Abstract
Yeast Rrp6p and its human counterpart, PM/Scl100, are exosome-associated proteins involved in the degradation of aberrant transcripts and processing of precursors to stable RNAs, such as the 5.8S rRNA, snRNAs, and snoRNAs. The activity of yeast Rrp6p is stimulated by the polyadenylation of its RNA substrates. We identified three RRP6-like proteins in Arabidopsis thaliana: AtRRP6L3 is restricted to the cytoplasm, whereas AtRRP6L1 and -2 have different intranuclear localizations. Both nuclear RRP6L proteins are functional, since AtRRP6L1 complements the temperature-sensitive phenotype of a yeast rrp6Δ strain and mutation of AtRRP6L2 leads to accumulation of an rRNA maturation by-product. This by-product corresponds to the excised 5′ part of the 18S-5.8S-25S rRNA precursor and accumulates as a polyadenylated transcript, suggesting that RRP6L2 is involved in poly(A)-mediated RNA degradation in plant nuclei. Interestingly, the rRNA maturation by-product is a substrate of AtRRP6L2 but not of AtRRP6L1. This result and the distinctive subcellular distribution of AtRRP6L1 to -3 indicate a specialization of RRP6-like proteins in Arabidopsis.
The exoribonucleolytic activity of the exosome is fundamental to many aspects of RNA metabolism. The exosome is involved in mRNA turnover and degradation of defective and cryptic transcripts, but also in processing of the 3′ extremities of a variety of noncoding RNAs and elimination of RISC-cleaved mRNA (for recent reviews, see references 27 and 46). While most exosome functions were initially characterized in the yeast Saccharomyces cerevisiae, related complexes are present in all higher eukaryotes investigated and in several Archaea (4, 9-11, 17, 26, 29, 35, 36).
Eukaryotic exosomes are composed of a core complex with which cytoplasm- and nucleus-specific subunits associate (reviewed in reference 38). One of these subunits, Rrp6p, a member of the RNase D family, associates with the nuclear exosome in yeast (2). Its human counterpart, PM/Scl-100, is predominantly nuclear but is also detected in the cytoplasm (7). Rrp6p plays a role in nuclear mRNA surveillance and in the degradation of rRNA maturation by-products or intergenic transcripts (30, 33, 47, 51). In addition, Rrp6p is involved in the final steps in processing several noncoding RNAs (1, 6).
In yeast, the TRAMP complex polyadenylates RNA substrates, which triggers their degradation by the nuclear exosome (30, 47, 51). In higher eukaryotes, evidence for polyadenylation of nuclear transcripts destined for degradation is emerging. Short poly(A) tails were detected upon cotranscriptional cleavage of human β-globin and murine serum albumin pre-mRNA (50). Human rRNA can also be polyadenylated at putative sites of endonucleolytic cleavage (44). In plants, polyadenylation of nuclear noncoding RNA also occurs, as polyadenylated transcripts of the low-abundance 5S rRNA spacer were reported in Nicotiana (20). During revision of our manuscript, a genome-wide search for exosome substrates revealed that a wide range of nuclear noncoding transcripts are polyadenylated in Arabidopsis thaliana (10).
We show here that three RRP6-like proteins (RRP6L1 to -3) are encoded by the A. thaliana genome and form two distinct subfamilies, one of which is specific to plants. Furthermore, each AtRRP6L protein has a specific subcellular distribution. RRP6L3 is cytoplasmic, whereas RRP6L1 and RRP6L2 have different intranuclear locations. RRP6L1 and RRP6L2 can be further distinguished based on yeast complementation assays and analysis of mutant plants. This analysis has revealed a specialization of the RRP6L1 and RRP6L2 proteins for two RNA substrates tested. Interestingly, we also observed polyadenylation of the excised 5′ external transcribed sequence (ETS), an rRNA maturation by-product that accumulates upon knockdown of RRP6L2, indicating that polyadenylation can mark a transcript destined for degradation by an RRP6-like protein in plant nuclei.
MATERIALS AND METHODS
Plant material.
All A. thaliana plants used in this study were of the Columbia ecotype (Col-0). Insertion mutant information was obtained from the SIGnAL website (http://signal.salk.edu). Transfer DNA (T-DNA) insertion lines were generated in the context of Gabi-KAT and Salk T-DNA programs and provided by Bernd Weisshaar (MPI for Plant Breeding Research, Cologne, Germany) or retrieved from NASC (http://arabidopsis.info/), respectively (3, 39, 41). Salk_004432 and Gabi_344G09 correspond to T-DNA insertion lines in At1g54440 and were named rrp6l1-1 and rrp6l1-2, respectively. Gabi_825G09 and Salk_113786 correspond to T-DNA insertion lines in At5g35910 and were named rrp6l2-1 and rrp6l2-2, respectively.
Sequence analysis.
Rice (Oryza sativa) and poplar (Populus trichocarpa) sequences were identified by BlastP and retrieved from the TIGR Rice Genome (http://www.tigr.org/tdb/e2k1/osa1) and the DOE Joint Genome Institute (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), respectively. To obtain full-length sequences for poplar proteins, predictions were improved using FGENESH+ software (data available upon request). Arabidopsis sequences initially identified in The Arabidopsis Information Resource (http://www.arabidopsis.org) were determined by translation of cloned cDNA sequences (GenBank accession numbers EU240662 to -4). PFAM (18) was used to determine protein domains. Sequence alignment was done with Muscle v3.52 (16). Phylogenetic analysis was performed using Phyml (23), following removal of poorly aligned and overly divergent positions using the Gblocks program (8). The unrooted tree was drawn with Treedyn (12).
