Abstract
The precise role of Cbl in epidermal growth factor (EGF) receptor (EGFR) endocytosis and trafficking remains to be fully uncovered. Here, we showed that mutant EGFR1044, which was truncated after residue 1044, did not associate with c-Cbl and was not ubiquitinated initially in response to EGF but was internalized with kinetics similar to those of wild-type EGFR. This finding indicates that c-Cbl-mediated ubiquitination is not required for EGF-induced EGFR endocytosis. We also showed that the previously identified internalization-deficient mutant receptor EGFR1010LL/AA bound to c-Cbl and was fully ubiquitinated in response to EGF, which indicates that c-Cbl binding and ubiquitination are not sufficient for EGFR internalization. We next investigated EGFR trafficking following EGFR internalization. We found that c-Cbl disassociation from EGFR occurred well in advance of EGFR degradation and that this event was concurrent with the selective dephosphorylation of EGFR at Y1045. This finding suggests that once EGFR is ubiquitinated, continual Cbl association is not required for EGFR degradation. Because EGFR1044 is ubiquitinated and degraded similarly to wild-type EGFR, we examined the role of another prominent Cbl homologue, Cbl-b, and found that Cbl-b was associated with both EGFR and EGFR1044. Further study showed that Cbl-b bound to EGFR at two regions: one in the C-terminal direction from residue 1044 and one in the N-terminal direction from residue 958. Moreover, Cbl-b association with EGFR rose markedly following a decrease in c-Cbl association, corresponding to a second peak of EGFR ubiquitination occurring later in EGFR trafficking. Using RNA interference to knock down both c-Cbl and Cbl-b, we were able to abolish EGFR downregulation. This knockdown had no affect on the rate of EGF-induced EGFR internalization. We found that the two Cbls accounted for total receptor ubiquitination and that while c-Cbl and Cbl-b are each alone sufficient to effect EGFR degradation, both are involved in the physiological, EGF-mediated process of receptor downregulation. Furthermore, these data ultimately reveal a previously unacknowledged temporal interplay of two major Cbl homologues with the trafficking of EGFR.
The mammalian Cbl protein family consists of the three homologues c-Cbl, Cbl-b, and Cbl-3, all of which associate with a wide variety of signaling proteins, including growth factor receptors and Src homology region 2 and 3 (SH2 and SH3) domain-containing proteins (reviewed in reference 50). Through these many associations, Cbl is able to regulate diverse signaling networks. One of the most extensively studied roles of Cbl is its function as a negative regulator of receptor tyrosine kinase (RTK) signaling (28, 35, 39, 55, 58). Two highly conserved amino-terminal domains contribute strongly to this regulatory function. First, Cbl's tyrosine kinase binding (TKB) domain recognizes phosphotyrosine residues and allows Cbl to interact directly with activated RTKs on the plasma membrane (15, 31, 54). Second, the RING finger domain recruits ubiquitin (Ub)-loaded enzymes (E2s), allowing Cbl to function as an E3 Ub ligase to direct the ubiquitination, and presumably the degradation, of its associated RTK (23, 27, 57, 59).
In many ways, the epidermal growth factor receptor (EGFR) serves as an archetypical model for growth factor-induced RTK signal transduction and trafficking (reviewed in references 5 and 43). Upon epidermal growth factor (EGF) binding, EGFR dimerizes, resulting in the activation of its intrinsic kinase and subsequent trans-autophosphorylation at multiple tyrosines along its carboxyl-terminal tail. These phosphotyrosines serve as tethering sites for the nucleation of protein complexes which transduce signals through downstream pathways, ultimately leading to important biological outcomes such as mitogenesis and survival (4, 34). Consequently, the deregulation of EGFR activity is a major factor in cellular transformation and cancer (40). The proper downregulation of EGFR and the attenuation of EGFR signaling are therefore critical in maintaining homeostasis. It has become increasingly evident that Cbl plays a prominent role in EGFR downregulation by virtue of its E3 Ub ligase activity (23, 28, 35). Upon EGF induction, Cbl is recruited to activated EGFR, associating directly via TKB domain binding with phosphotyrosine 1045 (pY1045) on EGFR (27). Cbl can also associate indirectly with activated EGFR through the adaptor protein Grb2, which binds the proline region of Cbl through the SH3 domain of Grb2 and pY1068 or pY1086 on EGFR through the SH2 domain of Grb2 (14). Following binding, Cbl is phosphorylated by EGFR, allowing the recruitment of Ub-loaded E2 proteins to the RING finger of Cbl and the subsequent ligation of monomeric Ub at multiple sites along the receptor (2, 23, 59). The event of multiple monoubiquitination, termed multiubiquitination, is thought to be a major determinant in EGFR downregulation, both during the initial sorting of activated EGFR into clathrin-coated pits and later during the sorting of the receptor into lysosomes for degradation (8, 17, 30, 33, 42, 45, 46).
Although it is generally agreed that Cbl acts to negatively regulate EGFR activity by promoting the intracellular trafficking and degradation of EGFR, it is still disputed whether Cbl binding or Cbl-mediated ubiquitination is altogether required for ligand-induced EGFR endocytosis (9, 30). Intuitively, the fact that Cbl binding and receptor ubiquitination occur at the plasma membrane seems to support a role for Cbl in EGFR endocytosis. Results from several studies also support a role for Cbl in EGFR internalization, either through Cbl's E3 Ub ligase activity or through its involvement in endocytic complexes (18, 19, 21, 22, 41, 44). Experiments employing mutations that either disrupted Cbl-containing complexes or hindered Cbl-EGFR interactions showed impaired rates of receptor internalization, though the complete inhibition of internalization was never observed (19, 21, 22, 44). Cbl was identified previously as a component of the CIN85-endophilin complex, which is thought to induce the negative membrane curvature required for clathrin-coated pit invagination and endocytosis (33, 41, 44, 47). Other findings indicate a role for EGFR multiubiquitination in endocytosis. The modification of certain plasma membrane proteins by multiubiquitination appears to be sufficient for their internalization (18, 20, 33). In addition, chimeric forms of EGFR fused to Ub to mimic a Cbl-multiubiquitinated receptor are constitutively internalized (18, 37).
On the other hand, many studies argue against a role for Cbl in EGFR internalization (7, 9, 16, 29, 30, 49). For example, the finding that receptor dimerization rather than kinase activation is sufficient to cause EGFR endocytosis suggests that Cbl tethering to EGFR—an event which requires the receptor to be phosphorylated—is dispensable for internalization (51, 52). It has also been demonstrated previously that Cbl overexpression does not significantly enhance EGFR internalization (28, 49). In experiments utilizing dominant-negative forms of Cbl, EGFR downregulation was severely inhibited, though the receptor was still observed to localize to internal vesicles (29). Moreover, studies using cells deficient in c-Cbl or conditionally defective in ubiquitination have revealed little or no impairment in EGFR internalization, although overall receptor downregulation was reduced (9). These studies went on to show that this reduction was caused by the defective sorting of EGFR into lysosomes. Evidence in support of this conclusion was provided by the finding that the ubiquitination of EGFR is necessary for its efficient transfer to the inner membranes of multivesicular bodies (30). Overall, these findings indicate a more downstream role for Cbl in EGFR downregulation than that at the level of EGFR internalization, namely, one at the level of late endosomal sorting and degradation.
The precise role played by Cbl during EGFR trafficking also remains to be clarified. While Cbl-mediated ubiquitination is likely a key factor in EGFR degradation, the necessity for continued Cbl-EGFR interaction in this process is uncertain (7, 16, 28). Many of the proteins involved in EGFR signaling and regulation are also targets of Cbl association and/or Cbl-mediated ubiquitination, including Grb2, phosphatidylinositol 3-kinase, Cdc42, and a wide array of non-RTKs (e.g., c-Src) (1, 3, 14, 24, 48, 56). While a number of these proteins are regulated by Cbl concurrently with EGFR (such as those directly in complex with the receptor), others are likely to be regulated by Cbl following disassociation from EGFR. It was recently demonstrated that after EGF-induced activation, Cool-1, while acting as a guanine nucleotide exchange factor for Cdc42, is able to sequester Cbl and decrease its association with EGFR, thereby regulating Cbl-EGFR interaction and thus Cbl-mediated EGFR ubiquitination (13, 56). Another significant observation is that Cbl undergoes EGF-induced autoubiquitination during its routing with RTKs (11, 54). An unresolved question is whether Cbl is degraded in lysosomes with the EGFR or in the proteosome. Cbl-b has been shown to be sorted with EGFR into later endocytic compartments and to coordinate EGFR degradation (11). Interestingly, results from these and related studies also reveal that the ubiquitination of both Cbl and EGFR is dependent on a functioning proteosome (10, 11). This finding is intriguing to say the least, as ubiquitinated EGFR is strongly believed to be degraded by lysosomal hydrolysis.
Studies of individual Cbl homologues seem to reveal a basic functional redundancy among them, and yet an even more complex picture is emerging (50). The possibility that c-Cbl and Cbl-b—structurally similar and widely coexpressed—may play distinct roles in EGFR regulation is supported by several recent findings (6, 10, 12). For example, overexpressed Cbl-b is more potent than c-Cbl in inhibiting EGF-mediated growth and in decreasing the output of signaling through the phosphatidylinositol 3-kinase-Akt pathway (12). Another finding showed that the carboxyl-terminal Ub-associated (UBA) domain of Cbl-b, but not that of c-Cbl, can bind ubiquitinated proteins (6). Furthermore, the overexpression of Cbl-b's UBA domain blocks EGFR degradation, likely acting in a dominant-negative fashion (6). Differential sorting routes for the Cbls may also exist, as suggested by the involvement of both lysosomal and proteasomal functioning in the degradation of Cbl and its receptor substrate. Together, these intriguing differences raise the question as to how the different Cbls interplay in EGFR downregulation.
In this study, we have demonstrated that Cbl interaction with EGFR and Cbl-mediated EGFR ubiquitination are neither necessary nor sufficient for EGF-mediated EGFR internalization. Cbl's negative regulation occurred at the level of EGFR degradation. Ubiquitination mediated by either c-Cbl or Cbl-b was sufficient to cause significant EGFR degradation, though both proteins were involved in the degradation. Furthermore, by utilizing RNA interference (RNAi) methods to knock down either Cbl protein, we uncovered two temporally distinct peaks of EGFR ubiquitination. Either peak corresponded to a high level of EGFR association with a particular Cbl protein. c-Cbl interaction with EGFR was strongest at an early stage of EGFR trafficking, while Cbl-b's peak association occurred later. In addition, we found specific dephosphorylation of pY1045 on EGFR to occur concomitantly with c-Cbl disassociation. We conclude that Cbl acts as a negative regulator of EGFR via Ub-mediated degradation, not internalization, and that there exists a temporal interplay of two Cbl isoforms with EGFR during the receptor's internal trafficking route.
MATERIALS AND METHODS
Antibodies and chemicals.
