Abstract
The Akt kinase is a key regulator of cell proliferation and survival. It is activated in part by PDK1-induced phosphorylation. Here we show that RalGDS, a Ras effector protein that activates Ral GTPases, has a second function that promotes Akt phosphorylation by PDK1 by bringing these two kinases together. In support of this conclusion is our finding that suppression of RalGDS expression in cells inhibits both epidermal growth factor and insulin-induced phosphorylation of Akt. Moreover, while PDK1 complexes with N-GDS, Akt complexes with the central region of RalGDS through an intermediary, JIP1. The biological significance of this newly discovered RalGDS function is highlighted by the observation that an N-terminally deleted mutant of RalGDS that retains the ability to activate Ral proteins but loses the ability to activate Akt also fails to promote cell proliferation. Thus, RalGDS forms a nexus that transduces growth factor signaling to both Ral GTPase and Akt-mediated signaling cascades.
The Akt kinase is a key mediator of the actions of many growth factors and hormones (10). Its effects on cells include regulation of metabolism, gene transcription, cell proliferation, angiogenesis, and apoptosis. Loss or gain of Akt activity contributes to a variety of diseases, including diabetes and cancer. Akt influences these processes by phosphorylating a broad spectrum of substrates, including MDM2, FOXO transcription factors, p27kip1, Bad, and GSK3. The three Akt isoforms, Akt1 to -3, belong to the AGC family of kinases, all of which are activated by the PDK1 kinase (for review, see reference 20). The accepted mode of Akt activation involves ligand-induced activation of phosphatidylinositol 3-kinase (PI3-kinase), which leads to the production of PIP2 and PIP3. These phospholipids interact with the PH domain of Akt to target the protein to PDK1 in the plasma membrane, where PDK1 phosphorylates Akt on threonine 308 (2). How specific PDK1 substrates are chosen for activation in response to specific stimuli is poorly understood. A secondary activation event occurs through phosphorylation of Akt at serine 473. Multiple kinases have been reported to be responsible for this event, the most recent of which is the rictor/mTor complex (29).
RalGDS is one of a family of guanine nucleotide exchange factors (GEFs) that activate Ral-GTPases in cells by promoting their switch from GDP-bound to GTP-bound states. A subset of these GEFs, including RalGDS, Rlf, Rgl, and Rgl3, are downstream effectors of Ras proteins, as active Ras binds to their C termini and targets them to Ral in cells (25). Recent studies on human cells and in knockout mice have highlighted the importance of this effector pathway in the Ras-mediated oncogenesis. For example, Ral exchange factors can replace Ras in assays designed to test the minimal number of genes that are required to transform primary human cells to a tumorigenic state (14, 26), while a RalGDS knockout suppresses tumor formation in a Ras-mediated tumor model (13).
Activation of RalGDS in cells also involves an enhancement of its intrinsic catalytic activity through a growth factor-induced interaction between RalGDS and the PDK1 kinase (32). Interestingly, the kinase activity of PDK1 is not involved. Instead, the N terminus of PDK1 associates with the noncatalytic N terminus of RalGDS, which relieves the latter's inhibitory effect on the GEF's catalytic domain. Moreover, PDK1 has been shown to promote lamellipodia formation in response to epidermal growth factor (EGF) by activating the Ral signaling cascade at the site of membrane protrusions (36).
To date, it has been assumed that all of the effects of RalGDS in cells, including its ability to promote oncogenesis, are through its ability to activate the highly similar (85% identity) Ral family members RalA and RalB (11). In their active GTP-bound states, RalA and RalB interact with a set of effectors that mediate their functions in cells. These include RalBP1 (RLIP) (4, 15, 23), the exocyst complex (3, 22, 24), and Zonab (12). Despite these similarities, RalA and RalB appear to play quite distinct roles in cells. For example, RalA is more effective than RalB in promoting basolateral membrane delivery (30), while RalB is more effective than RalA in promoting cell migration (27) and activation of the TBK1 kinase (5), even though both sets of events are mediated through exocyst components. In tumor cells, RalA appears to be more important for promoting anchorage-independent growth, while RalB appears to be more important for suppressing apoptosis and promoting metastasis (6). Finally, suppression of RalA activity appears to be a mode of action of the tumor suppressor PP2A (28). However, in many cases a Ral-GEF, like RalGDS, displays more potent activities than a constitutively activated version of Ral-GTPases, suggesting that Ral-GEFs are encoded with additional functions (19, 34).