Yeast complementation.
cDNAs for AtRRP6L1, -2, and -3 and yeast RRP6 were cloned into pRS 426-TDH (43) and transformed into wild-type (WT) (BY4742) and rrp6Δ (Euroscarf Y11777) yeast strains using the lithium acetate method. Following selection at 24°C, the cells were tested for growth at 24°C and 37°C.
Expression of GFP fusion proteins.
Full-length cDNAs of AtRRP6L1, -2, and -3 were obtained by reverse transcriptase (RT) PCR and fused to enhanced green fluorescent protein (EGFP) in N- and C-terminal orientations in pBinH, a pBinPLUS derivative (48) containing the cauliflower mosaic virus 35S promoter and terminator sequences and a hygromycin resistance cassette. Biolistic transformation of tobacco BY2 cells and analysis by confocal microscopy were as described previously (49). For stable transformants, Arabidopsis plants were transformed by the floral-dip method (13). Transformed plants were selected on agar plates containing Murashige and Skoog (MS) salts, 0.5% sucrose, and 50 μg/ml hygromycin. Stable transformants expressing GFP fusion proteins were sown on coverslips coated with MS salts, 0.5% sucrose, and 50 μg/ml hygromycin agar, and the root tips were examined 8 days after germination by confocal microscopy.
RNA extraction and analysis.
RNA was extracted using the Tri-reagent (Molecular Research Center) according to the manufacturer's manual. Polyadenylation sites were mapped by amplification of 3′ ends as described previously (37). The 5′ and 3′ ends of the ETS were determined by circular RT-PCR (cRT-PCR) as described previously (37). A complete list of primers used in this study is available upon request.
Smart cDNA and virtual Northern blots.
Full-length cDNAs were synthesized using the cDNA Smart synthesis kit (Clontech) following the manufacturer's protocol, except that different sequence tags at both the 3′ and 5′ ends of polyadenylated RNA were introduced during cDNA synthesis. Total cDNAs were amplified by PCR using the PCR Advantage II kit (Clontech) for 16 cycles (30 s at 94°C, 30 s at 60°C, and 6 min at 68°C). The resulting Smart PCR products (representing total full-length polyadenylated RNA) were separated on a 1.2% agarose gel-0.5× Tris-acetate-EDTA, blotted on a Hybond N+ membrane, and hybridized to [α-32P]dCTP-labeled DNA probes. This procedure is referred to as “virtual Northern” according to Franz et al. (19). Diluted Smart PCR products were used as a template for 3′ and 5′ rapid amplification of cDNA ends (RACE) PCR.
RESULTS
A small multigene family encodes RRP6-like proteins in plants.
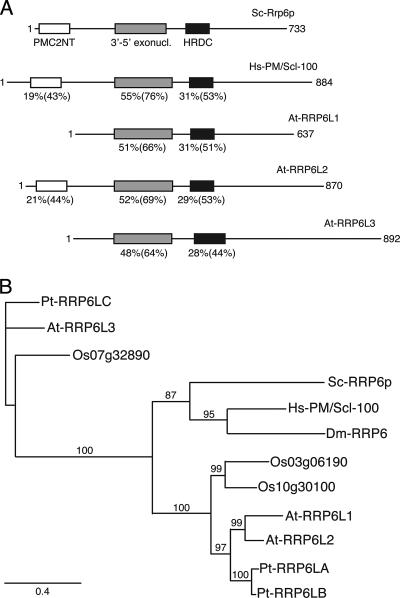
In a previous study aimed at identifying polyadenylated mitochondrial RNA degradation intermediates, we constructed an oligo(dT)-primed library from size-selected RNA isolated from A. thaliana seedlings (25). Interestingly, some clones in this library matched fragments of the polycistronic rRNA precursor in the nuclear genome. We therefore investigated a possible role of polyadenylation in the degradation of nuclear noncoding transcripts in A. thaliana, as described recently in yeast. Degradation of yeast nuclear noncoding RNAs depends on polyadenylation by the TRAMP complex and involves RRP6, a hydrolytic exoribonuclease associated with the nuclear exosome (30, 47, 51). While in yeast and humans, RRP6 is encoded by a single gene, the Arabidopsis genome contains three genes encoding RRP6-like proteins: At1g54440, At5g35910, and At2g32415, which we refer to as AtRRP6L1, AtRRP6L2, and AtRRP6L3, respectively. AtRRP6L1 to -3 encode proteins of 637, 870, and 892 amino acids, respectively. These three proteins are the only Arabidopsis proteins containing both the IPR002562 3′-5′ exoribonuclease and the IPR002121 HDRC motifs that characterize RRP6 proteins (Fig. 1A). The PMC2NT domain, present in the N terminus of S. cerevisiae Rrp6p, is found only in AtRRP6L2 (Fig. 1A).
FIG. 1.
Analysis of RRP6-like proteins. (A) Conservation of functional domains in RRP6 and RRP6-like proteins. Comparison of S. cerevisiae ScRrp6p and Homo sapiens HsPM/Scl-100 and the three A. thaliana RRP6-like proteins (AtRRP6L1, AtRRP6L2, and AtRRP6L3). Percent identity and, in parentheses, similarity with ScRrp6p are given below each domain, drawn as boxes. (B) Phylogenetic analysis of RRP6-like proteins presented as an unrooted maximum-likelihood tree. Bootstrap values above 70 (using 100 replications) are indicated along the branches. The scale bar indicates the evolutionary distance (amino acid substitutions per site). Pt, P. trichocarpa; At, A. thaliana; Os, O. sativa; Sc, S. cerevisiae; Ce, Caenorhabditis elegans; Hs H. sapiens; Dm, Drosophila melanogaster.
