT MRE11 (B), the MRE11-R/K-full mutant (C), and the MRE11-R/A-full mutant (D) with circular ssDNA is shown.
The MRE11 GAR motif is implicated in DNA binding.
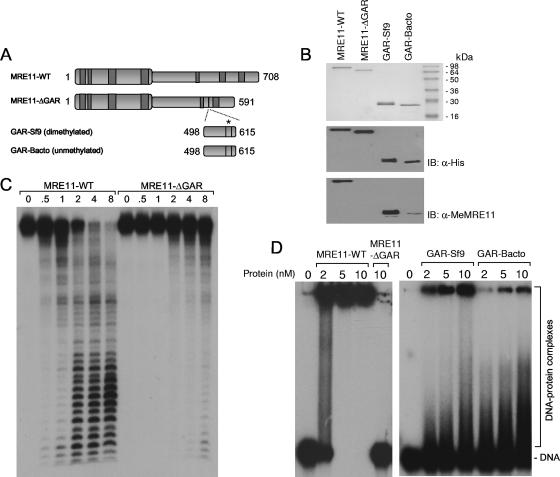
In light of these results, it was important to ask whether the GAR motif could be directly implicated in DNA binding in a methylation-dependent manner. To answer this question, we prepared MRE11 without the GAR region (MRE11-ΔGAR) or with only the GAR region (amino acids 498 to 615) comprised between the two putative DNA binding domains identified in budding yeast MRE11 (38). The GAR region was expressed in Sf9 insect cells and in E. coli, to produce methylated (GAR-Sf9) or unmethylated (GAR-Bacto) GAR motifs, respectively (Fig. 6 A and B). Western blotting with anti-MeMRE11 confirmed the absence of methylation in MRE11-ΔGAR and a decrease of methylation in GAR-Bacto compared to GAR-Sf9 (Fig. 6B). MRE11-ΔGAR multimerized (see Fig. S1A in the supplemental material) and interacted with NBS1 (see Fig. S1B in the supplemental material), suggesting that it was folded properly. MRE11-ΔGAR displayed a weak exonuclease activity compared to that of MRE11-WT, even if known nuclease and DNA binding domains are present (Fig. 6C). Furthermore, the DNA binding activity of MRE11-ΔGAR was reduced compared to that of WT MRE11. Also, GAR-Bacto was less efficient in binding DNA than was GAR-Sf9. Taken together, these results suggest that the regions encompassing the GAR motif and arginine methylation both contribute to MRE11 DNA binding (Fig. 6D).
FIG. 6.
Role of the methylated GAR motif in MRE11 DNA binding. (A) Schematic representation of purified MRE11 proteins. Full-length MRE11, MRE11 lacking amino acids 498 to 615, and the GAR domain purified from insect and human cells are depicted. The asterisk indicates the dimethylated or unmethylated GAR motif. (B) Coomassie blue staining of the purified proteins and Western blotting using anti-His and anti-MeMRE11 raised against the methylated GAR motif. (C) Exonuclease activity of WT MRE11 and MRE11-ΔGAR on dsDNA. The indicated amounts of WT or mutant MRE11 proteins were incubated with 100 nM of blunt-ended DNA. (D) DNA gel retardation assays of WT MRE11, MRE11-ΔGAR, GAR-Sf9, and GAR-Bacto on 100 nM of blunt-ended DNA.
Role of the GAR domain in MRE11 recruitment to DSBs.