In this paper, we show that RalGDS does, in fact, have another function. It contributes to Akt activation by growth factors by acting as a scaffold that brings PDK1 into the vicinity of Akt but not other PDK1 substrates. Surprisingly, RalGDS also participates in the complete activation of Akt by the additional phosphorylation of Akt at Ser473, which is not PDK1 dependent. Thus, RalGDS acts as a node that couples growth factors to two key signal transduction cascades, one mediated by Ral GTPases and the other by the Akt kinase.
MATERIALS AND METHODS
Cell culture and transfection.
COS-7 and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (HyClone). MCF10A cells were cultured in Dulbecco's modified Eagle's medium-F-12 supplemented with 5% horse serum (Gibco), 0.5 μg/ml hydrocortizone, 10 μg/ml insulin, 20 ng/ml EGF, and 0.5 μg/ml amphotericin B (Fungizone). MCF-7 cells were grown in minimal essential alpha medium with 5% fetal bovine serum. Transfection was performed using the Lipofectamine 2000 reagent (Invitrogen) following the manufacturer's instructions. Indicated cell lines were infected with retroviruses generated from the described retroviral vectors, and stable clones were obtained by selection with puromycin or neomycin.
Plasmid and cDNA.
The expression constructs for pMT3-Myc-RalGDS, pMT3-Myc-ΔN-RalGDS, pMT3-Myc-ΔC-RalGDS, and pMT3-Glu-RalGDS have been described previously (32). Retroviral constructs were gifts from C. Counter. pCMV-HA-AKT was a gift from P. Tsichlis, and pcDNA3-Flag-JIP1 was a gift from R. Buchsbaum.
Western blotting.
Cells were serum starved for 6 h and then pretreated with LY94002 or dimethyl sulfoxide for 0.5 h, and EGF or insulin was added as indicated. Cell were lysed in a radioimmunoprecipitation (RIPA) buffer (Tris-HCl, pH 7.5, 1% NP-40, 0.5% deoxycholate, 10 mM EDTA, 150 mM NaCl, 50 mM NaF) plus inhibitors. The protein concentration was determined by using the Micro BCA protein assay reagent kit (Pierce, Rockford, IL). An enhanced chemiluminescence detection assay was performed following the manufacturer's instruction (Perkin-Elmer, MA). Antibodies against Akt, phosphor-Akt (Thr308 and Ser473), phospho-ERK1/ERK2, phosphor-FOXO1 (Ser256), phosphor-GSK-3α/β(Ser21/9), GSK3β, and phosphor-protein kinase Cδ (PKCδ; Thr505) were purchased from Cell Signaling Technology (Danvers, MA). Antibodies against FOXO1, JIP1, and PKCδ were from Santa Cruz Biotechnology (Santa Cruz, CA). Glu antibodies were from Chemicon (Temecula, CA).
Immunoprecipitation.
Cells were transiently transfected with the indicated cDNA using Lipofectamine 2000 reagent (Invitrogen, CA). After 48 h, cells were lysed in RIPA-B buffer (Tris-HCl, pH 8.0, 1% NP-40, 50 mM NaCl, 50 mM NaF) plus inhibitors. Clarified lysates were incubated with an anti-Flag or anti-Myc antibody to immunoprecipitate the Flag-tagged or Myc-tagged proteins, followed by protein A-Sepharose (Invitrogen, CA). Pellets were washed three times with lysis buffer. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis and polyvinylidene difluoride transfer, proteins were detected by Western blot analysis using chemiluminescence.
Reverse transcription-PCR.
Total RNA was isolated using the RNeasy mini kit (Qiagen Inc., Valencia, CA) following the manufacturer's instructions. A 100-ng aliquot of the total RNA was used to perform reverse transcription-PCR (RT-PCR) with the Access RT-PCR system (Promega, Madison, WI). RalGDS was amplified using a sense primer (CCAGCTCCAGCACCAGTTCCAT) and antisense primer (GCAGCTCCCGGCTCAATGAGTA). The PCR results were analyzed with a 1% agarose gel. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control and amplified with a sense primer (CGGAGTCAACGGATTTGGTCGTAT) and antisense primer (AGCCTTCTCCATGGTGGTGAAGAC).
shRNA knockdown.
To knock down RalGDS, the short hairpin RNA (shRNA) vector was constructed by cloning the oligonucleotide 5′-GCCACTGTCACCCAGTTCA-3′ into the pSUPER.retro.neo vector. MCF10A cells were infected with retroviruses expressing this RalGDS shRNA or empty vectors, and a stable clone was obtained by selection with neomycin. The degree of RalGDS knockdown was determined by reverse transcription-PCR. To knock down JIP1, pSilencer 2.1-U6 hygro-JIP1i (gifts from Y. J. Lee) was used for clonal cell lines as described previously (31).
siRNA knockdown of RalGDS.