Rabbit anti-EGFR, anti-c-Cbl, anti-Cbl-b, mouse anti-pY99, anti-Ub, and goat anti-phospho-EGFR (pY1068 specific) were from Santa Cruz Biotech (Santa Cruz, CA). EGFR phosphotyrosine-specific rabbit anti-pY992 and mouse anti-pY1045 were obtained from Biosource (Medicorp) and U.S. Biological, respectively. Mouse anti-green fluorescent protein (anti-GFP) antibody used for immunoblotting and the pEYFP-N1 vector were from Clontech (Palo Alto, CA). Rabbit anti-GFP used for EGFR immunoprecipitation (IP) was a generous gift from L. Berthiaume. Alexa Fluor 647-labeled EGF and EGF were from Molecular Probes Inc. (Eugene, OR). Unless otherwise specified, all other chemicals were from Sigma (St. Louis, MO).
Plasmid construction.
The pEYFP-N1 expression plasmid containing the wild-type (wt) EGFR sequence was used as a template for the construction of all mutants employed in this study. For the creation of the EGFR Y1045F and LL/AA substitution mutations, the QuikChange XL site-directed mutagenesis kit from Stratagene was used. Briefly, primer pairs which mismatched the wt sequence at two or three central nucleotides were designed to accommodate the required substitution and used to amplify full-length plasmids containing the mutation. For the creation of the mutant forms truncated at residues 1044, 992, 973, and 957, upstream and downstream primers were designed to be complementary to the starting nucleotide sequence and the nucleotide sequence adjacent to the truncation site, respectively. Then the various EGFR truncation fragments with XhoI and KpnI restriction sites were amplified from the pEYFP/wtEGFR plasmid and subcloned into a new pEYFP vector.
Cell culture, transfection, and treatment.
293T and BT20 cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. 293T cells were transfected using calcium phosphate precipitation or using RNAi max Lipofectamine (Invitrogen, CA) reagent when cells were to be cotransfected with double-stranded RNA (dsRNA) and EGFR constructs. Following transfection, cells were serum starved overnight prior to treatment, and cycloheximide was added (at a final concentration of 0.5 μM) 30 min prior to treatment when the inhibition of protein synthesis was necessary. For EGF treatment, cells were precooled to 4°C and pulsed with EGF for 30 min. After being pulsed, cells were washed, given fresh serum-free medium, and transferred to 37°C to initiate the stimulation time course.
Subcellular fractionation.
Following treatment, 293T or BT20 cells were scraped into homogenization buffer (0.25 M sucrose, 20 mM Tris-HCl [pH 7], 1 mM MgCl2, 4 mM NaF, and 0.02% NaN3) containing 0.5 mM Na3VO4, 0.1 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride], 10 μg of aprotinin/ml, and 1 μM pepstatin A and homogenized by being ground 30 times in a glass-walled homogenizer. Homogenates were twice precleared of nuclei by centrifugation (at 100 × g for 5 min), and the resulting postnuclear supernatant was fractionated into total membrane and cytosolic fractions by centrifugation at 150,000 × g for 1 h. Total membrane fractions were resuspended in homogenization buffer and centrifuged again, along with cytosolic fractions, at the same speed as before. Fractions were then solubilized in 1% NP-40, assayed by the Bradford method for protein concentration, and then separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis.
IP and immunoblotting.
All methods described herein were carried out as described previously (53). Following treatment, 293T or BT20 cells were lysed in 1% NP-40 for 30 min and then assayed for protein concentration by the Bradford method. For IPs, lysates were first precleared with secondary IP immunoglobulin G (IgG; anti-rabbit protein A-conjugated IgG) and then incubated overnight with 1 μg of anti-GFP (rabbit) primary antibody per 1 mg of total protein. GFP (EGFR) precipitates were then incubated with 20 μg of anti-rabbit protein A-conjugated IgG for 3 h, centrifuged (at 1,000 × g for 1 min), and washed three times with phosphate-buffered saline, and the resulting pellet was resuspended in sample buffer containing SDS. Protein samples were separated by SDS-polyacrylamide gel electrophoresis: equal volumes of IP preparations (20 μl/lane) or equal amounts of lysates (20 μg/lane) were loaded onto 10% polyacrylamide gels. Following separation, proteins were transferred onto nitrocellulose and probed with primary antibodies. The primary antibodies were detected with a horseradish peroxidase-conjugated secondary antibody followed by enhanced chemiluminescence development (Pierce Chemical, Rockford, IL) and light detection on Fuji Super RX film (Tokyo, Japan). For graphical analysis, subsaturated band exposures were scanned using a GS-800 calibrated densitometer and quantified using Quantity One software (Bio-Rad).
Flow cytometry.
EGFR internalization was assayed by flow cytometry by a method modified from that previously described by Duan et al. (9). Briefly, cells at ∼75% confluence were serum starved overnight and incubated with cold Alexa Fluor 647-labeled EGF at 4°C for 30 min. Excess ligand was removed, and cells were incubated at 37°C for the times indicated in the figures to allow internalization. Following each time point, cells were washed with an acid solution (0.2 M acetic acid, 0.5 M NaCl, pH 2.8) to remove uninternalized EGF, and then the fluorescence emission from internalized EGF was detected by flow cytometry.
RNAi.
Strong candidate sequences for the coding regions for both c-Cbl and Cbl-b were chosen by algorithms provided by Invitrogen and used to design small interfering RNA (siRNA) duplexes for use in all RNAi experiments performed. Candidate c-Cbl and Cbl-b siRNAs were tested in 293T and BT20 cells, along with a nonspecific control siRNA, to assess optimal times of knockdown and concentrations of dsRNA. An RNAi max transfection kit was employed for the cotransfection of cells with dsRNA and EGFR constructs (Invitrogen, CA). Significant (>90%) knockdown was obtained following 48 h of transfection using 100 pmol of c-Cbl dsRNA and 200 pmol of Cbl-b dsRNA. Neither Cbl siRNA duplex, nor the negative siRNA duplex, showed nonspecific interference effects.
RESULTS
c-Cbl interaction with EGFR and c-Cbl-mediated ubiquitination of EGFR are neither required nor sufficient for EGFR ligand-induced internalization.
We first employed three different EGFR mutant forms, including the Y1045F mutant receptor [EGFR(Y1045F)], the form truncated from the C terminus to position 1044 (EGFR1044), and the receptor with the 1010LL/AA mutation (EGFR1010LL/AA), along with wt EGFR, to study the role of c-Cbl interaction and the c-Cbl-mediated ubiquitination of EGFR in the internalization of EGFR following EGF stimulation (Fig. 1). EGFR(Y1045F) was newly constructed with a single phenylalanine substitution at tyrosine 1045. Both EGFR1044 and EGFR1010LL/AA, which contains two alanines replacing L1010 and L1011, were constructed previously. EGFR and the mutant proteins were tagged with yellow fluorescent protein (YFP).
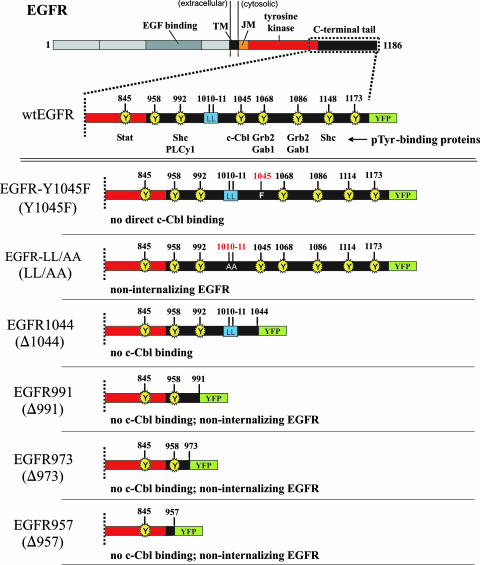
FIG. 1.
EGFR constructs employed in this study. Each construct was tagged at its C terminus with YFP. EGFR(Y1045F) contains a single phenylalanine substitution at tyrosine 1045 which has been shown to abolish its ability to interact directly with c-Cbl (27). EGFR1044, an EGFR truncated after residue 1044, should conceivably lack both direct c-Cbl interaction (via pY1045) and indirect interaction at pY1068 and pY1086 via Grb2. The EGFR-LL/AA mutant, in which alanines replace two critical leucines at positions 1010 and 1011, has been previously reported to be deficient in ligand-induced internalization (52). EGFR991, EGFR973, and EGFR957 are EGFRs truncated from their C termini to positions 991, 973, and 957, respectively, and were constructed previously (52). TM, transmembrane domain; JM, juxtamembrane domain; PLCy1, phospholipase C-γ1; pTyr, phosphotyrosine.
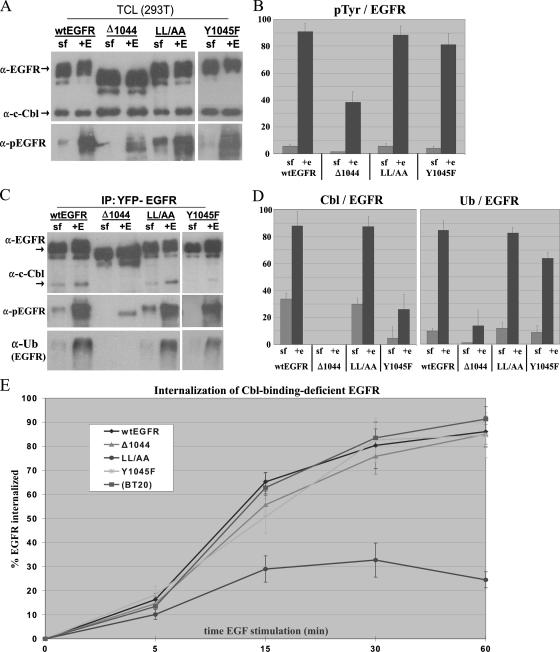
293T cells were transiently transfected with sequences encoding YFP-tagged wt EGFR, EGFR(Y1045F), EGFR1044, and EGFR1010LL/AA, serum starved overnight, and then either treated with 100 ng of EGF/ml for 10 min or left untreated. We showed by immunoblotting that wt EGFR and all mutant EGFRs were expressed at similar levels (Fig. 2A). No endogenous EGFR was detected—this result was expected as 293T cells express very low levels of the receptor. c-Cbl was expressed at similar levels in cells transfected with various mutant constructs (Fig. 2A). Immunoblotting for phosphotyrosine demonstrated that EGFR was activated by EGF to similar levels in EGFR(Y1045F)- and EGFR1010LL/AA-expressing cells and to a lesser extent in cells expressing EGFR1044. Densitometric analysis of phosphotyrosine immunoblots showed that EGFR1044 phosphorylation was approximately 45% the level of wt EGFR phosphorylation (Fig. 2B); this result is likely due to the lack of phosphorylated tyrosines downstream of residue 1044.
FIG. 2.