We searched for RRP6-like sequences in other plant genomes and found that both rice and poplar also contain three genes encoding RRP6-like proteins. Phylogenetic analysis revealed that these sequences form two strongly supported groups (Fig. 1B). The first group contains AtRRP6L1 and AtRRP6L2, and the second group contains AtRRP6L3. The presence of AtRRP6L3-like genes in rice, poplar, and Arabidopsis indicates an early divergence in plants from a common ancestor. While RRP6L3-related proteins form a plant-specific subgroup, the AtRRP6L1/2 group clusters with yeast and animal RRP6-like proteins. In each of the three plant species, RRP6L1 and RRP6L2 cluster as a pair (Fig. 1B). In addition, the positions of introns are clearly conserved in an intra- rather than interspecific manner (data not shown). Therefore, we propose that the RRP6L1/2 gene duplication occurred independently in these three plant species rather than anterior to the speciation of rice, poplar, and Arabidopsis.
It is interesting to note that both the rice and Arabidopsis genomes encode RRP6-like proteins containing or lacking the N-terminal PMC2NT domain.
In conclusion, three RRP6-like proteins are encoded in the genomes of rice, poplar, and Arabidopsis. Two of these genes, represented by AtRRP6L1 and -2, are the closest homologues to yeast and human RRP6, whereas the AtRRP6L3 type appears to be specific to plants.
Subcellular distribution of RRP6-like proteins in plants.
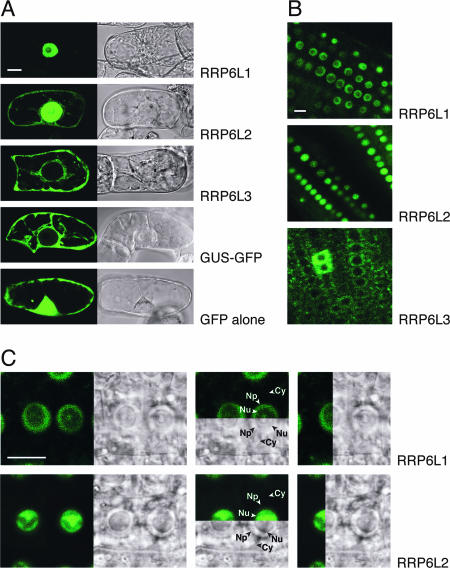
Both AtRRP6L1 and AtRRP6L2 are predicted to be nuclear by the PSORT program (http://psort.ims.u-tokyo.ac.jp), while no specific localization is predicted for AtRRP6L3. To address this issue experimentally, we expressed N- and C-terminal GFP fusion proteins of AtRRP6L1, -2, and -3 in tobacco BY2 cells. The results were identical for the two orientations of the fusion proteins. AtRRP6L3 fusion proteins were excluded from the nucleus and located exclusively in the cytosol (Fig. 2A). In contrast, both AtRRP6L1 and -2 fused to GFP accumulated predominantly in the nucleus, although a faint cytoplasmic signal was observed for AtRRP6L2 (Fig. 2A). These results indicate that, in contrast to AtRRP6L3, both AtRRP6L1 and -2 can be targeted to the nucleus. To further study the distribution of AtRRP6-like proteins, we produced stably transformed Arabidopsis plants. The results obtained confirmed the cytoplasmic localization of AtRRP6L3 and the nuclear localization of both AtRRP6L1 and -2 (Fig. 2B). In root tip cells, where nucleoli are particularly large, the intranuclear distributions of RRP6L1 and RRP6L2 could be distinguished. The AtRRP6L1 fusion protein accumulated in both the nucleoplasm and the nucleolar vacuole (Fig. 2C, top). In contrast, AtRRP6L2 accumulated predominantly in nucleoli, and the signal was weaker in the nucleoplasm (Fig. 2C, bottom).
FIG. 2.
Subcellular distribution of RRP6-like proteins. (A) GFP fluorescence (left) and Nomarski (right) images of tobacco BY2 cells transiently expressing EGFP and EGFP fusion proteins. β-Glucuronidase-GFP (GUS-GFP) is a large protein that cannot enter the nuclear compartment by passive diffusion. (B) Root tips of stably transformed Arabidopsis plants expressing RRP6L-EGFP fusion proteins. (C) Enlarged view of root tips of transformed Arabidopsis plants showing the intranuclear distribution of fusion proteins. Comparison of fluorescence and Nomarski panels shows that RRP6L1-EGFP (top) is mainly in the nucleoplasm and in the nucleolar vacuole. RRP6L2-EGFP is detected mainly inside the nucleolus (bottom). Cy, cytoplasm; Np, nucleoplasm; Nu, nucleolus. Size bar = 10 μm.
Hence, AtRRP6L1 and -2 are targeted to the nucleus, whereas AtRRP6L3 is restricted to the cytoplasm. The subcellular distribution thus again differentiates AtRRP6L3 from AtRRP6L1 and -2. In addition, AtRRP6L1 and -2 have distinct intranuclear distributions.
AtRRP6L1 complements the growth defect of a yeast rrp6Δ strain.
