Abstract
ADP-ribosylation is a reversible posttranslational modification mediated by poly-ADP-ribose polymerase (PARP). The results of recent studies demonstrate that ADP-ribosylation contributes to transcription regulation. Here, we report that transcription factor NFAT binds to and is ADP-ribosylated by PARP-1 in an activation-dependent manner. Mechanistically, ADP-ribosylation increases NFAT DNA binding. Functionally, NFAT-mediated interleukin-2 (IL-2) expression was reduced in T cells upon genetic ablation or pharmacological inhibition of PARP-1. Parp-1−/− T cells also exhibit reduced expression of other NFAT-dependent cytokines, such as IL-4. Together, these results demonstrate that ADP-ribosylation mediated by PARP-1 provides a molecular switch to positively regulate NFAT-dependent cytokine gene transcription. These results also imply that, similar to the effect of calcineurin inhibition, PARP-1 inhibition may be beneficial in modulating immune functions.
ADP-ribosylation is a reversible posttranslational modification that transfers ADP-ribose from NAD+ to Glu, Asp, and/or Arg amino acids of target proteins (18). Similar to ubiquitination, ADP-ribosylation modifies target proteins to various masses due to the assorted chain lengths of the ADP-ribose. ADP-ribosylation is inhibited by the NAD+ analog 3-aminobenzamide and, more specifically, by PJ-34 (45). Poly-ADP-ribose polymerase-1 (PARP-1) is a nuclear enzyme that accounts for the bulk of ADP-ribosylation in vivo (43). Indeed, only ∼10% of PARP activity remains in Parp-1−/− cells upon DNA damage. In addition to its role in DNA damage repair, the results of recent studies demonstrate that PARP-1 contributes to gene transcription regulation (26, 40).
Transcription factor NFAT is the master regulator of interleukin-2 (IL-2) gene transcription (24, 42). In resting cells, NFAT resides in the cytosol. The nuclear accumulation of NFAT is regulated by calcineurin-mediated dephosphorylation (9, 15, 23). The immunosuppressant drugs cyclosporine A (CsA) and tacrolimus (FK506) inhibit calcineurin and abrogate NFAT activation. Indeed, understanding the mechanism of NFAT activation has contributed to the great advances in transplantation surgery (27). Given that immunosuppressant therapy using CsA or FK506 causes neuro- and nephrotoxicity (1, 19), further understanding of the molecular basis of NFAT activation will provide alternate therapeutic targets for the treatment of transplant patients.
Once in the nucleus, NFAT interacts with coregulators to achieve optimal NFAT activation (11, 21, 28). These NFAT coregulators include Fos-Jun, C/EBPs, and Fox3p, which form a composite transcription complex to regulate NFAT-mediated gene transcription. In addition, transcription coactivator CREB-binding protein/p300 and class II histone deacetylases are recruited to modulate NFAT-mediated transcription (3, 12, 16, 48). Here, we report that PARP-1 binds to and ADP-ribosylates NFAT. The ADP-ribosylation mediated by PARP-1 provides a molecular switch to positively regulate NFAT-dependent cytokine gene transcription, including the transcription of IL-2. Hence, PARP-1 inhibition may be beneficial in modulating immune functions.
MATERIALS AND METHODS
Mice.
Animal experiments were performed in accordance with the guidelines of the Albert Einstein College of Medicine Institute of Animal Studies. Parp-1−/− mice were obtained from the Jackson Laboratory and were back-crossed with C57BL/6 mice at least nine times before use. Back-crossed Parp-1−/− mice were used for the isolation of mouse embryonic fibroblasts (MEFs), as well as primary T cells.
Reagents.
Antibodies for NFATc4 (sc13036), NFATc2 (sc7295 and sc7296), fibrillin (sc7540), PARP-1 (sc7150, sc8007, and sc25780), CD3 (553295), and CD28 (553058) were obtained from Santa Cruz Biotech or Pharmingen. Tubulin antibody was obtained from the monoclonal antibody facility at the University of Iowa. The PARP-1 inhibitors PJ-34 and 3-aminobenzamide, calcineurin inhibitor CsA, recombinant PARP-1, and NAD+ were obtained from Calbiochem, Sigma, and/or Fisher Scientific. [32P]NAD+ was obtained from Amersham.
Cell culture.
Primary MEF cells were isolated from embryonic-day-13.5 embryos after trypsin digestion as described previously (49). Primary MEFs were used for experiments within the first three passages. Naïve CD4+ T cells were isolated from lymph nodes and spleens of 4- to 6-week-old control C57BL/6 and Parp-1−/− mice as described previously (29). Th1 cells were differentiated for 7 days in the presence of mouse IL-12 (10 ng/ml), anti-mouse IL-4 (10 μg/ml), and mouse IL-2 (10 units/ml). Cells were challenged with ionomycin (2 μM) and phorbol ester (100 nM) or anti-mouse CD3 (0.12 to 0.5 μg/ml) plus anti-mouse CD28 (0.12 to 0.5 μg/ml) as indicated in the figures. MEFs, as well as COS and HEK293 cells, were cultured in Dulbecco modified Eagle medium. Jurkat T cells were cultured in RPMI medium. All media were supplemented with 10% fetal calf serum, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) (Invitrogen). Cells were transfected by using Lipofectamine (Invitrogen).
Binding assays.
Biotinylated NFAT DNA binding elements (biotin-ATTACAGGGAAAATATTGCCACACTGTCTC) were incubated with tissue extracts prepared from heart expressing constitutively active calcineurin (31). Therefore, endogenously expressed activated NFAT and its associated proteins were investigated. DNA binding precipitation was performed as described previously (49), and NFAT-associated proteins were visualized by Coomassie blue staining. NFAT-associated proteins were excised and subjected to in-gel digestion and proteomic analysis. Chromatin immunoprecipitation assays were performed on cross-linked DNA isolated from Jurkat T cells using NFATc2 (sc7296), PARP-1 (sc7150), and isotype-matching immunoglobulin G antibodies. Primers for the amplification of IL-2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter were, for IL-2, 5′-AGCTGACATGTAAGAAGCAATCT-3′ and 5′-TGGTTTCCTGTTTCAGAATGG-3′ and, for GAPDH, 5′-GGCTCTCTGCTCCTCCCTGTTCC-3′ and 5′-CAATGAAGGGGTCGTTGATGGC-3′. Coimmunoprecipitations were carried out using antibodies against NFATc4 (sc13036) and PARP-1 (sc7150), and enhanced chemiluminescence was performed to visualize bound NFAT and PARP-1. Gel mobility shift assays were performed as described previously (49) using nuclear extracts prepared from control C57BL/6 and Parp-1−/− cells or Jurkat T cells treated or not treated with PJ-34. Gel shift assays using recombinant proteins were performed after in vitro ADP-ribosylation of the NFATc2 DNA binding domain (DBD) (130 ng/reaction mixture) by PARP-1 (2 ng/reaction mixture; Sigma) in the presence and absence of NAD+ (0.1 μM) and/or PJ-34 (5 μM; Calbiochem). The percentage of DNA binding was determined by dividing the amount of shifted probe (NFAT-DNA complex) by the total amount of probe input (free probe plus shifted probe).
ADP-ribosylation assays.