The effects of EGF-induced EGFR association with Cbl and EGFR ubiquitination on EGFR internalization. 293T cells were transiently transfected with a sequence encoding YFP-tagged wt EGFR, EGFR1044 (Δ1044), EGFR1010LL/AA (LL/AA), or EGFR(Y1045F) (Y1045F). Following overnight removal of serum, cells were treated for 10 min with 100 ng of EGF/ml (+E) or left untreated (sf). (A and C) TCLs were immunoblotted directly (A) or first immunoprecipitated with anti-YFP and then immunoblotted with the indicated antibodies (C). α-pEGFR, anti-phospho-EGFR. (B and D) Graphical representation of immunoblot data. The IP and immunoblot analyses presented in panels A and C were performed in triplicate, and the corresponding band intensities were quantified by densitometry. (B) Graphical representation of the anti-phospho-EGFR/anti-EGFR band intensity ratio taken from TCL immunoblots in panel A. pTyr, phosphotyrosine. (D) Graphical representation of anti-Cbl/anti-EGFR and anti-Ub/anti-EGFR band intensity ratios taken from the immunoblots in panel C. All data were normalized to EGFR and arbitrarily fit to a scale of 0 to 100. (E) Quantitative analysis of EGF-mediated internalization of various EGFR mutant forms by flow cytometry. 293T cells were transiently transfected with a sequence encoding YFP-tagged wt EGFR, EGFR1044, EGFR1010LL/AA, or EGFR(Y1045F). BT20 cells were used to assess the internalization of endogenous EGFR. Following overnight removal of serum, cells were treated with 100 ng of Alexa Fluor 647-labeled EGF/ml for the times indicated and their rates of internalization were measured by flow cytometry. Data are the means of results from at least three experiments performed in triplicate.
In order to assess the state of Cbl interaction with and Cbl-mediated ubiquitination of the various EGFR mutant forms, we subjected the same cell lysates analyzed as described above to IP with an anti-YFP antibody and probed for c-Cbl and Ub (Fig. 2C and D). Both wt EGFR and EGFR1010LL/AA interacted with c-Cbl, and the two forms were ubiquitinated to similar extents following EGF stimulation (Fig. 2C). c-Cbl association with EGFR(Y1045F) was significantly reduced compared to that with wt EGFR (to ∼30% of the level of Cbl-wt EGFR association). However, the level of EGFR(Y1045F) ubiquitination was unexpectedly high—approximately 75% of the level of wt EGFR ubiquitination. No c-Cbl association with EGFR1044 was observed, and EGFR1044 was not ubiquitinated in response to EGF.
We then evaluated the EGF-induced internalization of these mutant receptors in 293T cells transiently transfected with sequences encoding YFP-tagged wt EGFR or mutant forms by flow cytometry. We also assessed the rate of internalization of endogenous EGFR in BT20 cells. As shown in Fig. 2E, after 30 min of stimulation, ∼80% of overexpressed wt and endogenous EGFRs were internalized. Interestingly, both EGFR(Y1045F) and EGFR1044 were internalized to the same extent as wt EGFR. These results demonstrate that neither c-Cbl association with EGFR nor Cbl-mediated ubiquitination of EGFR is required for EGF-mediated receptor internalization. On the other hand, EGFR1010LL/AA, as expected, showed only minimal internalization, which indicates that neither c-Cbl association with EGFR nor Cbl-mediated ubiquitination of EGFR is sufficient for EGF-mediated receptor internalization.
Characterization of Cbl-EGFR interaction during EGFR trafficking.
Despite Cbl's established role as a downregulator of EGFR signaling, our results suggested that Cbl does not exert its negative regulatory role at the level of EGFR internalization. Reports of other investigations have proposed a more downstream role for Cbl in EGFR trafficking (22, 38, 45). We decided to characterize Cbl-EGFR interaction during the receptor's internal trafficking following ligand stimulation.
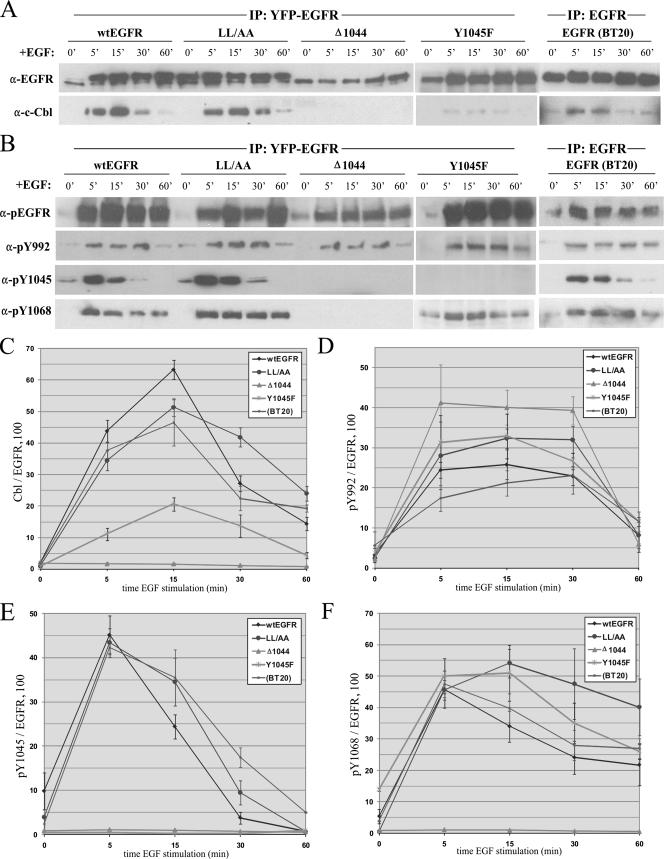
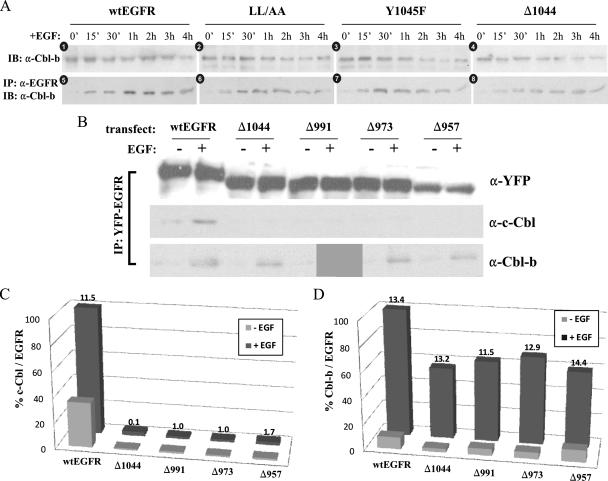
To directly assess c-Cbl-EGFR interaction during EGFR trafficking, we performed IP experiments. 293T cells transfected with a sequence encoding YFP-tagged wt EGFR, EGFR1010LL/AA, EGFR1044, or EGFR(Y1045F) were stimulated with EGF over the same 1-h time course used for the experiments described above. At each time point, cell lysates were immunoprecipitated using anti-YFP antibody and immunoblotted with antibodies to EGFR and c-Cbl (Fig. 3A). c-Cbl association with wt EGFR was strongest within the first 15 min of stimulation with EGF (Fig. 3C). After this time, the level of c-Cbl interaction decreased rapidly and was virtually undetectable by 60 min of EGF stimulation. In EGFR1010LL/AA-expressing cells, c-Cbl disassociation was delayed approximately 15 min, while EGFR1044-expressing cells showed no c-Cbl interaction. The initial interaction of EGFR(Y1045F) with c-Cbl displayed dynamics similar to those of the interaction of wt EGFR with c-Cbl, though the overall level of c-Cbl association with this mutant form was substantially lower than that of c-Cbl association with the wt (Fig. 3C). To ensure that our observations were not limited to 293T cells exogenously expressing an exogenous EGFR, we performed the same experiments with BT20 cells by using a monoclonal EGFR antibody to immunoprecipitate the endogenous receptor (Fig. 3A, rightmost panels). Immunoblotting for c-Cbl revealed that the endogenous EGFR interacted with c-Cbl similarly to wt EGFR in 293T cells. It is also important that, in these experiments, no degradation of EGFR by 1 h was observed. To eliminate the possibility that the repletion of EGFR levels by new protein synthesis was interfering with the interpretation of our data, we performed the same experiments in the presence of 50 μg of cycloheximide/μl but found no difference in the levels of EGFR or in the pattern of c-Cbl interaction (data not shown).
FIG. 3.
c-Cbl-EGFR association during EGF-mediated EGFR trafficking and its relationship with EGFR phosphorylation at Y1045. 293T cells were transiently transfected with a sequence encoding YFP-tagged wt EGFR, EGFR1044 (Δ1044), EGFR1010LL/AA (LL/AA), or EGFR(Y1045F) (Y1045F), serum starved overnight, and then treated with 100 ng of EGF/ml for the times indicated. (A and B) Cell lysates were immunoprecipitated using anti-YFP and then immunoblotted with anti-EGFR and anti-Cbl (A) or antibodies to EGFR phosphotyrosines (B). BT20 cells were treated in parallel to assess the association of Cbl with endogenous EGFR. 0′, 0 min; α-pEGFR, anti-phospho-EGFR. (C to F) Quantitative graphical analyses of c-Cbl association with EGFR (C) and phosphorylation of specific EGFR tyrosines (D to F). Immunoblot analyses depicted in panels A and B were performed in triplicate, and the band intensities were quantified by densitometry. Each datum point represents a percentage ratio of band intensities, normalized to EGFR.
Our results demonstrate that a significant portion of c-Cbl disassociated from EGFR after between 15 and 30 min of EGF stimulation, a time well in advance of receptor degradation. Indeed, we observed no appreciable level of EGFR degradation over the first hour of EGF stimulation (Fig. 3A), nor was the loss of c-Cbl association due to the loss of c-Cbl itself, as c-Cbl levels had not significantly changed even after 2 h of EGF stimulation, when EGFR degradation begins to occur (Fig. 4A, panels 1 and 2). Together, these findings suggested that c-Cbl disassociation preceded EGFR degradation and prompted us to investigate specific mechanisms of c-Cbl disassociation.
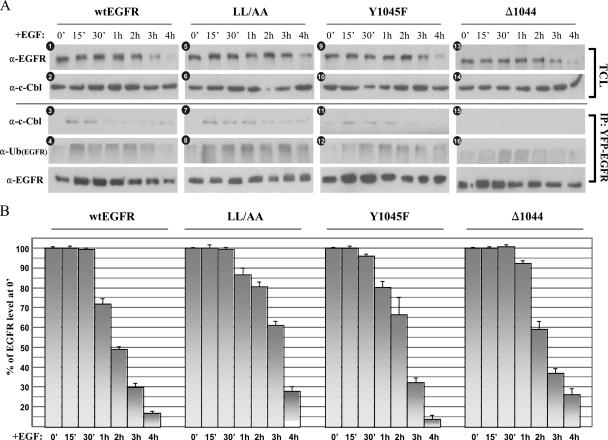
FIG. 4.