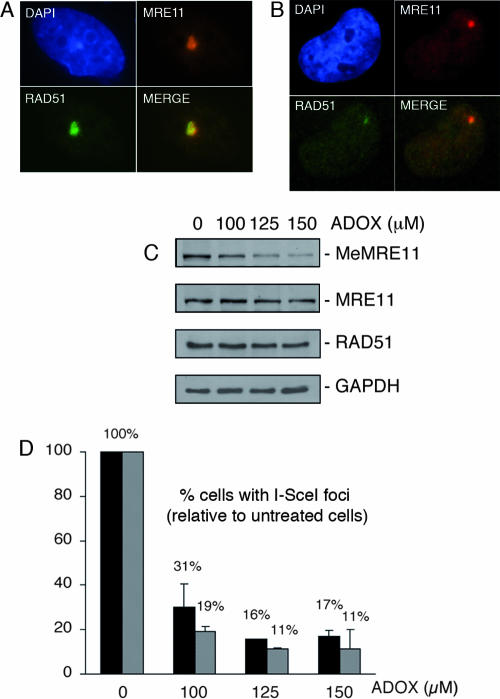
Once DNA damage occurs, the MRN complex localizes to DSBs, forms nuclear foci, and remains at these sites until DNA damage is repaired (23). If methylation of MRE11 occurs prior to DNA repair, there may be a general DNA repair defect when methylation is inhibited. To test this possibility, we used a cell line where a unique DSB can be produced in vivo following transfection with I-SceI (29). Localization to a unique DSB was monitored in mock-treated (Fig. 7A) or methylation-inhibitor-treated cells (Fig. 7B). At a 100-μM concentration of ADOX, MRE11 and RAD51 DNA damage foci were reduced to 31% and 19% of the level of the control in the absence of arginine methylation (Fig. 7D). The reduction in MRE11 and RAD51 focus formation was not due to a general decrease in protein concentration but rather to a decrease in arginine-methylated proteins, such as MRE11 (Fig. 7C). These results suggest that arginine methylation is necessary to target MRE11 to DSBs and activate homologous recombination.
FIG. 7.
(A and B) The localization of MRE11 to a unique DSB in vivo is dependent on arginine methylation. (A) MRE11 and RAD51 focus formation on a unique DSB in vivo. DR95 cells were transfected with pCBASce, and immunofluorescence was conducted with the indicated antibodies. Micrographs depict DNA stained with DAPI (4′,6′-diamidino-2-phenylindole) (blue), anti-MRE11 (red), or anti-RAD51 (green). The merge picture is an overlay of the green and red channels. (B) MRE11 and RAD51 focus formation is reduced in ADOX-treated cells. The left side shows a picture of DR95 cells treated for 8 h with 125 μM ADOX, transfected with pCBASce, and incubated with ADOX for another 16 h. Immunofluorescence analysis was conducted with the indicated antibodies. Micrographs depict DNA stained with DAPI (blue), anti-MRE11 (red), or anti-RAD51 (green). The right side shows (merge picture) an overlay of the green and red channels. (C) MeMRE11, MRE11, RAD51, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) protein levels following ADOX treatment (0 to 150 μM). (D) Quantification of the percentage of cells showing an I-SceI-induced MRE11 (black bars) or RAD51 (hatched bars) focus following ADOX treatment (0 to 150 μM) relative to untreated cells.
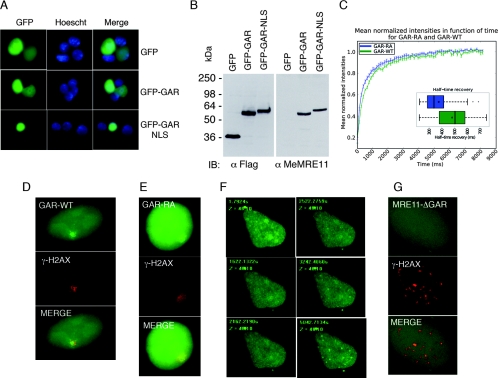
Based on these observations, we reasoned that perhaps the methylated GAR motif is necessary to target MRE11 to chromatin and nuclear foci following DNA damage. The GAR domain was fused to GFP or an NLS-containing version of GFP. Fluorescence microscopy revealed that GFP-GAR was localized throughout the cytoplasm and nucleus, whereas GFP-GAR-NLS was found to be nuclear (Fig. 8A). Both forms were methylated on arginines (Fig. 8B). In order to gain insights into the binding of the GAR domain to nuclear structures, FRAP was performed. FRAP is a technique widely used in cell biology to observe the dynamics of biological systems, including the diffusion of nuclear components. In order to gather information on the dynamics of the movement of the GAR domain in vivo, FRAP analysis was performed on GAR-WT and GAR-RA fused to GFP-NLS in the absence of DNA damage. The average half time recovery distribution of the GAR-WT was 485.7 ms, while that for the GAR-RA mutant was 379.1 ms (P value of 0.0245). Since methylation of the GAR domain influences DNA binding and the localization to nuclear structures (6), this result suggests that the decreased mobility of GAR-WT compared to that of GAR-RA is due to the influence of arginine methylation on GAR binding to chromosomal DNA. Next, cells were transfected with GAR-WT and GAR-RA fused to GFP-NLS and laser-induced DSBs were created in a unique region of the nucleus. GAR-WT focus formation colocalized with γ-H2AX (Fig. 8D). Similarly, GAR-RA also formed a unique focus, albeit at a lower level (Fig. 8E). Live-cell microscopy established that after treatment with etoposide, nuclear foci formed by GAR-WT peaked 1 h 30 min after treatment (Fig. 8E). GFP alone (data not shown) or an MRE11-ΔGAR mutant did not form any specific foci after etoposide treatment (Fig. 8G). These results show that the GAR domain of MRE11 is essential for determining the localization of MRE11 following DNA damage.