COS cells transiently transfected with NFATc4 were labeled with [32P]NAD+. The transfected cells were rendered permeable to NAD+ by using hypotonic buffer as described previously (46). The in vivo incorporation of ADP-ribose onto transfected NFATc4 and endogenous PARP-1 was examined by immunoprecipitation and subsequent autoradiography. The in vitro ADP-ribosylation on recombinant NFATc2 DBD (13 to 130 ng/reaction mixture) was performed by using purified PARP-1 (2 ng/reaction mixture; Sigma) and NAD+ (0.1 μM to 500 μM) as described previously (30). PARP inhibitor PJ-34 (5 μM; Calbiochem) or 3-aminobenzamide (3 mM; Fisher Scientific) was preincubated with PARP-1 as indicated in the figures. The ADP-ribosylation of NFATc2 DBD and PARP-1 was examined by immunoblotting analysis and enhanced chemiluminescence. The in vitro ADP-ribosylation using [32P]NAD+ was visualized by autoradiography.
IL-2 determination.
The amount of secreted IL-2 protein was determined by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's protocol (Raybiotech). Quantitative PCR amplification was performed to assess the expression of IL-2 transcript (the primers for IL-2 were 5′-ATGTACAGGATGCAACTCCTG-3′ and 5′-CAAGTTAGTGTTGAGATGATGC-3′ and those for GAPDH were 5′-ACCTGACCTGCCGTCTAGAA-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′).
Statistical analysis.
Experiments were conducted at least three times, and all values are reported as the means ± standard errors of the means. Statistical analyses were performed by using the Student t test, and statistical significance was determined at values of P <0.05.
RESULTS
Identification of PARP-1 as an NFAT-interacting protein.
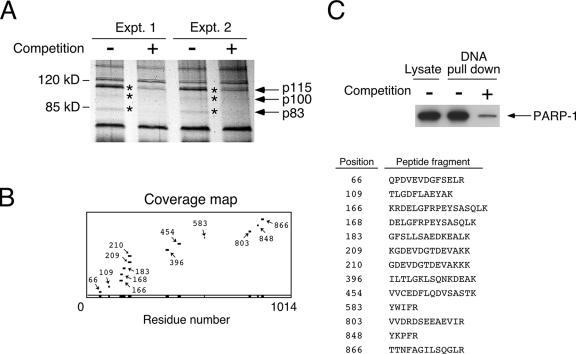
To identify novel coregulators of NFAT, we performed DNA affinity isolation using biotinylated NFAT binding elements to avidly pull down activated endogenous NFAT and its associated proteins (Fig. 1A). Protein staining indicated that several specific NFAT-interacting proteins were present in NFAT-DNA precipitates. Competition with nonbiotinylated NFAT binding oligonucleotides reduced the binding of these proteins. Mass spectrometry analysis identified the 115-kDa NFAT-interacting protein as PARP-1 (Fig. 1B). The wide spread of PARP-1 peptide fragments and the presence of conjoining peptides indicated the likelihood of PARP-1 in the DNA precipitates. Subsequent immunoblotting analysis validated the presence of PARP-1 in the DNA precipitates; PARP-1 was sensitive to competition with nonbiotinylated NFAT binding oligonucleotides (Fig. 1C). These data demonstrate that PARP-1 was present in the NFAT-DNA precipitates.
FIG. 1.
Identification of PARP-1 as an NFAT-interacting protein. (A) DNA affinity isolation of NFAT-interacting proteins using biotinylated NFAT DNA binding elements. Binding of proteins (p115, p100, and p83; marked by asterisks) was ascertained by competition with nonbiotinylated NFAT oligonucleotides and revealed by protein staining. Two representative results of the binding assays are shown (Expt. 1 and Expt. 2). Molecular size markers are shown on the left. (B) The 115-kDa NFAT-interacting protein was excised and subjected to tryptic digestion and matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis. The peptide mass obtained (with coverage of at least 12%) was searched against the NCBI protein database using ProFound software. With confidence in the 99th percentile, computation analysis unambiguously predicted that the 115-kDa NFAT-interacting protein is PARP-1. (C) DNA affinity isolation was performed using NFAT DNA binding elements. The presence of PARP-1 in the NFAT-DNA precipitates was detected by immunoblotting analysis. Competition with nonbiotinylated NFAT oligonucleotides was used to ascertain specificity. +, present; −, absent.
PARP-1 binds to the REL DBD of NFAT.
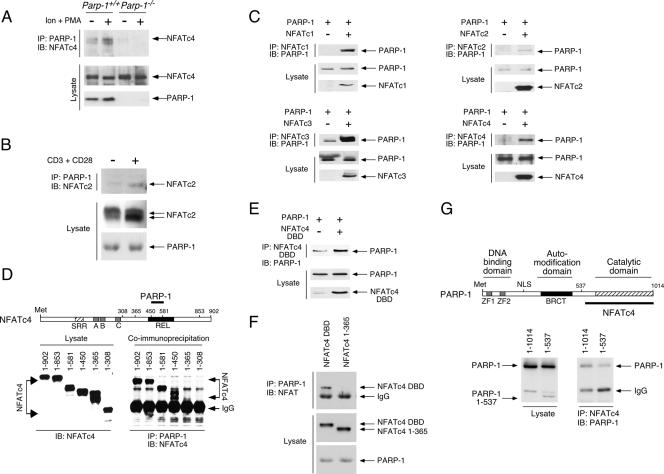
Given that PARP-1 can bind nonspecifically to DNA, we investigated whether protein-protein interaction was involved in order to ascertain PARP-1-NFAT association. We performed coimmunoprecipitation assays and demonstrated the presence of endogenous NFATc4 in the PARP-1 precipitates (Fig. 2A). Notably, NFATc4-PARP-1 interaction was potentiated by NFAT activation using the calcium ionophore ionomycin and the phorbol ester phorbol myristate acetate (PMA), which elicits NFAT nuclear translocation. Interaction between NFATc2 and PARP-1 upon T-cell activation was also observed (Fig. 2B), suggesting that PARP-1 interacts with the conserved NFAT domain. Indeed, the interaction between PARP-1 and NFAT (NFATc1, NFATc2, NFATc3, and NFATc4) was further confirmed by coexpression and subsequent coimmunoprecipitation to demonstrate the presence of PARP-1 in NFAT precipitates (Fig. 2C). We also mapped the PARP-1 binding domain on NFAT by reversed coimmunoprecipitation to detect NFAT in PARP-1 precipitates (Fig. 2D). Deletion analysis revealed that PARP-1 interacted with the REL DBD of NFATc4. The interaction between PARP-1 and the NFATc4 DBD was further confirmed by coexpression and subsequent coimmunoprecipitation (Fig. 2E and F). On the other hand, the COOH-terminal catalytic domain of PARP-1 was required for NFATc4 interaction (Fig. 2G). Together, these data demonstrate that the COOH-terminal catalytic domain of PARP-1 binds to the REL DBD of NFAT.
FIG. 2.