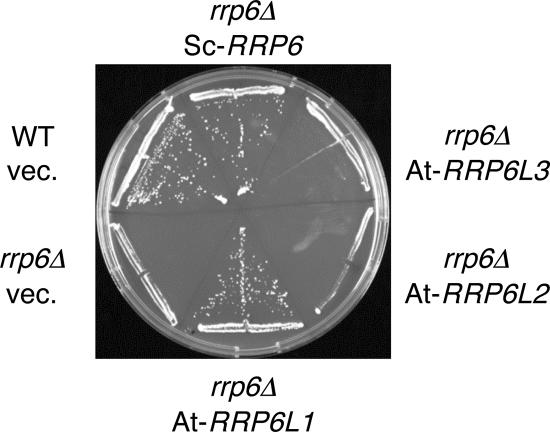
To determine whether Arabidopsis RRP6-like proteins can functionally replace S. cerevisiae Rrp6p, we expressed AtRRP6L1, -2, and -3 in the temperature-sensitive rrp6Δ yeast strain (2). Expression of transgenes was confirmed by RT-PCR (not shown). AtRRP6L3, cytoplasmic in plants, did not support the growth of the rrp6Δ strain at the nonpermissive temperature (Fig. 3). In contrast, AtRRP6L1 was able to complement the thermosensitive growth phenotype of rrp6Δ yeast (Fig. 3). This demonstrates that AtRRP6L1 is a functional protein that can perform at least one of the biological roles of Rrp6p in yeast. AtRRP6L1 lacks the N-terminal PMC2NT domain. In yeast Rrp6p, this domain is not required to complement the growth phenotype of the rrp6Δ strain (45). Surprisingly, RRP6L2, which is the closest homologue of yeast Rrp6p, did not restore the growth phenotype of the rrp6Δ strain, although the expression and stability of the protein were sufficient, as determined by Western analysis (not shown). Even though RRP6L2 does not complement the growth phenotype of the rrp6Δ strain, we show below that RRPL2 and yeast Rrp6p perform similar functions in rRNA metabolism.
FIG. 3.
AtRRP6L1 complements yeast rrp6Δ. Growth at the nonpermissive temperature (37°C) of WT and rrp6Δ yeast strains harboring empty vector (vec.) or vectors encoding either S. cerevisiae Rrp6p (ScRRP6) or the indicated Arabidopsis RRP6-like proteins.
Characterization of mutant plants.
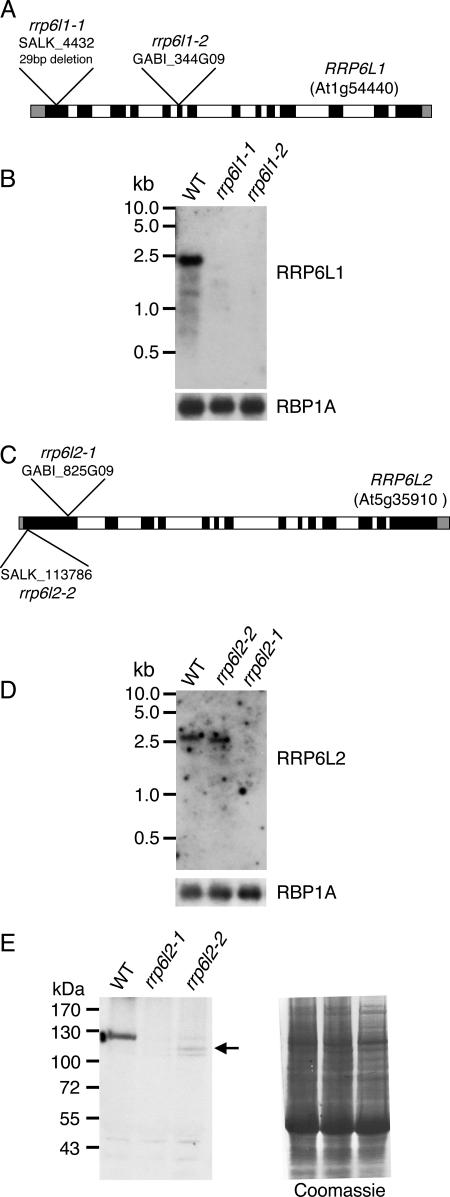
Since RRP6L1 and -2 are putatively nuclear proteins, we wanted to test whether they could be involved in the turnover of polyadenylated noncoding RNAs transcribed in the nucleus. To this end, two independent T-DNA insertion lines for AtRRP6L1 (rrp6l1-1 and rrp6l1-2) and for AtRRP6L2 (rrp6l2-1 and rrp6l2-2) were characterized. In rrp6l1-1, the T-DNA is inserted into the first exon of RRP6L1 (Fig. 4A). This insertion event led to a deletion of 29 bp of the coding region. Line rrp6l1-2 harbors a T-DNA in the sixth exon of RRP6L1. No RRP6L1 mRNA was detected in either rrp6l1 mutant by virtual Northern blots (see Materials and Methods) (Fig. 4B), indicating that both T-DNA insertions severely affect RRP6L1 expression. In both rrp6l2-1 and rrp6l2-2, the T-DNA is inserted in the first exon of RRP6L2 (Fig. 4C). No RRP6L2 mRNA was detected in rrp6l2-1. In rrp6l2-2, the mRNA was slightly reduced in size but accumulated to a WT level (Fig. 4D). We therefore mapped mRNA extremities by 5′ and 3′ RACE. This analysis revealed that rrp6l2-2 mRNA consists of the entire RRP6L2 mRNA, in which the complete 5′ untranslated region plus 29 nucleotides (nt) of the RRP6L2 open reading frame is replaced by 16 nt encoded by the T-DNA. The resulting chimeric rrp6l2-2 mRNA is 2,839 nt compared to 2,936 nt for WT RRP6L2 mRNA. The rrp6l2-2 mRNA potentially codes for a protein lacking the 86 N-terminal amino acids compared to WT RRP6L2. This truncated protein lacks about 60% of the PMC2NT domain, which corresponds to amino acids 35 to 123 of the WT RRP6L2.
FIG. 4.
