Abstract
The mechanisms underlying the circadian control of gene expression in peripheral tissues and influencing many biological pathways are poorly defined. Factor VII (FVII), the protease triggering blood coagulation, represents a valuable model to address this issue in liver since its plasma levels oscillate in a circadian manner and its promoter contains E-boxes, which are putative DNA-binding sites for CLOCK-BMAL1 and NPAS2-BMAL1 heterodimers and hallmarks of circadian regulation. The peaks of FVII mRNA levels in livers of wild-type mice preceded those in plasma, indicating a transcriptional regulation, and were abolished in Clock−/−; Npas2−/− mice, thus demonstrating a role for CLOCK and NPAS2 circadian transcription factors. The investigation of Npas2−/− and ClockΔ19/Δ19 mice, which express functionally defective heterodimers, revealed robust rhythms of FVII expression in both animal models, suggesting a redundant role for NPAS2 and CLOCK. The molecular bases of these observations were established through reporter gene assays. FVII transactivation activities of the NPAS2-BMAL1 and CLOCK-BMAL1 heterodimers were (i) comparable (a fourfold increase), (ii) dampened by the negative circadian regulators PER2 and CRY1, and (iii) abolished upon E-box mutagenesis. Our data provide the first evidence in peripheral oscillators for an overlapping role of CLOCK and NPAS2 in the regulation of circadianly controlled genes.
Circadian clocks are endogenous oscillators that drive biochemical, physiological, and behavioral processes in organisms (36), and the alteration of which is associated with human diseases (17, 26, 42). It is well documented that the circadian rhythms of mammals are controlled by a master circadian pacemaker located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus (38) as well as by peripheral oscillators located in most tissues (43).
The basic circadian molecular clockworks consist of interacting positive and negative transcriptional/translational feedback loops (11). The positive loop involves CLOCK-BMAL1 and NPAS2-BMAL1 heterodimers, three basic helix-loop-helix transcriptional activators that bind to E-box elements (CACGTG) located in the regulatory regions of the Period (Per1, Per2, and Per3) and Cryptochrome (Cry1 and Cry2) genes (3, 13, 16, 24). CRY and PER proteins form oligomers that, once transported into the nucleus, repress their own transcription by inhibiting CLOCK-BMAL1 or NPAS2-BMAL1 (25). Furthermore, other transcription factors (CIPC, DEC1, DEC2, and Fbxl3) play important roles in the negative-feedback loop (15, 19, 44). The positive and negative limbs are interlaced: CLOCK-BMAL1 heterodimers indirectly regulate a rhythm in Bmal1 transcription; the nuclear orphan receptor genes Rev-erbα and Rora are coordinately activated by CLOCK-BMAL1 to produce proteins that compete for the same promoter element but have opposing actions on Bmal1 transcription (1, 32). Oscillations in gene expression resulting from this feedback loop have period lengths of approximately 24 h, thus giving rise to overt circadian rhythms (36).
Very recently, the role of CLOCK and NPAS2 has been elegantly investigated in the central and peripheral oscillators. Genetic and behavioral evidences in mice indicate that CLOCK and NPAS2 have partially redundant functions within the SCN (7). Whether this compensatory mechanism controls the gene expression in cells of peripheral oscillators is strongly debated (8, 9), particularly for circadian gene outputs. To address this issue, we took advantage of the key protease synthesized by hepatocytes that triggers blood coagulation (33), factor VII (FVII), whose levels in plasma oscillate in a circadian manner (6, 31). Through studies with transgenic mice and cellular and molecular models, we investigated the role of cardinal clock components in circadian transcription in liver oscillators.
MATERIALS AND METHODS
Animal and experimental design.
Experiments were performed with C57BL/6J wild-type (n = 48; Charles River Laboratory, Milano, Italy), ClockΔ19/Δ19 (n = 8; Jackson Laboratory, Bar Harbor, ME), Npas2−/− (n = 20), Clock−/−; Npas2−/− (n = 14) mice and littermate controls (wild type; n = 7 to 16 for each mutant). Food and water were supplied ad libitum. Mice were kept in a 12-h-light-12-h-dark cycle (LD 12:12; lights-on at 0600) or in total darkness (DD). It is conventional to divide the 24-hour LD cycle into 24-hour Zeitgeber time (ZT) units and indicate the time of lights-on as ZT0 and the time of lights-off as ZT12. Sampling was performed at ZT0, -4, -6, -8, -12, -16, -18, and -20. The 24-hour circadian cycle is divided into 24-hour circadian time (CT) units. As a reference point, in DD, the phase that would normally (i.e., in LD 12:12) coincide with the onset of darkness, which is called CT12, is conventionally used. The part of the cycle covering CT0 to CT12 is then called subjective day, and the part covering CT12 to CT24 is called subjective night. Because the free-running period of circadian rhythms in mice does not change from 24 h during the first days in DD, ZTs and CTs mark the same phases and can be used to identify the same time points of sampling between the LD and DD tests. Sampling was performed in DD at projected CT0, -2, -4, -6, -8, -10, -12, -14, -16, -18, and -20. In DD, a weak red light (<0.1 lx) was used during the sampling. Blood samples were collected in 0.38% trisodium citrate. After centrifugation at 2,500 × g for 10 min, the plasma was separated and stored at −80°C. The livers were quickly removed, frozen on dry ice, and stored at −80°C until processing.
