Abstract
The Forkhead transcription factor FoxM1 is an important regulator of gene expression during the G2 phase. Here, we show that FoxM1 transcriptional activity is kept low during G1/S through the action of its N-terminal autoinhibitory domain. We found that cyclin A/cdk complexes are required to phosphorylate and activate FoxM1 during G2 phase. Deletion of the N-terminal autoinhibitory region of FoxM1 generates a mutant of FoxM1 (ΔN-FoxM1) that is active throughout the cell cycle and no longer depends on cyclin A for its activation. Mutation of two cyclin A/cdk sites in the C-terminal transactivation domain leads to inactivation of full-length FoxM1 but does not affect the transcriptional activity of the ΔN-FoxM1 mutant. We show that the intramolecular interaction of the N- and C-terminal domains depends on two RXL/LXL motifs in the C terminus of FoxM1. Mutation of these domains leads to a similar gain of function as deletion of the N-terminal repressor domain. Based on these observations we propose a model in which FoxM1 is kept inactive during the G1/S transition through the action of the N-terminal autorepressor domain, while phosphorylation by cyclin A/cdk complexes during G2 results in relief of inhibition by the N terminus, allowing activation of FoxM1-mediated gene transcription.
FoxM1 is a transcription factor of the Forkhead family. It is also known in the literature as Trident (in the mouse) (3), HFH-11 (in humans) (18), WIN (in the rat) (17), or MPP-2 (partial human cDNA) (13). FoxM1 is expressed ubiquitously in all embryonic tissues, while in adults its expression is only observed in actively proliferating tissues (3, 18). Disruption of the mouse FoxM1 gene results in various organ defects due to the lack of progenitor cell proliferation (2-5, 12). Moreover, FoxM1 controls expression of a subset of genes in the G2/M-specific gene cluster, among which are essential regulators of mitosis (5, 11). Consistently, the inhibition of FoxM1-mediated gene expression results in pleiotropic cell cycle defects, including severe delay in mitotic entry, chromosome missegregation, and polyploidization (5, 11, 16). In addition, FoxM1-deficient primary mouse embryonic fibroblasts (MEFs) display dramatic changes in chromosome numbers and premature senescence, suggesting that, in addition to promoting cell cycle progression, FoxM1 is also required to maintain chromosomal stability (5).
FoxM1 protein levels vary during cell cycle progression. Both FoxM1 mRNA and protein levels are barely detectable in quiescent cells, whereas they increase in cells stimulated to reenter the cell cycle (3). FoxM1 expression reaches maximum levels in late G1 or early S phase and is sustained at these levels throughout G2 and mitosis (3). While the earliest findings have reported that phosphorylation of FoxM1 occurs mainly during mitosis (3), others have shown that phosphorylation of FoxM1 is initiated by cyclin-cdk complexes in early G1 and continues during the G2 and M phases of the cell cycle (8). FoxM1 associates with cyclin E-cdk2 complexes in the G1 and S phases of the cell cycle, whereas it preferentially binds the cyclin B-cdk1 complex in the G2 phase (8). Moreover, Raf/MEK/mitogen-activated protein kinase signaling can stimulate FoxM1 nuclear translocation through phosphorylation in late S phase (7). This suggest that phosphorylation of FoxM1 involves multiple regulatory kinases at different stages of the cell cycle. In addition, FoxM1 interacts with the cell cycle-inhibitory pocket protein pRb and the cdk-activating phosphatase Cdc25B in G1 and in G1/S, respectively (8). These proteins are important cell cycle regulators and might regulate FoxM1 transcriptional activity via their effects on cdk activity. Remarkably, although FoxM1 is expressed as early as late G1, many of its target genes are only induced in G2 (5). Therefore, FoxM1 activity must somehow be inhibited during early S phase and mitotic exit in order to prevent the unscheduled expression of mitotic proteins in S and G1. However, the mechanism responsible for this level of regulation is poorly understood. Here, we set out to study this aspect of FoxM1 function through analysis of the regulation of FoxM1 at the posttranscriptional level during an ongoing cell cycle. We show that the N-terminal region of FoxM1, shown to act as an autorepressor domain (9, 14), is involved in cell cycle-dependent inactivation of FoxM1 during G1 and S phases. Increased FoxM1 transcriptional activity occurs specifically during G2 and is dependent on active cyclin A/cdk complexes. We show that a truncated form of FoxM1 protein that is lacking the N-terminal autorepressor domain (ΔN-FoxM1) is constitutively active throughout the cell cycle and no longer requires cyclin A for its activation. Similarly, mutating two RXL/LXL motifs in the C terminus of FoxM1 disrupts the intramolecular interaction between N- and C-terminal domains, causing this mutant to behave like ΔN-FoxM1. Our data strongly suggest that cyclin A/cdk-mediated phosphorylation of FoxM1 is involved in relieving the repressive function that resides within the N-terminal region of FoxM1. These findings provide a new mechanism of regulation of transcriptional activity of FoxM1 during the cell cycle.
MATERIALS AND METHODS
Cell culture, transfections, and drugs.
U2OS cells, 293-T cells, immortalized MEFs, and FoxM1-ER-inducible cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics. Thymidine, nocodazole, and MG132 were added at final concentrations of 2.5 mM, 250 ng/ml, and 5 μM, respectively, and were all purchased from Sigma. Cells were transfected with plasmid DNA using the standard calcium phosphate transfection protocol. Small inhibitory RNA (siRNA) oligonucleotides were transfected with HiPerFect (Qiagen) following the manufacturer's protocol.
Antibodies.
Rabbit anti-Cdk2 (sc-163), mouse anti-Cdk1 (sc-54), rabbit anti-cyclin A (sc-751), mouse anti-cyclinB1 (sc-245), goat antiactin (sc-1616), and rabbit anti-FoxM1 MPP-C20 (sc-502), MPP-K19 (sc-500), and MPP-H300 (sc-13016) antibodies were all purchased from Santa Cruz Biotechnology. Mouse anti-flag-M2 (f3165), anti-flag-agarose beads (A2220), and mouse anti-α-tubulin (t5168) were from Sigma. HA-11 affinity matrix (AFC-101P) and mouse anti-hemagglutinin (anti-HA; mms-101p) were purchased from Covance. Rabbit anti-CENP-F (ab5) and antiphosphothreonine (9381) were from Abcam and Cell Signaling, respectively. The following secondary antibodies were used: peroxidase-conjugated goat anti-rabbit, goat anti-mouse, and rabbit anti-goat antibodies were from DAKO, and chicken anti-mouse/Alexa 488 and goat anti-rabbit/Alexa 568 were from Molecular Probes.
Plasmids and oligonucleotides.