FIG. 8.
The MRE11 GAR domain is arginine methylated in vivo and localized to nuclear foci following DNA damage. (A) Plasmids expressing GFP, GFP-GAR, or GFP-GAR-NLS were transfected into 293T cells, and their localization was monitored by fluorescence microscopy. (B) Western blots of the different GFP constructs expressed in 293T cells. The expression of GFP or GFP fusions was monitored using anti-Flag (α-Flag), and the methylation status was monitored by by using anti-MeMRE11 (α-MeMRE11). IB, immunoblot. (C) FRAP analysis of GAR WT (green) and mutant GAR-RA (blue) fused to GFP. HeLa cells were transfected with a WT or mutant GAR expression vector. Twenty-four hours after the transfection, a region of interest in the nucleus was photobleached, and images were then taken at the indicated time points using an Olympus laser confocal microscope. The relative fluorescence intensities in the bleached areas of the WT and mutant GAR were plotted. A box-and-whisker diagram graphically depicts the half time recovery of the GAR-WT and GAR-RA proteins. The average and median are represented with a square and a line, respectively. GAR-WT (D) and GAR-RA (E) (green) colocalizes with γ-H2AX (red) at laser-induced DSBs. The merge picture is an overlay of the green and red channels. (F) HeLa DR95 cells expressing GAR-WT were treated with etoposide (50 μM) and DNA-damage-induced focus formation was monitored by live-cell microscopy over time. (G) MRE11-ΔGAR (green) does not form foci after etoposide treatment. γ-H2AX focus formation (red) and the merge picture of the green and red channel are depicted.
DISCUSSION
In the present study, we characterized the effect of arginine methylation on MRE11 biochemical functions. To date, MRE11 and 53BP1 are the only two proteins involved in DNA damage signaling and repair that are methylated on arginines by PRMT1 (1, 4). Although PRMT1 methylates nine arginines in the GAR motif of MRE11, in vitro analyses revealed that only the first six of nine arginines are important for the regulation of exonuclease and endonuclease activities. The replacement of the arginines of RI by alanines had a drastic effect on MRE11 activities. These effects were partially restored when positively charged lysines were introduced. We infer that both arginines and the positive charges on arginines contribute to optimal MRE11 biochemical activities. These mutants also helped us to assess the effect of this region on DNA binding. Although DNA binding was reduced in R/A-RI and R/A-full mutants, an enhanced endonuclease activity was observed. Electron microscopy gave us some clues to explain this result; the MRE11-WT protein distribution along DNA was normal, but R/A-RI and R/A-full mutants formed large aggregates on ssDNA molecules. These aggregates may facilitate catalytic activity and were observed also on blunt and 3′-tailed DNA (data not shown). In order to get insights into the multimeric state of the MRE11 arginine mutants, the native molecular weight of the R/A full mutant was also compared to that of the WT protein by gel filtration analysis (see Fig. S1A in the supplemental material). As previously published, WT MRE11 was dimeric in solution (18), but the R/A full mutant formed dimers and complexes containing a variable number of molecules. Hence, we conclude from these observations that protein-protein interactions between MRE11 molecules and the local concentration of MRE11 on DNA DSBs might affect the endonuclease or exonuclease activities used to process broken DNA.