All treatments were conducted under the guidelines established by the Italian Ministry of Health and by the Institutional Animal Care and Use Committee of the Morehouse School of Medicine, the University of Texas Southwestern Medical Center, and the University of Massachusetts Medical School.
Real-time quantitative reverse transcription (RT)-PCR analysis.
Total RNA was isolated from mouse liver using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
For quantitative real-time RT-PCR, the fluorogenic forward primer and the corresponding unlabeled reverse primer were designed using LUX Designer software online. The target sequence was based on mouse coagulation FVII mRNA (GenBank accession no. NM_010172). Oligonucleotide sequences were as follows: 5′-cacagttGACTTTGAGGGTCGGAACTGtGFAM-3′ (forward; lowercase letters indicate complementary bases for hairpin formation with the 3′ end and FAM is 6-carboxyfluorescein) and 5′-GCGGCTGCTGGAGTTTCTTT-3′ (reverse). The PCR product size is 225 bp. As an internal control, the mouse GAPDH (glyceraldehyde 3-phosphate dehydrogenase)-certified LUX JOE-labeled primers (Invitrogen, Carlsbad, CA) were used (PCR product size, 200 bp).
The real-time RT-PCRs were carried out using the Platinum quantitative RT-PCR ThermoScript one-step system (Invitrogen, Carlsbad, CA). Reactions were set up in accordance with the manufacturers' instructions and performed on an ABI PRISM 7700 sequence detection system (Applied Biosystems, Warrington, United Kingdom). Briefly, target and internal control mRNA sequences were independently amplified. The reaction was carried out in 25 μl in a single tube containing 3 mM MgSO4, 0.2 mM of each deoxynucleoside triphosphate, 140 nM of each primer, 20 U of RNaseOUT RNase inhibitor (Invitrogen, Carlsbad, CA), 1 μl ROX reference dye, 0.5 μl ThermoScript Plus-Platinum Taq enzyme mix, and 500 ng RNA template. Cycling parameters were 50°C for 30 min and 95°C for 5 min to activate Taq DNA polymerase, followed by 40 cycles of 95°C for 15 s, 60°C for 45 s, and 72°C for 30 s.
The comparable amplification efficiencies of FVII and GAPDH transcripts prompted us to exploit the comparative threshold cycle method (CT; the fractional cycle number at which the amount of amplified copies reaches a fixed threshold). Normalized ΔCT values from each time point were then subtracted from the calibrator sample ΔCT value (ΔΔCT) to determine the relative abundance values (2−ΔΔCT).
Time-series data were analyzed by one-way analysis of variance (ANOVA) or by Kruskal-Wallis one-way ANOVA to determine significant differences. Differences between mouse strains were tested by two-way ANOVA. A comparison of <0.05 was considered statistically significant.
Evaluation of FVII activity in plasma.
The activity of FVII was tested by activated factor X (FXa) generation assays measuring the ability of FVII to activate its physiologic substrate FX. As previously described (31), FXa generation was measured in mouse plasma diluted (1:20) in human FVII-depleted plasma (Dade Behring, Marburg, Germany) to selectively assess the contribution of mouse FVII. Upon the triggering of coagulation by an excess of tissue factor (Innovin; Dade Behring, Marburg, Germany), the generation of FXa was evaluated by measuring its activity toward a specific fluorogenic substrate (methylsulfonyl-d-cyclohexylalanyl-Gly-Arg-7-amino-4-methylcoumarin acetate; American Diagnostica Inc., Stamford, CT) over time.
The initial rate of activity was expressed as relative fluorescence units (RFU) per second. One-way and two-way ANOVAs were used to determine significant differences (P < 0.05). Bonferroni's test was applied for post hoc comparisons.
Expression vectors.
To create the pFVII-Luc vector, the 1,087-bp sequence preceding the translation ATG initiation codon of mouse FVII gene was cloned upstream of the firefly luciferase coding sequence (Luc+) in the pGL3 basic vector (Promega, Madison, WI). The region was amplified from mouse liver DNA obtained through phenol chloroform extraction. To permit cloning in the pGL3 polylinker, the HindIII sites (in bold) were introduced within primers (forward, 5′-CGCGTAAGCTTACGCAGGAGCCAGCCAAG-3′; reverse, 5′-ATGCCAAGCTTGATGAGACTGGCTTCTGGC-3′). PCRs were subjected to 35 cycles of the following: 94°C for 30 s, 57°C for 30 s, and 72°C for 40 s. After cloning, the putative promoter sequence was validated by DNA sequencing. Cloning provided us with a vector containing the 1,087-bp sequence oriented in the opposite direction (pFVII-Luc-inv), which was exploited as an additional negative control in the experiments.
E-boxes of the FVII promoter were mutated at bp −1055 to −1050 (E1 mutation, CACGTG to TCGCTC) and at bp −484 to −479 (E2 mutation, CACGTG to GCTAGT) using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Constructs were validated by direct sequencing.
Expression vectors for the mouse circadian transcription factors Clock (pcDNA3-mClock), Bmal1 (pcDNA3-mBmal1), Npas2 (pcDNA3-mNpas2), Per2 (pcDNA3-mPer2), and Cry1 (pcDNA3-mCry1) have been previously described (24, 35). The pSV-β-galactosidase (Promega, Madison, WI), driving the expression of the galactosidase, was used as the control plasmid (pBKCMVLgal).
Cell culture, transfection, and reporter assays.