The 6xDB and CENP-F luciferase reporters have been described previously (5). HA-Nt-FoxM1, glutathione S-transferase (GST), GST-Nt-FoxM1, and GST-Ct-FoxM1 constructs were from W. Korver, and corresponding proteins were purified from bacterial cultures. Full-length human FoxM1 and ΔN-FoxM1 mutant were gifts from B. Luscher. FoxM1AAA (T600A/T611A/S638A), FoxM1T600A, FoxM1T611A, FoxM1T600A/T611A, FoxM1R716A, and FoxM1 R716A/L722A were obtained by site-directed mutagenesis in both full-length FoxM1 and ΔN-FoxM1. RNA interference (RNAi)-insensitive expression constructs were also obtained by site-directed mutagenesis. Correctly mutated plasmids were identified through direct sequence analysis. Expression constructs for cyclinB1, empty pSuper, and pS-cyclinB1 were described previously (5). Short hairpin RNA (shRNA)-targeting vectors for cyclin A (pS-cycA) and for cdk2 (pS-cdk2) and cdk1 (pS-cdk1) were gifts from M. van Vugt and R. Wolthuis, respectively. Expression plasmids for cyclin A, DNcdk2, and DNcdk1 were kind gifts from S. van den Heuvel. Double-stranded RNA oligonucleotides used for cyclin A2 RNAi were purchased from Ambion.
Reporter assays.
Cells were transfected using the standard calcium phosphate transfection protocol. Luciferase activity was determined 48 h after transfection, using the dual luciferase kit (Promega) according to the manufacturer's instructions. Relative luciferase activity was expressed as the ratio of firefly luciferase activity to control Renilla luciferase activity.
Western blot analysis, immunoblotting, and kinase assays.
Western blot analysis was performed as described elsewhere (5). For immunoblotting and in vitro kinase assays, cells were lysed in ELB buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM EDTA, and 0.1% NP-40) and 100 μg of protein was used for immunoprecipitation with the appropriate antibody coupled to protein A/G-agarose beads. For immunoblotting, the precipitates were extensively washed, resuspended in sample buffer, boiled, and analyzed in Western blot assays. For kinase assays, the immunoprecipitates were extensively washed and incubated in kinase buffer (50 mM HEPES pH 7.5, 5 mM MgCl2, 2.5 mM MnCl2, 1 mM dithiothreitol [DTT]) with 1 μl of substrate (histone H1, GST, GST-Nt-FoxM1, or GST-Ct-FoxM1), 50 μM cold ATP, and 2.5 μCi [γ-32P]ATP for 30 min at 30°C. Samples were then denatured in sample buffer, boiled, and loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Results were visualized by Coomassie blue staining of the gel followed by autoradiography. For cyclin A/cdk2-dependent disruption of the N-/C-terminal complexes of FoxM1, washed immunoprecipitates were incubated with 100 ng of active recombinant cyclin A/cdk2 (Cell Signaling) for 30 min prior to analysis of the complexes in immunoblot assays.
RT-PCR and chromatin immunoprecipitation.
Total RNA was isolated by using either TRIzol reagent or the Qiagen RNeasy kit, according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) and chromatin immunoprecipitations were performed as previously described (5).
FACS and immunofluorescence analysis.
Fluorescence-activated cell sorter (FACS) and immunofluorescence analyses were performed as described previously (5).
Tryptic phosphopeptide analysis.
For analysis of in vitro phosphorylation by cyclin A/cdk2, FoxM1 and respective phospho site mutants were expressed in 293T cells and immunoprecipitated using anti-FoxM1 antibodies. The resulting immunocomplexes were incubated in kinase buffer (50 mM HEPES pH 7.5, 10 mM Mg-acetate, 1 mM DTT) in the presence of 50 μM ATP and 5 μCi [γ-32P]ATP, with or without 100 ng of recombinant cyclin A/cdk2 (Cell Signaling), for 30 min at 30°C. Phosphorylated FoxM1 was processed for two-dimensional phosphopeptide mapping. For analysis of in vivo phosphorylation of FoxM1, endogenous FoxM1 in control or cyclin A-depleted U2OS cells was labeled using 2 mCi [32P]orthophosphate per 10-cm dish. Phosphorylated FoxM1 was immunoprecipitated, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and electroblotted to nitrocellulose. After exposing the blot to X-ray film, bands of interest were cut out for tryptic phosphopeptide analysis. Tryptic phosphopeptide analysis was performed as described earlier (1).
Phosphopeptide analysis by mass spectrometry.
Epitope-tagged FoxM1 was expressed in U2OS cells and 10 15-cm dishes were harvested, after which FoxM1 was isolated by immunoprecipitation. Subsequently, immunoprecipitated proteins were reduced in 10 mM DTT and alkylated in 50 mM iodoacetamide, followed by digestion using sequencing-grade trypsin (Roche Diagnostics, Ingelheim, Germany) overnight at a protein/protease ratio of 50:1. Phosphopeptide enrichment was performed as previously described (10). A home-built nanoflow vented column system, comprised of an LC-Packings Ultimate quaternary solvent delivery system, a thermostatted FAMOS autosampler, and a Switchos six-port switching module (LC-Packings, Amsterdam, The Netherlands), coupled on-line to a linear ion trap-Fourier transform ion cyclotron resonance mass spectrometer (Thermo Electron, Bremen, Germany), was used for all analyses. Spectra were processed using the Bioworks 3.3 software (Thermo, Bremen, Germany), and the subsequent data analysis was conducted using the Mascot (version 2.2) software platform (Matrix Science, London, United Kingdom). The IPI human database (version 3.36; containing 69,012 sequences and 29,002,682 residues) was searched with trypsin, allowing two missed cleavages, carbamidomethyl (C) as fixed modification and oxidation (M), N-acetylation (protein N terminus), deamidation (NQ), phosphorylation (ST), and phosphorylation (Y) as variable modifications. The peptide tolerance was set to 10 ppm, and tandem mass spectrometry tolerances to 0.9-Da phosphorylated peptides with MASCOT scores of 50 or higher were considered to be reliable, whereas the remaining phosphorylated peptides were confirmed manually.
RESULTS
FoxM1 transcriptional activity is restricted to the G2/M phases of the cell cycle.