RalGDS in MCF-7 cells was suppressed with SMARTpool small interfering RNAs (siRNAs) from Dharmacon (Chicago, IL). A 50 nM concentration of RalGDS siRNA or a nontargeted scrambled negative control siRNA was delivered into cells using Oligofectamine (Invitrogen) following the manufacturer's instructions. The degree of RalGDS knockdown was determined by reverse transcription-PCR. Experiments were repeated with one of the oligos in this mixture (5′-GCAGAAAGGACTCAAGATT-3′).
RESULTS
Suppression of RalGDS expression blocks growth factor activation of Akt.
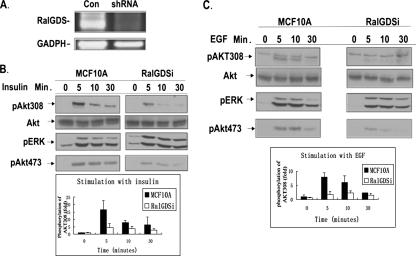
We previously showed that PDK1 stimulates the catalytic activity of RalGDS by forming a complex with it in response to growth factor activation (32). Here we tested whether RalGDS can influence PDK1 function by assessing how depletion of RalGDS from cells affects growth factor activation of a major target of PDK1, Akt. To this end, the mammary epithelial cell line MCF-10A was infected with a retrovirus, pSuper.retro.neo GDSi, which expressed an shRNA predicted to block RalGDS expression. After G418 selection, a cell line stably expressing the shRNA was selected that displayed a >80% reduction in RalGDS mRNA compared to control cells, as detected by RT-PCR. In contrast, no significant effect on the mRNA for GADPH was observed (Fig. 1A).
FIG. 1.
Knockdown of RalGDS suppresses activation of Akt by growth factors in MCF10A cells. (A) Expression levels of RalGDS RNA in MCF10A cells stably expressing RalGDS shRNA or empty vector detected by RT-PCR. (B) MCF10A cells stably expressing either RalGDS shRNA or empty vector were serum starved for 6 h and then stimulated with insulin (1 μM) for the indicated times, and cell lysates were assayed for pAkt308, pAkt473, pERK, and total Akt. pAkt308 band intensities were quantified with NIH ImageJ 1.34s, and the data represent the averages of data from three independent experiments (+ standard deviations). (C) MCF10A cells were stimulated with EGF (5 ng/ml) after being starved for 6 h and then analyzed as in panel B.
These cells and cells infected with empty virus were then serum starved for 6 h and then stimulated with either insulin (Fig. 1B) or EGF (Fig. 1C) for various times, and the activation of Akt was assessed using phospho-specific antibodies directed against the PDK1 target site on Akt, Thr308. These experiments showed that depletion of RalGDS in these cells was associated with a >70% decrease in both insulin-induced (Fig. 1B) and EGF-induced (Fig. 1C) phosphorylation of threonine 308 (top row) compared to control cells, without an effect on total Akt (second row). RalGDS depletion did not block all signaling pathways regulated by these ligands, since insulin- or EGF-induced activation of Erk mitogen-activated protein kinase was not inhibited in these cells (third row). Interestingly, RalGDS depletion also blocked Akt phosphorylation at serine 473 (fourth row), which is not phosphorylated by PDK1, but rather the rictor/mTor complex, which is also PI3 kinase dependent.
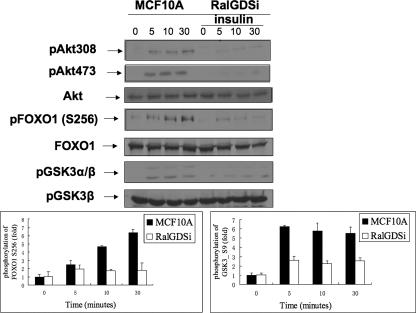
Akt can phosphorylate a variety of downstream target proteins, including FOXO1 and GSK3. Analysis of cell lysates of RalGDS-depleted MCF-10A cells with antibodies specific for sites on these AKT targets showed that, just as Akt phosphorylation was blocked (Fig. 2, first and second lanes), insulin-induced phosphorylation of both Foxo1 (Fig. 2, fourth lane) and GSK3 (Fig. 2, sixth lane) was also suppressed in RalGDS-depleted MCF-10A cells compared to control cells.
FIG. 2.
Signaling downstream from Akt is suppressed by RalGDS knockdown in MCF10A cells. MCF10A cells stably expressing empty vector or RalGDS shRNA were serum starved for 6 h and then stimulated with insulin (1 μM) for the indicated times, and cell lysates were assayed for pAkt308, pAkt473, total Akt, pFOXO1 (Ser256), total FOXO1, pGSK3α/β (Ser21/9), and total GSK3β. pFOXO1 and pGSK3β band intensities were quantified with NIH ImageJ 1.34s, and the data represent the averages of data from three independent experiments.