Experiments were conducted for mouse hepatoma cell lines (Hepa1-6) grown in Dulbecco's modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% inactivated fetal bovine serum (Invitrogen, Carlsbad, CA). The day before transfection, cells were split and seeded in 35-mm tissue culture dishes.
Cells at approximately 70% confluence were cotransfected with 200 ng of pFVII-Luc (or of each mutagenized vector), 2 μg of plasmids encoding circadian transcription factors (CLOCK, BMAL1, NPAS2, PER2, and CRY1), and 350 ng of pBKCMVLgal to normalize transfection efficiency. To keep constant the amount of transfected DNA, a gutted pcDNA3 vector was used.
After a 54-h incubation, cells were lysed and luciferase activity was measured using a luciferase assay system (Promega, Madison, WI). Cell lysates were also assessed for l-galactosidase activity. Luciferase activity was normalized against the l-galactosidase activity.
Each experiment was performed at least four times. One-way ANOVA was used to determine significant differences (P < 0.05) and Bonferroni's test was used in post hoc analyses.
RESULTS
Liver mRNA FVII expression and plasma FVII levels.
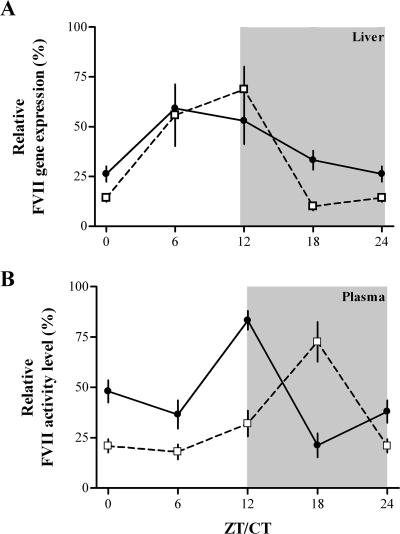
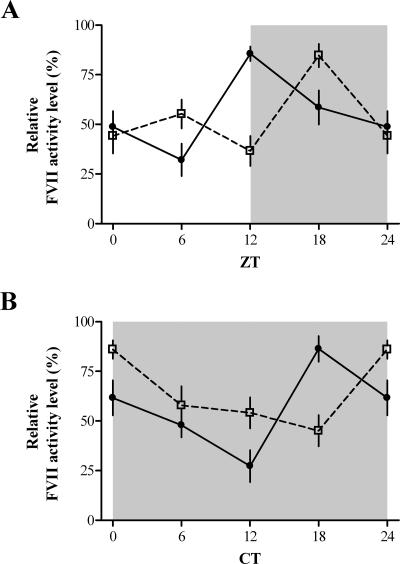
To investigate whether the circadian oscillations of FVII levels previously observed in mouse plasma correlate with cyclic transcription of the FVII gene, we evaluated by using quantitative real-time RT-PCR the FVII mRNA levels in the livers from a group of 48 wild-type C57BL/6J mice. The hepatic expression of FVII mRNA showed a statistically significant rhythmicity both in LD (n = 24) and DD (n = 24) [one-way ANOVA; LD, F(3,20) = 3.2, P < 0.05; DD, F(3,20) = 6.9, P < 0.003] (Fig. 1A). FVII mRNA levels peaked at ZT6 and CT12 in LD and DD, respectively (Fig. 1A). Differences in the patterns between LD and DD conditions were not significant (two-way ANOVA; F(1,40) = 0.75, P > 0.3).
FIG. 1.
Temporal variations of liver FVII mRNA expression (A) and of plasma FVII activity (B) levels in mice. Mice (n = 6 for each time point) exposed to LD or DD conditions were sampled at the indicated ZT/CT. Values were normalized with respect to the maximum value (100%) measured for each condition. The mean ± the standard error of the mean (SEM) at each time point is shown. Statistical analysis was carried out using one-way and two-way ANOVAs. Gray areas indicate dark phases. LD, filled circles; DD, empty squares.
The parallel analysis of plasma samples from the same animals revealed a significant rhythmicity of FVII activity levels in both LD and DD (one-way ANOVA; LD, F(3,20) = 20.1, P < 0.0001; DD, F(3,20) = 10.8, P < 0.001) (Fig. 1B), with peaks approximately 6 h later than those of the mRNA levels.
FVII expression in circadian rhythm mutant mice.
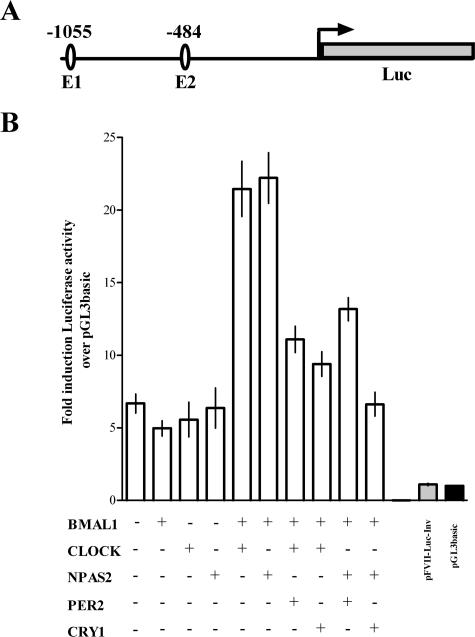
The mouse FVII promoter contains two perfect E-box elements (CACGTG) at positions −1055 and −484 (see Fig. 5A), putative DNA-binding sites for CLOCK-BMAL1 and NPAS2-BMAL1 heterodimers. These sequence features, together with the circadian nature of FVII mRNA level variations, prompted us to evaluate in vivo the role of the cardinal clock components CLOCK and NPAS2 in the generation of FVII rhythms.