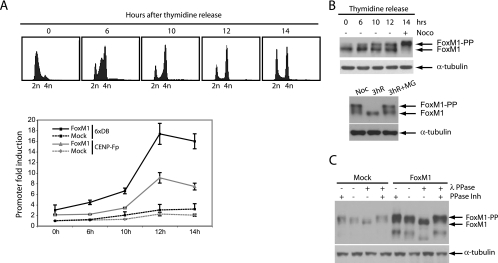
Our recent data have shown that most known FoxM1 target genes are not induced until the cell reaches G2 phase (5), although FoxM1 itself is expressed from S phase onwards. Thus, additional levels of regulation must exist to control proper timing of FoxM1 activation during G2. In order to examine regulation of FoxM1 transcriptional activity throughout the cell cycle, we analyzed the transactivation of luciferase reporters containing the 6XDB Forkhead-binding motif or CENP-F promoter, a specific FoxM1-responsive gene (5), at different phases of the cell cycle. For this purpose, FoxM1 was transiently expressed in human U2OS osteosarcoma cells from a cytomegalovirus promoter-driven expression vector to ensure cell cycle-independent FoxM1 expression throughout the cell cycle (Fig. 1A). We found that FoxM1 activity was low in cells synchronized at the G1/S transition and increased only as the cells entered the G2 phase after release from the G1/S block (10 to 12 h after release) (Fig. 1A), whereas expression of FoxM1 was relatively constant (Fig. 1B). Induction of FoxM1 activity in G2 was not due to enhanced DNA binding, as chromatin immunoprecipitation assays showed that FoxM1 is already bound to its target promoters in G1/S (see Fig. S1A in the supplemental material). Interestingly, while the majority of cells appeared to have completed S phase at 10 h in these experiments, luciferase activity peaked some 2 h later (Fig. 1A). However, analysis of luciferase mRNA showed that mRNA levels are readily induced at 10 h after the release (see Fig. S1B in the supplemental material), suggesting that this delay is a consequence of slow accumulation of the luciferase enzyme. These data suggest that FoxM1 activation occurs as cells progress to G2 phase, even if it is constitutively expressed throughout the cell cycle. Transcriptional activation of FoxM1 correlates well with the appearance of a slower-migrating band 10 h after release (Fig. 1B, upper panel). This shifted band was also present in extracts prepared from cells trapped in G2/M by nocodazole treatment (Fig. 1B, upper panel), whereas it disappeared in cells that reentered the G1 phase after 3 h of release from the nocodazole block (Fig. 1B, lower panel). We further showed that this shifted band corresponds to a phosphorylated form of FoxM1, since phosphatase treatment prevented the shift (Fig. 1C).
FIG. 1.
Cell cycle-dependent transcriptional activation of FoxM1. (A) U2OS osteosarcoma cells were cotransfected with 6xDB or CENP-F luciferase reporter, synchronized at the G1/S transition by 24-h administration of thymidine, and released from the block for the indicated times. DNA profiles of cell cultures after release from the G1/S block (top panels) were determined by FACS using propidium iodide staining. Transactivation of the 6xDB construct and of the CENP-F promoter (CENP-Fp) by endogenous FoxM1 (Mock) or by ectopic FoxM1 was measured in a dual luciferase assay. In all experiments relative luciferase reporter activity was expressed as the ratio of firefly luciferase activity to control Renilla luciferase activity. (B) U2OS cells were blocked at the G1/S transition by thymidine treatment and released from the thymidine block at the indicated times. Endogenous FoxM1 protein levels in these cells were viewed by Western blotting (upper panel). Nocodazole-blocked cells (mitotic shake-off) were released from the nocodazole block after 3 h in the absence or presence of the proteasome inhibitor MG132. Endogenous FoxM1 protein levels in these cells were viewed by Western blotting (lower panel). (C) G2 cell lysates (12 h after release from the thymidine block) were prepared from U2OS cells transfected with either mock or FoxM1 expression vectors and subjected to lambda phosphatase (λPPase) treatment in the absence or presence of phosphatase inhibitors (PPase Inh). Endogenous and ectopically expressed FoxM1 proteins were viewed by Western blotting.
FoxM1 transcriptional activity is enhanced by cyclin A/cdk function.
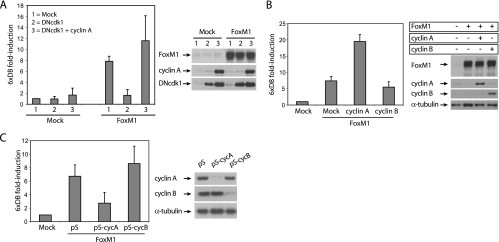
Previous data from our lab showed that FoxM1 activity is highest in G2/M-arrested cells (5). Treatment of cells with nocodazole, which blocks cell cycle progression in G2/M, enhanced FoxM1 activity, whereas removal of serum arrests cells in G0 with very low FoxM1 transcriptional activity (5). In addition, expression of the cdk inhibitors p16 and p21, or a dominant negative form of cdk2 (DNcdk2), caused a dramatic reduction in FoxM1 activity (5). Similarly, expression of a dominant negative form of cdk1 (DNcdk1) also abolished FoxM1 transcriptional activity (Fig. 2A). Interestingly, the effect of DNcdk1 was reverted when cyclin A was coexpressed (Fig. 2A), suggesting that the inhibitory effect of DNcdk1 on FoxM1 activity is due to titration of the endogenous cyclin A. Indeed, we found that maximum activity of FoxM1 coincided with maximum cyclin A-associated kinase activity 12 h after release from the G1/S thymidine block (see Fig. S1C in the supplemental material). Consistently, exogenous expression of cyclin A, but not cyclin B, enhanced FoxM1 transcriptional activity (Fig. 2B, left panel), suggesting that FoxM1-dependent transactivation might be specifically mediated via cyclin A-associated kinases.
FIG. 2.
Regulation of FoxM1 transcriptional activity by cyclin A/cdk's. (A) Luciferase assay in U2OS cells transiently cotransfected with control, or FoxM1 expression vectors and 6xDB luciferase reporter in combination with DNcdk1, alone or in combination with ectopically expressed cyclin A (left panel). The expression of all constructs was confirmed by Western blotting (right panel). (B and C) Transactivation of 6xDB by FoxM1 was measured in U2OS cells that were either transfected with empty vector (mock), cyclin A- or cyclin B-expressing vectors (B), or with shRNA-targeting vectors against cyclin A (pS-cycA) or cyclin B (pS-cyB) (C). Presented data are the averages of three independent experiments performed in duplicate. Expression levels of all constructs were viewed by Western blotting.