Although human MRE11 has been studied for many years, a complete structure-function analysis has not been performed. An analysis of budding yeast Mre11 protein revealed that residues 1 to 292 contained a phosphoesterase homology domain, residues 407 to 421 are required for DNA binding, and residues 643 to 692 at the C terminus of the protein contained a second DNA binding site (38). According to pfam04152, the presumed DNA binding domain of human MRE11 is between amino acids 250 and 461. Efforts to purify unmethylated MRE11, either from E. coli or Sf9 cells treated with methylation inhibitors, were unsuccessful due to the degradation of the protein in both systems. To overcome this problem and study the direct implication of the GAR motif in MRE11 DNA binding activity, we expressed and purified only the GAR region (amino acids 498 to 615), which lies outside the putative DNA binding domain of MRE11. Protein expression in insect cells produced a protein dimethylated asymmetrically, while expression in bacteria produced a soluble unmethylated protein. Interestingly, we observed that methylation of this region changed the elution profile on gel filtration analysis, suggesting a conformational change (see Fig. S1A in the supplemental material). Moreover, we showed that arginine methylation of the MRE11 GAR motif influences DNA binding directly. This result was consistent with the DNA binding patterns of our R/A and R/K MRE11 mutants, but without any amino acid substitutions. Similar analyses performed with the MRE11-ΔGAR protein, lacking amino acids 498 to 615, corroborated these results. MRE11-ΔGAR includes both putative DNA binding domains of MRE11 but displays a weak DNA binding activity in gel retardation assays. Furthermore, MRE11-ΔGAR exonuclease activity is impaired compared to that of full-length MRE11. Our results demonstrate a direct implication of the MRE11 GAR motif in DNA binding that impacts indirectly on nuclease activity. We infer that the GAR region contains a new DNA binding domain for human MRE11.
The function of the GAR region was also monitored using live-cell microscopy and FRAP analysis. First we observed that constructs expressing GFP-GAR with or without an NLS were methylated in vivo. This result is consistent with the observation that PRMT1 is ubiquitously expressed and localized within the cytoplasm and the nucleus (36). However, our data suggest that although colocalization of PRMT1 and MRE11 occurs in promyelytic nuclear bodies (6), MRE11 methylation could also occur outside nuclear bodies. Since MRE11 (but not MRN) interacts with PRMT1, we propose that methylation by PRMT1 occurs before the incorporation of MRE11 into the MRN complex. When DSBs were introduced by laser or etoposide, the GFP-GAR-NLS, but not GFP-MRE11-ΔGAR, formed distinct nuclear foci that colocalized with γ-H2AX. The remaining capability of GFP-GAR-RA to form foci is consistent with the fact that we still observed DNA binding for the unmethylated GAR-Bacto protein compared to methylated GAR-Sf9. Taken together, these results suggest that the GAR domain is essential to control the accumulation of MRE11 on DNA or chromatin, while arginine methylation on the domain may not be essential in the targeting. The increased diffusion rate of the GAR-RA construct suggests that arginine methylation may be required to retain the protein on chromatin in a more efficient manner. Chromatin binding could occur through direct binding to DNA or protein-protein interactions. For instance, proteins with a Tudor domain are well known to bind methylated arginine or lysine residues. We have shown that the GAR domain could be accessible for such interactions. Proteins binding the MRE11 GAR motif have been identified in our laboratory (J.-Y. Masson, unpublished observations).