FIG. 5.
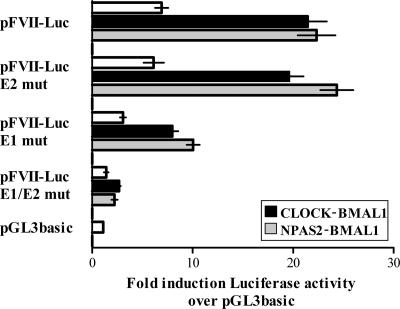
Transcriptional regulation of mouse FVII promoter by circadian factors. (A) Schematic representation of the reporter gene construct. E-box elements (E1 and E2) in the 5′ regulatory region (1,087 bp) of the mouse FVII gene are represented by ellipses. The numbering refers to the translation start site (arrow). (B) Relative luciferase activity in Hepa1-6 cells transfected with pFVII-Luc and different combinations of circadian transcription factors (BMAL1, CLOCK, NPAS2, PER2, and CRY1). The abscissa displays mean (±SEM) induction levels of luciferase activity over that of the negative control (pGL3basic).
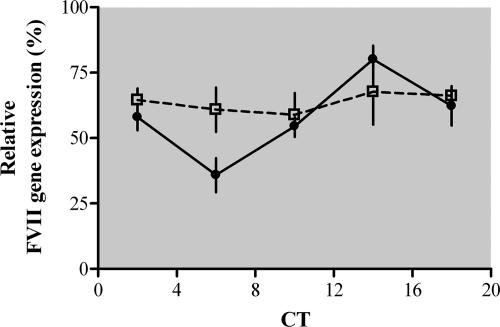
The Clock−/−; Npas2−/− double mutant mice, kept under DD conditions to avoid direct-light effects, provided us with a valuable model to address this issue. Differently from wild-type mice, which displayed robust rhythms of FVII mRNA levels in liver (n = 16; one-way ANOVA; F(4,29) = 7.91, P < 0.0002) (Fig. 2), the Clock−/−; Npas2−/− mice did not show appreciable temporal variations in FVII expression (n = 14; one-way ANOVA; F(4,29) = 0.18, P > 0.9) (Fig. 2). However, daily mean FVII expression levels in Clock−/−; Npas2−/− mice were not significantly different from those detected in wild-type mice (two-way ANOVA; F(1,50) = 1.48, P > 0.2), highlighting the importance of the other cis/trans elements responsible for the constitutive expression of mouse FVII gene (39).
FIG. 2.
Liver FVII mRNA expression levels in Clock−/−; Npas2−/− mice. Clock−/−; Npas2−/− (empty squares) and wild-type (filled circles) mice (n = 14 to 16 for each genotype) were sampled at the indicated CT on the first day in DD. FVII expression levels were normalized with respect to the maximum value (100%) measured for each strain. The mean ± SEM at each time point is shown. Statistical analysis was carried out using one-way and two-way ANOVAs. The gray area indicates the dark phase.
Results in Clock−/−; Npas2−/− mice prompted us to examine the specific contribution of CLOCK and NPAS2 to circadian FVII rhythms in Npas2−/− and ClockΔ19/Δ19 mutant mice, respectively.
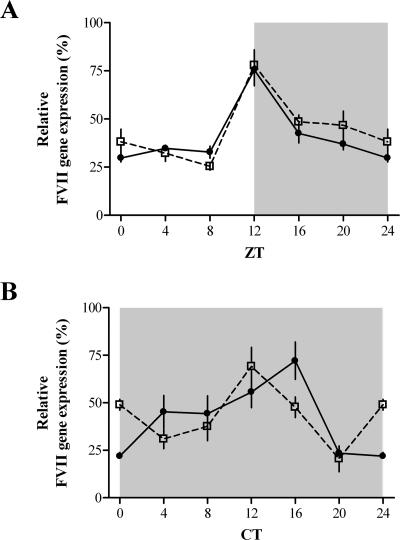
Npas2−/− mice express an inactive NPAS2 variant lacking the basic helix-loop-helix domain (12, 35). The expression of FVII mRNA in the livers of Npas2−/− mice kept under LD and DD conditions showed a statistically significant rhythmicity with peaks at ZT-/CT12 (n = 10 for each lighting condition; Kruskal-Wallis one-way ANOVA; LD, P < 0.005; DD, P < 0.003) (Fig. 3). As expected, liver FVII mRNA expressions in wild-type mice were rhythmic under both lighting conditions (n = 10 for each condition; Kruskal-Wallis one-way ANOVA; P < 0.0001) (Fig. 3).
FIG. 3.
Liver FVII mRNA expression levels in Npas2−/− mice. Npas2−/− (empty squares) and wild-type (filled circles) mice (n = 10 for each genotype) under LD (A) and DD (B) conditions were sampled at the indicated ZT/CT. FVII expression levels were normalized with respect to the maximum value (100%) measured for each genotype/condition. The mean ± SEM at each time point is shown. Statistical analysis was carried out using the Kruskal-Wallis one-way ANOVA. Gray areas indicate dark phases.