PARP-1 binds to the REL DBD of NFAT. (A) Parp-1+/+ and Parp-1−/− MEFs were treated (+) or not treated (−) with ionomycin and the phorbol ester PMA (Ion + PMA) for 30 min. Nuclear extracts were prepared and immunoprecipitated (IP) with PARP-1 antibody. The presence of NFATc4 in the PARP-1 precipitates was analyzed by immunoblotting analysis (IB). (B) In vitro-differentiated Th1 cells were stimulated or not stimulated with anti-mouse CD3 and anti-mouse CD28 for 2 h. Extracts were prepared and immunoprecipitated (IP) with PARP-1 antibody. The presence of NFATc2 in the PARP-1 precipitates was analyzed by immunoblotting analysis (IB). (C) Cell extracts prepared from COS cells transiently transfected with PARP-1 and/or NFAT (NFATc1, NFATc2, NFATc3, or NFATc4) were immunoprecipitated (IP) with NFAT antibody. The presence of PARP-1 in the NFAT precipitates was analyzed by immunoblotting analysis (IB). (D) Deletion analysis revealed that PARP-1 binds to the NFATc4 REL DBD. COS cells were transiently transfected with PARP-1 and/or various NFATc4 proteins (residues 1 to 902, 1 to 853, 1 to 581, 1 to 450, 1 to 365, and 1 to 308). Interaction between PARP-1 and NFATc4 proteins was examined by immunoprecipitation (IP) with PARP-1 antibody and analyzed by immunoblotting analysis (IB). SRR, Ser-rich region. (E, F) Cell extracts prepared from COS cells transiently transfected with PARP-1 and/or NFATc4 DBD (NFATc4 344-749) were immunoprecipitated (IP) with NFAT antibody (panel E). The presence of PARP-1 in NFAT precipitates was analyzed by immunoblotting analysis (IB). Reversed immunoprecipitation to detect NFATc4 DBD in PARP-1 precipitates was also performed (panel F). (G) Deletion analysis revealed that the COOH-terminal catalytic domain of PARP-1 is required to bind to NFATc4. COS cells were transiently transfected with NFATc4 and PARP-1 (1-1014) or ΔCOOH-PARP-1 (PARP-1 1-537). Interaction between PARP-1 and NFATc4 proteins was examined by immunoprecipitation (IP) with NFATc4 antibody and analyzed by immunoblotting analysis (IB). NLS, nuclear localization signal; +, present; −, absent; IgG, immunoglobulin G.
PARP-1 ADP-ribosylates NFAT DBD.
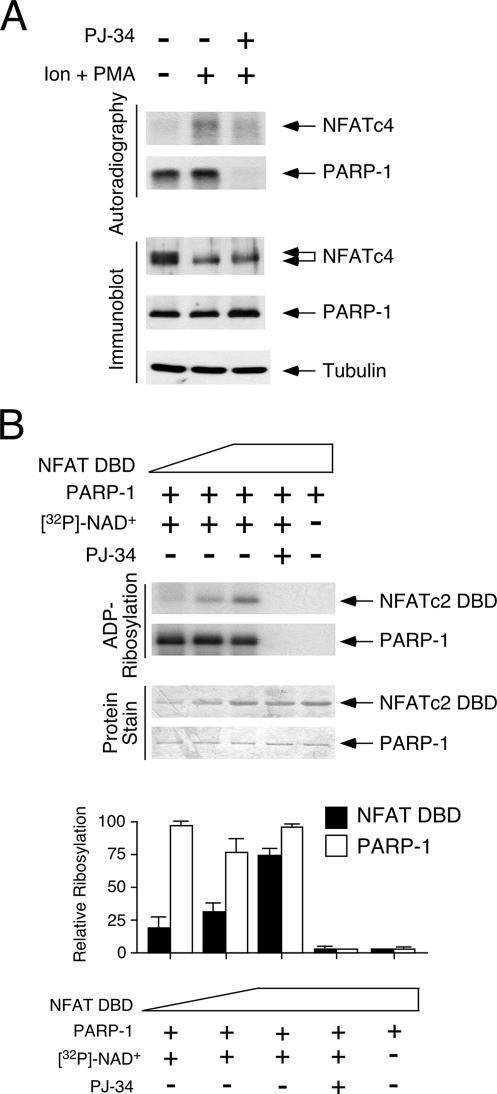
Next, we asked whether the binding of PARP-1 to the DBD promoted the ADP-ribosylation of NFAT. To test this hypothesis, we performed in vivo-labeling analysis in the presence of [32P]NAD+. The results of the in vivo-labeling studies demonstrated the incorporation of [32P]ADP-ribose onto NFATc4 and PARP-1. Notably, ADP-ribosylated NFATc4 was found only in the presence of ionomycin and the phorbol ester PMA (Fig. 3A), indicating that the ADP-ribosylation of NFATc4 is activation dependent. PARP inhibitor PJ-34, however, reduced the ADP-ribosylation of NFATc4 and PARP-1.
FIG. 3.
PARP-1 ADP-ribosylates NFAT DBD. (A) COS cells transiently transfected with NFATc4 were incubated with [32P]NAD+ for 2 h before stimulation with ionomycin and phorbol ester PMA (Ion + PMA). NFATc4 and PARP-1 in cell lysate were immunoprecipitated, and the incorporation of [32P]ADP-ribose was examined by autoradiography. The specificity of PARP-1-mediated ADP-ribosylation was ascertained by using PARP inhibitor PJ-34. Immunoblotting analysis indicated similar expression levels of NFAT and PARP-1 in the cell lysate. (B) In the presence of [32P]NAD+, increasing amounts of recombinant NFATc2 DBD protein were ADP-ribosylated by purified PARP-1 in vitro (top panel). The specificity of PARP-1-mediated ADP-ribosylation was ascertained by using PARP inhibitor PJ-34 or in the absence of NAD+. The extent of [32P]ADP-ribose incorporation in NFATc2 DBD and PARP-1 was determined by using a PhosphorImager, and the results are presented in the bottom panel. +, present; −, absent.
To demonstrate direct ADP-ribosylation of NFAT by PARP-1, we performed in vitro ADP-ribosylation assays using recombinant NFATc2 DBD and purified PARP-1 proteins. The results of the ADP-ribosylation assays demonstrated a dose-dependent increase in the incorporation of ADP-ribose into the NFATc2 DBD (Fig. 3B). A similar extent of auto-ADP-ribosylation of PARP-1 was used as a control. ADP-ribosylation on the NFATc2 DBD and PARP-1, however, was sensitive to the PARP inhibitor PJ-34 or the absence of NAD+. Notably, the extent of ADP-ribosylation was higher in PARP-1 than in the NFATc2 DBD, a result which was in agreement with the high intrinsic autoribosylation of PARP-1 (32). Together, these data demonstrate that PARP-1 catalyzes the transfer of ADP-ribose onto NFAT and itself using NAD+ as a substrate.
Conserved Glu residue on NFAT DBD is ADP-ribosylated by PARP-1.
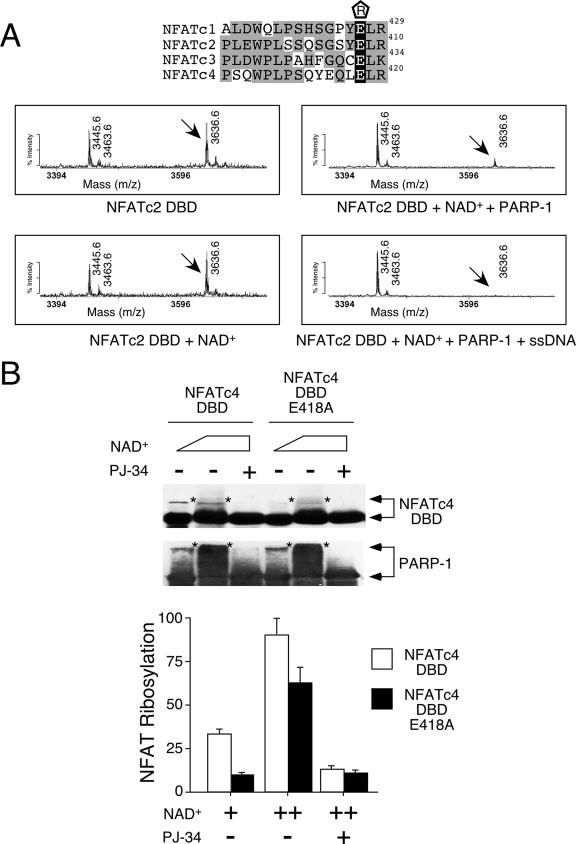
Few reports have identified target sites of PARP-1 acceptor proteins (18). We next mapped the ADP-ribosylation site on the NFAT DBD by using mass spectrometry (Fig. 4A). Unlike phosphorylation, ADP-ribosylation modifies target proteins to various masses due to the assorted chain lengths of the poly-ADP-ribose. Although similar to ubiquitination, which modifies target proteins to various extents, the moieties of ADP-ribosylation, ADP-ribose, are indigestible by trypsin, and the large carbohydrate chain can impede the detection of the modified peptide in the mass spectrometer. Hence, we determined the amounts of loss of specific NFAT peptides upon ADP-ribosylation. In comparison to its intensity in the mass spectrum obtained from the NFATc2 DBD, the intensity of peptide 3636.6 was reduced in the presence of PARP-1. Peptide 3636.6 completely disappeared upon the activation of PARP-1 using single-stranded DNA. The subsequent sequencing of peptide 3636.6 identified Glu408 of NFATc2 as a possible ADP-ribosylation target. Based on previous structural analysis (7), Glu408 is located away from the DNA contact and is exposed, providing accessibility for ADP-ribosylation. Notably, the Glu corresponding to Glu408 of NFATc2 is conserved in other NFAT members (Glu427 of NFATc1, Glu432 of NFATc3, and Glu418 of NFATc4) (Fig. 4A).