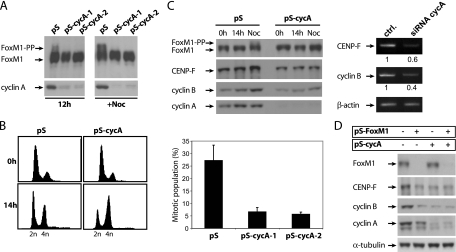
To further confirm the requirement for cyclin A in transactivation by FoxM1, expression of endogenous cyclin A was repressed by an shRNA-targeting vector. Specific removal of cyclin A, but not cyclin B, through RNAi-mediated depletion was able to cause a strong reduction in FoxM1 activity, similar to what is observed after expression of a dominant negative version of cdk1 (Fig. 2C). In addition, cyclin A-associated kinases appear to be required for in vivo phosphorylation of endogenous FoxM1, as depletion of cyclin A with two different shRNA-targeting vectors resulted in a strong reduction of phosphorylated forms of FoxM1 in G2 phase (Fig. 3A; see also Fig. S2A in the supplemental material). In addition, removal of cyclin A caused cells to accumulate in G2 and resulted in a strong reduction in the mitotic population after nocodazole treatment (Fig. 3B), similar to what we have observed following depletion of FoxM1 (5). Down-regulation of cyclin A also led to a reduction in the expression of two well-known target genes of FoxM1 during G2/M progression, CENP-F and cyclin B, at both protein and mRNA levels (Fig. 3C). This was observed both in G2 cells released from a thymidine block and in nocodazole-arrested cells (Fig. 3C). Codepletion of cyclin A and FoxM1 does not lead to a further reduction of FoxM1 target genes (Fig. 3D), indicating that cyclin A and FoxM1 act together, rather than independently, to regulate these genes. Also, while depletion of cyclin A caused a clear reduction in CENP-F and cyclin B mRNA levels in wild-type MEFs, this effect was much less pronounced in FoxM1-deficient MEFs (see Fig. S2B in the supplemental material). These data suggest that cyclin A acts through FoxM1 to activate transcription of genes required for proper G2/M progression.
FIG. 3.
Cyclin A is required for FoxM1 target gene expression and for proper G2/M progression. (A) Western blot analysis of endogenous FoxM1 protein in human U2OS cells transfected with empty pSuper or two different pS-cycA-targeting constructs. Cells were blocked at the G1/S transition by 24-h administration of thymidine and released from the thymidine block for 12 h or 14 h in the presence of nocodazole. (B) U2OS cells cotransfected with spectrin-green fluorescent protein (GFP) plasmid and empty pSuper or pS-cycA targeting constructs were synchronized at G1/S transition by thymidine treatment and released from the block for 14 h in the absence (left graphs) or presence of nocodazole (right graph). The left graphs show FACS analysis of DNA profiles of spectrin-GFP-positive cells using propidium iodide staining. The right panel shows the quantification of the mitotic cell population in nocodazole-treated cells using phospho-histone H3 staining (pH 3). (C) Western blot analysis of FoxM1 target gene (CENP-F and cyclin B1) expression in U2OS cells expressing pS or pS-cycA targeting vectors blocked at the G1/S transition by 24-h administration of thymidine and released from the thymidine block for 14 h in the presence or absence of nocodazole (left panel). RT-PCR analysis of CENP-F and cyclin B1 mRNA levels in U2OS cells transfected with control siRNA or siRNA oligonucleotides targeting cyclin A and released from a thymidine block for 12 h (right panel). (D) Western blot analysis of FoxM1 target gene expression in human U2OS cells that were either transfected with empty pSuper RNAi vector (pS), with an RNAi-expressing construct targeting FoxM1 (pS-FoxM1), or with an RNAi-expressing construct targeting cyclin A (pS-cycA) or were cotransfected with both pS-FoxM1 and pS-cyclin A targeting constructs.
Deletion of the N-terminal region of FoxM1 generates a constitutively active form of FoxM1.
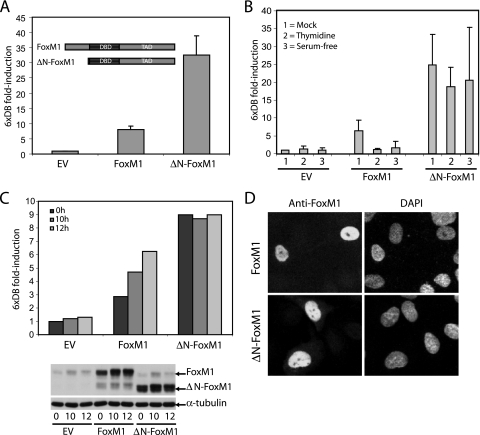
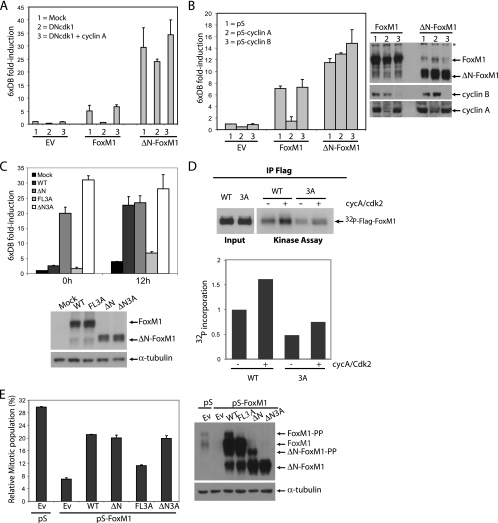
Removal of the N-terminal domain, right up to the start of the DNA-binding domain (ΔN-FoxM1), results in a protein with increased activity (Fig. 4A). To define the contribution of the N terminus of FoxM1 to the cell cycle-dependent regulation of its transcriptional activity, we examined the activity of this truncated mutant during different stages of the cell cycle. In contrast to the full-length protein, transactivation by the ΔN-FoxM1 variant was hardly affected upon synchronization of cells by serum starvation or by the addition of thymidine (Fig. 4B). Likewise, the activity of this mutant remained constantly high throughout the cell cycle, whereas full-length FoxM1 activity only increased during the G2 phase (10 h to 12 h after release from the G1/S block) (Fig. 4C). This is not due to differences in the subcellular localization or protein expression, since both full-length FoxM1 and ΔN-FoxM1 localize to the nucleus and show similar expression levels throughout the cell cycle (Fig. 4C and D; see also Fig. S3 in the supplemental material). These data show that ΔN-FoxM1 behaves as a constitutively active form of FoxM1 whose activity is independent of cell cycle progression. As cyclin A-containing complexes directly phosphorylate FoxM1 and enhance its transcriptional activity, we next examined the effect of cyclin A on the transcriptional activity of the ΔN-FoxM1 mutant. As shown in Fig. 2A, the activity of full-length FoxM1 was dramatically reduced in cells expressing DNcdk1 (by 90%) and fully restored by overexpression of cyclin A. In contrast, the activity of the ΔN-FoxM1 mutant was only slightly affected by DNcdk1 (less than ∼20%) (Fig. 5A). Furthermore, depletion of cyclin A by shRNA had no apparent effect on the activity of ΔN-FoxM1, while it strongly decreased the activity of full-length FoxM1 (Fig. 5B). The difference in cyclin A dependence between the wild type and ΔN-FoxM1 was observed over a range of FoxM1 concentrations, indicating that the observed difference was not an artifact of saturation of our assay system (see Fig. S4A in the supplemental material). Our observations strongly suggest that cyclin A/cdk complexes activate FoxM1 during the G2 phase primarily through relief of repression mediated by its autoinhibitory N-terminal domain. However, the fact that ΔNFoxM1 transcriptional activity was slightly inhibited by DNcdk1 points to the possibility that an additional mechanism(s) may contribute to cyclin A/cdk-mediated activation of FoxM1.