It was previously shown that point mutations in the forkhead-associated domain and deletion of amino acids 682 to 693 on human NBS1, corresponding to the MRE11 binding domain, resulted in a failure to observe MRE11 focus formation following gamma irradiation (31). However, the deletion of amino acids 682 to 693 of human NBS1 resulted in an about threefold decrease in homologous recombination, although NBS1 foci were still formed. This result suggests that MRE11 localization to DNA damage sites, rather than NBS1 localization, could be a key event for homologous recombination (31). Based on these observations and the data presented in this paper, we infer that there are at least two important ways to target MRE11 to DNA damage sites, one depending on the N and C termini of human NBS1 and the second depending on the GAR domain. The inactivation of one of these regions in NBS1 or MRE11 results in a decrease in MRE11 focus formation.
We have found no evidence for DNA-damage-induced MRE11 methylation in the GAR region. However, our data suggest that this modification is necessary for a proper cellular response to DNA damage. Recently, histone H3 methylation on lysine 79 has been shown to be required for the recruitment of 53BP1 to sites of DNA damage (14). In this case also, the methylation of lysine 79 was not increased in response to DNA damage. Although these observations suggest constitutive arginine methylation on MRE11 and 53BP1, this process might be more dynamic and transient than anticipated. A new alternative pathway for the removal of a methyl group from arginines has been reported. Deimination by peptidylarginine deiminase 4 converts unmodified and monomethylated arginine to citrulline at the tails of histones H3 and H4. Citrulline deposition on histones appears to be short lived (10). Although a specific enzyme responsible for dimethylarginine demethylation has yet to be discovered, a similar level of control could be foreseeable in the future.
We previously reported that cells containing hypomethylated MRE11 displayed intra-S-phase DNA damage checkpoint defects (6). When WT and PRMT1−/− embryonic stem cells were exposed to etoposide, PRMT1−/− cells progressed more slowly through the S phase. We wanted to extend this study and verify whether DNA repair was impaired under these conditions. If MRE11 functions are dependent on methylation, the formation of RAD51 foci should be reduced in these cells. In order to study DNA repair at the resolution of a single lesion, we used the DR95 cell line (29). We observed that pretreatment of the cells with ADOX decreased the single-focus formation of MRE11 and RAD51. Our interpretation is that MRE11 arginine methylation is an initial step in preparing MRE11 and the MRN complex for DSB repair. Hence, the inhibition of arginine methylation decreases RAD51 focus formation, because homologous recombination is dependent on the resection of the DSB by MRN and its recruitment to the break is impaired under these conditions. These results confirm that arginine methylation on MRE11 is important for DNA repair.
Genetic instability associated with defective DNA repair and checkpoint signaling is a key feature of cancer cells. Ataxia-telangiectasia-like disease, cause by mutations in MRE11, was identified in four patients presenting the clinical features of ataxia telangiectasia, including progressive cerebellar degeneration, increased levels of chromosome aberrations, and increased sensitivity to ionizing radiation at the cellular and chromosomal levels (34). Very interestingly, one in-frame stop codon found in two patients with ataxia-telangiectasia-like disorder resulted in the truncation of MRE11 to 571 amino acids (28). This mutation occurs directly in the GAR motif, changing arginine 572 to a stop codon having for consequence nonsense-mediated mRNA decay. In addition, in a screening of 159 primary tumors, a G-to-A transition, changing arginine 572 to glutamine in lymphoma, was found (12). These results may highlight the importance of the MRE11 GAR motif in disease progression and cancer formation.
Supplementary Material
Acknowledgments
We thank Alan Anderson, Mark Bedford, Louise Simard, Raymund Wellinger, and Jacques Côté for critically reviewing the manuscript and Josée Lavoie for GFP constructs. We thank Eric Paquet for statistical analysis of the FRAP data, Carl St-Pierre of the Unité d'Imagerie Cellulaire for technical help, and Isabelle Brodeur for preliminary results.
U.D. and A.R. are recipients of an FRSQ doctoral scholarship and a CIHR doctoral scholarship, respectively. J.Y.M. holds a CIHR New Investigator Award and S.R. is a CIHR investigator. This research was supported by funds from the NCIC (017121) to J.Y.M. and CIHR (MOP-67070) to S.R.