FIG. 4.
Conserved Glu residue on NFAT DBD is ADP-ribosylated by PARP-1. (A) Recombinant NFATc2 DBD was incubated with NAD+ in the presence and absence of purified PARP-1 and subjected to tryptic digestion and matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis. The effect of PARP-1 activator single-stranded DNA (ssDNA) was also determined. Target peptide 3636.6 was sequenced by tandem mass spectrometry to confirm its identity. The conserved Glu residues of NFAT members are highlighted in black in the sequence alignment above, with their positions given at the right. “R” enclosed in a pentagon above the sequences indicates ADP-ribosylation. Gray highlighting indicates similar amino acid sequences on the DBDs of different NFAT members. (B) Wild-type or Ala418 NFATc4 DBD expressed in COS cells was immunoprecipitated and incubated with increasing amounts of NAD+ in the presence of purified PARP-1. After incubation, NFAT DBD and PARP-1 were analyzed by immunoblotting analysis. The extent of ADP-ribosylation was revealed by the reduction in electrophoretic mobility of NFAT DBD and PARP-1 (top panel, indicated by asterisks). PARP-1 inhibitor PJ-34 was used to demonstrate specificity. The extent of [32P]ADP-ribose incorporation in NFATc4 DBD and PARP-1 was determined by using a PhosphorImager, and the results are presented in the bottom panel. +, present; ++, higher concentration present; −, absent.
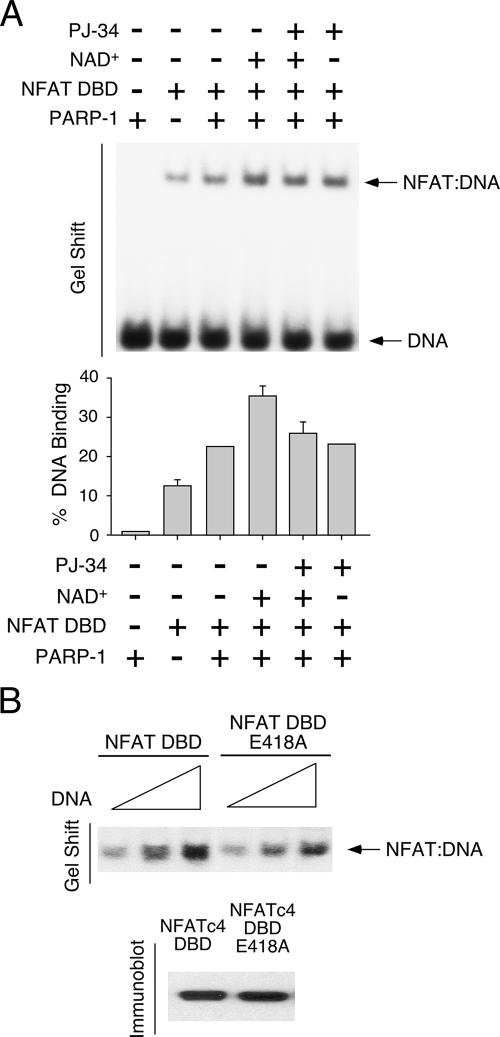
To ascertain the ADP-ribosylation site on NFAT, we performed site-directed mutagenesis to replace the conserved Glu with Ala. Since the recombinant E408A NFATc2 DBD was not soluble, we expressed wild-type and Ala418 NFATc4 DBD in COS cells, immunoprecipitated them, and tested to determine whether ADP-ribosylation was affected by using recombinant PARP-1 (Fig. 4B). The results of in vitro ADP-ribosylation assays demonstrated reduced ADP-ribosylation upon Ala replacement of Glu418 of NFATc4. Ala418 NFATc4 remained ADP-ribosylated at high concentrations of NAD+, suggesting the presence of additional target sites. Together, these data demonstrate that PARP-1 ribosylates Glu418 of NFATc4.
PARP-1 regulates NFAT DNA binding.
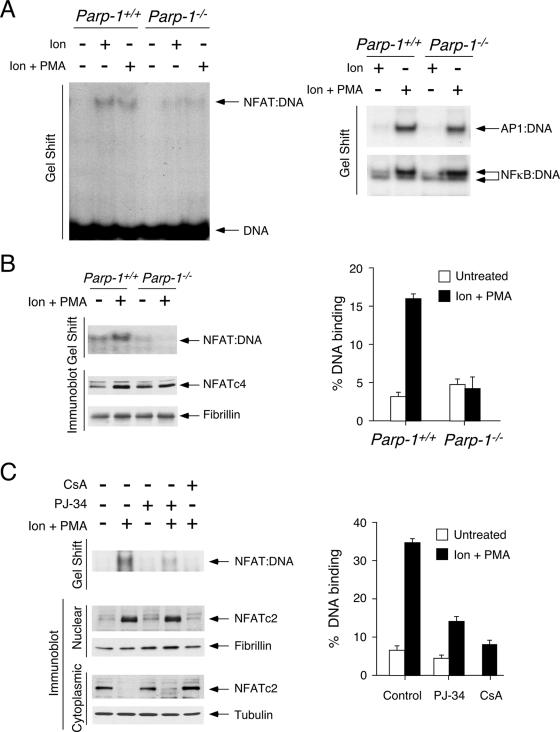
Given that the interaction between PARP-1 and NFAT requires the DBD (Fig. 2D and E), ADP-ribosylation at the REL DBD might modulate NFAT DNA binding. We performed gel mobility shift assays to test this hypothesis. The formation of the NFAT-DNA complex was evident upon stimulation with ionomycin or ionomycin plus PMA using nuclear extract isolated from Parp-1+/+ T cells (Fig. 5A). The formation of the NFAT-DNA complex, however, was reduced in the absence of PARP-1 using nuclear extract isolated from Parp-1−/− T cells. The DNA binding of AP-1 and NF-κB was similar, indicating the specific effect on NFAT in activated Parp-1−/− T cells. The results of gel mobility shift assays also demonstrated reduced NFAT DNA binding in Parp-1−/− MEFs (Fig. 5B). A similar amount of nuclear NFATc4 was detected in Parp-1−/− and Parp-1+/+ MEFs, however, indicating that the subcellular distribution of NFATc4 was not affected by the ablation of PARP-1.
FIG. 5.