Effects of EGF-induced EGFR association with c-Cbl and EGFR ubiquitination on the degradation of EGFR. 293T cells were transiently transfected with a sequence encoding YFP-tagged wt EGFR, EGFR1010LL/AA (LL/AA), EGFR(Y1045F) (Y1045F), or EGFR1044 (Δ1044). Cells were serum starved overnight and then treated with 100 ng of EGF/ml for the times indicated. (A) TCLs were either immunoblotted directly or first immunoprecipitated with anti-YFP and then immunoblotted with anti-EGFR (α-EGFR), anti-c-Cbl, or anti-Ub. The immunoblots shown are representative of at least three independent experiments. α-Ub(EGFR), anti-ubiquitinated EGFR. (B) Analysis of wt EGFR and mutant EGFR degradation over 4-h time course of EGF stimulation. Anti-EGFR blots of TCLs as shown in panel A were prepared in triplicate and densitometrically quantified, and the results were normalized to the anti-EGFR band at 0 min (0′). Data are means (± standard deviations [SD]).
We examined whether c-Cbl disassociation resulted from specific alterations to EGFR. We screened the immunoprecipitates from the analysis presented in Fig. 3A with a panel of antibodies specific to major EGFR phosphotyrosines (anti-pY992, anti-pY1045, and anti-pY1068) and an antibody that recognizes all phosphotyrosines on EGFR (anti-pEGFR) (Fig. 3B). Interestingly, the level of Y1045 phosphorylation of either transiently expressed or endogenous EGFR significantly decreased after 15 min of stimulation with EGF, and this decrease was contemporaneous with c-Cbl disassociation (Fig. 3E). Moreover, Y1045 dephosphorylation of EGFR1010LL/AA was delayed compared with that of wt EGFR in 293T cells, and the delay was comparable with the delay of c-Cbl disassociation observed for EGFR1010LL/AA (Fig. 3C and E). The dephosphorylation at Y1045 was not a result of total receptor dephosphorylation, as immunoblotting for total phosphotyrosine showed no change in overall EGFR hyperphosphorylation over 1 h of EGF stimulation (Fig. 3B, top-lane panels). Furthermore, dephosphorylation at pY1045 appeared to be specific, since the levels of Y992 and Y1068 phosphorylation did not fall as sharply as that of Y1045 phosphorylation (compare Fig. 3D and F with E). Y992 also appeared to be specifically dephosphorylated, though this dephosphorylation occurred much later than Y1045 dephosphorylation (Fig. 3D). Although pY1045 is the major Cbl docking site, Cbl can also interact with EGFR though Grb2 at pY1068 on the receptor, though we did not observe significant dephosphorylation at this site during c-Cbl disassociation from EGFR. Together, these results suggest a potential mechanism for Cbl disassociation involving specific dephosphorylation of the EGFR at Y1045.
Cbl-mediated ubiquitination and degradation of EGFR.
To elucidate the overall role of Cbl in EGFR downregulation, we first investigated wt EGFR levels by Western blotting of total cell lysates (TCLs) over an extended time course of EGF stimulation in order to profile the receptor's degradation (Fig. 4A, panel 1, and B). In wt EGFR sequence-transfected 293T cells, we observed significant receptor degradation after 2 h of EGF stimulation. By 4 h of EGF stimulation, wt EGFR levels were virtually undetectable while c-Cbl levels remained unchanged (Fig. 4A, panel 2). Similar patterns of degradation of the endogenous receptor and c-Cbl in BT20 cells were observed (see Fig. 9). All these experiments were performed in the presence of cycloheximide in order to inhibit new protein synthesis; parallel experiments performed without inhibition still showed EGFR degradation after 2 h of EGF stimulation, though the effect was less pronounced (data not shown). We next assessed the state of wt EGFR ubiquitination over the same extended time course. As we were unable to confidently assess the ubiquitination of EGFR by using our EGFR antibody alone, we instead immunoprecipitated EGFR and probed the precipitates with the same antibody used for the analyses presented in Fig. 2 that recognizes both monomeric ubiquitin and polyubiquitin (Fig. 4A, panel 4). Receptor ubiquitination rose strongly to reach a peak around 15 min after the beginning of EGF stimulation, after which ubiquitination levels fell by ∼30%. To our surprise, a small rise in receptor ubiquitination appeared after 30 min of EGF stimulation, peaking sometime between 1 and 2 h of EGF stimulation, a time when c-Cbl interaction was already very low, as shown in the corresponding c-Cbl immunoblot of immunoprecipitated EGFR (Fig. 4A, panel 3).
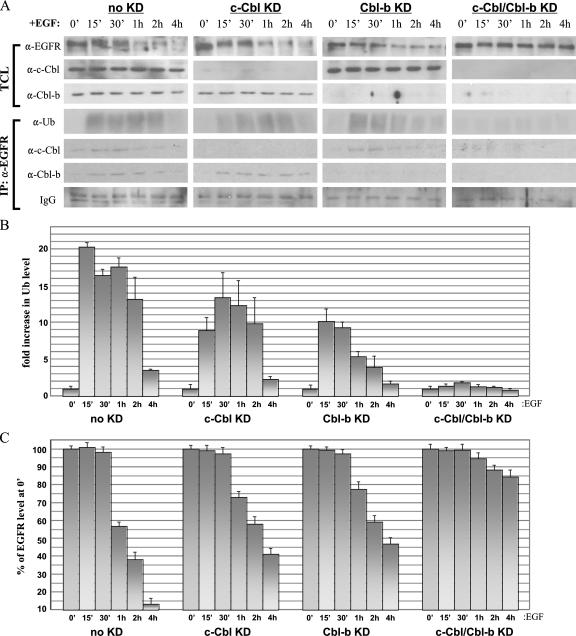
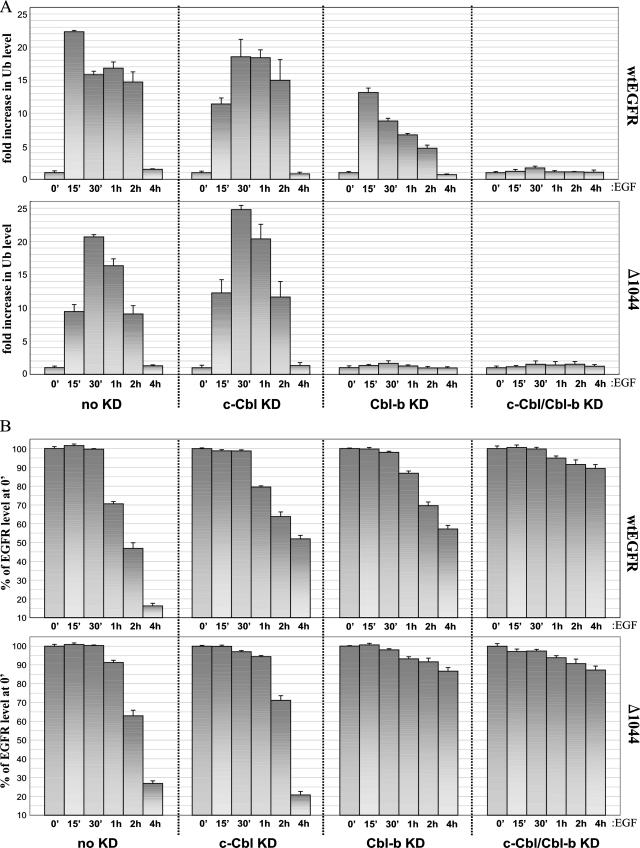
FIG. 9.
Effects of c-Cbl and/or Cbl-b knockdown on EGFR ubiquitination and degradation in BT20 cells. (A) BT20 cells were transfected with one of the following RNAi duplexes: 100 pmol of nonspecific control dsRNA (no knockdown [no KD]), 100 pmol of c-Cbl dsRNA (c-Cbl KD), 100 pmol of Cbl-b dsRNA (Cbl-b KD), or 100 pmol each of c-Cbl and Cbl-b dsRNAs (c-Cbl/Cbl-b KD). Thirty-six hours after transfection, cells were serum starved overnight and then treated with 100 ng of EGF/ml for the times indicated. TCLs were either immunoblotted directly or first immunoprecipitated with anti-YFP and then immunoblotted with the indicated antibodies. α-EGFR, anti-EGFR. (B) Graphical analysis of Cbl-mediated ubiquitination in BT20 cells. Anti-Ub blotting analyses of EGFR immunoprecipitates as presented in panel A were performed in triplicate, the corresponding band intensities were quantified by densitometry, and the results were normalized to the anti-Ub band at 0 min (0′). (C) Graphical analysis of EGFR degradation in BT20 cells. Anti-EGFR blots of TCLs as shown in panel A were prepared in triplicate and densitometrically quantified, and the results were normalized to the anti-EGFR band at 0 min.
We next performed parallel experiments using 293T cells transfected with a sequence encoding EGFR1010LL/AA or EGFR(Y1045F) (Fig. 4). The profile of EGFR(Y1045F) degradation was similar to that of wt EGFR degradation, though EGFR1010LL/AA degradation occurred later than wt EGFR degradation (Fig. 4A, panels 1 and 5, and B). This delay in EGFR1010LL/AA degradation is not surprising; the inability of EGFR1010LL/AA to undergo ligand-induced endocytosis would mean that it relied primarily on constitutive endocytosis mechanisms to accumulate internally in endosomes. We then looked at the ubiquitination of the EGFR1010LL/AA and EGFR(Y1045F) mutant forms (Fig. 4A, panels 8 and 12). The initial ubiquitination level of EGFR(Y1045F) was expectedly lower than that of wt EGFR, but the level of ubiquitination increased significantly after 15 min of EGF stimulation to peak at around 1 h of EGF stimulation. The delayed rise in the ubiquitination level was unanticipated since the degree of c-Cbl association was already very low at this time. This result extended the previously observed disparity between the unexpectedly high level of EGFR(Y1045F) ubiquitination relative to the level of c-Cbl association (Fig. 2B). When we looked at EGFR1010LL/AA ubiquitination following EGF stimulation, we observed a pattern qualitatively similar to that of wt EGFR ubiquitination, although the first rise in EGFR1010LL/AA ubiquitination occurred slowly (over the first hour of EGF stimulation) and then, following a small fall in ubiquitination, the level rose again at ∼3 h of EGF stimulation. We also noticed that the overall intensity of EGFR1010LL/AA ubiquitination was greater than that of wt EGFR ubiquitination (Fig. 4A, panel 8). A longer time of c-Cbl retention with the plasma membrane-bound receptor can only partially explain this higher level of ubiquitination, as the later wave of ubiquitination occurred well after c-Cbl disassociation.
When we transfected 293T cells with the construct encoding the truncated EGFR1044, we were surprised to find that this mutant form was not only significantly degraded but was also ubiquitinated in response to EGF (Fig. 4A, panels 13 and 16), despite the complete lack of c-Cbl interaction (Fig. 4A, panel 15). Earlier we observed low levels of EGFR1044 ubiquitination following 10 min of EGF stimulation (Fig. 2D). Here, we showed that over the extended EGF stimulation time course, EGFR1044 was undergoing significant ubiquitination, peaking at 1 h of EGF stimulation.
EGFR association with Cbl-b in response to EGF.