FIG. 4.
Deletion of the N-terminal region of FoxM1 leads to a hyperactive mutant that is constitutively active throughout the cell cycle. (A) Transactivation of the 6xDB luciferase reporter was measured in U2OS cells expressing full-length FoxM1 or ΔN-FoxM1. (B) Transactivation of 6xDB by FoxM1 or ΔN-FoxM1 in U2OS cells blocked in G1/S by thymidine treatment or by serum starvation (serum free). (C) 6xDB luciferase reporter transactivation by full-length FoxM1 or ΔN-FoxM1 in U2OS cells that were blocked at the G1/S transition by thymidine treatment and released from the thymidine block for the indicated times. Endogenous as well as ectopically expressed FoxM1 or ΔN-FoxM1 protein levels were viewed by Western blotting. (D) Nuclear localization of full-length FoxM1 and ΔN-FoxM1 introduced in U2OS cells, as visualized by anti-FoxM1 and 4′,6′-diamidino-2-phenylindole (DAPI) staining using fluorescence microscopy.
FIG. 5.
The N-terminal-truncated FoxM1 mutant is insensitive to inactivation of cyclin A/cdk complexes. (A) 6xDB luciferase reporter transactivation by full-length FoxM1 or ΔN-FoxM1 in U2OS cells expressing DNcdk1 alone or together with cyclin A. (B) Transactivation of 6xDB by FoxM1 or ΔN-FoxM1 was measured in U2OS cells at 12 h after release from the G1/S block. U2OS cells were either transfected with empty pSuper RNAi vector (1) or with an RNAi-expressing construct targeting cyclin A (2) or cyclin B (3). Endogenous cyclin A and cyclin B as well as ectopically expressed FoxM1 or ΔN-FoxM1 protein levels were viewed by Western blotting. *, nonspecific band. (C) Transactivation of 6xDB by the indicated constructs was measured in U2OS cells synchronized at the G1/S transition by 24-h administration of thymidine (0 h) and at 12 h after release from the G1/S block. Expression of all constructs was determined by Western blotting. (D) In vitro phosphorylation of the Flag-tagged wild type and 3A FoxM1 mutant. The flag-tagged wild-type and 3A proteins were transfected in 293T cells and immunoprecipitated using anti-flag antibody. The immunoprecipitates were then used in kinase assays with a radioactive label in the absence or presence of active purified cycA/cdk2 complex (Cell Signaling). Kinase activity was viewed by autoradiography (upper panel). Quantification of 32P incorporation is shown (lower panel). (E) Quantification of the mitotic cell fraction in U2OS cells transfected with pS or pS-FoxM1 targeting vector in combination with RNAi-insensitive forms of the indicated proteins. The percentage of the mitotic cell population was measured using pH 3 staining 16 h after release from the G1/S transition in the presence of nocodazole. Expression of all constructs was determined by Western blotting.
The N terminus of FoxM1 contains two conserved cdk phosphorylation (S/T)P sites at amino acid serine 4 (S4P) and serine 35 (S35P), and at least the latter appears to be phosphorylated in vivo (see Fig. S5E in the supplemental material). To determine the contribution of these sites in G2-dependent activation of FoxM1, we mutated both sites to alanine and examined the transcriptional activity of this mutant throughout the cell cycle. Both wild-type FoxM1 and the FoxM1S4A/S35A mutant show a similar pattern of activation as cells progress from G1/S to the G2 phase of the cell cycle (data not shown), indicating that additional residues in FoxM1 serve as substrates for cyclin A/cdk complexes to mediate release of the N-terminal inhibitory domain. Additional cdk phosphorylation sites (T600/T611/S638; amino acid numbering in the FoxM1C variant) in the C-terminal transactivation domain (TAD) may contribute to FoxM1 activation by cyclin/cdk complexes (6). Therefore, we mutated all three sites into alanine in both full-length FoxM1 and ΔN-FoxM1 and assessed their transcriptional activities throughout the cell cycle. We found that mutation of T600/T611/S638 sites into alanine strongly reduced transcriptional activation of full-length FoxM1 (FL3A) during the G2 phase (Fig. 5C). However, mutation of these sites in ΔN-FoxM1 (ΔN3A) had no effect on its activity (Fig. 5C), although both FL3A and ΔN3A showed strongly decreased phosphorylation (see Fig. S4B and C in the supplemental material). Similarly, the 3A FoxM1 mutant was less phosphorylated by active recombinant cyclin A/cdk2 in vitro (Fig. 5D). Finally, we assessed the functionality of these mutants by testing their ability to promote G2/M progression in the absence of endogenous FoxM1. U2OS cells were cotransfected with the FoxM1 shRNA-targeting vector and RNAi-insensitive expression plasmids encoding the various proteins. Cells lacking FoxM1 fail to enter mitosis and show a significant delay in G2, resulting in a low mitotic index in the presence of nocodazole (5) (Fig. 5E). As expected, full-length FoxM1, ΔN-FoxM1, and ΔN3A mutants were able to reverse the mitotic entry defect in FoxM1-depleted cells to a large extent, while the FL3A mutant failed to do so significantly (Fig. 5E, left panel). Collectively, these data suggest that cyclin A/cdk-dependent phosphorylation of the C-terminal TAD is required for relief of inhibition by the N-terminal autorepressor domain to promote proper G2/M progression.
Phosphorylation of FoxM1 by cyclin A/cdk complexes.
Because exogenous expression of cyclin A induces a strong activation of FoxM1, we examined whether FoxM1 protein was a direct substrate for cyclin A-containing cdk complexes. We performed in vitro kinase assays using GST-FoxM1 variants as a substrate. We found that cyclin A/cdk complexes can strongly phosphorylate both N-terminal and C-terminal domains of FoxM1 in vitro (see Fig. S5A in the supplemental material). To verify if cyclin A/cdk complexes can phosphorylate the T600/T611/S638 residues, we first analyzed if all sites were relevant to cell cycle-dependent regulation of FoxM1. Mutating single residues within this triad did not result in loss of FoxM1 activation in G2 (not shown), but a double T600/T611 mutant fully reproduced the functional effects seen with the triple mutant (see Fig. S5B in the supplemental material), indicating that the S638 site is not relevant for this aspect of FoxM1 regulation. To test if T600 and T611 can be phosphorylated by cyclin A/cdk2, we immunoprecipitated ectopically expressed single and double T600/T611 mutants of FoxM1 and incubated these immunocomplexes with active recombinant cyclin A/cdk2 complexes. In vitro-phosphorylated proteins were subsequently analyzed by tryptic phosphopeptide mapping. As shown in Fig. S5C of the supplemental material, several peptides in FoxM1 are phosphorylated by cyclin A/cdk2, two of which appear to correspond to a T600- and a T611-containing peptide (see Fig. S5C in the supplemental material), as they are missing in the peptide maps of the corresponding T600/T611 single or double alanine mutants. Of these, the T611 peptide runs at the expected mobility as predicted with Mobility software, while the T600 peptide displays a different running behavior, possibly as a result of incomplete cleavage of the protein.