PARP-1 regulates NFAT DNA binding. (A) Parp-1+/+ and Parp-1−/− Th1 cells were treated or not treated with ionomycin (Ion) or ionomycin and PMA (Ion + PMA), and prepared nuclear extracts were incubated with 32P-labeled oligonucleotides containing binding sites for NFAT, AP-1, or NF-κB. Gel mobility shift assays were performed to determine the formation of protein-DNA complexes. (B) Nuclear extracts prepared from Parp-1+/+ and Parp-1−/− MEFs were incubated with 32P-labeled NFAT. Gel mobility shift assays demonstrated reduced formation of NFAT-DNA complex in Parp-1−/− extracts. Immunoblotting analysis indicated similar amounts of NFAT proteins in the nuclear extracts. The expression level of nuclear protein fibrillin was used as loading control. The amounts of NFAT-DNA complexes were quantified by using a PhosphorImager. (C) Jurkat T cells were treated or not treated with PARP inhibitor PJ-34 for 2 h before stimulation with ionomycin and phorbol ester PMA (Ion + PMA) for 30 min. Gel mobility shift assays demonstrated reduced formation of NFAT-DNA complexes in PJ-34-treated nuclear extracts. Immunoblot analysis indicated similar amounts of NFAT in cytoplasmic or nuclear fraction in cells treated or not treated with PJ-34. CsA was used as control to block nuclear translocation of NFAT and, hence, formation of NFAT-DNA complex. The amounts of NFAT-DNA complexes were quantified by using a PhosphorImager. +, present; −, absent.
We further confirmed the role of ADP-ribosylation on NFAT DNA binding, but not subcellular distribution, by using PARP inhibitor PJ-34 (Fig. 5C). Jurkat T cells were pretreated or not with PJ-34 and challenged with and without ionomycin and PMA, and the cytoplasmic and nuclear fractions were prepared for immunoblotting and gel mobility shift assays. The results of the immunoblotting analysis indicate that the calcineurin-dependent nuclear translocation of NFATc2 was not affected by PARP inhibition (Fig. 5C). In the presence and absence of PJ-34, similar amounts of NFATc2 were found in the cytosolic fraction at resting state and in the nuclear extracts upon activation (Fig. 5C). The formation of the NFAT-DNA complex, however, was reduced upon PARP inhibition (Fig. 5C). The results for the control indicated that the inhibition of calcineurin function by using CsA blocked NFATc2 nuclear translocation and, thus, abolished the formation of the NFAT-DNA complex. These data demonstrate that the inhibition of PARP activity with PJ-34 reduced the DNA binding, but not the subcellular distribution, of NFAT.
ADP-ribosylation regulates NFAT DNA binding.
Next, we asked whether the ADP-ribosylation mediated by PARP-1 affects the NFAT DNA binding (Fig. 6A). Recombinant NFATc2 DBD was incubated with 32P-labeled NFAT DNA binding element in the presence and absence of purified PARP-1 and/or NAD+. The results of gel mobility shift assays indicate that purified PARP-1 did not form a complex with the NFAT DNA binding element. The presence of PARP-1 protein, however, increased NFAT DNA binding. Notably, incubation with NAD+ further increased the formation of NFAT-DNA complexes. Treatment with PJ-34, however, abolished the effect of PARP-1. In addition, wild-type and Ala418 NFATc4 DBD were expressed in COS cells and tested to determine whether ADP-ribosylation affected DNA binding. The results of gel mobility shift assays indicated that Ala replacement of Glu418 reduced NFATc4 DNA binding (Fig. 6B), supporting the idea that ADP-ribosylation potentiates NFAT DNA binding. Together, these data demonstrate that ADP-ribosylation promotes NFAT-DNA interaction.
FIG. 6.
ADP-ribosylation regulates NFAT DNA binding. (A) Recombinant NFATc2 DBD was incubated with 32P-labeled NFAT DNA binding element in the presence and absence of purified PARP-1 proteins for gel mobility shift assays. Effect of NAD+ and PJ-34 on the formation of NFAT-DNA complex was also examined. The amounts of NFAT-DNA complexes were quantified by using a PhosphorImager. (B) Wild-type or Ala418 NFATc4 DBD expressed in COS cells was incubated with various amounts of 32P-labeled NFAT DNA binding element (100, 150, and 200 fmol) for gel mobility shift assays. Similar amounts of wild-type or Ala418 NFATc4 DBD in gel shift assays were also shown. +, present; −, absent.
PARP-1 contributes to NFAT-dependent gene transcription.
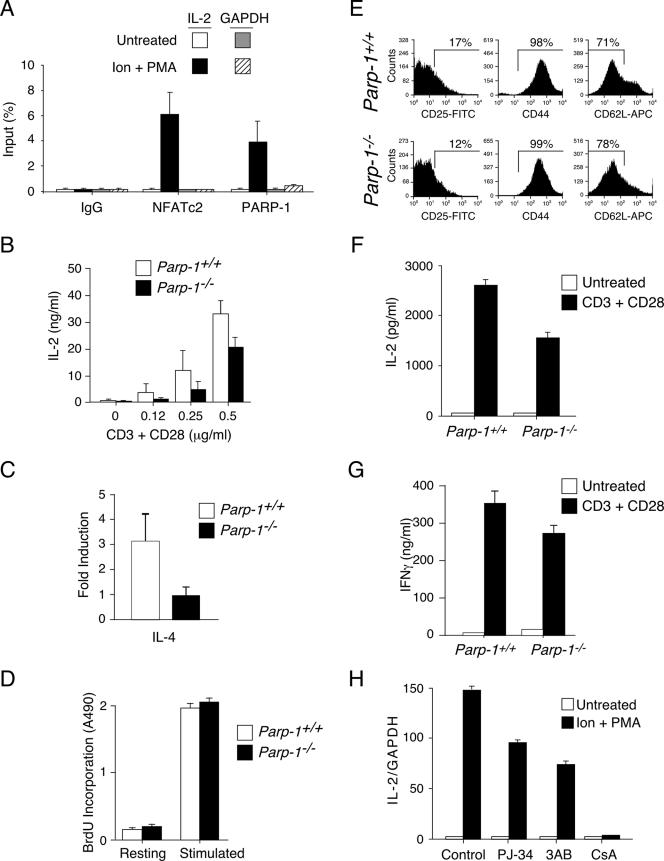
The results of previous studies establish that NFAT is the master regulator of IL-2 gene transcription (24, 42). Targeted disruption of NFAT or the administration of CsA to inhibit NFAT activation reduces IL-2 expression (8, 14, 24, 29, 34, 38, 42). Therefore, NFAT is required for IL-2 gene transcription. We next asked whether PARP-1 is associated with the IL-2 transcription loci. We addressed this question by performing chromatin immunoprecipitation assays using NFATc2 and PARP-1 antibodies (Fig. 7A). The results of quantitative PCR amplification revealed activation-dependent recruitment of NFATc2 to the IL-2 transcription loci. Similarly, PARP-1 was recruited to the same IL-2 transcription loci upon activation. These data demonstrate that PARP-1 and NFATc2 are recruited to the IL-2 transcription loci upon T-cell activation.
FIG. 7.