ClockΔ19/Δ19 mice express a CLOCK protein variant devoid of the residues encoded by exon 19, which renders CLOCK-BMAL1 heterodimers functionally defective (22, 41). Plasma FVII levels in these mice (n = 8) were robustly rhythmic in both LD and DD (one-way ANOVA; LD, F(3,21) = 9.5, P < 0.001; DD, F(3,21) = 4.6, P < 0.01), with peaks delayed by 6 h compared to those observed in wild-type mice (n = 7; one-way ANOVA; LD, F(3,18) = 15.1, P < 0.0001; DD, F(3,18) = 7.3, P < 0.003) (Fig. 4). Differences in daily levels between genotypes were not significant for either LD or DD (two-way ANOVA; LD, F(1,42) = 3.6, P > 0.1; DD, F(1,42) = 0.2, P > 0.6).
FIG. 4.
FVII activity levels in ClockΔ19/Δ19 mice. ClockΔ19/Δ19 (empty squares) and wild-type (filled circles) mice (n = 8 for each time point) exposed to LD (A) and DD (B) conditions were sampled at different ZT/CT. Each point represents the mean ± SEM of the logarithm of FVII activity expressed as RFU per second. FVII levels were normalized with respect to the maximum value (100%) measured for each genotype/condition. Statistical analysis was carried out using one-way and two-way ANOVAs. The gray areas indicate dark phases.
Transactivation of mouse FVII promoter by circadian clock components.
To investigate the molecular mechanisms underlying our observations in mutant mice, we performed reporter gene assays. This approach was also aimed at corroborating findings in ClockΔ19/Δ19 mice, in which a residual activity of CLOCK-BMAL1 could be present (22, 41). For these studies, we created a luciferase reporter construct that contains the 5′ regulatory region of mouse FVII gene (1,087 bp) including the E-box elements (Fig. 5A) and that was previously shown to possess promoter activity (39). Because of the liver-specific expression of FVII, the mouse hepatoma cell line Hepa1-6 was used for reporter expression analysis.
Normalized luciferase activity measured in lysates from cells transfected with the pFVII-Luc was approximately sixfold higher (F(2,36) = 8.3, P < 0.001) than that in cells transfected with the gutted vector (pGL3basic) or pFVII-Luc-inv, which were used as negative controls (Fig. 5B).
The cotransfection of pFVII-Luc with each single pcDNA3-mBmal1, pcDNA3-mClock, or pcDNA3-mNpas2 vector did not result in a significant transactivation (F(3,30) = 0.71, P > 0.2) (Fig. 5B). In contrast, the cotransfection of pFVII-Luc with the combination of pcDNA3-mClock-pcDNA3-mBmal1 or of pcDNA3-mNpas2-pcDNA3-mBmal1 resulted in an approximately fourfold increase in FVII promoter activity, which was significantly higher than that in the absence of transactivators (F(2,24) = 8.71, P < 0.001) (Fig. 5B). The transactivation activities of pcDNA3-mClock-pcDNA3-mBmal1 or of pcDNA3-mNpas2-pcDNA3-mBmal1 were not significantly different (Bonferroni's post hoc test, P > 0.05).
The expression of PER2 or CRY1, the key components of the negative feedback loop, was also investigated. The cotransfection of either pcDNA3-mPer2 or pcDNA3-mCry1 strongly inhibited CLOCK-BMAL1-dependent (F(2,22) = 18.57, P < 0.0001) or NPAS2:BMAL1-dependent (F(2,25) = 24.78, P < 0.0001) transactivation (Fig. 5B), in accordance with the circadian nature of FVII gene expression.
To unequivocally demonstrate the role of E-boxes in the circadian regulation of mouse FVII transcription, the sequence elements were abrogated in the pFVII-Luc vector by site-directed mutagenesis. The mutation of both E-boxes abolished both CLOCK-BMAL1- and NPAS2-BMAL1-dependent transactivation (F(3,22) = 28.6, P < 0.0001; F(3,24) = 30.4, P < 0.0001, respectively) (Fig. 6). The effect of the abolition of each single E-box was also sought. The mutation of the proximal E-box element (E2 in Fig. 5A) had no significant effects on CLOCK-BMAL1- or NPAS2-BMAL1-mediated transcription (F(3,30) = 0.57, P > 0.6) (Fig. 6). Differently, the mutation of the distal enhancer (E1 in Fig. 5A) drastically reduced FVII transactivation by both heterodimers (F(3,32) = 14.8, P < 0.0001) (Fig. 6). This observation might indicate the interaction of CLOCK-BMAL1 or NPAS2-BMAL1 with other components involved in FVII promoter transcription.
FIG. 6.
Functional role of E-boxes in the transcriptional regulation of mouse FVII promoter. The relative luciferase activity in Hepa1-6 cells transfected with pFVII-Luc variants (E1, E2, or E1/E2 mutation [mut]) and different combinations of circadian transcription factors (CLOCK, BMAL1, and NPAS2). The abscissa displays mean (±SEM) induction levels of luciferase activity over that of pGL3basic.
DISCUSSION
Hepatocytes synthesize, process, and secrete an impressive number of molecules whose levels are regulated by transcriptional, posttranscriptional, and posttranslational mechanisms (2, 34). The circadian control of expression plays a role—with potential implications in pathophysiology (17, 26, 42)—for a number of genes, among them the FVII gene, chosen as the output gene model for liver. FVII level variations in humans are associated with a bleeding tendency (27) or an increased risk for cardiovascular disease (4, 14, 28).