In addition, we analyzed the phosphopeptide map of in vivo-phosphorylated endogenous FoxM1 obtained from [32P]orthophosphate-labeled control or cyclin A-depleted U2OS cells. This showed that T611, and possibly also T600 to a lesser extent, is also phosphorylated in vivo in a cyclin A-dependent manner (see Fig. S5D in the supplemental material). Phosphorylation of T611 in vivo was further confirmed by mass spectrometry using exogenously expressed FoxM1 isolated from U2OS cells (see Fig. S5E in the supplemental material). These data show that FoxM1 is phosphorylated on T611 and possibly T600, in a cyclin A-dependent manner, both in vitro and in vivo.
The N-terminal domain can repress FoxM1 activity in trans and is inactivated through cyclin A/Cdk2-mediated phosphorylation.
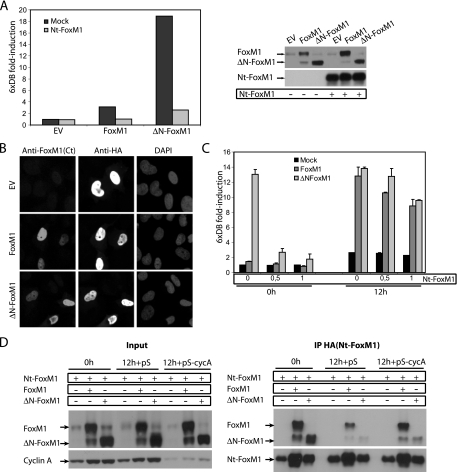
To demonstrate that the N terminus of FoxM1 carries repressor activity, we next assessed FoxM1 transcriptional activity in cells expressing the FoxM1 N-terminal fragment. We found that transactivation by the ΔN-FoxM1 mutant was dramatically reduced in the presence of the N-terminal fragment during G1/S (Fig. 6A). The N-terminal fragment of FoxM1 appears to localize predominantly in the nucleus and did not interfere with the subcellular localization of full-length FoxM1 and ΔN-FoxM1 at either the G1/S block or during G2 (Fig. 6B; see also Fig. S3 in the supplemental material). Since the N-terminal repressive action was cell cycle controlled, we speculated that expression of the N-terminal fragment would not affect FoxM1 activity during the G2 phase. To test this, we examined the effect of the N-terminal fragment on FoxM1 activity in G1/S and G2, respectively. At doses that repress ΔN-FoxM1 activity in G1/S phase (0 h), the N-terminal fragment had no significant effect on transcriptional activity of the full-length FoxM1 or ΔN-FoxM1 mutant in G2 cells (12 h) (Fig. 6C). Yet, at higher doses, the N-terminal fragment was able to prevent transcriptional activation of both full-length FoxM1 and ΔN-FoxM1 during the G2 phase (data not shown). These data indicate that the N-terminal region of FoxM1 is a potent autorepressor domain that acts to restrict FoxM1 activity in the G1 and S phases. Cyclin A/cdk-dependent phosphorylation of the C-terminal TAD appears to be required for relief of autoinhibition by the N-terminal domain. Therefore, the N terminus may exert its inhibitory action through direct binding to the C-terminal TAD of FoxM1. To examine the possible interaction between the N-terminal domain and FoxM1, we performed immunoprecipitation experiments in U2OS cells expressing full-length FoxM1 or ΔN-FoxM1. We found that the N-terminal fragment of FoxM1 strongly interacts with both full-length FoxM1 and ΔN-FoxM1 at G1/S (Fig. 6D). Interestingly, the interaction was strongly reduced in G2 cells, but this reduction was prevented upon depletion of cyclin A (Fig. 6D). These data indicate that cyclin A-dependent kinase activity relieves FoxM1 autorepression by disrupting the interaction between the N-terminal and C-terminal domains.
FIG. 6.
The N-terminal domain represses FoxM1 transcriptional activity during G1/S through interaction with the C-terminal transactivation domain. (A) Transactivation of the 6xDB luciferase reporter was measured in U2OS cells transfected with empty vector or full-length FoxM1- or ΔN-FoxM1-expressing vectors in the absence or presence of an exogenously expressed HA-tagged N-terminal fragment of FoxM1 (HA-Nt-FoxM1) at the G1/S transition. Expression of endogenous and ectopically expressed FoxM1 proteins was viewed by Western blotting with anti-FoxM1 antibody. (B) Subcellular localizations of full-length FoxM1 and ΔN-FoxM1 were visualized in U2OS cells transiently expressing the HA-Nt-FoxM1 fragment by using an anti-FoxM1 antibody that recognizes the C-terminal domain of the protein in combination with HA and 4′,6′-diamidino-2-phenylindole (DAPI) staining using fluorescence microscopy. (C) Transactivation of the 6xDB luciferase reporter was measured in U2OS cells transfected with empty vector or vectors expressing full-length FoxM1 or ΔN-FoxM1 in the absence or presence of increasing amounts of HA-Nt-FoxM1 fragment (0, 0.5, and 1 μg, respectively) at the indicated times after release from the thymidine block. Presented data are the averages of four independent experiments performed in duplicate. (D) Immunoprecipitation of the HA-Nt-FoxM1 fragment transiently expressed in U2OS cells in combination with full-length FoxM1 or ΔN-FoxM1. U2OS cells were either transfected with empty pSuper RNAi vector (pS) or with an RNAi-expressing construct targeting cyclin A. Cells were synchronized at the G1/S transition by 24-h administration of thymidine (0 h) and released from the G1/S block for 12 h. Expression of all constructs was detected by Western blotting.
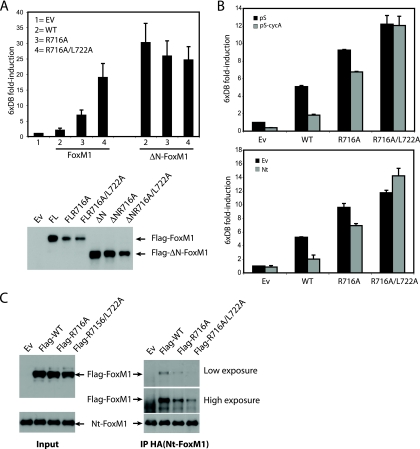
Interestingly, mutation of two RXL/LXL motifs in the C-terminal domain of FoxM1 (R716A-X-L718A and L722A-X-L724A) caused hyperactivation of FoxM1 in G1/S, similar to deletion of the N-terminal domain, while the same mutations did not further affect the activity of ΔN-FoxM1 (Fig. 7A). Also, the activity of the RXL/LXL double mutant was not affected by depletion of cyclin A (Fig. 7B, left panel) or by expression of the N-terminal fragment (Fig. 7B, right panel). This suggests that the RXL/LXL motifs within the C-terminal domain are required for interaction with the N-terminal repressor domain. To test this directly, we coexpressed both wild-type and RXL/LXL single or double mutants and tested their ability to interact with the N-terminal domain in transfected cells. Indeed, the double RXL/LXL mutant failed to interact with the N-terminal mutant (Fig. 7C), consistent with a model in which binding of the N-terminal domain to the C-terminal domain inhibits FoxM1 activity.