Our above-described results strongly indicated that another E3 Ub ligase, in addition to c-Cbl, was acting to affect the ubiquitination and degradation of EGFR. Another major Cbl isoform, Cbl-b, was a strong candidate for investigation, given that its size, N-terminal domains, and EGFR regulatory functions are all very similar to those of c-Cbl (6, 10, 12). We therefore obtained an antibody specific to Cbl-b and first determined the total levels of Cbl-b by Western blotting of the lysates and immunoprecipitates prepared for the analyses presented in Fig. 4A (Fig. 5A). In 293T cells transfected with sequences encoding wt EGFR or each of the mutant derivatives, Cbl-b was significantly expressed, although unlike c-Cbl levels, total Cbl-b levels did not remain constant during the EGF stimulation time course (Fig. 5A, top panels). We were unable to explain the small degree of fluctuation observed in Cbl-b levels at later times of EGF stimulation, although the general trend revealed a decrease of the protein by 3 to 4 h of EGF stimulation, contemporaneous with EGFR degradation. Performing the same experiments without cycloheximide did not alter this observed decline (data not shown).
FIG. 5.
EGF-induced association between EGFR and Cbl-b. (A) EGF-induced association between EGFR and Cbl-b. 293T cells were transiently transfected with a sequence encoding YFP-tagged wt EGFR, EGFR1010LL/AA (LL/AA), EGFR(Y1045F) (Y1045F), or EGFR1044 (Δ1044). Cells were serum starved overnight and then treated with 100 ng of EGF/ml for the times indicated. TCLs and anti-EGFR (α-EGFR) immunoprecipitates described in the legend to Fig. 4A were immunoblotted (IB) with an antibody to Cbl-b. The immunoblots shown are representative of at least three independent experiments. 0′, 0 min. (B) EGF-induced association between c-Cbl or Cbl-b and various truncated EGFRs. 293T cells were transiently transfected with a sequence encoding YFP-tagged wt EGFR, EGFR1044, EGFR991 (Δ991), EGFR973 (Δ973), or EGFR957 (Δ957). Cells were serum starved overnight and then treated with 100 ng of EGF/ml for 20 min. TCLs were first immunoprecipitated with anti-YFP and then immunoblotted with anti-EGFR, anti-c-Cbl, or anti-Cbl-b antibodies. +, with EGF; −, without EGF. (C and D) Quantitative analysis of c-Cbl (C) and Cbl-b (D) interaction with wt EGFR and the EGFR truncation constructs listed above. Western blots of EGFR immunoprecipitates as shown in panel B were prepared in triplicate and densitometrically quantified. Datum points represent percentage ratios of band intensities (calculated as Cbl band intensity/EGFR band intensity × 100), normalized to wt EGFR following EGF stimulation. Values given at the top of “+EGF” bars are SD values.
We next tested Cbl-b association with EGFR by co-IP. The EGFR immunoprecipitates were immunoblotted with anti-Cbl-b (Fig. 5A). As seen in the bottom panels in Fig. 5A, Cbl-b interacted with wt EGFR, EGFR1010LL/AA, EGFR(Y1045F), and EGFR1044 following EGF stimulation. Significantly, in all transfectants, Cbl-b association with EGFR was weak at early time points of EGF stimulation. This delayed association of Cbl-b following EGF stimulation was also observed for the endogenous receptor in BT20 immunoprecipitates of EGFR (see Fig. 9A). Most striking was that Cbl-b associated with the EGFR1044 truncation form (Fig. 5A, panel 8). Although Cbl-b's association with the truncated EGFR1044 form was weaker than its association with the other EGFRs, the association remained EGF dependent. While IP experiments showed a strong association between EGFR and Cbl-b even at 4 h of EGF stimulation, this result reflects only the association between undegraded EGFR and Cbl-b. As shown in Fig. 4, at 4 h, the majority of EGFR (Fig. 4A, panels 1, 5, 9, and 13) and Cbl-b (Fig. 5A) had already degraded. Overall, these findings demonstrate that unlike that of c-Cbl, the association of Cbl-b with EGFR following EGF stimulation is delayed and prolonged.
To further explore this apparently differential association of Cbl-b and c-Cbl with the EGFR, we examined which region of EGFR is responsible for binding Cbl-b. 293T cells were transfected with sequences encoding full-length wt EGFR or one of a series of EGFR truncation mutant forms, including EGFR1044, EGFR truncated at residue 991 (EGFR991), EGFR truncated at residue 973 (EGFR973), and EGFR truncated at residue 957 (EGFR957) (as shown in Fig. 1). The association of c-Cbl and Cbl-b with wt EGFR and the various EGFR mutant forms was evaluated by co-IP with or without EGF stimulation (Fig. 5B). As anticipated, both Cbls associated with the full-length receptor, though c-Cbl failed to interact with any of the truncation mutant receptors, thus demonstrating that c-Cbl interaction with EGFR is restricted to the region in the C-terminal direction from residue 1044 (Fig. 5B and C). Cbl-b, on the other hand, associated to similar extents with all EGFR truncation forms investigated, at a level approximately 50% of that with the full-length receptor (Fig. 5B and D). This result suggests that EGFR sequences in the C-terminal direction from 1044 remain important in mediating interaction with Cbl-b. Further truncation of EGFR to positions 991, 973, and 957 did not reduce the association of Cbl-b with EGFR, indicating that (i) sequences between positions 1044 and 958 are not essential in Cbl-b association and (ii) the EGFR intracellular domain in the N-terminal direction from position 958 contributes significantly (approximately 50%) to the EGF-mediated association of Cbl-b with the receptor.
Neither c-Cbl nor Cbl-b is required for EGF-induced EGFR internalization.
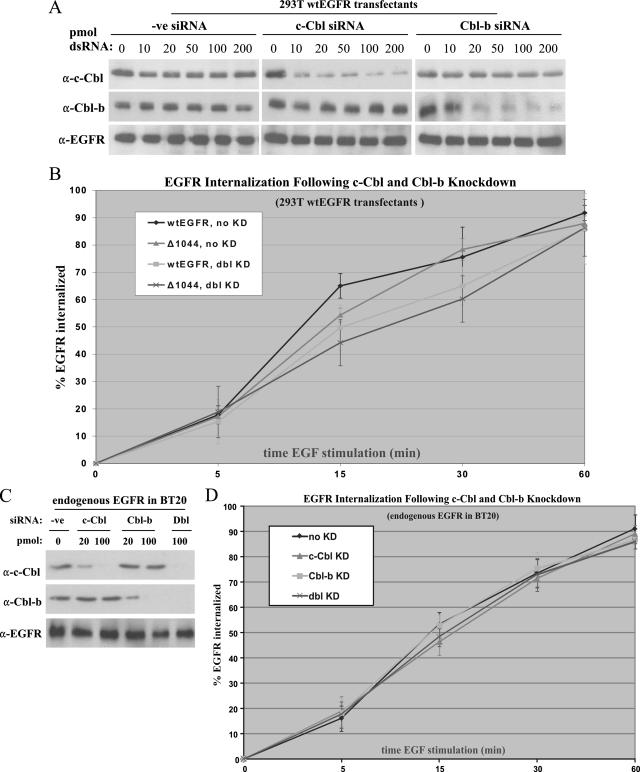
To conclusively define the roles of c-Cbl and Cbl-b in EGFR ubiquitination, internalization, and degradation, we employed RNAi experiments (Fig. 6). Strong candidate sequences in the coding regions for both Cbls were chosen and used to design siRNA duplexes for use in all our subsequent RNAi experiments. To first assess the degree of mRNA knockdown, 293T cells were cotransfected with candidate c-Cbl and Cbl-b siRNAs, along with a nonspecific control siRNA, and a sequence encoding wt EGFR. The immunoblots in Fig. 6A demonstrate significant and Cbl-specific knockdown (>90%) at 48 h posttransfection. The highest level of Cbl knockdown was observed using 100 pmol of c-Cbl dsRNA or 200 pmol of Cbl-b dsRNA. Neither Cbl siRNA duplex, nor the control siRNA duplex, showed nonspecific interference effects.
FIG. 6.
RNAi-mediated knockdown of c-Cbl or Cbl-b does not affect EGFR internalization. (A) 293T cells were transfected with a sequence encoding wt EGFR and the indicated duplex RNA (dsRNA) at various amounts for 48 h. Optimal amounts of dsRNA for knockdown were 100 pmol for c-Cbl and 200 pmol for Cbl-b. A control duplex (-ve siRNA) was employed to assess nonspecific effects on protein expression. α-c-Cbl, anti-c-Cbl. (B) Quantitative analysis by flow cytometry of EGF-mediated internalization following c-Cbl or Cbl-b knockdown in 293T cells transfected with a sequence encoding wt EGFR or EGFR1044 (Δ1044). Data are the means of results (± SD) from three independent experiments. KD, knockdown; dbl KD, double knockdown. (C) BT20 cells were transfected with the indicated types and amounts of dsRNA for 36 h. The optimal amount of dsRNA for effective knockdown was 100 pmol for both c-Cbl and Cbl-b. A control duplex (-ve siRNA) was employed to assess nonspecific effects on protein expression. Dbl, combination of c-Cbl siRNA and Cbl-b siRNA. (D) Quantitative analysis by flow cytometry of EGF-mediated internalization following c-Cbl and/or Cbl-b knockdown in BT20 cells. Data are the means of results (± SD) from three independent experiments.
Using the optimized conditions established from these experiments, we employed RNAi to reinvestigate the role of Cbl in EGFR internalization, now in the context of both Cbls. 293T cells were transfected with a sequence encoding wt EGFR or EGFR1044 along with siRNA duplexes corresponding to both Cbls or the control siRNA. Following 48 h of knockdown, in which the levels of both Cbls were reduced >90% as seen in Fig. 6A, cells were serum starved overnight and treated with 100 ng of fluorescently labeled EGF/ml for various times. EGFR internalization was then analyzed by flow cytometry (Fig. 6B). Overall, the c-Cbl-Cbl-b double knockdown had little effect on EGF-mediated internalization of either wt EGFR or EGFR1044. Initial internalization rates were nearly identical for all transfectants (between 15 and 20% of EGFR was internalized at 5 min of EGF stimulation); although there was a noticeable dip in receptor internalization rates for our double-knockdown transfectants compared with those for the no-knockdown transfectants during the middle of the stimulation time course, this decrease was statistically insignificant (P > 0.05 for all data pairs at 15 and 30 min of EGF stimulation). Nevertheless, by 60 min of EGF stimulation, the internalization levels for normal and Cbl-deficient transfectants were virtually identical.
To validate our results in the context of endogenous EGFR internalization, we knocked down c-Cbl and/or Cbl-b in BT20 cells by the same methods described above. As shown in Fig. 6C, significant knockdown of either Cbl was obtained following transfection with 100 pmol of the corresponding dsRNA duplex. EGFR internalization in BT20 cells was then examined by flow cytometry (Fig. 6D). The knockdown of either c-Cbl, Cbl-b, or both Cbls had no significant effect on the internalization of EGFR following EGF stimulation. Overall, these results suggest that Cbl knockdown does not significantly alter EGF-mediated EGFR internalization.
Either c-Cbl or Cbl-b is sufficient for EGF-mediated ubiquitination and the degradation of EGFR.