Characterization of rrpl1 and -2 TDNA insertion mutants. (A) Diagram of the intron-exon structure of RRP6L1. Exons are in black, introns in white. The 5′ and 3′ untranslated regions are drawn as gray blocks. T-DNA insertion sites for rrp6l1-1 and -2 are shown. In rrp6l1-1, the T-DNA insertion removed 29 nt from the genomic sequence. (B) Detection of RRP6L1 mRNA in WT and mutant plants by virtual Northern analysis, as described in Materials and Methods. A probe for RBP1A mRNA was used as a loading control. (C) Organi-zation and T-DNA insertion sites in RRP6L2. (D) Virtual Northern blot analysis of RRP6L2 mRNA in WT and mutant plants showed a size difference between transcripts in WT and rrp6b-2 mutants. (E) Western blots of proteins extracted from WT, rrp6l21-1, or rrp6l2-2 seedlings were probed with antibodies against AtRRP6L2 (left). The truncated protein encoded by the mutant rrp6b-2 allele is indicated by an arrow. As a loading control, the membrane was stained with Coomassie blue (right).
Both rrp6l2 mutants were further characterized by Western blotting using antibodies raised against RRP6L2. We did not detect any RRP6L2 protein in rrp6l2-1 and observed a faint signal that could correspond to the truncated protein in rrp6l2-2 (Fig. 4E). We therefore cannot exclude the possibility that low levels of a truncated RRP6L2 can be produced from rrp6l2-2 mRNA. However, these results indicate that RRP6L2 expression is severely decreased in rrp6l2-2 and probably abolished in rrp6l2-1.
All four single mutants were indistinguishable from WT plants in terms of growth and development under standard growth conditions. Similarly, both homozygous double mutants, rrp6l1-2 rrp6l2-1 and rrp6l1-2 rrp6l2-2, did not show any obvious growth or developmental phenotype.
An rRNA maturation by-product accumulates in rrp6l2, but not in rrp6l1, mutants.
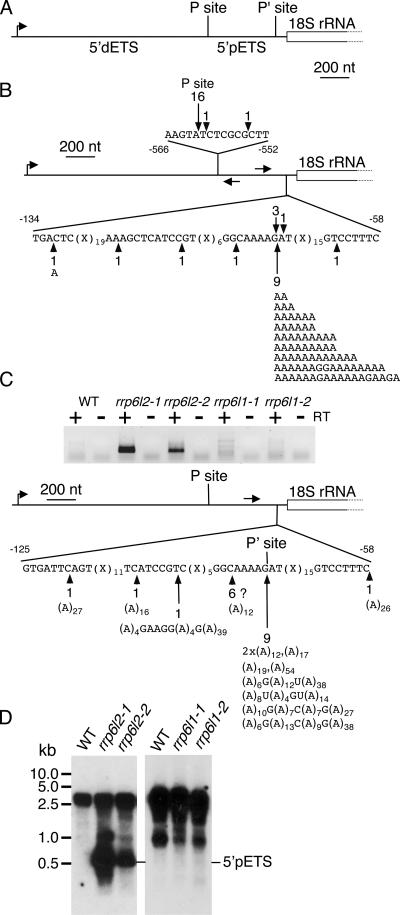
We tested these insertion mutants for the accumulation of an rRNA maturation by-product, which is a typical molecular phenotype of yeast rrp6 mutant strains (2). One of the earliest processing events of rRNA maturation is an endonucleolytic cleavage in the 5′ ETS. In S. cerevisiae, this cleavage takes place at site A0, located about 90 nt upstream of the 18S rRNA. The excised ETS accumulates in the absence of functional Rrp6p. In Arabidopsis, the complete 5′ ETS is relatively large (about 1.8 kb). The primary processing site (P site) is located 1,275 nt downstream of the transcription initiation site and about 560 nt upstream of the 5′ extremity of the 18S rRNA (40). To our knowledge, a second processing site, equivalent to the A0 site in yeast, has not been characterized in Arabidopsis. Cleavage at such a site will release a plant rRNA maturation by-product akin to the yeast 5′ ETS. We will refer to this product as the 5′ pETS (5′-proximal ETS) to distinguish it from the distal 5′ ETS corresponding to the sequence between the transcription initiation site and the P site (Fig. 5A).
FIG. 5.
A polyadenylated rRNA maturation by-product accumulates in rrp6l2 mutants. (A) Diagram showing the 5′ region of the polycistronic rRNA transcript of A. thaliana. Distal (5′ dETS) and proximal (5′ pETS) RNA segments and processing sites P and P′, respectively, are indicated. The promoter is shown by a bent arrow. (B) Mapping of 5′ and 3′ ends of the 5′ pETS by cRT-PCR. Total RNA from rrp6l2-1 plants was self-ligated by T4 RNA ligase, and cDNA was synthesized using a gene-specific reverse primer. The same primer was combined with a gene-specific forward primer to amplify joined 5′ and 3′ ends by PCR. Primers are indicated on the diagram. 5′ ends are shown above the diagram, 3′ ends are shown below, and nonencoded nucleotides at the 3′ ends of 5′ pETS transcripts are indicated. (C) Characterization of the 5′ pETS by 3′ RACE. Oligo(dT)12-primed cDNA was synthesized from total RNA from WT or mutant plants. 3′ ends were then amplified by PCR using a gene-specific forward primer (arrow above the diagram) and a reverse primer specific for the oligo(dT) primer adapter sequence. The PCR products were analyzed by electrophoresis (top). The PCR products obtained from rrp6l2 samples were cloned and sequenced to map polyadenylation sites (bottom). The locations and frequencies of polyadenylation sites are indicated on the sequence, and the sizes and nucleotide compositions of poly(A) tails are given below. Clones obtained in an A-rich region may correspond to artifacts and are indicated by a question mark. (D) Accumulation of the 5′ pETS in WT and rrp6l2 (left) or WT and rrp6l1 (right) plants as determined by virtual Northern blotting. Full-length oligo(dT)-primed cDNA was amplified by 16 PCR cycles, separated on an agarose gel, blotted, and hybridized to a DNA probe corresponding to the 5′ pETS.