FIG. 7.
Autoinhibition of FoxM1 transcriptional activity requires binding of the N-terminal domain to RXL/LXL motifs in the C-terminal transactivation domain. (A) Transactivation of 6xDB luciferase reporter was measured in U2OS cells transfected with empty, wild-type full-length FoxM1, or ΔN-FoxM1 vector or mutants carrying the R716A single mutation or R716A/L722A double mutation at the G1/S transition (right graph). Expression of all constructs was detected by Western blotting (lower panel). (B) Transactivation of the 6xDB luciferase reporter was measured in U2OS cells transfected with empty vector, wild-type full-length FoxM1, full-length FoxM1R716A single, or FoxM1R716A/L722A double mutants in U2OS cells cotransfected with empty pSuper or pS-cycA targeting constructs (left panel) or cotransfected with empty or HA-Nt-FoxM1-expressing vector (lower panel). (C) Immunoprecipitation of the HA-Nt-FoxM1 fragment in 293T cells transiently coexpressed with wild-type FoxM1 or FoxM1R716A single or FoxM1R716A/L722A double mutants. Protein levels of all constructs were viewed by Western blotting.
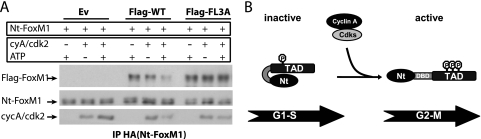
Because expression of the N-terminal fragment of FoxM1 reduced transcriptional activation of full-length FoxM1 as efficiently as depletion of cyclin A, we postulated that phosphorylation of FoxM1 by cyclin A/cdk complexes might cause release of inhibition by the N terminus. To test this hypothesis, we investigated whether active cyclin A/cdk2 complexes could disrupt the interaction between the N and C terminus of wild-type FoxM1, but not of the 3A mutant. To this end, immunoprecipitated complexes containing the N-terminal fragment and FoxM1 protein were incubated in vitro with active cyclin A/cdk2 in the presence or absence of ATP. Incubation of the wild-type FoxM1-Nt intermolecular complexes with active cyclin A/cdk2 resulted in efficient disruption of the interaction (Fig. 8A). This disruption requires ATP, as mere addition of cyclin A/cdk2 was insufficient to release the N terminus. Moreover, incubation of cyclin A/cdk2 with the 3A FoxM1 mutant lacking the cdk phosphorylation sites had no effect on its interaction with the N terminus (Fig. 8A). Taken together, these data suggest that cyclin A/cdk2 phosphorylates the C-terminal TAD of FoxM1 to relieve the autoinhibition by the N-terminal repressor domain, allowing transcriptional activation of FoxM1 in G2 (Fig. 8B).
FIG. 8.
Autoinhibition of the FoxM1 transcriptional activity is released upon phosphorylation by active cyclin A/cdk2 complexes during G2. (A) The immunoprecipitates containing HA-Nt-FoxM1 and wild-type Flag-FoxM1 (Flag-WT) or HA-Nt-FoxM1 and Flag-FoxM13A mutant (Flag-FL3A) were incubated with commercial active cyclin A/cdk2 in the presence or absence of ATP. FoxM1 and cdk2 protein levels were viewed by Western blotting. (B) Model illustrating the cell cycle-dependent regulation of FoxM1 transcriptional activity. During G1/S, FoxM1 activity is repressed by its own N-terminal domain, most likely via direct interaction with the C-terminal TAD. As cells progress to the G2 phase, phosphorylation by cyclin A/cdk complexes promotes the inactivation of the N-terminal autorepressor domain by disrupting the interaction between the N terminus and the C-terminal TAD, allowing activation of the FoxM1 protein.
DISCUSSION
In this study, we aimed to gain more insight into how FoxM1 activity is regulated during an ongoing cell cycle. Our data point to the existence of a cyclin A-dependent mechanism that controls transactivation by FoxM1, allowing a tight restriction of FoxM1 transactivation to the G2 phase of the cell cycle.
We provide evidence that although FoxM1 can already be expressed in late G1 and in S phase and can bind to its endogenous promoters, its transcriptional activity is kept low until entry into G2. Our data indicate that cyclin A, but not cyclin B, plays an important role in the regulation of FoxM1 transcriptional activity during G2. Ectopic expression of cyclin A greatly increases FoxM1 transcriptional activity, while removal of cyclin A through RNAi-mediated depletion leads to a strong reduction in FoxM1 activity, similar to what is seen after expression of dominant negative versions of cdk1 and cdk2. More importantly, cyclin A depletion results in a defect in G2/M progression and a reduction in FoxM1 target gene expression, similar to what is seen in FoxM1-deficient cells. Previously, cyclin B/cdk2 was shown to phosphorylate FoxM1 (8), but the exact contribution of this phosphorylation to FoxM1 activation remains to be determined. Cyclin E/cdk2 was also shown to phosphorylate and activate FoxM1 at G1/S (6, 15). A similar observation was reported by Major and coworkers, who showed that the cyclin E/cdk2 complex binds and phosphorylates the C-terminal region of FoxM1 in order to recruit the transcriptional coactivator p300 (8). Accordingly, a minor shifted form of FoxM1 is seen in thymidine-blocked cells (Fig. 1), suggesting that phosphorylation of FoxM1 is a multistep process that starts as early as late G1. However, because cyclin E/cdk2 activity is limited to the G1/S transition of the cell cycle, it may not be sufficient for full activation of FoxM1 but could prime FoxM1 for eventual activation by cyclin A/cdk complexes in G2. Indeed, the major cyclin A-dependent shift in FoxM1 mobility that is observed as cells progress from S to G2 suggests that cyclin A plays a major role in promoting FoxM1 phosphorylation. Accordingly, we also found that FoxM1 is in vitro phosphorylated by cyclin A/cdk2 complexes and that the endogenous protein is in vivo phosphorylated during G2 in a cyclin A-dependent manner. On the basis of our observations, it is difficult to discriminate which cyclin A/cdk complex is most crucial for FoxM1 activation during G2 phase, as both cdk1 and cdk2 complexes could act redundantly.