Footnotes
Published ahead of print on 19 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, M. M., B. Wang, Z. Xia, J. C. Morales, X. Lu, L. A. Donehower, D. A. Bochar, S. J. Elledge, and P. B. Carpenter. 2005. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle 41854-1861. [DOI] [PubMed] [Google Scholar]
- 2.Allfrey, V. G., R. Faulkner, and A. E. Mirsky. 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51786-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18263-272. [DOI] [PubMed] [Google Scholar]
- 4.Boisvert, F. M., J. Cote, M. C. Boulanger, and S. Richard. 2003. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 21319-1330. [DOI] [PubMed] [Google Scholar]
- 5.Boisvert, F. M., U. Dery, J. Y. Masson, and S. Richard. 2005. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boisvert, F. M., M. J. Hendzel, J. Y. Masson, and S. Richard. 2005. Methylation of MRE11 regulates its nuclear compartmentalization. Cell Cycle 4981-989. [DOI] [PubMed] [Google Scholar]
- 7.Boisvert, F. M., A. Rhie, S. Richard, and A. J. Doherty. 2005. The GAR motif of 53BP1 is arginine methylated by PRMT1 and is necessary for 53BP1 DNA binding activity. Cell Cycle 41834-1841. [DOI] [PubMed] [Google Scholar]
- 8.Comb, D. G., N. Sarkar, and C. J. Pinzino. 1966. The methylation of lysine residues in protein. J. Biol. Chem. 2411857-1862. [PubMed] [Google Scholar]
- 9.Connelly, J. C., and D. R. Leach. 2002. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27410-418. [DOI] [PubMed] [Google Scholar]
- 10.Cuthbert, G. L., S. Daujat, A. W. Snowden, H. Erdjument-Bromage, T. Hagiwara, M. Yamada, R. Schneider, P. D. Gregory, P. Tempst, A. J. Bannister, and T. Kouzarides. 2004. Histone deimination antagonizes arginine methylation. Cell 118545-553. [DOI] [PubMed] [Google Scholar]
- 11.El-Andaloussi, N., T. Valovka, M. Toueille, R. Steinacher, F. Focke, P. Gehrig, M. Covic, P. O. Hassa, P. Schar, U. Hubscher, and M. O. Hottiger. 2006. Arginine methylation regulates DNA polymerase beta. Mol. Cell 2251-62. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda, M., and K. Mikoshiba. 1997. The function of inositol high polyphosphate binding proteins. Bioessays 19593-603. [DOI] [PubMed] [Google Scholar]
- 13.Gary, J. D., and S. Clarke. 1998. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 6165-131. [DOI] [PubMed] [Google Scholar]
- 14.Huyen, Y., O. Zgheib, R. A. Ditullio, Jr., V. G. Gorgoulis, P. Zacharatos, T. J. Petty, E. A. Sheston, H. S. Mellert, E. S. Stavridi, and T. D. Halazonetis. 2004. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432406-411. [DOI] [PubMed] [Google Scholar]
- 15.Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre, and J. E. Haber. 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 143414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jazayeri, A., J. Falck, C. Lukas, J. Bartek, G. C. Smith, J. Lukas, and S. P. Jackson. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 837-45. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, J., H. Tauchi, S. Sakamoto, A. Nakamura, K. Morishima, S. Matsuura, T. Kobayashi, K. Tamai, K. Tanimoto, and K. Komatsu. 2002. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 121846-1851. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J. H., R. Ghirlando, V. Bhaskara, M. R. Hoffmeyer, J. Gu, and T. T. Paull. 2003. Regulation of Mre11/Rad50 by Nbs1: effects on nucleotide-dependent DNA binding and association with ataxia-telangiectasia-like disorder mutant complexes. J. Biol. Chem. 27845171-45181. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. H., and T. T. Paull. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 30493-96. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner, and J. E. Haber. 1998. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94399-409. [DOI] [PubMed] [Google Scholar]
- 21.Lim, D. S., S. T. Kim, B. Xu, R. S. Maser, J. Y. Lin, J. H. J. Petrini, and M. B. Kastan. 2000. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature 404613-617. [DOI] [PubMed] [Google Scholar]