PARP-1 contributes to NFAT-dependent gene transcription. (A) Recruitment of PARP-1 to IL-2 transcription loci upon T-cell activation. Chromatin immunoprecipitation was performed with Jurkat T cells to demonstrate binding of NFATc2 and PARP-1 in the IL-2 transcription loci upon stimulation with ionomycin and PMA (Ion + PMA). Immunoprecipitation with isotype-matched immunoglobulin G (IgG) was used as control. (B to D) Naïve CD4+ T cells isolated from Parp-1+/+ and Parp-1−/− mice were stimulated or not with anti-mouse CD3 and anti-mouse CD28 (0.12 to 0.5 μg/ml) for 12 h. The amounts of IL-2 secreted were determined by ELISA and are presented in panel B. Quantitative PCR was performed to determine the level of expression of IL-4 in naïve T cells upon stimulation. The expression of IL-4 was normalized to that of GAPDH, and the level of induction is presented in panel C. The extent of T-cell proliferation before (Resting) and after stimulation is indicated in panel D. BrdU, bromodeoxyuridine. (E to G) In vitro-differentiated Th1 cells isolated from Parp-1+/+ and Parp-1−/− mice were stained with antibodies against CD25, CD44, and CD62L and analyzed by fluorescence-activated cell sorting (E). FITC, fluorescein isothiocyanate. Parp-1+/+ and Parp-1−/− Th1 cells were stimulated or not stimulated for IL-2 (F) and IFN-γ (G) production with anti-mouse CD3 and anti-mouse CD28 for 24 h, and production levels were determined by ELISA. (H) Jurkat T cells were stimulated with ionomycin and PMA (Ion + PMA) in the presence or absence of PARP-1 inhibitors (PJ-34 or 3-aminobenzamide [3AB]) for 1 h before isolation of total RNA for cDNA synthesis. Quantitative PCR was performed to determine the expression of IL-2 transcripts. The level of expression of GAPDH was used as control. The effect of CsA on IL-2 expression is also shown.
Given that PARP-1, which accounts for the majority of ADP-ribosylation in vivo (43), is recruited to the IL-2 gene loci (Fig. 7A) and that ADP-ribosylation increased NFAT DNA binding (Fig. 5), we asked whether naïve CD4+ T cells isolated from Parp-1−/− mice exhibited defects in IL-2 expression. The results of ELISA assays indicated that T-cell activation elicited by anti-mouse CD3 and anti-mouse CD28 increased IL-2 production in Parp-1+/+ naïve CD4+ T cells in a dose-dependent manner (Fig. 7B). The production of IL-2, however, was reduced in Parp-1−/− naïve CD4+ T cells. Reduced IL-2 production was also found in splenocytes freshly isolated from Parp-1−/− mice (data not shown). In addition to IL-2, NFAT regulates other cytokines in T cells, including IL-4 and gamma interferon (IFN-γ). Similar to that of IL-2, IL-4 and IFN-γ expression levels are reduced in NFAT-null mice or upon CsA treatment (5, 20, 38, 39, 44). Indeed, the induction of IL-4 mRNA was reduced in Parp-1−/− naïve CD4+ T cells (Fig. 7C). The extent of T-cell proliferation in Parp-1+/+ and Parp-1−/− naïve CD4+ T cells, however, was similar after stimulation (Fig. 7D), indicating that the difference in IL-2 and IL-4 production was intrinsic and was not due to changes in T-cell numbers in Parp-1+/+ and Parp-1−/− mice. These data indicate that PARP-1 contributes to NFAT-dependent gene transcription.
Next, we extended the role of PARP-1 in NFAT-dependent gene transcription in immune cells by using in vitro-differentiated Th1 cells. The results of cell-sorting analysis indicated that the expression of Th1 cell markers was similar in Parp-1+/+ and Parp-1−/− Th1 cells (Fig. 7E). The expression of IL-2, however, was reduced in Parp-1−/− Th1 cells (Fig. 7F). The expression of the Th1 cytokine IFN-γ was also reduced in Parp-1−/− Th1 cells (Fig. 7G). These data support the idea that PARP-1 contributes to NFAT-dependent gene transcription.
In addition to the genetic approach in determining IL-2 gene expression in Parp-1+/+ and Parp-1−/− cells, we further ascertained the role of ADP-ribosylation in IL-2 gene expression by using PARP inhibitors (Fig. 7H). The results of quantitative PCR analysis reveal that the expression of IL-2 mRNA in Jurkat T cells was increased upon stimulation with ionomycin and PMA. The administration of PARP inhibitor PJ-34 or 3-aminobenzamide reduced IL-2 gene expression. The results for the control indicated that the administration of CsA blocked IL-2 expression. These data confirm that PARP-1 positively regulates IL-2 gene expression.
DISCUSSION
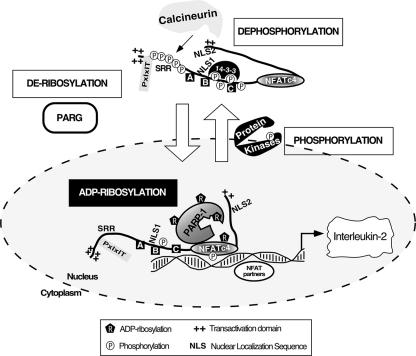
In this report, we have demonstrated that PARP-1 binds to and ADP-ribosylates NFAT in an activation-dependent manner. The results of genetic-ablation and pharmacological-inhibition approaches demonstrate that ADP-ribosylation regulates NFAT DNA binding and IL-2 gene expression. Hence, in addition to phosphorylation, ADP-ribosylation provides another layer of regulation of NFAT-dependent gene transcription. Notably, similar to phosphorylation, ADP-ribosylation is reversible, and the hydrolysis of ADP-ribose is mediated by PARG (4). Interestingly, cytoplasmic PARG accounts for most of the cellular PARG activity (18). Given the compartmentalization of the molecules involved in NFAT phosphorylation and ADP-ribosylation, it is tempting to speculate that in the cytosol, calcineurin mediates dephosphorylation and promotes NFAT nuclear accumulation, where nuclear NFAT is ADP-ribosylated by PARP-1 to increase avidity toward DNA (Fig. 8). Upon transcription termination, ADP-ribosylated NFAT is phosphorylated by nuclear NFAT kinases and returns to the cytosol, where PARG promotes the hydrolysis of ADP-ribose. One question remaining is whether dephosphorylation is a prerequisite of subsequent ADP-ribosylation of NFAT, especially since calcium and extracellular signal-regulated kinase also contribute to PARP-1 activation (10, 22). Conversely, whether phosphorylation is necessary in order to recruit PARG to hydrolyze the ADP-ribose on NFAT remains elusive. This causal relationship is intriguing and may provide fine-tuning of NFAT-mediated gene transcription.
FIG. 8.
Schematic illustration of PARP-1 in NFAT signaling pathway. At resting state, basal phosphorylation is required to maintain cytosolic localization of NFAT. Upon an increase in intracellular calcium, activated calcineurin dephosphorylates NFAT to promote nuclear translocation. Once in the nucleus, NFAT interacts with NFAT partners to mediate gene transcription. Here, we demonstrate that PARP-1 acts as coregulator of NFAT. Interaction with PARP-1 promotes ADP-ribosylation of NFAT, which increases avidity of NFAT DNA binding, to regulate gene transcription. Transcription termination requires rephosphorylation of NFAT, which is mediated by multiple protein kinases. Similar to phosphorylation, ADP-ribosylation is reversible, and hydrolysis of ADP-ribose is mediated by PARG. Given that cytoplasmic PARG accounts for most of the cellular PARG activity, it is tempting to speculate that compartmentalization of the molecules involved in NFAT phosphorylation (protein kinases and calcineurin phosphatase) and ADP-ribosylation (PARP-1 and PARG) provides fine-tuning of NFAT-mediated gene transcription. SRR, Ser-rich region; boxes A, B, and C, Ser-Pro motifs; 14-3-3, phospho-Ser binding protein; PxIxIT, calcineurin docking site.