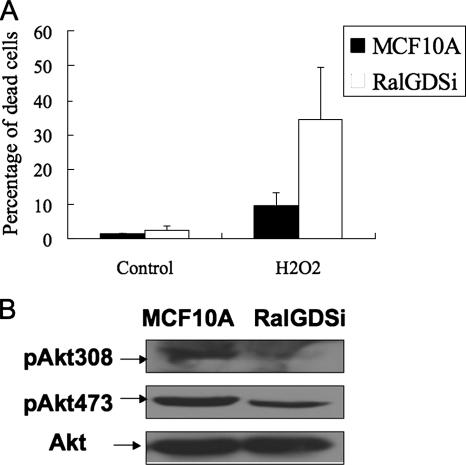
To confirm that the suppression of Akt activity by RalGDS depletion has biological effects consistent with the known role of Akt in cell viability, control MCF10A cells and MCF10A cells with depleted RalGDS were both treated with H2O2 (800 μM) for 3 h, a stimulus known to produce death in these cells (1). Cells were then stained with trypan blue, and the number of dead cells was quantified. H2O2 treatment induced a higher death rate in RalGDS-depleted cells (35%) compared to control cells (9.7%) (Fig. 3A). Figure 3B shows that RalGDS depletion led to suppressed Akt activity in growing cells, just as it blocked growth factor activation of Akt in serum-starved cells (Fig. 1). Thus, RalGDS plays a significant physiological role in cell survival.
FIG. 3.
RalGDS knockdown sensitizes cells to H2O2-induced cell death. (A) MCF10A cells were treated with H2O2 (800 μM) for 3 h, followed by a phosphate-buffered saline wash, and then stained with a mixed solution of 0.4% trypan blue and phosphate-buffered saline (1:1) for 5 min. Stained cells were observed with a microscope and quantified. The data represent the averages of data from two independent experiments, each performed in triplicate (+ standard errors of the mean). (B) Extracts of MCF10A cells and RalGDS-depleted MCF10A cells (RalGDSi) growing in 10% serum were assayed for Akt activity by immunoblotting with either pAkt308 or pAkt473 antibodies.
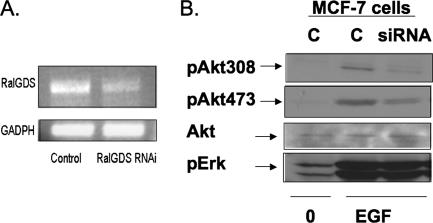
The role of RalGDS in Akt regulation was confirmed in another cell type and with a different mode of RalGDS depletion, by transient-transfection experiments of siRNA (targeted to part of the RNA different from that used in shRNA experiments previously) into MCF-7 breast cancer cells. RT-PCR performed 2 days after transfection showed that RalGDS expression was depleted ∼70% compared to cells transfected with control siRNA (Fig. 4A). Again, analysis of cell lysates with phospho-specific Akt antibodies showed that RalGDS depletion was associated with the suppression of EGF-induced phosphorylation of both Thr308 and Ser473 in Akt, while RalGDS depletion had no effect on the phosphorylation of Erk (Fig. 4B).
FIG. 4.
Akt activation by EGF is suppressed in RalGDS-depleted MCF-7 cells. (A) Expression levels of RalGDS detected by RT-PCR in MCF-7 cells transfected with RalGDS siRNA or control siRNA. (B) MCF-7 cells transfected with RalGDS siRNA or control siRNA were serum starved for 6 h and then stimulated with EGF (5 ng/ml) for 5 min, and cell lysates were assayed for pAkt308, pAkt473, total Akt, and pERK. Results are representative of experiments performed at least three times.
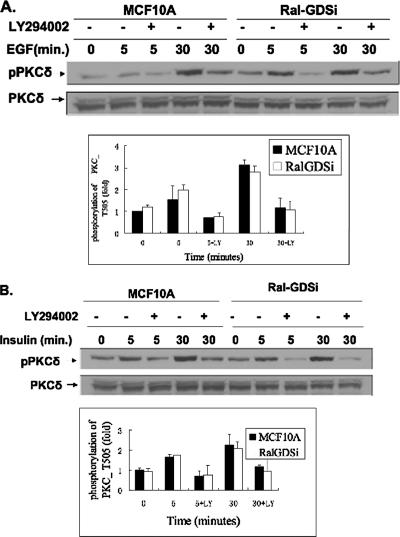
RalGDS knockdown does not affect growth factor activation of another PDK1 substrate, PKCδ.