- 22.Lukas, C., F. Melander, M. Stucki, J. Falck, S. Bekker-Jensen, M. Goldberg, Y. Lerenthal, S. P. Jackson, J. Bartek, and J. Lukas. 2004. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 232674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelms, B. E., R. S. Maser, J. F. MacKay, M. G. Lagally, and J. H. J. Petrini. 1998. In situ visualization of DNA double-strand break repair in human fibroblasts. Science 280590-592. [DOI] [PubMed] [Google Scholar]
- 24.Paik, W. K., and S. Kim. 1968. Protein methylase I. Purification and properties of the enzyme. J. Biol. Chem. 2432108-2114. [PubMed] [Google Scholar]
- 25.Paull, T. T., and M. Gellert. 1999. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 131276-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paull, T. T., and M. Gellert. 1998. The 3′-exonuclease to 5′-exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell 1969-979. [DOI] [PubMed] [Google Scholar]
- 27.Phair, R. D., S. A. Gorski, and T. Misteli. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375393-414. [DOI] [PubMed] [Google Scholar]
- 28.Pitts, S. A., H. S. Kullar, T. Stankovic, G. S. Stewart, J. I. Last, T. Bedenham, S. J. Armstrong, M. Piane, L. Chessa, A. M. Taylor, and P. J. Byrd. 2001. hMRE11: genomic structure and a null mutation identified in a transcript protected from nonsense-mediated mRNA decay. Hum. Mol. Genet. 101155-1162. [DOI] [PubMed] [Google Scholar]
- 29.Rodrigue, A., M. Lafrance, M.-C. Gauthier, D. McDonald, M. Hendzel, S. C. West, M. Jasin, and J. Y. Masson. 2006. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 25222-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto, S., K. Iijima, D. Mochizuki, K. Nakamura, K. Teshigawara, J. Kobayashi, S. Matsuura, H. Tauchi, and K. Komatsu. 2007. Homologous recombination repair is regulated by domains at the N- and C-terminus of NBS1 and is dissociated with ATM functions. Oncogene 266002-6009. [DOI] [PubMed] [Google Scholar]
- 32.Sartori, A. A., C. Lukas, J. Coates, M. Mistrik, S. Fu, J. Bartek, R. Baer, J. Lukas, and S. P. Jackson. 2007. Human CtIP promotes DNA end resection. Nature 450509-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sogo, J., A. Stasiak, W. De Bernadin, R. Losa, and T. Koller. 1987. Negative staining of proteins and filaments, p. 61-79. In J. Sommerville and U. Scheer (ed.), Electron microscopy in molecular biology. IRL Press, Oxford, United Kingdom.
- 34.Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan, N. G. J. Jaspers, A. Raams, P. J. Byrd, J. H. J. Petrini, and A. M. R. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an Ataxia telangiectasia-like disorder. Cell 99577-587. [DOI] [PubMed] [Google Scholar]
- 35.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 36.Tang, J., J. D. Gary, S. Clarke, and H. R. Herschman. 1998. PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 27316935-16945. [DOI] [PubMed] [Google Scholar]
- 37.Trujillo, K. M., S. S. F. Yuan, E. Y. H. P. Lee, and P. Sung. 1998. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 27321447-21450. [DOI] [PubMed] [Google Scholar]
- 38.Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa, and T. Ogawa. 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95705-716. [DOI] [PubMed] [Google Scholar]
- 39.Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman, and Y. Shiloh. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 225612-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varon, R., C. Vissinga, M. Platzer, K. M. Cerosaletti, K. H. Chrzanowska, K. Saar, G. Beckmann, E. Seemanova, P. R. Cooper, N. J. Nowak, M. Stumm, C. M. R. Weemaes, R. A. Gatti, R. K. Wilson, M. Digweed, A. Rosenthal, K. Sperling, P. Concannon, and A. Reis. 1998. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell 93467-476. [DOI] [PubMed] [Google Scholar]
- 41.Zhao, S., W. Renthal, and E. Y. Lee. 2002. Functional analysis of FHA and BRCT domains of NBS1 in chromatin association and DNA damage responses. Nucleic Acids Res. 304815-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.