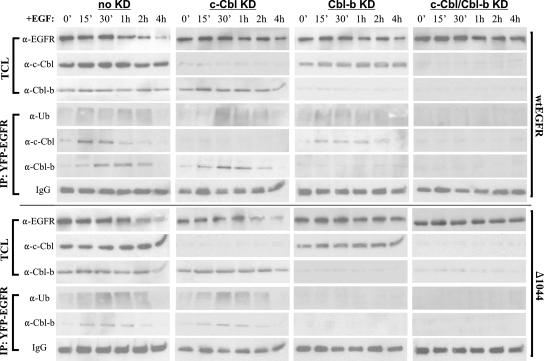
Since the levels of EGFR internalization by 1 h of EGF stimulation were nearly identical in all our transfectants regardless of the presence or absence of Cbl, we remained convinced that Cbl's function in EGFR downregulation was to induce receptor degradation later during trafficking. We next employed RNAi to address the absolute requirement for Cbl interaction and Cbl-mediated ubiquitination in EGFR downregulation. 293T cells cotransfected with a sequence encoding either YFP-tagged wt EGFR or EGFR1044 and control siRNA (no knockdown), c-Cbl siRNA (c-Cbl knockdown), Cbl-b siRNA (Cbl-b knockdown), or c-Cbl and Cbl-b siRNAs (double knockdown) were treated with EGF and analyzed over a 4-h time course by Western blotting (Fig. 7). TCLs from each time point were either immunoblotted directly or first immunoprecipitated with anti-YFP and then immunoblotted with various antibodies. The results from our no-knockdown transfectants (expressing a nonspecific siRNA) fully agreed with the data presented in Fig. 6. EGF-stimulated wt EGFR underwent rapid initial ubiquitination coincident with a rise in c-Cbl-EGFR interaction (at ∼15 min of EGF stimulation), while a later rise in ubiquitination, following a modest decline in ubiquitination, was coincident with a strong rise in Cbl-b-EGFR interaction (at ∼1 h of EGF stimulation). EGFR1044, on the other hand, showed only a single rise in ubiquitination, and this rise occurred only after 15 min of EGF stimulation, corresponding to the peak of EGFR1044 interaction with Cbl-b. Despite the difference in ubiquitination patterns between these receptors, both wt EGFR and EGFR1044 were significantly degraded by 4 h of EGF stimulation. We next turned to the immunoblots from our Cbl knockdown transfectants. Probing TCLs of the c-Cbl and Cbl-b knockdown cells revealed the disappearance of >90% of both homologues. Furthermore, neither Cbl protein could be appreciably detected in the corresponding EGFR immunoprecipitates, thus minimizing the possibility that even low levels of Cbl could affect EGFR downregulation. In c-Cbl knockdown cells, the ubiquitination of wt EGFR was delayed and appeared to be strongest when Cbl-b interaction was at its peak (at ∼30 min of EGF stimulation). In contrast, wt EGFR expressed in Cbl-b knockdown cells showed only an initial rise in ubiquitination following EGF stimulation. Despite these differences, significant degradation of wt EGFR (∼50%) occurred by 4 h of EGF stimulation in both c-Cbl knockdown and Cbl-b knockdown cells. When we extended our EGF stimulation time course in these single knockdown experiments, we did eventually observe even higher levels of degradation, by ∼6 h of EGF stimulation (data not shown), indicating that either Cbl homologue alone is sufficient to effect receptor degradation. Most importantly, both the ubiquitination and the degradation of wt EGFR in the double-knockdown cells were eliminated. When we looked at the immunoblots from EGFR1044 c-Cbl knockdown cells, the pattern of receptor ubiquitination and degradation was closely similar to that for cells with no knockdown. This result is not surprising, as c-Cbl cannot directly influence the truncated receptor. Under Cbl-b knockdown conditions, however, both receptor ubiquitination and degradation were virtually abolished, revealing that Cbl-b was alone necessary for both events in the case of EGFR1044.
FIG. 7.
Effects of c-Cbl and/or Cbl-b knockdown on EGFR ubiquitination and degradation in 293T cells. 293T cells were transfected with a sequence encoding YFP-tagged wt EGFR or EGFR1044 (Δ1044) and one of the following RNAi duplexes: 200 pmol of nonspecific control dsRNA (no knockdown [no KD]), 100 pmol of c-Cbl dsRNA (c-Cbl KD), 200 pmol of Cbl-b dsRNA (Cbl-b KD), or 100 pmol of c-Cbl dsRNA and 200 pmol of Cbl-b dsRNA (c-Cbl/Cbl-b KD). Forty-eight hours after transfection, cells were serum starved overnight and then treated with 100 ng of EGF/ml for the times indicated. TCLs were either immunoblotted directly or first immunoprecipitated with anti-YFP and then immunoblotted with the indicated antibodies. 0′, 0 min; α-EGFR, anti-EGFR.
We wanted to present the data in Fig. 7 in a comparative and consistent manner. To this end, we performed all anti-EGFR and anti-Ub immunoblot analyses presented in Fig. 7 in triplicate and quantitated band intensities by densitometry. In order to normalize the data for graphical analysis, samples corresponding to 0 min of EGF stimulation with no knockdown were run alongside each anti-Ub immunoblot of immunoprecipitated EGFR (Fig. 8A) and each anti-EGFR immunoblot (Fig. 8B). Figure 8A compares the ubiquitination levels of wt EGFR and EGFR1044. Under conditions of no knockdown, the ubiquitination pattern of wt EGFR was seen as an initial strong rise, peaking at 15 min of EGF stimulation, followed by a ∼25% decline in the signal and then another small rise of 5 to 10% at 1 h of EGF stimulation. Although the degree of receptor deubiquitination following the initial rise was clearly evident—ubiquitination levels at 30 min had fallen by 30% from those at 15 min of EGF stimulation—the meaningfulness of the second rise and/or augmentation of EGFR ubiquitination can be statistically disputed (Fig. 8A, top left graph: P > 0.05 for comparison of samples at 30 min and 1 h). It is possible that different receptors in the same sample were being ubiquitinated by c-Cbl and Cbl-b concurrently, which would serve to blunt the distinction between early and late ubiquitination events. In the case of either single Cbl knockdown, however, this overlap was eliminated and early and late ubiquitination events could be demarcated. By comparing the c-Cbl and Cbl-b knockdown graphs, it can be seen that the early rise in ubiquitination was ∼20% stronger than the later rise. It is also significant that the peak level of ubiquitination in either single Cbl knockdown sample was lower than the ubiquitination level seen under no-knockdown conditions. This result may likely indicate that a small fraction of Cbl-b ubiquitinates EGFR at early times of EGF stimulation and a small fraction of c-Cbl continues to ubiquitinate EGFR at later times. Interestingly, the trend of Cbl-b-mediated EGFR1044 ubiquitination showed a pronounced rise kinetically similar to the initial rise in wt EGFR ubiquitination following EGF stimulation. These subtle differences aside, it can be clearly seen that both Cbls account for total EGFR ubiquitination, as the elimination of both homologues virtually abolishes the Ub signal (Fig. 8A, top right graph). Furthermore, as shown by the top right graph of Fig. 8B, both Cbls participate in EGFR degradation, while eliminating either Cbl serves only to delay this event. Thus, both Cbls account for total EGFR ubiquitination and degradation.
FIG. 8.
Graphical analysis of Cbl-mediated ubiquitination and EGFR degradation. 293T cells were transfected and treated as described in the legend to Fig. 7. (A) Analysis of Cbl-mediated ubiquitination. Anti-Ub blotting analyses of EGFR immunoprecipitates as presented in the figure were performed in triplicate, the corresponding band intensities were quantified by densitometry, and the results were normalized to the anti-Ub band at 0 min (0′). (B) Analysis of EGFR degradation. Anti-EGFR blots of TCLs as shown in the figure were prepared in triplicate and densitometrically quantified, and the results were normalized to the anti-EGFR band at 0 min. Δ1044, EGFR1044; KD, knockdown.
Lastly, we wanted to confirm the requirement for c-Cbl and Cbl-b in the downregulation of endogenous EGFR, as well as to assess the relevance of this phenomenon in another cell type. To this end, we employed the same methods described above to analyze the roles of both c-Cbl and Cbl-b in endogenous EGFR ubiquitination and degradation in BT20 cells (Fig. 9), which express levels of EGFR similar to those in our 293T transfectants (compare Fig. 7 and 9A, top left panels). BT20 cells showed a pattern of EGF-mediated EGFR ubiquitination (Fig. 8A, top left graph, and 9B, left graph) and degradation (Fig. 8B, top left graph, and 9C, left graph) nearly identical to that in 293T cells expressing wt EGFR. Near-complete knockdown (>90%) of either Cbl or both Cbls in BT20 cells led to the same perturbations in EGFR ubiquitination and degradation that occurred in 293T cells. Significantly, the knockdown of both Cbls in BT20 cells led to the elimination of both EGFR ubiquitination and degradation (Fig. 9B and C, rightmost graphs).
Taken together, our results reveal that (i) Cbl downregulates EGFR through ubiquitination and (ii) while c-Cbl and Cbl-b are each alone sufficient to effect EGFR degradation, both are involved in the physiological, EGF-mediated process of receptor downregulation.
DISCUSSION
The Cbls are oncoproteins by virtue of their prominent role in regulating signal transduction (reviewed in reference 50). Understanding how they regulate upstream receptors such as EGFR is vital to delineating exactly how they contribute to cancer. In this investigation, we have conclusively demonstrated that Cbl plays its role in EGFR downregulation at the level of Ub-mediated degradation and not internalization. In order to determine whether Cbl interaction with and Cbl-mediated ubiquitination of EGFR are required for the ligand-induced internalization of EGFR, we employed various mutant EGFRs with distinct characteristics (Fig. 1). These included an internalization-deficient EGFR (EGFR1010LL/AA) and two mutant forms deficient for the binding of Cbl: EGFR(Y1045F), which lacks direct c-Cbl binding, and EGFR1044, which lacks any c-Cbl binding. Both Western analysis and kinetic analysis of receptor internalization showed that EGFR internalization does not depend on the interaction of EGFR with c-Cbl- or c-Cbl-mediated ubiquitination (Fig. 2). Specifically, we showed that EGFR1044 was not associated with c-Cbl and its ubiquitination was not detectable at 10 min of EGF stimulation but that EGFR1044 was internalized similarly to wt EGFR in response to EGF. These results demonstrate that neither c-Cbl association with EGFR nor Cbl-mediated ubiquitination of EGFR is required for EGF-induced EGFR internalization. On the other hand, EGFR1010LL/AA was associated with c-Cbl and ubiquitinated to the same extent as wt EGFR; however, it showed only minimal internalization, which indicates that neither c-Cbl association with EGFR nor Cbl-mediated ubiquitination of EGFR is sufficient for EGF-induced EGFR internalization.