To test whether phosphorylation of all PDK1 substrates is dependent upon RalGDS, phosphorylation was assessed using an antibody specific for the site on PKCδ that is phosphorylated by PDK1. Figure 5 shows that EGF-induced (Fig. 5A) and insulin-induced (Fig. 5B) phosphorylation of PKCδ was not blocked in MCF-10A cells lacking RalGDS compared to control MCF-10A cells. Treatment of RalGDS-depleted cells with LY294002, a PI3-kinase inhibitor, did block PKCδ phosphorylation on this site, confirming that both EGF- and insulin-induced phosphorylation of PKCδ is dependent upon PI3-kinase signaling. Thus, these results show that RalGDS is required for PDK1 activation of Akt, but not all of its substrates. They also argue that the inhibition of Akt phosphorylation on both Thr308 and Ser473 by RalGDS knockdown is not due to suppression of overall PI3-kinase signaling.
FIG. 5.
RalGDS knockdown does not affect growth factor activation of another PDK1 substrate, PKCδ. (A) MCF10A cells stably expressing RalGDS shRNA or empty vector were serum starved for 6 h and pretreated with either LY294002 for 1 h or control medium, and then cells were stimulated with EGF (5 ng/ml) for the indicated times. Cell lysates were then assayed for pPKCδ (Thr505) and total PKCδ. pPKCδ band intensities were quantified with NIH ImageJ 1.34s, and the data represent the averages of data from three independent experiments. (B) MCF10A cells pretreated as for panel A were stimulated with insulin (1 μM) for the indicated times, and cell lysates were assayed for pPKCδ (Thr505) and total PKCδ. pPKCδ band intensities were quantified with NIH ImageJ 1.34s, and the data represent the averages of data from three independent experiments.
RalGDS forms a complex with the Akt-binding protein JIP1.
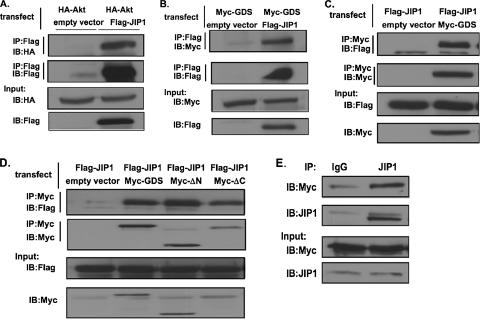
Since we found that RalGDS preferentially promotes the activation of Akt by PDK1, we searched for a protein that might link RalGDS specifically to Akt. JIP1 was a reasonable candidate because it has already been shown to bind to Akt and facilitate its activation by phosphorylation at both Thr308 and S473 (17). Moreover, Ral signaling has already been shown to promote signaling to Jnk (9), a kinase whose activity JIP1 was first shown to influence. Figure 6A reproduces the earlier finding that JIP1 binds Akt by showing that immunoprecipitates of Flag-JIP1 precipitated hemagglutinin (HA)-Akt when both proteins were transfected into COS-7 cells.
FIG. 6.
RalGDS forms a complex with JIP1 in cells. (A) HA-Akt was transfected into COS-7 cells along with empty vector or vector expressing Flag-JIP1. JIP1 was immunoprecipitated (IP) with anti-Flag antibodies and then immunoblotted (IB) with anti-HA antibody to detect Akt (first row). Alternatively, Flag-JIP1 was immunoprecipitated and blotted with anti-Flag antibody (second row). Finally, lysates were immunoblotted with anti-HA antibody (third row) and anti-Flag antibody (fourth row). (B) Myc-RalGDS (Myc-GDS) was transfected into COS-7 cells along with either empty vector or vector expressing Flag-JIP1. JIP1 was immunoprecipitated with anti-Flag antibodies and then immunoblotted with anti-Myc antibody to detect RalGDS (first row). Alternatively, Flag-Jip1 was immunoprecipitated and blotted with anti-Flag antibody (second row). Samples were also blotted for Myc-RalGDS (third row) and Flag-JIP1 (fourth row). (C) Myc-RalGDS or empty vector was transfected into COS-7 cells along with Flag-JIP1. RalGDS was immunoprecipitated with anti-Myc antibodies and then immunoblotted with anti-Flag antibody to detect JIP1 (first row). Alternatively, Myc-RalGDS was immunoprecipitated and blotted with Myc antibody (second row). Finally, Flag-JIP and Myc-RalGDS were immunoblotted (third and fourth rows). (D) Flag-JIP1 was transfected into COS-7 cells along with empty vector or vector expressing Myc-RalGDS, Myc-RalGDSΔN (Myc-ΔN), or Myc-RalGDSΔC (Myc-ΔC). RalGDS or mutants were immunoprecipitated with anti-Myc antibodies, and the presence of JIP1 in the immunoprecipitates was assayed by immunoblotting with anti-Flag antibody (first row). Alternatively, Myc-RalGDS was immunoprecipitated and blotted with Myc antibody (second row). Expression levels of JIP and GDS in lysates were also evaluated (third and fourth rows). (E) Lysates of MCF10A cells stably expressing Myc-RalGDS were immunoprecipitated with either control mouse immunoglobulin G (IgG) or JIP1 antibodies and then samples were immunoblotted with anti-Myc (first row) or anti-JIP1 (second row) antibodies. Extracts were also blotted directly with anti-Myc antibodies to measure Myc-RalGDS levels (third row) and with anti-JIP1 antibodies to measure JIP1 levels (fourth row). Results are representative of experiments performed at least twice.