To characterize the putative 5′ pETS in A. thaliana, we performed cRT-PCR to determine both the 5′ and 3′ extremities. Briefly, the 3′ and 5′ ends were ligated prior to cDNA synthesis, and PCR amplification of the region spanning the joined ends was done. The 5′ pETS was successfully amplified when cDNA from rrp6l2 plants, but not from WT or rrp6l1 samples, was used. The 5′ pETS amplified from rrp6l2 was cloned. Most 5′ extremities (16/18 clones) mapped to the P site (Fig. 5B). The 3′ ends mapped about 80 nt upstream of the 18S rRNA (Fig. 5B). Interestingly, nonencoded adenosines were present in 55% of the clones analyzed. Poly(A) tails were up to 18 nt in length, with an average size of 9 to 10 nt. These lengths do not necessarily reflect the in vivo RNA population, since cRT-PCR may preferentially amplify RNA with shorter poly(A) tails. However, these results unequivocally show that this rRNA maturation by-product can be polyadenylated in rrp6l2 plants, since oligo(dT) primers were not used in cRT-PCR.
We further analyzed the polyadenylation status of the 5′ pETS by 3′ RACE. Oligo(dT)-primed cDNA was produced from WT and rrp6l RNA, followed by 3′ RACE with a sense primer located 210 nt upstream of the 18S rRNA. We clearly detected polyadenylated 5′ pETS in rrp6l2-1 and to a lesser extent in rrp6l2-2, while only a faint smear was observed for WT and rrp6l1 samples (Fig. 5C). Sequence analysis of the cloned PCR products from the WT sample revealed that most were PCR artifacts; only 1 in 50 clones actually corresponded to the polyadenylated 5′ pETS. This suggests that the 5′ pETS can be polyadenylated in WT plants, but the level is extremely low. In contrast, all sequences from rrp6l2 mutants corresponded to polyadenylated 5′ pETS. The positions and frequencies of polyadenylation sites are presented for rrp6l2-1 (Fig. 5C). Some clones mapped to regions devoid of A's and had poly(A) tails longer than the oligo(dT)12 used for cDNA synthesis. These clones represent unambiguous polyadenylation sites. For six clones (Fig. 5C), we cannot exclude artificial priming by a stretch of four A's. However, nine clones mapped precisely to the 3′ site previously determined by cRT-PCR, immediately downstream of this A-rich region (AAAAG). The fact that all of these clones contained the G nucleotide indicates that cDNA synthesis was not primed by the genomically encoded A stretch. Remarkably, the poly(A) tails were slightly heteropolymeric, i.e., they contained several non-A nucleotides. These data confirm that the 5′ pETS is polyadenylated in rrp6l2 mutants.
The 5′ pETS was detected as a weak signal in rrp6l2 mutants, but not in WT or rrp6l1 plants, by using Northern hybridization of total RNA. To enhance the sensitivity and confirm the polyadenylation of the 5′ pETS, we performed virtual Northern blotting, a PCR-based method designed for quantitative comparison of polyadenylated RNA samples. Briefly, full-length oligo(dT)-primed cDNAs were amplified by a limited number of PCR cycles, separated on agarose gels, blotted, and hybridized to radiolabeled probes (19). Several RNA species were detected using a 5′ pETS DNA probe (Fig. 5D). The larger transcripts were rRNA precursor transcripts that were present in all samples. The smaller RNA, which was the size of the 5′ pETS (450 nt), accumulated in both rrp6l2 mutants (Fig. 5D), indicating that AtRRP6L2 is involved in the degradation of the 5′ pETS. Accumulation of this RNA was not observed in either of the rrp6l1 mutants or in the WT. Similarly, a 5.8S rRNA precursor transcript accumulated only upon downregulation of RRP6L2, and not RRP6L1, as determined by Northern blot analysis (data not shown). No increase in accumulation of the 5′ pETS and the 5.8S rRNA precursor was observed in rrp6l1-2 rrp6l2-1 and rrp6l1-2 rrp6l2-2 double mutants (data not shown). These results strongly suggest that both the 5′ pETS and the 5.8S rRNA precursor are substrates for AtRRP6L2, but not for AtRRP6L1.
DISCUSSION
While the compositions and functions of exosome complexes in yeast and humans have been investigated in depth, the plant core exosome was characterized only very recently (10). These initial data reveal some interesting features. First, AtRRP41 is a true phosphorolytic subunit, as judged by the conservation of essential residues and its in vitro activity (11, 15). If RRP41 retains its activity upon complex assembly, the plant exosome core complex would therefore have an exoribonucleolytic activity similar to those of archaeal exosomes (32), while yeast and human core subunits are catalytically inactive (15, 31). Second, at least three components of plant exosomes are not encoded by single genes: RRP40, RRP44, and RRP45. In the case of RRP45 proteins, single loss-of-function mutants were viable while simultaneous downregulation of both proteins was lethal, indicating a certain degree of redundancy (26). However, loss of AtRRP45b, but not of AtRRP45a, specifically affected cuticular-wax biosynthesis, suggesting that their functional overlap is only partial. These results illustrate that isoforms of core components of the exosome can impact a specific cellular process. In the present study, we show that plants as diverse as Arabidopsis, poplar, and rice contain three RRP6-like proteins, in contrast to single RRP6 proteins in yeast, Drosophila, trypanosomes, and human. Although their association with the exosome remains to be investigated, our results show that two of these proteins can perform a function of the yeast Rrp6p, since RRP6L1 complements the growth phenotype of the yeast mutant and RRP6L2 is involved in the degradation of the 5′ pETS. Importantly, RRP6L proteins show a distinctive subcellular distribution, which suggests a specialization of RRP6L proteins in Arabidopsis.