Our investigations with liver mRNA demonstrated that rhythms of FVII observed in mouse plasma are due to control at the transcriptional level. This finding together with the presence of two functional E-boxes in the FVII promoter, shown through site-directed mutagenesis, provided us with a valuable experimental tool to investigate the role of CLOCK and NPAS2 circadian transcription factors, paralog proteins that form transcriptionally active complexes with BMAL1 on E-boxes (13, 18, 25, 35). Their contribution to variations of FVII expression levels in vivo was clearly demonstrated in Clock−/−; Npas2−/− mutant mice, in which the rhythms of FVII mRNA in liver were abolished, even in the first day under DD conditions. Overall, this observation indicates that FVII gene transcription is strongly influenced by the molecular circadian machinery, which is known to be disrupted in Clock−/−; Npas2−/− mice (7).
Npas2−/− and ClockΔ19/Δ19 mutant mice, expressing functionally defective heterodimers (22, 35, 41), were exploited to dissect in vivo the contribution of the single transcription factors to the generation of FVII rhythms.
The impairment of NPAS2 function in Npas2−/− mice has profound physiologic consequences and results in sleep disturbance (10, 12). NPAS2 is required for the circadian expression of mPer2 mRNA in the forebrain (35), and it is expressed in a circadian manner in peripheral oscillators such as the liver (9, 20). Interestingly, we detected normal circadian FVII rhythms in the livers of Npas2−/− mice, thus indicating that NPAS2 is not crucial for the circadian expression of the FVII gene.
Robust circadian FVII rhythms were also observed in ClockΔ19/Δ19 mice. Although not mimicking a completely CLOCK-null condition, these mice show arrhythmicity (21, 30, 41) and markedly blunted molecular rhythms in the SCN (5, 37), thus indicating a major impairment of CLOCK functions. Therefore, our findings in ClockΔ19/Δ19 mice support the hypothesis that CLOCK is also dispensable for the circadian control of FVII levels. The changes in peaks observed in this mouse strain could depend on the altered expression of other clock genes (29) or on alterations in metabolism (40). Particularly, ClockΔ19/Δ19 mice show attenuated diurnal feeding rhythms and develop metabolic syndromes, and we previously demonstrated that feeding is a major determinant of temporal FVII level variations (31).
The overlapping roles of CLOCK and NPAS2 in the circadian control of mouse FVII expression in vivo were further corroborated by reporter gene assays with the mouse FVII promoter, revealing comparable transactivation efficiencies for CLOCK-BMAL1 and NPAS2-BMAL1.
The redundant roles of CLOCK and NPAS2 in vivo have recently been shown in the SCN as well as in the liver for clock genes such as Per1 and Per2 (7, 9). It must be mentioned that the rhythmicity of mPer2 expression in liver explants from Clock−/−; mPer2Luciferase mice is abolished, suggesting the inability of NPAS2 alone to maintain circadian rhythm variations in cultured cells (8). This observation might indicate that the circadian expression of liver genes in Clock−/− mice is most likely being driven by oscillating systemic cues, such as hormones and metabolites, which are directly or indirectly controlled by the SCN. However, investigations using mice with a conditionally active liver clock demonstrated that the circadian transcription of most liver genes, including the FVII gene (data available at http://www.ebi.ac.uk/arrayexpress; accession number E-MEXP-842), depend exclusively on functional hepatocyte clocks (23).
Taken together, our in vivo, cellular, and molecular observations of the components underlying the circadian rhythm control of FVII expression provide the first evidence in peripheral oscillators of overlapping roles of CLOCK and NPAS2 in the regulation of a circadianly controlled gene.
Acknowledgments
The work was supported by grants from the University of Ferrara (M.P., C.B., A.F., and F.B.), the Fondazione Cassa di Risparmio di Cento (I.C., A.F., and C.B.), Telethon (GP05214; M.P. and F.B.), and the National Institutes of Health NS-43459 (G.T.).
We are grateful to Urs Albrecht (University of Fribourg, Switzerland), Pablo Sdrenko (Max Planck Institute for Molecular Endocrinology, Hannover, Germany), Steven L. McKnight (University of Texas Southwestern Medical Center, Dallas, TX), Steven M. Reppert (University of Massachusetts Medical School, Worcester, MA), and Michael H. Hastings (Medical Research Council, Cambridge, United Kingdom), who kindly provided expression vectors for the mouse circadian transcription factors. We also thank Steven L. McKnight (University of Texas Southwestern Medical Center) and Jason P. DeBruyne (University of Massachusetts Medical School) for providing us with samples from Npas2−/− and Clock−/−; Npas2−/− mutant mice, respectively.
Footnotes
Published ahead of print on 3 March 2008.