We then turned our query to downstream stages of EGFR trafficking and revealed that c-Cbl disassociated from EGFR well in advance of EGFR degradation (Fig. 3). Interestingly, c-Cbl disassociation from EGFR1010LL/AA was delayed; this mutant form was functionally similar to wt EGFR, though incapable of internalizing, therefore suggesting that the subcellular environment may be important to c-Cbl's disassociation mechanism (Fig. 3). Since EGFR1010LL/AA is restricted mostly to the plasma membrane, it may have poor access to machinery responsible for c-Cbl's disassociation. Interestingly, the loss of c-Cbl binding to EGFR corresponded to the specific dephosphorylation of Y1045, shedding light on a potential mechanism for Cbl's disassociation. Whether this dephosphorylation event is the cause or the consequence of Cbl disassociation remains to be clarified (Fig. 3). Several studies have also shown that Cbl is recruited by soluble factors at later times (>15 min) of EGF stimulation (1, 3, 13, 14, 48, 56). We next looked at receptor ubiquitination and degradation over a longer time course of EGF stimulation. Every EGFR, including the truncated EGFR1044, was degraded and ubiquitinated (Fig. 4). Since EGFR1044 should completely lack c-Cbl binding and c-Cbl-mediated ubiquitination, we searched for alternate E3 Ub ligases that might be responsible. The strongest candidates were in fact the two other major Cbl isoforms: Cbl-b and Cbl-3. Both homologues bind EGFR and regulate its signaling in an EGF-dependent fashion, though only Cbl-b's Ub ligase activity has been demonstrated previously (10, 12, 25, 26, 50). Although Cbl-3 possesses the TKB and RING finger domains necessary to function as an E3 ligase, its expression profile among different tissues is limited and it lacks the C-terminal tail possessed by c-Cbl and Cbl-b, which contains a wide range of regulatory sites, including multiple SH3 domain binding motifs and a UBA domain (25, 50). It therefore seemed to us that Cbl-b was the stronger candidate to investigate.
Our analysis indeed revealed that Cbl-b was working in conjunction with c-Cbl to bind and ubiquitinate wt EGFR, EGFR1010LL/AA, and EGFR(Y1045F), while Cbl-b alone was acting on EGFR1044. c-Cbl and Cbl-b functions were not simply redundant, as each appeared to act on EGFR at temporally distinct times during EGF-mediated trafficking (Fig. 4 and 5). By investigating these events over an extended time course of EGF stimulation, we uncovered an interesting interplay between c-Cbl and Cbl-b in which interaction with EGFR regulated the ubiquitination and degradation of EGFR. Within 15 min of EGF stimulation, EGFR interacted primarily with c-Cbl, which mediated the receptor's rapid initial ubiquitination (Fig. 4). During this early stage of rapid EGFR ubiquitination, Cbl-b binding with the receptor appeared to be inhibited. This result may be due to location constraints, competition with c-Cbl for binding sites of the receptor, and/or some unknown requirement for additional EGFR modifications. Following peak c-Cbl association and the initial rise in receptor ubiquitination, EGFR was partially deubiquitinated (Ub levels dropped ∼30% after between 15 and 30 min of EGF stimulation). This drop was likely due to the action of the Ub-specific protease Y, previously shown to be a key deubiquitinating enzyme for EGFR (36). The onset of the next stage (the following ∼30 min of EGF-induced trafficking) was marked by several contemporaneous events, including c-Cbl disassociation from EGFR, specific dephosphorylation at Y1045, and a strong rise in Cbl-b association with EGFR. It is the latter event that most likely accounts for the second smaller rise in EGFR ubiquitination observed at ∼ 1 h of EGF stimulation.
We also investigated which region of EGFR is responsible for binding Cbl-b (Fig. 5B to D). Two regions of EGFR, one in the C-terminal direction from position 1044 and one in the N-terminal direction from position 958, were important in mediating the association between EGFR and Cbl-b. It is not surprising that the region in the C-terminal direction from position 1044 contributed to a substantial amount (∼50%) of Cbl-b interaction, as this region contains the major phosphotyrosine docking sites of Cbl (Y1045, Y1068, and Y1086). On the other hand, we do not know how the EGFR region in the N-terminal direction from position 957 mediates the interaction with Cbl-b. c-Cbl and Cbl-b are thought to share the same phosphotyrosine binding sites on EGFR, though it appears that Cbl-b, and not c-Cbl, can bind at one or more other sites upstream of position 1044 (50).
Why c-Cbl is incapable of binding the same upstream sites as Cbl-b remains unclear, given the high functional and sequence homology between these isoforms. A possible explanation may be derived from the previously reported differences in the binding capacities of Cbl-b's and c-Cbl's C-terminal UBA domains, the former of which is unable to interact with ubiquitinated proteins while the latter interacts with a wide variety of ubiquitinated proteins (6). Furthermore, the overexpression of Cbl-b's UBA was shown previously to act in a dominant-negative manner to inhibit EGFR degradation, suggesting that Cbl-b directly interacts with EGFR via Ub moieties (6). Whether our reported Cbl-b association upstream of EGFR residue 957 is Ub dependent or phosphotyrosine dependent is, however, unclear, though multiple ubiquitination sites in the EGFR kinase domain upstream of residue 957 have been reported previously (6). Nonetheless, we have demonstrated that Cbl-b association is EGF dependent and that Cbl-b itself is still capable of acting as an E3 ligase for EGFR at this site, as our EGFR1044, which completely lacks c-Cbl binding, was still ubiquitinated following EGF stimulation (Fig. 4). Even in light of the reported differences of c-Cbl and Cbl-b associations with EGFR, it is important not to overinterpret the interplay of these two Ub ligases as a simple sequential process involving a single receptor or receptor population. It is equally possible that following EGF stimulation, each Cbl isoform acts on a separate spatiotemporal pool of EGFR to effect its degradation, and these processes may kinetically differ.
In order to conclusively establish the overall role of Cbl in EGFR downregulation, we employed RNAi experiments to knock down either or both Cbl homologues (Fig. 6 to 9). Our data showed that either c-Cbl or Cbl-b was capable of ubiquitinating EGFR and was sufficient for complete EGFR degradation. In the absence of both Cbls, however, EGFR was neither ubiquitinated nor degraded. On the other hand, the double knockdown of both Cbls did not significantly affect EGF-mediated internalization in both 293T cells exogenously expressing wt EGFR and BT20 cells with endogenous EGFR (Fig. 6). Although the double Cbl knockdown experiments clearly demonstrated the involvement of both Cbls in EGFR ubiquitination and degradation and the requirement for at least one of these proteins, the existence of another E3 Ub ligase(s), acting as an intermediary or auxiliary ligase for EGFR, cannot be ruled out, especially in light of the possibility that Cbl-b interacts with EGFR upstream of its C-terminal tail through Ub. Whatever the case may be, we have conclusively demonstrated that the ubiquitination of EGFR, whether mediated by c-Cbl, Cbl-b, or both together, leads to the degradation of EGFR.
In conclusion, we have found that Cbl acts as a negative regulator of EGFR at the level of EGFR degradation, not EGFR internalization. Based on the results of RNAi knockdown of both c-Cbl and Cbl-b, either c-Cbl or Cbl-b is sufficient to ubiquitinate and cause the degradation of EGFR, and the presence of at least one of these proteins is necessary, as the elimination of both homologues abolished EGFR ubiquitination and, consequently, prevented it degradation. Through detailed analysis of EGFR trafficking, we have also unveiled a temporal interplay of these two Cbls with EGFR during the internal EGFR trafficking route. It remains to be seen whether this downregulatory dualism of Cbl is shared among cell types and other receptor systems. Given the multitude of Cbl substrates known and the complexity of their regulatory networks, it seems to be a distinct possibility.
Acknowledgments
This work was supported in part by grants from the Canadian Institutes of Health Research and the Alberta Heritage Foundation for Medical Research. Z.W. is an Alberta Heritage Foundation for Medical Research Senior Scholar.
We would like to thank L. Berthiaume for providing us with the anti-GFP antibody used for IP.
Footnotes
Published ahead of print on 3 March 2008.
REFERENCES
- 1.Bao, J., G. Gur, and Y. Yarden. 2003. Src promotes destruction of c-Cbl: implications for oncogenic synergy between Src and growth factor receptors. Proc. Natl. Acad. Sci. USA 1002438-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowtell, D. D., and W. Y. Langdon. 1995. The protein product of the c-cbl oncogene rapidly complexes with the EGF receptor and is tyrosine phosphorylated following EGF stimulation. Oncogene 111561-1567. [PubMed] [Google Scholar]
- 3.Buday, L., A. Khwaja, S. Sipeki, A. Farago, and J. Downward. 1996. Interactions of Cbl with two adapter proteins, Grb2 and Crk, upon T cell activation. J. Biol. Chem. 2716159-6163. [DOI] [PubMed] [Google Scholar]
- 4.Burgering, B. M., and P. J. Coffer. 1995. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376599-602. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, G. 1987. Receptors for epidermal growth factor and other polypeptide mitogens. Annu. Rev. Biochem. 56881-914. [DOI] [PubMed] [Google Scholar]
- 6.Davies, G. C., S. A. Ettenberg, A. O. Coats, M. Mussante, S. Ravichandran, J. Collins, M. M. Nau, and S. Lipkowitz. 2004. Cbl-b interacts with ubiquitinated proteins; differential functions of the UBA domains of c-Cbl and Cbl-b. Oncogene 237104-7115. [DOI] [PubMed] [Google Scholar]
- 7.de Melker, A. A., G. van der Horst, J. Calafat, H. Jansen, and J. Borst. 2001. c-Cbl ubiquitinates the EGF receptor at the plasma membrane and remains receptor associated throughout the endocytic route. J. Cell Sci. 1142167-2178. [DOI] [PubMed] [Google Scholar]
- 8.de Melker, A. A., G. van der Horst, and J. Borst. 2004. Ubiquitin ligase activity of c-Cbl guides the epidermal growth factor receptor into clathrin-coated pits by two distinct modes of Eps15 recruitment. J. Biol. Chem. 27955465-55473. [DOI] [PubMed] [Google Scholar]
- 9.Duan, L., Y. Miura, M. Dimri, B. Majumder, I. L. Dodge, A. L. Reddi, A. Ghosh, N. Fernandes, P. Zhou, K. Mullane-Robinson, N. Rao, S. Donoghue, R. A. Rogers, D. Bowtell, M. Naramura, H. Gu, V. Band, and H. Band. 2003. Cbl-mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J. Biol. Chem. 27828950-28960. [DOI] [PubMed] [Google Scholar]