Phylogenetic analysis clearly distinguished AtRRP6L1 and -2, which group with human and yeast proteins, from AtRRP6L3, which seems to be a plant-specific gene. Interestingly, AtRRP6L3-GFP fusion proteins were found exclusively in the cytoplasm in both tobacco BY2 cells and A. thaliana. While yeast Rrp6p is restricted to the nucleus (2), the protein was found in both the cytoplasm and nucleus of trypanosomes (24). Interestingly, in human and Drosophila, a small fraction of PM/Scl-100 and dRRP6, respectively, was also detected in the cytoplasm (7, 22). Taken together, these results suggest that RRP6-related proteins could have a function in the cytoplasm in higher eukaryotes, including plants. It remains to be determined whether the biological role of RRP6L3 is similar to that of cytoplasmic RRP6 proteins in animals and humans or whether it has a plant-specific function that could also be independent of the exosome.
Only plant mutants downregulated for AtRRP6L2 accumulated the 5′ pETS, a maturation by-product of rRNA synthesis and a classical substrate for yeast Rrp6p. A role of AtRRP6L2 in the degradation of plant ETS is supported by its domain architecture and its subcellular distribution. AtRRP6L2 is the only RRP6-like protein in Arabidopsis, which contains the N-terminal PMC2NT domain. This domain mediates the interaction of yeast Rrp6p with the RNA binding protein Rrp47 (45). The absence of Rrp47p impairs both 3′ processing of stable RNA and degradation of the 5′ ETS (34). Thus, the N-terminal region of RRP6L2 could be necessary for degradation of the ETS in plants by interaction with a cofactor analogous to Rrp47. In fact, a protein with significant homology to Rrp47p is encoded by At5g25080. In addition, the intranuclear distribution of the AtRRP6L2 fusion proteins coincides with the localization of the 5′ ETS in nucleoli (42).
The ancient role of polyadenylation in marking transcripts destined for degradation is conserved from bacteria to higher eukaryotes, including humans, Drosophila, and plants, as well as in some organelles (5, 10, 14, 21, 28, 30, 36, 44, 47, 50, 51). Our results suggest that RRP6L2 participates in this process in Arabidopsis nuclei. The fact that the 5′ pETS that accumulates upon knockdown of AtRRP6L2 is polyadenylated is consistent with recent results in yeast, which show that several nuclear transcripts can be polyadenylated by the TRAMP complex and are subsequently degraded by Rrp6p (30, 47, 51). The fact that only RRP6L2 is able to degrade the 5′ pETS, combined with the distinct subcellular distribution of RRP6L proteins, suggests that RRP6-like proteins are specialized, or at least not fully redundant, in plants. These results reveal an intriguing complexity compared to other organisms studied to date.
Acknowledgments
This work was funded by the Centre National de la Recherche Scientifique (CNRS) (France), a fellowship of the Deutsche Forschungsgemeinschaft to H.L., and a Ph.D. fellowship from the French Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche (MNER) (France) to S.H.
We thank Uli Mühlenhoff (Marburg) and Jean-Luc Evrard (Strasbourg) for kindly providing the yeast and GFP cloning vectors, respectively.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 185399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., E. Petfalski, A. Podtelejnikov, M. Mann, D. Tollervey, and P. Mitchell. 1999. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 132148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, J. M., A. N. Stepanova, T. J. Leisse, C. J. Kim, H. Chen, P. Shinn, D. K. Stevenson, J. Zimmerman, P. Barajas, R. Cheuk, C. Gadrinab, C. Heller, A. Jeske, E. Koesema, C. C. Meyers, H. Parker, L. Prednis, Y. Ansari, N. Choy, H. Deen, M. Geralt, N. Hazari, E. Hom, M. Karnes, C. Mulholland, R. Ndubaku, I. Schmidt, P. Guzman, L. Aguilar-Henonin, M. Schmid, D. Weigel, D. E. Carter, T. Marchand, E. Risseeuw, D. Brogden, A. Zeko, W. L. Crosby, C. C. Berry, and J. R. Ecker. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301653-657. [DOI] [PubMed] [Google Scholar]
- 4.Andrulis, E. D., J. Werner, A. Nazarian, H. Erdjument-Bromage, P. Tempst, and J. T. Lis. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420837-841. [DOI] [PubMed] [Google Scholar]
- 5.Bollenbach, T. J., G. Schuster, and D. B. Stern. 2004. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 78305-337. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, M. W., K. T. Burkard, and J. S. Butler. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 27313255-13263. [DOI] [PubMed] [Google Scholar]
- 7.Brouwer, R., C. Allmang, R. Raijmakers, Y. van Aarssen, W. V. Egberts, E. Petfalski, W. J. van Venrooij, D. Tollervey, and G. J. Pruijn. 2001. Three novel components of the human exosome. J. Biol. Chem. 2766177-6184. [DOI] [PubMed] [Google Scholar]
- 8.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17540-552. [DOI] [PubMed] [Google Scholar]
- 9.Chekanova, J. A., J. A. Dutko, I. S. Mian, and D. A. Belostotsky. 2002. Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3′ → 5′ exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Res. 30695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chekanova, J. A., B. D. Gregory, S. V. Reverdatto, H. Chen, R. Kumar, T. Hooker, J. Yazaki, P. Li, N. Skiba, Q. Peng, J. Alonso, V. Brukhin, U. Grossniklaus, J. R. Ecker, and D. A. Belostotsky. 2007. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 1311340-1353. [DOI] [PubMed] [Google Scholar]
- 11.Chekanova, J. A., R. J. Shaw, M. A. Wills, and D. A. Belostotsky. 2000. Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem. 27533158-33166. [DOI] [PubMed] [Google Scholar]
- 12.Chevenet, F., C. Brun, A. L. Banuls, B. Jacq, and R. Christen. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clough, S. J., and A. F. Bent. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16735-743. [DOI] [PubMed] [Google Scholar]