The ability of RalGDS to bind to JIP1 was then tested in this same system. Immunoprecipitates of Flag-JIP1 from these cells contained Myc-RalGDS, and immunoprecipitates of Myc-RalGDS contained Flag-JIP1 (Fig. 6B and C). We showed previously that PDK1 forms a complex with the N terminus of RalGDS, and so we tested whether JIP1 binds to the same region of the protein by including an N-terminally deleted mutant RalGDS, ΔN-RalGDS, in this assay. Figure 6D shows that ΔN-RalGDS bound JIP1 as effectively as full-length protein, indicating that PDK1 and JIP1 form complexes with different parts of the protein. A mutant with a deleted C-terminal Ras-binding domain also bound to JIP1 to a comparable degree (Fig. 6D), indicating that JIP1 binds to the middle of the protein in a region that contains the catalytic domain. Finally, Myc-RalGDS also bound to endogenous JIP1 that was immunoprecipitated from MCF-10A cells (Fig. 6E).
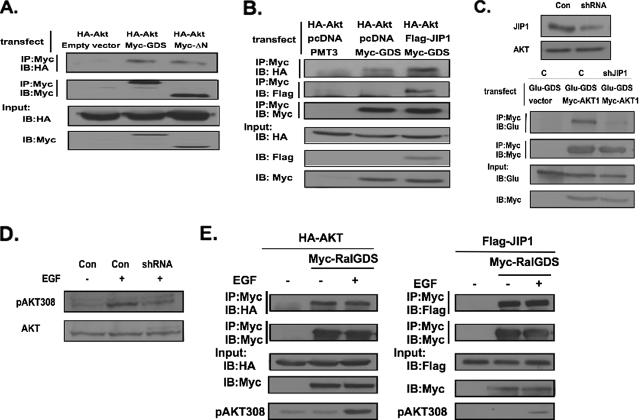
Finally, we tested whether RalGDS forms a complex with Akt in cells in a JIP1-dependent manner. First, Myc-RalGDS was cotransfected with HA-Akt into COS-7 cells. Immunoprecipitation of RalGDS led to coprecipitation of Akt (Fig. 7A). Like JIP1, Akt binding was also detected when the PDK1-binding N terminus of RalGDS was deleted, consistent with the fact that JIP1 bridges Akt to RalGDS (Fig. 7A). This notion was also supported by the observation that the efficiency of Akt binding to RalGDS was significantly less than that of JIP1. Moreover, Akt binding to RalGDS was enhanced when JIP1 was overexpressed in cells by transfection (Fig. 7B).
FIG. 7.