We found that a truncated mutant of FoxM1 that lacks the N-terminal domain is a hyperactive transcriptional regulator, consistent with recent results from others (9, 14). Here, we show that transcriptional activation of this mutant no longer depends on cyclin A/cdk activity, as neither overexpression of dominant negative forms of cdk's nor depletion of cyclin A could inhibit the activity of this mutant. These data imply that the N-terminal repressor domain is regulated in a cell cycle-dependent fashion, requiring cyclin A/cdk-dependent inactivation in G2. We found that phosphorylation of the C-terminal TAD (T600 and T611 residues) is required for G2-specific activation of FoxM1. Substitution of these residues to alanine clearly reduced the level of FoxM1 phosphorylation by cyclin A/cdk complexes and prevented activation of full-length FoxM1 in G2, while it did not affect transcriptional activity of ΔN-FoxM1. These data strongly suggest that cyclin A/cdk-mediated phosphorylation of the C-terminal TAD is required for relief of the inhibitory function of the N terminus.
Recently, it was shown that the N- and C-terminal domains of FoxM1 can form a direct complex (14), suggesting that inhibition by the N terminus occurs through direct interaction of this domain with the C-terminal transactivation domain, thereby preventing transcriptional activation of FoxM1. Importantly, our data indicate that this molecular interaction within FoxM1 can be disrupted by active cyclin A/cdk complexes, allowing for full activation of FoxM1. Moreover, point mutations in two conserved RXL/LXL sites in the TAD of FoxM1 (R716/L718 and L722/724) strongly reduced the interaction with the N-terminal repressor domain of FoxM1 and behaved as hyperactive mutants. Similar to the N-terminal deletion mutant, mutation of the RXL/LXL motifs eliminates the requirement of cyclin A/cdk for FoxM1 activation, indicating that these RXL/LXL motifs mediate the binding between the N terminus and the TAD of FoxM1. In contrast, mutation of another LXL motif present in the TAD of FoxM1 (L656A), which has been recently shown to be required for FoxM1B activation (9), did not show any significant effect on FoxM1C transactivation (data not shown), suggesting that the different FoxM1 isoforms require different modes of regulation.
Taken together, our findings uncover a crucial role for cyclin A/cdk complexes in optimal activation of FoxM1 during the G2 phase of the cell cycle. These data are most consistent with a model in which FoxM1 activity is kept low due to repression by its own N-terminal domain through direct binding to the C-terminal TAD. This binding might also allow docking of cellular components, yet unknown, that further suppress FoxM1 activity. Through constitutive cyclin A/cdk action, and once sufficient levels of phosphorylated FoxM1 protein are achieved, repression by the N terminus is relieved, allowing full activation of FoxM1.
Supplementary Material
Acknowledgments
We thank O. Kranenburg for critically reviewing the manuscript. We thank Lisa Caballero, Jill Meisenhelder, and Tony Hunter for help with the Mobility software. We also thank all members of the Medema laboratory for valuable discussions.
This work was supported by the Dutch Cancer Society (NKI200-2192 and UU2007-3826), The Netherlands Organization of Health Research and Development (918.46.616), and The Netherlands Proteomics Center.
Footnotes
Published ahead of print on 19 February 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Boyle, W. J., P. van der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201110-149. [DOI] [PubMed] [Google Scholar]
- 2.Kim, I. M., S. Ramakrishna, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2005. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 28022278-22286. [DOI] [PubMed] [Google Scholar]
- 3.Korver, W., J. Roose, and H. Clevers. 1997. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 251715-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krupczak-Hollis, K., X. Wang, V. V. Kalinichenko, G. A. Gusarova, I. C. Wang, M. B. Dennewitz, H. M. Yoder, H. Kiyokawa, K. H. Kaestner, and R. H. Costa. 2004. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev. Biol. 27674-88. [DOI] [PubMed] [Google Scholar]
- 5.Laoukili, J., M. R. Kooistra, A. Bras, J. Kauw, R. M. Kerkhoven, A. Morrison, H. Clevers, and R. H. Medema. 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7126-136. [DOI] [PubMed] [Google Scholar]
- 6.Luscher-Firzlaff, J. M., R. Lilischkis, and B. Luscher. 2006. Regulation of the transcription factor FOXM1c by cyclin E/CDK2. FEBS Lett. 5801716-1722. [DOI] [PubMed] [Google Scholar]
- 7.Ma, R. Y., T. H. Tong, A. M. Cheung, A. C. Tsang, W. Y. Leung, and K. M. Yao. 2005. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J. Cell Sci. 118795-806. [DOI] [PubMed] [Google Scholar]
- 8.Major, M. L., R. Lepe, and R. H. Costa. 2004. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol. Cell. Biol. 242649-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park, H. J., Z. Wang, R. H. Costa, A. Tyner, L. F. Lau, and P. Raychaudhuri. 24 September 2007, posting date. An N-terminal inhibitory domain modulates activity of FoxM1 during cell cycle. Oncogene doi: 10.1038/sj.onc.1210814. [DOI] [PMC free article] [PubMed]
- 10.Pinkse, M. W., S. Mohammed, J. W. Gouw, B. van Breukelen, H. R. Vos, and A. J. Heck. 2007. Highly robust, automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J. Proteome Res. 7687-697. [DOI] [PubMed] [Google Scholar]
- 11.Wang, I. C., Y. J. Chen, D. Hughes, V. Petrovic, M. L. Major, H. J. Park, Y. Tan, T. Ackerson, and R. H. Costa. 2005. Forkhead Box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 2510875-10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, X., H. Kiyokawa, M. B. Dennewitz, and R. H. Costa. 2002. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. USA 9916881-16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westendorf, J. M., P. N. Rao, and L. Gerace. 1994. Cloning of cDNAs for M phase phosphoproteins recognized by the MPM2 monoclonal antibody and determination of the phosphorylated epitope. Proc. Natl. Acad. Sci. USA 91714-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wierstra, I., and J. Alves. 2006. Despite its strong transactivation domain, transcription factor FOXM1c is kept almost inactive by two different inhibitory domains. Biol. Chem. 387963-976. [DOI] [PubMed] [Google Scholar]
- 15.Wierstra, I., and J. Alves. 2006. FOXM1c is activated by cyclin E/Cdk2, cyclin A/Cdk2, and cyclin A/Cdk1, but repressed by GSK-3α. Biochem. Biophys. Res. Commun. 34899-108. [DOI] [PubMed] [Google Scholar]
- 16.Wonsey, D. R., and M. T. Follettie. 2005. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 655181-5189. [DOI] [PubMed] [Google Scholar]
- 17.Yao, K. M., M. Sha, Z. Lu, and G. G. Wong. 1997. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J. Biol. Chem. 27219827-19836. [DOI] [PubMed] [Google Scholar]
- 18.Ye, H., T. F. Kelly, U. Samadani, L. Lim, S. Rubio, D. G. Overdier, K. A. Roebuck, and R. H. Costa. 1997. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 171626-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.