- 14.Dreyfus, M., and P. Regnier. 2002. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Cell 111611-613. [DOI] [PubMed] [Google Scholar]
- 15.Dziembowski, A., E. Lorentzen, E. Conti, and B. Seraphin. 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 1415-22. [DOI] [PubMed] [Google Scholar]
- 16.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estevez, A. M., T. Kempf, and C. Clayton. 2001. The exosome of Trypanosoma brucei. EMBO J. 203831-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franz, O., I. Bruchhaus, and T. Roeder. 1999. Verification of differential gene transcription using virtual northern blotting. Nucleic Acids Res. 27e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulnecek, J., and A. Kovarik. 2007. Low abundant spacer 5S rRNA transcripts are frequently polyadenylated in Nicotiana. Mol. Genet. Genomics 278565-573. [DOI] [PubMed] [Google Scholar]
- 21.Gagliardi, D., P. P. Stepien, R. J. Temperley, R. N. Lightowlers, and Z. M. Chrzanowska-Lightowlers. 2004. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 20260-267. [DOI] [PubMed] [Google Scholar]
- 22.Graham, A. C., D. L. Kiss, and E. D. Andrulis. 2006. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol. Biol. Cell 171399-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52696-704. [DOI] [PubMed] [Google Scholar]
- 24.Haile, S., M. Cristodero, C. Clayton, and A. M. Estevez. 2007. The subcellular localisation of trypanosome RRP6 and its association with the exosome. Mol. Biochem. Parasitol. 15152-58. [DOI] [PubMed] [Google Scholar]
- 25.Holec, S., H. Lange, K. Kuhn, M. Alioua, T. Borner, and D. Gagliardi. 2006. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 262869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooker, T. S., P. Lam, H. Zheng, and L. Kunst. 2007. A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19904-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houseley, J., J. LaCava, and D. Tollervey. 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7529-539. [DOI] [PubMed] [Google Scholar]
- 28.Ibrahim, F., J. Rohr, W. J. Jeong, J. Hesson, and H. Cerutti. 2006. Untemplated oligoadenylation promotes degradation of RISC-cleaved transcripts. Science 3141893. [DOI] [PubMed] [Google Scholar]
- 29.Koonin, E. V., Y. I. Wolf, and L. Aravind. 2001. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Res. 11240-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121713-724. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Q., J. C. Greimann, and C. D. Lima. 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 1271223-1237. [DOI] [PubMed] [Google Scholar]
- 32.Lorentzen, E., P. Walter, S. Fribourg, E. Evguenieva-Hackenberg, G. Klug, and E. Conti. 2005. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 12575-581. [DOI] [PubMed] [Google Scholar]
- 33.Milligan, L., C. Torchet, C. Allmang, T. Shipman, and D. Tollervey. 2005. A nuclear surveillance pathway for mRNAs with defective polyadenylation. Mol. Cell. Biol. 259996-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell, P., E. Petfalski, R. Houalla, A. Podtelejnikov, M. Mann, and D. Tollervey. 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 236982-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91457-466. [DOI] [PubMed] [Google Scholar]
- 36.Orban, T. I., and E. Izaurralde. 2005. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrin, R., H. Lange, J. M. Grienenberger, and D. Gagliardi. 2004. AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 325174-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raijmakers, R., G. Schilders, and G. J. Pruijn. 2004. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur. J. Cell Biol. 83175-183. [DOI] [PubMed] [Google Scholar]
- 39.Rosso, M. G., Y. Li, N. Strizhov, B. Reiss, K. Dekker, and B. Weisshaar. 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53247-259. [DOI] [PubMed] [Google Scholar]
- 40.Saez-Vasquez, J., D. Caparros-Ruiz, F. Barneche, and M. Echeverria. 2004. A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol. Cell. Biol. 247284-7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholl, R. L., S. T. May, and D. H. Ware. 2000. Seed and molecular resources for Arabidopsis. Plant Physiol. 1241477-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaw, P. J., M. I. Highett, A. F. Beven, and E. G. Jordan. 1995. The nucleolar architecture of polymerase I transcription and processing. EMBO J. 142896-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slomovic, S., D. Laufer, D. Geiger, and G. Schuster. 2006. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 342966-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stead, J. A., J. L. Costello, M. J. Livingstone, and P. Mitchell. 2007. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 355556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanacova, S., and R. Stefl. 2007. The exosome and RNA quality control in the nucleus. EMBO Rep. 8651-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Engelen, F. A., J. W. Molthoff, A. J. Conner, J. P. Nap, A. Pereira, and W. J. Stiekema. 1995. pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res. 4288-290. [DOI] [PubMed] [Google Scholar]
- 49.Vetter, G., J. M. Hily, E. Klein, L. Schmidlin, M. Haas, T. Merkle, and D. Gilmer. 2004. Nucleo-cytoplasmic shuttling of the beet necrotic yellow vein virus RNA-3-encoded p25 protein. J. Gen. Virol. 852459-2469. [DOI] [PubMed] [Google Scholar]
- 50.West, S., N. Gromak, C. J. Norbury, and N. J. Proudfoot. 2006. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell 21437-443. [DOI] [PubMed] [Google Scholar]
- 51.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121725-737. [DOI] [PubMed] [Google Scholar]