RalGDS forms a complex with Akt. (A) HA-Akt was transfected into COS-7 cells along with Myc-RalGDS or Myc-RalGDSΔN. Myc-RalGDS or Myc-RalGDSΔN was immunoprecipitated (IP) with anti-Myc antibodies and then immunoblotted (IB) with anti-HA antibody to detect Akt (first row) and immunoblotted with anti-Myc antibodies to detect Myc-GDS (second row). HA-Akt and Myc-GDS expression levels in lysates are shown in the third and fourth rows. (B) HA-Akt was transfected into COS-7 cells along with Myc-RalGDS, Flag-JIP1, and empty vector. RalGDS was immunoprecipitated with anti-Myc antibodies and then immunoblotted with either anti-HA antibody to detect Akt (first row), anti-Flag antibody to detect JIP1(second row), or anti-Myc antibody to detect GDS (third row). HA-Akt, Flag-JIP1, and Myc-GDS expression levels are shown in fourth, fifth, and sixth rows. (C) Top: Extracts of COS-7 cells (Con) or COS-7 cells constitutively expressing shRNAi against JIP1 (shRNA) were immunoblotted with anti-JIP1 antibodies or total Akt antibodies. Bottom: COS-7 cells or COS-7 cells expressing JIP1 shRNA were transfected with Glu-RalGDS, and either empty vector or vector expressing Myc-Akt. Myc-Akt was immunoprecipitated, and the sample was immunoblotted with anti-Glu-GDS antibodies (first row) or My-AKT antibodies (second row). Lysates were also blotted with anti-Glu-GDS antibodies (third row) or anti-Myc antibodies (fourth row). (D) COS-7 cells or COS-7 cells expressing JIP1 shRNA were serum starved and then stimulated with EGF for 5 min. Lysates were immunoblotted with pAKT T308-specific antibodies. (E) HA-Akt or Flag-JIP1 was transfected into COS-7 cells along with Myc-RalGDS. Cells were serum starved for 6 h and then stimulated with EGF (5 ng/ml) for 5 min. Cell lysates were immunoprecipitated with anti-Myc antibody and then immunoblotted with anti-HA antibody to detect Akt and with anti-Flag antibody to detect JIP1. Samples were also blotted with HA, Myc, or Flag antibodies to detect expression levels of proteins in cells. Samples were also immunoblotted with pAKT antibodies to confirm EGF stimulation was effective. Data are representative of experiments performed at least twice.
More direct confirmation of this model is supported by the finding that the depletion of JIP1 from COS-7 cells, by expression of an shRNA shown previously to specifically block its expression in cells (31) (Fig. 7C, top), suppressed Akt binding to RalGDS (Fig. 7C, bottom). Importantly, suppression of JIP1 in these cells also suppressed PDK1-mediated phosphorylation of Akt assessed by immunoblotting of cell lysates with pThr308 Akt-specific antibodies (Fig. 7D). We previously found that PDK1 binding to the N terminus of RalGDS was stimulated by EGF. To test whether Akt or JIP1 binding to RalGDS was regulated in a similar manner, COS-7 cells were cotransfected with Myc-RalGDS and HA-Akt or Flag-JIP1. Cells were serum starved and then stimulated with EGF (Fig. 7E). Neither of the binding activities was affected by EGF. Thus, JIP1 and Akt appear to be constitutively associated with RalGDS, whereas PDK1 becomes associated independently with the protein upon growth factor stimulation, which presumably accounts for elevated Akt activation by PDK1.
Expression of RalGDS but not an N-terminally deleted RalGDS enhances Akt activation and promotes NIH 3T3 cell proliferation.
There is a growing appreciation that RalGDS makes a significant contribution to cellular growth control and oncogenesis through its role as an effector of Ras and a mediator of Ral protein activation. However, most experiments have shown that expression of RalGDS is more potent in this context than expression of constitutively activated Ral GTPase, suggesting that RalGDS has another function in cellular growth control. To test whether activation of Akt by RalGDS may be involved in the ability of RalGDS to promote cell proliferation, we compared the ability of RalGDS, which can bind PDK1, with the N-terminally deleted RalGDS, ΔN-RalGDS, which cannot, for their abilities to promote Akt activity and cell proliferation when overexpressed in NIH 3T3 cells.
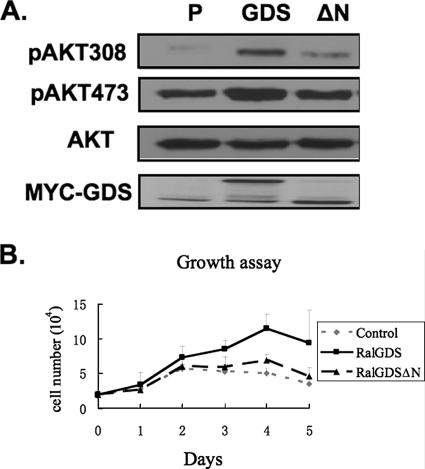
Compared to control cells expressing an empty vector growing in serum, RalGDS cells displayed elevated levels of activated Akt as assessed by T308 and S473 phosphorylation. In contrast, no elevation of Akt was detected in ΔN-RalGDS cells, consistent with the fact that this form of the protein fails to bind PDK1 (Fig. 8A).
FIG. 8.
Enhanced cell proliferation and elevated Akt activation both depend upon the N terminus of RalGDS. (A) Pools of NIH 3T3 cells stably expressing empty vector, Myc-RalGDS (GDS), or Myc-RalGDSΔN (ΔN) were lysed and assayed for pAkt308, pAkt473, total Akt, and Myc-GDS. (B) Pools of NIH 3T3 cells stably expressing empty vector, Myc-RalGDS, or Myc-RalGDSΔN were plated at 2 × 104 cells/ml in a 12-well plate in low serum medium (0.25%). Cell numbers were determined at the indicated times. The data represent the averages of data from three independent experiments.