REFERENCES
- 1.Akashi, M., and T. Takumi. 2005. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 12441-448. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, U. 2006. Molecular orchestration of the hepatic circadian symphony. Genome Biol. 7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunger, M. K., L. D. Wilsbacher, S. M. Moran, C. Clendenin, L. A. Radcliffe, J. B. Hogenesch, M. C. Simon, J. S. Takahashi, and C. A. Bradfield. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 1031009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campo, G., M. Valgimigli, P. Ferraresi, P. Malagutti, M. Baroni, C. Arcozzi, D. Gemmati, G. Percoco, G. Parrinello, R. Ferrari, and F. Bernardi. 2006. Tissue factor and coagulation factor VII levels during acute myocardial infarction: association with genotype and adverse events. Arterioscler. Thromb. Vasc. Biol. 262800-2806. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, M. Y., C. M. Bullock, C. Li, A. G. Lee, J. C. Bermak, J. Belluzzi, D. R. Weaver, F. M. Leslie, and Q. Y. Zhou. 2002. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature 417405-410. [DOI] [PubMed] [Google Scholar]
- 6.Colognesi, I., V. Pasquali, A. Foà, P. Renzi, F. Bernardi, C. Bertolucci, and M. Pinotti. 2007. Temporal variations of coagulation factor VII activity in mice are influenced by lighting regime. Chronobiol. Int. 24305-313. [DOI] [PubMed] [Google Scholar]
- 7.DeBruyne, J. P., D. R. Weaver, and S. M. Reppert. 2007. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat. Neurosci. 10:543-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeBruyne, J. P., D. R. Weaver, and S. M. Reppert. 2007. Peripheral circadian oscillators require CLOCK. Curr. Biol. 17R538-R539. [DOI] [PubMed] [Google Scholar]
- 9.DeBruyne, J. P., E. Noton, C. M. Lambert, E. S. Maywood, D. R. Weaver, and S. M. Reppert. 2006. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50465-477. [DOI] [PubMed] [Google Scholar]
- 10.Dudley, C. A., C. Erbel-Sieler, S. J. Estill, M. Reick, P. Franken, S. Pitts, and S. L. McKnight. 2003. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science 301379-383. [DOI] [PubMed] [Google Scholar]
- 11.Gachon, F., E. Nagoshi, S. A. Brown, J. Ripperger, and U. Schibler. 2004. The mammalian circadian timing system: from gene expression to physiology. Chromosoma 113103-112. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, J. A., D. Zhang, S. J. Estill, C. Michnoff, J. Rutter, M. Reick, K. Scott, R. Diaz-Arrastia, and S. L. McKnight. 2000. Impaired cued and contextual memory in NPAS2-deficient mice. Science 2882226-2230. [DOI] [PubMed] [Google Scholar]
- 13.Gekakis, N., D. Staknis, H. B. Nguyen, F. C. Davis, L. D. Wilsbacher, D. P. King, J. S. Takahashi, and C. J. Weitz. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 2801564-1569. [DOI] [PubMed] [Google Scholar]
- 14.Girelli, D., C. Russo, P. Ferraresi, O. Olivieri, M. Pinotti, S. Friso, F. Manzato, A. Mazzucco, F. Bernardi, and R. Corrocher. 2000. Polymorphisms in the factor VII gene and the risk of myocardial infarction in patients with coronary artery disease. N. Engl. J. Med. 343774-780. [DOI] [PubMed] [Google Scholar]
- 15.Godinho, S. I., E. S. Maywood, L. Shaw, V. Tucci, A. R. Barnard, L. Busino, M. Pagano, R. Kendall, M. M. Quwailid, M. R. Romero, J. O'Neill, J. E. Chesham, D. Brooker, Z. Lalanne, M. H. Hastings, and P. M. Nolan. 2007. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316897-900. [DOI] [PubMed] [Google Scholar]
- 16.Griffin, E. A., D. Staknis, and C. J. Weitz. 1999. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286768-771. [DOI] [PubMed] [Google Scholar]
- 17.Hastings, M. H., A. B. Reddy, and E. S. Maywood. 2003. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 4649-661. [DOI] [PubMed] [Google Scholar]
- 18.Hogenesch, J. B., Y. Z. Gu, S. Jain, and C. A. Bradfield. 1998. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 955474-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honma, S., T. Kawamoto, Y. Takagi, K. Fujimoto, F. Sato, M. Noshiro, Y. Kato, and K. Honma. 2002. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419841-844. [DOI] [PubMed] [Google Scholar]
- 20.Kaasik, K., and C. C. Lee. 2004. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430467-471. [DOI] [PubMed] [Google Scholar]
- 21.Kennaway, D. J., A. Voultsios, T. J. Varcoe, and R. W. Moyer. 2003. Melatonin and activity rhythm responses to light pulses in mice with the Clock mutation. Am. J. Physiol. Regul. Integr. Comp. Physiol. 284R1231-R1240. [DOI] [PubMed] [Google Scholar]
- 22.King, D. P., Y. Zhao, A. M. Sangoram, L. D. Wilsbacher, M. Tanaka, M. P. Antoch, T. D. Steeves, M. H. Vitaterna, J. M. Kornhauser, P. L. Lowrey, F. W. Turek, and J. S. Takahashi. 1997. Positional cloning of the mouse circadian clock gene. Cell 89641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornmann, B., O. Schaad, H. Bujard, J. S. Takahashi, and U. Schibler. 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 5e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kume, K., M. J. Zylka, S. Sriram, L. P. Shearman, D. R. Weaver, X. Jin, E. S. Maywood, M. H. Hastings, and S. M. Reppert. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98193-205. [DOI] [PubMed] [Google Scholar]