Optimal NFAT-mediated gene transcription requires the integration of two signaling pathways—activation of the calcium/calcineurin phosphatase and Ras/Raf/mitogen-activated protein kinase (MAPK) signaling. In addition to DNA damage, calcium and Ras/Raf/MAPK signaling have recently been shown to activate PARP-1. The calcium induction of PARP-1 is mediated by calmodulin-dependent protein kinase II, which phosphorylates PARP-1 (25). The extracellular signal-regulated kinase group of the MAPK family also phosphorylates PARP-1 (10). Given the activation-dependent recruitment of PARP-1 to NFAT transcription loci, it is tempting to speculate that the integration of calcium and Ras/Raf/MAPK coordinates the recruitment of PARP-1 in NFAT-mediated gene transcription.
We have demonstrated that PARP-1 ADP-ribosylates conserved Glu residues on NFAT (Glu427 of NFATc1, Glu408 of NFATc2, Glu432 of NFATc3, and Glu418 of NFATc4). Based on previous structural analysis (7), the Glu residue is exposed and lies in the NH2-terminal half of the NFAT DBD. Interestingly, there is a large surface at the NH2-terminal half of the NFAT DBD to make contact with an NFAT partner, such as Fos-Jun. Given that the NFAT partner forms a ternary complex with NFAT, ADP-ribosylation of NFAT at this interface may provide additional charge-charge interaction and/or hydrophobic stacking to stabilize the DNA binding. Alternatively, the ADP-ribose may ensure DNA binding by stabilizing the conformation of dephosphorylated NFAT, which is likely reconfigured to expose the nuclear localization sequence for nuclear import. Nonetheless, extended incorporation and branching of the ADP-ribose polymers may interfere with the ternary complex formation and disrupt the cooperation between NFAT and NFAT partners. Indeed, we found that the exogenous expression of PARP-1 inhibits transient NFAT reporter gene transcription and DNA binding (data not shown). Hence, understanding the physiological relevancy of PARP-1 will require investigation of the endogenous expression of specific gene targets by using PARP-1 inhibitors or Parp-1+/+ and Parp-1−/− mice.
The NFAT group of transcription factors was first identified as a critical component in cytokine gene expression upon T-cell activation (11, 21). In additional to the established role of NFAT in cytokine gene regulation in immune cells (33-37, 41, 47), targeted disruption of the calcineurin-regulated NFAT members has further illuminated the role of NFAT in multiple biological processes, including cardiac morphogenesis (6, 13, 31) and neural pathfinding (17). Recently, we have extended the role of NFAT in adipokine gene transcription in adipocytes. Similarly, PARP-1 contributes to multiple aspects of pathophysiological regulation, including inflammatory disease, ischemia-reperfusion injury, diabetes pathogenesis, arthritis, myocardial injury, etc. (18, 40, 45). Given their diverse roles in multiple tissues, future studies using NFAT- and PARP-null mice to investigate the transcription cooperation between NFAT and PARP-1 are warranted.
In conclusion, we have demonstrated that the ADP-ribosylation mediated by PARP-1 provides a molecular switch to positively regulate NFAT-dependent cytokine gene transcription. The integration of ADP-ribosylation (PARP versus PARG) and phosphorylation (NFAT kinases versus calcineurin) would affect the duration of nuclear localization, avidity of DNA binding, and/or recruitment of other transcription cofactors in NFAT-dependent gene transcription. Such a combinatorial effect may implement a dosage-dependent regulation on the expression of distinct NFAT targets in response to different extracellular stimuli (2, 50).
Acknowledgments
We thank members of our laboratories for their critical reading of the manuscript. We are grateful for the assistance from the Laboratory of Macromolecular Analysis and Proteomics (AECOM) on mass spectrometry analysis.
O.A.O. and N. S.-N. are trainees sponsored by NIH training grants 1F31-GM66607 and 5T32-GM07491, respectively. This research is supported, in part, by grants from the National Institutes of Health (C.-W.C. and F. M.), American Diabetes Association (C.-W.C.), and American Heart Association (C.-W.C.).
Footnotes
Published ahead of print on 25 February 2008.
REFERENCES
- 1.Andoh, T. F., and W. M. Bennett. 1998. Chronic cyclosporine nephrotoxicity. Curr. Opin. Nephrol. Hypertens. 7265-270. [DOI] [PubMed] [Google Scholar]
- 2.Arron, J. R., M. M. Winslow, A. Polleri, C. P. Chang, H. Wu, X. Gao, J. R. Neilson, L. Chen, J. J. Heit, S. K. Kim, N. Yamasaki, T. Miyakawa, U. Francke, I. A. Graef, and G. R. Crabtree. 2006. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 441595-600. [DOI] [PubMed] [Google Scholar]
- 3.Baksh, S., H. R. Widlund, A. A. Frazer-Abel, J. Du, S. Fosmire, D. E. Fisher, J. A. DeCaprio, J. F. Modiano, and S. J. Burakoff. 2002. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol. Cell 101071-1081. [DOI] [PubMed] [Google Scholar]
- 4.Bonicalzi, M. E., J. F. Haince, A. Droit, and G. G. Poirier. 2005. Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell. Mol. Life Sci. 62739-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, P. M., J. Pimm, V. Ramassar, and P. F. Halloran. 1996. Identification of a calcium-inducible, cyclosporine sensitive element in the IFN-gamma promoter that is a potential NFAT binding site. Transplantation 61933-939. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C. P., J. R. Neilson, J. H. Bayle, J. E. Gestwicki, A. Kuo, K. Stankunas, I. A. Graef, and G. R. Crabtree. 2004. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 118649-663. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., J. N. Glover, P. G. Hogan, A. Rao, and S. C. Harrison. 1998. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 39242-48. [DOI] [PubMed] [Google Scholar]
- 8.Chow, C. W., M. Rincon, and R. J. Davis. 1999. Requirement for transcription factor NFAT in interleukin-2 expression. Mol. Cell. Biol. 192300-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clipstone, N. A., and G. R. Crabtree. 1992. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 357695-697. [DOI] [PubMed] [Google Scholar]
- 10.Cohen-Armon, M., L. Visochek, D. Rozensal, A. Kalal, I. Geistrikh, R. Klein, S. Bendetz-Nezer, Z. Yao, and R. Seger. 2007. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: a link to histone acetylation. Mol. Cell 25297-308. [DOI] [PubMed] [Google Scholar]
- 11.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.)S67-S79. [DOI] [PubMed] [Google Scholar]
- 12.Dai, Y. S., J. Xu, and J. D. Molkentin. 2005. The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment. Mol. Cell. Biol. 259936-9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Pompa, J. L., L. A. Timmerman, H. Takimoto, H. Yoshida, A. J. Elia, E. Samper, J. Potter, A. Wakeham, L. Marengere, B. L. Langille, G. R. Crabtree, and T. W. Mak. 1998. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392182-186. [DOI] [PubMed] [Google Scholar]
- 14.Emmel, E. A., C. L. Verweij, D. B. Durand, K. M. Higgins, E. Lacy, and G. R. Crabtree. 1989. Cyclosporin A specifically inhibits function of nuclear proteins involved in T cell activation. Science 2461617-1620. [DOI] [PubMed] [Google Scholar]