- 10.Ettenberg, S. A., Y. R. Rubinstein, P. Banerjee, M. M. Nau, M. M. Keane, and S. Lipkowitz. 1999. cbl-b inhibits EGF-receptor-induced apoptosis by enhancing ubiquitination and degradation of activated receptors. Mol. Cell Biol. Res. Commun. 2111-118. [DOI] [PubMed] [Google Scholar]
- 11.Ettenberg, S. A., A. Magnifico, M. Cuello, M. M. Nau, Y. R. Rubinstein, Y. Yarden, A. M. Weissman, and S. Lipkowitz. 2001. Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J. Biol. Chem. 27627677-27684. [DOI] [PubMed] [Google Scholar]
- 12.Ettenberg, S. A., M. M. Keane, M. M. Nau, M. Frankel, L. M. Wang, J. H. Pierce, and S. Lipkowitz. 1999. cbl-b inhibits epidermal growth factor receptor signaling. Oncogene 181855-1866. [DOI] [PubMed] [Google Scholar]
- 13.Feng, Q., D. Baird, X. Peng, J. Wang, T. Ly, J. L. Guan, and R. A. Cerione. 2006. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat. Cell Biol. 8945-956. [DOI] [PubMed] [Google Scholar]
- 14.Fukazawa, T., S. Miyake, V. Band, and H. Band. 1996. Tyrosine phosphorylation of Cbl upon epidermal growth factor (EGF) stimulation and its association with EGF receptor and downstream signaling proteins. J. Biol. Chem. 27114554-14559. [DOI] [PubMed] [Google Scholar]
- 15.Galisteo, M. L., I. Dikic, A. G. Batzer, W. Y. Langdon, and J. Schlessinger. 1995. Tyrosine phosphorylation of the c-cbl proto-oncogene protein product and association with epidermal growth factor (EGF) receptor upon EGF stimulation. J. Biol. Chem. 27020242-20245. [DOI] [PubMed] [Google Scholar]
- 16.Grovdal, L. M., E. Stang, A. Sorkin, and I. H. Madshus. 2004. Direct interaction of Cbl with pTyr 1045 of the EGF receptor (EGFR) is required to sort the EGFR to lysosomes for degradation. Exp. Cell Res. 300388-395. [DOI] [PubMed] [Google Scholar]
- 17.Haglund, K., P. P. Di Fiore, and I. Dikic. 2003. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem. Sci. 28598-603. [DOI] [PubMed] [Google Scholar]
- 18.Haglund, K., S. Sigismund, S. Polo, I. Szymkiewicz, P. P. Di Fiore, and I. Dikic. 2003. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5461-466. [DOI] [PubMed] [Google Scholar]
- 19.Haglund, K., N. Shimokawa, I. Szymkiewicz, and I. Dikic. 2002. Cbl-directed monoubiquitination of CIN85 is involved in regulation of ligand-induced degradation of EGF receptors. Proc. Natl. Acad. Sci. USA 9912191-12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84277-287. [DOI] [PubMed] [Google Scholar]
- 21.Huang, F., and A. Sorkin. 2005. Growth factor receptor binding protein 2-mediated recruitment of the RING domain of Cbl to the epidermal growth factor receptor is essential and sufficient to support receptor endocytosis. Mol. Biol. Cell 161268-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, X., F. Huang, A. Marusyk, and A. Sorkin. 2003. Grb2 regulates internalization of EGF receptors through clathrin-coated pits. Mol. Biol. Cell 14858-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joazeiro, C. A., S. S. Wing, H. Huang, J. D. Leverson, T. Hunter, and Y. C. Liu. 1999. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286309-312. [DOI] [PubMed] [Google Scholar]
- 24.Kassenbrock, C. K., S. Hunter, P. Garl, G. L. Johnson, and S. M. Anderson. 2002. Inhibition of Src family kinases blocks epidermal growth factor (EGF)-induced activation of Akt, phosphorylation of c-Cbl, and ubiquitination of the EGF receptor. J. Biol. Chem. 27724967-24975. [DOI] [PubMed] [Google Scholar]
- 25.Keane, M. M., S. A. Ettenberg, M. M. Nau, P. Banerjee, M. Cuello, J. Penninger, and S. Lipkowitz. 1999. cbl-3: a new mammalian cbl family protein. Oncogene 183365-3375. [DOI] [PubMed] [Google Scholar]
- 26.Keane, M. M., O. M. Rivero-Lezcano, J. A. Mitchell, K. C. Robbins, and S. Lipkowitz. 1995. Cloning and characterization of cbl-b: a SH3 binding protein with homology to the c-cbl proto-oncogene. Oncogene 102367-2377. [PubMed] [Google Scholar]
- 27.Levkowitz, G., H. Waterman, S. A. Ettenberg, M. Katz, A. Y. Tsygankov, I. Alroy, S. Lavi, K. Iwai, Y. Reiss, A. Ciechanover, S. Lipkowitz, and Y. Yarden. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 41029-1040. [DOI] [PubMed] [Google Scholar]
- 28.Levkowitz, G., H. Waterman, E. Zamir, Z. Kam, S. Oved, W. Y. Langdon, L. Beguinot, B. Geiger, and Y. Yarden. 1998. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 123663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lill, N. L., P. Douillard, R. A. Awwad, S. Ota, M. L. Lupher, Jr., S. Miyake, N. Meissner-Lula, V. W. Hsu, and H. Band. 2000. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J. Biol. Chem. 275367-377. [DOI] [PubMed] [Google Scholar]
- 30.Longva, K. E., F. D. Blystad, E. Stang, A. M. Larsen, L. E. Johannessen, and I. H. Madshus. 2002. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupher, M. L., Jr., K. A. Reedquist, S. Miyake, W. Y. Langdon, and H. Band. 1996. A novel phosphotyrosine-binding domain in the N-terminal transforming region of Cbl interacts directly and selectively with ZAP-70 in T cells. J. Biol. Chem. 27124063-24068. [DOI] [PubMed] [Google Scholar]
- 32.Reference deleted.
- 33.Marmor, M. D., and Y. Yarden. 2004. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 232057-2070. [DOI] [PubMed] [Google Scholar]
- 34.Marshall, C. J. 1996. Cell signalling. Raf gets it together. Nature 383127-128. [DOI] [PubMed] [Google Scholar]
- 35.Miyake, S., M. L. Lupher, Jr., B. Druker, and H. Band. 1988. The tyrosine kinase regulator Cbl enhances the ubiquitination and degradation of the platelet-derived growth factor receptor alpha. Proc. Natl. Acad. Sci. USA 957927-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizuno, E., T. Iura, A. Mukai, T. Yoshimori, N. Kitamura, and M. Komada. 2005. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell 165163-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosesson, Y., K. Shtiegman, M. Katz, Y. Zwang, G. Vereb, J. Szollosi, and Y. Yarden. 2003. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J. Biol. Chem. 27821323-21326. [DOI] [PubMed] [Google Scholar]
- 38.Opresko, L. K., C. P. Chang, B. H. Will, P. M. Burke, G. N. Gill, and H. S. Wiley. 1995. Endocytosis and lysosomal targeting of epidermal growth factor receptors are mediated by distinct sequences independent of the tyrosine kinase domain. J. Biol. Chem. 2704325-4333. [DOI] [PubMed] [Google Scholar]
- 39.Ota, Y., and L. E. Samelson. 1997. The product of the proto-oncogene c-cbl: a negative regulator of the Syk tyrosine kinase. Science 276418-420. [DOI] [PubMed] [Google Scholar]
- 40.Peles, E., and Y. Yarden. 1993. Neu and its ligands: from an oncogene to neural factors. Bioessays 15815-824. [DOI] [PubMed] [Google Scholar]
- 41.Petrelli, A., G. F. Gilestro, S. Lanzardo, P. M. Comoglio, N. Migone, and S. Giordano. 2002. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature 416187-190. [DOI] [PubMed] [Google Scholar]
- 42.Ravid, T., J. M. Heidinger, P. Gee, E. M. Khan, and T. Goldkorn. 2004. c-Cbl-mediated ubiquitinylation is required for epidermal growth factor receptor exit from the early endosomes. J. Biol. Chem. 27937153-37162. [DOI] [PubMed] [Google Scholar]
- 43.Schlessinger, J., and A. Ullrich. 1992. Growth factor signaling by receptor tyrosine kinases. Neuron 9383-391. [DOI] [PubMed] [Google Scholar]
- 44.Soubeyran, P., K. Kowanetz, I. Szymkiewicz, W. Y. Langdon, and I. Dikic. 2002. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature 416183-187. (Erratum, 417:102.) [DOI] [PubMed] [Google Scholar]
- 45.Stang, E., F. D. Blystad, M. Kazazic, V. Bertelsen, T. Brodahl, C. Raiborg, H. Stenmark, and I. H. Madshus. 2004. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol. Biol. Cell 153591-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stang, E., L. E. Johannessen, S. L. Knardal, and I. H. Madshus. 2000. Polyubiquitination of the epidermal growth factor receptor occurs at the plasma membrane upon ligand-induced activation. J. Biol. Chem. 27513940-13947. [DOI] [PubMed] [Google Scholar]
- 47.Szymkiewicz, I., K. Kowanetz, P. Soubeyran, A. Dinarina, S. Lipkowitz, and I. Dikic. 2002. CIN85 participates in Cbl-b-mediated down-regulation of receptor tyrosine kinases. J. Biol. Chem. 27739666-39672. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, S., L. Neff, R. Baron, and J. B. Levy. 1995. Tyrosine phosphorylation and translocation of the c-cbl protein after activation of tyrosine kinase signaling pathways. J. Biol. Chem. 27014347-14351. [DOI] [PubMed] [Google Scholar]
- 49.Thien, C. B., F. Walker, and W. Y. Langdon. 2001. RING finger mutations that abolish c-Cbl-directed polyubiquitination and downregulation of the EGF receptor are insufficient for cell transformation. Mol. Cell 7355-365. [DOI] [PubMed] [Google Scholar]
- 50.Thien, C. B., and W. Y. Langdon. 2001. Cbl: many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2294-307. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Q., G. Villeneuve, and Z. Wang. 2005. Control of epidermal growth factor receptor endocytosis by receptor dimerization, rather than receptor kinase activation. EMBO Rep. 6942-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Q., F. Zhu, and Z. Wang. 2007. Identification of EGF receptor C-terminal sequences 1005-1017 and di-leucine motif (1010)LL(1011) as essential in EGF receptor endocytosis. Exp. Cell Res. 3133349-3363. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Y., S. Pennock, X. Chen, and Z. Wang. 2002. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol. Cell. Biol. 227279-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, Y., Y. G. Yeung, W. Y. Langdon, and E. R. Stanley. 1996. c-Cbl is transiently tyrosine-phosphorylated, ubiquitinated, and membrane-targeted following CSF-1 stimulation of macrophages. J. Biol. Chem. 27117-20. [DOI] [PubMed] [Google Scholar]
- 55.Waterman, H., G. Levkowitz, I. Alroy, and Y. Yarden. 1999. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J. Biol. Chem. 27422151-22154. [DOI] [PubMed] [Google Scholar]
- 56.Wu, W. J., S. Tu, and R. A. Cerione. 2003. Activated Cdc42 sequesters c-Cbl and prevents EGF receptor degradation. Cell 114715-725. [DOI] [PubMed] [Google Scholar]
- 57.Yokouchi, M., T. Kondo, A. Houghton, M. Bartkiewicz, W. C. Horne, H. Zhang, A. Yoshimura, and R. Baron. 1999. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J. Biol. Chem. 27431707-31712. [DOI] [PubMed] [Google Scholar]
- 58.Yoon, C. H., J. Lee, G. D. Jongeward, and P. W. Sternberg. 1995. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science 2691102-1105. [DOI] [PubMed] [Google Scholar]
- 59.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102533-539. [DOI] [PubMed] [Google Scholar]