- 25.Kwon, I., J. Lee, S. H. Chang, N. C. Jung, B. J. Lee, G. H. Son, K. Kim, and K. H. Lee. 2006. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol. Cell. Biol. 267318-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansour, H. A., J. Wood, T. Logue, K. V. Chowdari, M. Dayal, D. J. Kupfer, T. H. Monk, B. Devlin, and V. L. Nimgaonkar. 2006. Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. 5150-157. [DOI] [PubMed] [Google Scholar]
- 27.Mariani, G., F. H. Herrmann, A. Dolce, A. Batorova, D. Etro, F. Peyvandi, K. Wulff, J. F. Schved, G. Auerswald, J. Ingerslev, F. Benardi, and the International Factor VII Deficiency Study Group. 2005. Clinical phenotypes and factor VII genotype in congenital factor VII deficiency. Thromb. Haemost. 93481-487. [DOI] [PubMed] [Google Scholar]
- 28.Meade, T. W., S. Mellows, M. Brozovic, G. J. Miller, R. R. Chakrabarti, W. R. North, A. P. Haines, Y. Stirling, J. D. Imeson, and S. G. Thompson. 1986. Haemostatic function and ischaemic heart disease: principal results of the Northwick Park Heart Study. Lancet ii533-537. [DOI] [PubMed] [Google Scholar]
- 29.Noshiro, M., M. Furukawa, S. Honma, T. Kawamoto, T. Hamada, K. Honma, and Y. Kato. 2005. Tissue-specific disruption of rhythmic expression of Dec1 and Dec2 in clock mutant mice. J. Biol. Rhythms 20404-418. [DOI] [PubMed] [Google Scholar]
- 30.Oishi, K., K. Miyazaki, and N. Ishida. 2002. Functional CLOCK is not involved in the entrainment of peripheral clocks to the restricted feeding: entrainable expression of mPer2 and mBmal1 mRNAs in the heart of Clock mutant mice on Jcl:ICR background. Biochem. Biophys. Res. Commun. 298198-202. [DOI] [PubMed] [Google Scholar]
- 31.Pinotti, M., C. Bertolucci, F. Portaluppi, I. Colognesi, E. Frigato, A. Foà, and F. Bernardi. 2005. Daily and circadian rhythms of tissue factor pathway inhibitor and factor VII activity. Arterioscler. Thromb. Vasc. Biol. 25646-649. [DOI] [PubMed] [Google Scholar]
- 32.Preitner, N., F. Damiola, L. Lopez-Molina, J. Zakany, D. Duboule, U. Albrecht, and U. Schibler. 2002. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110251-260. [DOI] [PubMed] [Google Scholar]
- 33.Rapaport, S. I., and L. V. Rao. 1995. The tissue factor pathway: how it has become a “prima ballerina”. Thromb. Haemost. 747-17. [PubMed] [Google Scholar]
- 34.Reddy, A. B., N. A. Karp, E. S. Maywood, E. A. Sage, M. Deery, J. S. O'Neill, G. K. Wong, J. Chesham, M. Odell, K. S. Lilley, C. P. Kyriacou, and M. H. Hastings. 2006. Circadian orchestration of the hepatic proteome. Curr. Biol. 161107-1115. [DOI] [PubMed] [Google Scholar]
- 35.Reick, M., J. A. Garcia, C. Dudley, and S. L. McKnight. 2001. NPAS2: an analog of clock operative in the mammalian forebrain. Science 293506-509. [DOI] [PubMed] [Google Scholar]
- 36.Reppert, S. M., and D. R. Weaver. 2001. Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63647-676. [DOI] [PubMed] [Google Scholar]
- 37.Ripperger, J. A., L. P. Shearman, S. M. Reppert, and U. Schibler. 2000. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14679-689. [PMC free article] [PubMed] [Google Scholar]
- 38.Saper, C. B., T. E. Scammell, and J. Lu. 2005. Hypothalamic regulation of sleep and circadian rhythms. Nature 4371257-1263. [DOI] [PubMed] [Google Scholar]
- 39.Stauffer, D. R., B. N. Chukwumezie, J. A. Wilberding, E. D. Rosen, and F. J. Castellino. 1998. Characterization of transcriptional regulatory elements in the promoter region of the murine blood coagulation factor VII gene. J. Biol. Chem. 2732277-2287. [DOI] [PubMed] [Google Scholar]
- 40.Turek, F. W., C. Joshu, A. Kohsaka, E. Lin, G. Ivanova, E. McDearmon, A. Laposky, S. Losee-Olson, A. Easton, D. R. Jensen, R. H. Eckel, J. S. Takahashi, and J. Bass. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 3081043-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vitaterna, M. H., D. P. King, A. M. Chang, J. M. Kornhauser, P. L. Lowrey, J. D. McDonald, W. F. Dove, L. H. Pinto, F. W. Turek, and J. S. Takahashi. 1994. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, H., D. F. Fischer, A. Kalsbeek, M. L. Garidou-Boof, J. van der Vliet, C. van Heijningen, R. Y. Liu, J. N. Zhou, and D. F. Swaab. 2006. Pineal clock gene oscillation is disturbed in Alzheimer's disease, due to functional disconnection from the “master clock.” FASEB J. 201874-1876. [DOI] [PubMed] [Google Scholar]
- 43.Yoo, S. H., S. Yamazaki, P. L. Lowrey, K. Shimomura, C. H. Ko, E. D. Buhr, S. M. Siepka, H. K. Hong, W. J. Oh, O. J. Yoo, M. Menaker, and J. S. Takahashi. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 1015339-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, W. N., N. Malinin, F. C. Yang, D. Staknis, N. Gekakis, B. Maier, S. Reischl, A. Kramer, and C. J. Weitz. 2007. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat. Cell Biol. 9268-275. [DOI] [PubMed] [Google Scholar]