- 15.Fruman, D. A., C. B. Klee, B. E. Bierer, and S. J. Burakoff. 1992. Calcineurin phosphatase activity in T lymphocytes is inhibited by FK 506 and cyclosporin A. Proc. Natl. Acad. Sci. USA 893686-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Rodriguez, C., and A. Rao. 1998. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 1872031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graef, I. A., F. Wang, F. Charron, L. Chen, J. Neilson, M. Tessier-Lavigne, and G. R. Crabtree. 2003. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell 113657-670. [DOI] [PubMed] [Google Scholar]
- 18.Hassa, P. O., S. S. Haenni, M. Elser, and M. O. Hottiger. 2006. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 70789-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauben, M. 1996. Cyclosporine neurotoxicity. Pharmacotherapy 16576-583. [PubMed] [Google Scholar]
- 20.Hodge, M. R., A. M. Ranger, F. Charles de la Brousse, T. Hoey, M. J. Grusby, and L. H. Glimcher. 1996. Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 4397-405. [DOI] [PubMed] [Google Scholar]
- 21.Hogan, P. G., L. Chen, J. Nardone, and A. Rao. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 172205-2232. [DOI] [PubMed] [Google Scholar]
- 22.Homburg, S., L. Visochek, N. Moran, F. Dantzer, E. Priel, E. Asculai, D. Schwartz, V. Rotter, N. Dekel, and M. Cohen-Armon. 2000. A fast signal-induced activation of poly(ADP-ribose) polymerase: a novel downstream target of phospholipase c. J. Cell Biol. 150293-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain, J., P. G. McCaffrey, Z. Miner, T. K. Kerppola, J. N. Lambert, G. L. Verdine, T. Curran, and A. Rao. 1993. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature 365352-355. [DOI] [PubMed] [Google Scholar]
- 24.Jain, J., P. G. McCaffrey, V. E. Valge-Archer, and A. Rao. 1992. Nuclear factor of activated T cells contains Fos and Jun. Nature 356801-804. [DOI] [PubMed] [Google Scholar]
- 25.Ju, B. G., D. Solum, E. J. Song, K. J. Lee, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 2004. Activating the PARP-1 sensor component of the Groucho/ TLE1 corepressor complex mediates a CaMKinase IIdelta-dependent neurogenic gene activation pathway. Cell 119815-829. [DOI] [PubMed] [Google Scholar]
- 26.Kim, M. Y., T. Zhang, and W. L. Kraus. 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 191951-1967. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J., J. D. Farmer, Jr., W. S. Lane, J. Friedman, I. Weissman, and S. L. Schreiber. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66807-815. [DOI] [PubMed] [Google Scholar]
- 28.Macian, F. 2005. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 5472-484. [DOI] [PubMed] [Google Scholar]
- 29.Macian, F., F. Garcia-Cozar, S. H. Im, H. F. Horton, M. C. Byrne, and A. Rao. 2002. Transcriptional mechanisms underlying lymphocyte tolerance. Cell 109719-731. [DOI] [PubMed] [Google Scholar]
- 30.Mendoza-Alvarez, H., and R. Alvarez-Gonzalez. 2001. Regulation of p53 sequence-specific DNA-binding by covalent poly(ADP-ribosyl)ation. J. Biol. Chem. 27636425-36430. [DOI] [PubMed] [Google Scholar]
- 31.Molkentin, J. D., J. R. Lu, C. L. Antos, B. Markham, J. Richardson, J. Robbins, S. R. Grant, and E. N. Olson. 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogata, N., K. Ueda, M. Kawaichi, and O. Hayaishi. 1981. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Biol. Chem. 2564135-4137. [PubMed] [Google Scholar]
- 33.Oukka, M., I. C. Ho, F. C. de la Brousse, T. Hoey, M. J. Grusby, and L. H. Glimcher. 1998. The transcription factor NFAT4 is involved in the generation and survival of T cells. Immunity 9295-304. [DOI] [PubMed] [Google Scholar]
- 34.Peng, S. L., A. J. Gerth, A. M. Ranger, and L. H. Glimcher. 2001. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 1413-20. [DOI] [PubMed] [Google Scholar]
- 35.Ranger, A. M., L. C. Gerstenfeld, J. Wang, T. Kon, H. Bae, E. M. Gravallese, M. J. Glimcher, and L. H. Glimcher. 2000. The nuclear factor of activated T cells (NFAT) transcription factor NFATp (NFATc2) is a repressor of chondrogenesis. J. Exp. Med. 1919-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranger, A. M., M. Oukka, J. Rengarajan, and L. H. Glimcher. 1998. Inhibitory function of two NFAT family members in lymphoid homeostasis and Th2 development. Immunity 9627-635. [DOI] [PubMed] [Google Scholar]
- 37.Rengarajan, J., B. Tang, and L. H. Glimcher. 2002. NFATc2 and NFATc3 regulate T(H)2 differentiation and modulate TCR-responsiveness of naive T(H)cells. Nat. Immunol. 348-54. [DOI] [PubMed] [Google Scholar]
- 38.Rooney, J. W., M. R. Hodge, P. G. McCaffrey, A. Rao, and L. H. Glimcher. 1994. A common factor regulates both Th1- and Th2-specific cytokine gene expression. EMBO J. 13625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooney, J. W., T. Hoey, and L. H. Glimcher. 1995. Coordinate and cooperative roles for NF-AT and AP-1 in the regulation of the murine IL-4 gene. Immunity 2473-483. [DOI] [PubMed] [Google Scholar]
- 40.Schreiber, V., F. Dantzer, J. C. Ame, and G. de Murcia. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7517-528. [DOI] [PubMed] [Google Scholar]
- 41.Schuh, K., B. Kneitz, J. Heyer, F. Siebelt, C. Fischer, E. Jankevics, E. Rude, E. Schmitt, A. Schimpl, and E. Serfling. 1997. NF-ATp plays a prominent role in the transcriptional induction of Th2-type lymphokines. Immunol. Lett. 57171-175. [DOI] [PubMed] [Google Scholar]
- 42.Shaw, J. P., P. J. Utz, D. B. Durand, J. J. Toole, E. A. Emmel, and G. R. Crabtree. 1988. Identification of a putative regulator of early T cell activation genes. Science 241202-205. [DOI] [PubMed] [Google Scholar]
- 43.Shieh, W. M., J. C. Ame, M. V. Wilson, Z. Q. Wang, D. W. Koh, M. K. Jacobson, and E. L. Jacobson. 1998. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J. Biol. Chem. 27330069-30072. [DOI] [PubMed] [Google Scholar]
- 44.Szabo, S. J., J. S. Gold, T. L. Murphy, and K. M. Murphy. 1993. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc. Mol. Cell. Biol. 134793-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Virag, L., and C. Szabo. 2002. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 54375-429. [DOI] [PubMed] [Google Scholar]
- 46.Walker, J., and C. K. Pearson. 1981. NAD+, ADP-ribosylation and transcription in permeabilized mammalian cells. Biochem. J. 199813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xanthoudakis, S., J. P. Viola, K. T. Shaw, C. Luo, J. D. Wallace, P. T. Bozza, D. C. Luk, T. Curran, and A. Rao. 1996. An enhanced immune response in mice lacking the transcription factor NFAT1. Science 272892-895. [DOI] [PubMed] [Google Scholar]
- 48.Yang, T., R. J. Davis, and C. W. Chow. 2001. Requirement of two NFATc4 transactivation domains for CBP potentiation. J. Biol. Chem. 27639569-39576. [DOI] [PubMed] [Google Scholar]
- 49.Yang, T. T., Q. Xiong, I. A. Graef, G. R. Crabtree, and C. W. Chow. 2005. Recruitment of the extracellular signal-regulated kinase/ribosomal S6 kinase signaling pathway to the NFATc4 transcription activation complex. Mol. Cell. Biol. 25907-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, X. Y., T. T. Yang, W. Schubert, S. M. Factor, and C. W. Chow. 2007. Dosage-dependent transcriptional regulation by the calcineurin/NFAT signaling in developing myocardium transition. Dev. Biol. 303825-837. [DOI] [PubMed] [Google Scholar]