Then, the cells were compared for their ability to proliferate in low serum levels. In this assay, RalGDS cells showed an enhanced proliferation rate compared to control cells. In contrast, ΔN-RalGDS cells, which showed basal levels of Akt activity, proliferated no better than controls cells. This was despite the fact that we showed previously that ΔN-RalGDS activates Ral proteins in cells better than RalGDS, because the N terminus plays a negative regulatory role on the GEF activity of the protein (11). We performed the same experiment in MCF10A cells, but RalGDS expression only activated Akt to a small degree and had little effect on cell proliferation under these conditions. This could be because RalGDS levels are not as rate limiting in MCF10 as NIH 3T3 cells or because we could not express RalGDS in MCF10 cells at high enough levels to see this phenotype. Thus, a comparison between wild-type and mutant RalGDS could not be made for these cells (data not shown). Nevertheless, the findings argue that the ability of RalGDS to promote Akt activation plays an important part in the protein's ability to influence normal and oncogenic cell proliferation.
DISCUSSION
This study reveals RalGDS as a new signaling component involved in the regulation of Akt activation by growth factors and hormones. This conclusion is based on the observation that depletion of RalGDS in both nontransformed and transformed mammary epithelial cells severely blocks activation of Akt by EGF and insulin stimulation. In contrast, other signaling cascades downstream from these ligands, such as the Ras pathway that activates Erk mitogen-activated protein kinase, are not affected. Moreover, overexpression of RalGDS enhances Akt activity.
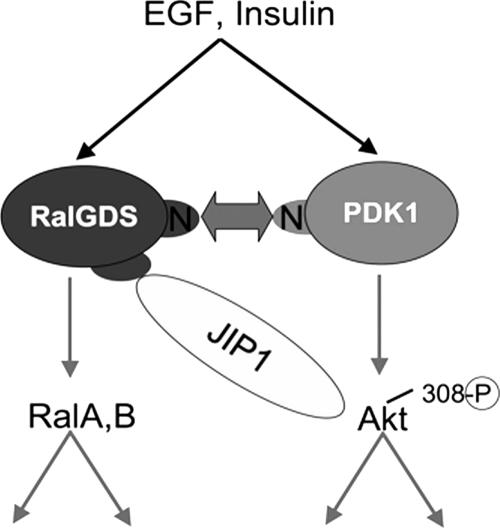
We previously showed that PDK1, a kinase that activates Akt by phosphorylation at threonine 308, has the additional function of stimulating the catalytic activity of RalGDS (32). This occurs independently of the protein's kinase activity. Instead, the noncatalytic N terminus of PDK1 forms a growth factor-induced complex with the N terminus of RalGDS, relieving its inhibitory effect on the catalytic activity of the exchange factor. Those findings raised the possibility that signal transduction might flow in the opposite direction as well, such that RalGDS influences the kinase function of PDK1 (Fig. 9). The data provided here confirm this hypothesis by showing that RalGDS promotes the ability of PDK1 to activate Akt. Moreover, it shows that RalGDS regulates the specificity of PDK1 signaling in that it promotes the activation of one PDK1 substrate, Akt, but not another, PKCδ. Specificity mediated by RalGDS appears to involve its function as a scaffold. We show here that in addition to its previously identified PDK1 association domain in its N terminus, RalGDS also has an independent site in the middle of the protein that complexes with Akt. This scaffold model is consistent with recent fluorescence resonance energy transfer data that showed that RalGDS, PDK1, and Akt associate together in lamellipodia, where Ral and Akt are both activated upon EGF stimulation (36). Moreover, an analogous scaffold function has been ascribed to MAKAPa, which preferentially promotes PDK1 activation of p90RSK in the cytoplasm but not of Akt in the plasma membrane (21).
FIG. 9.
Two-way signal transduction between RalGDS and PDK1. Previously, we showed that PDK1 activates the intrinsic catalytic activity of RalGDS by forming a complex between its N terminus and the N terminus of RalGDS. Now we show that signal transduction travels in the other direction because RalGDS promotes the phosphorylation of Akt at T308 by PDK1 through the scaffold function of JIP1, which binds both Akt and RalGDS. The model suggests direct binding, but since these experiments were done in COS-7 cells it could be indirect. RalGDS also promotes the phosphorylation of Akt at S473, but the mechanism involved is different and not yet known. Thus, RalGDS transduces two important signaling cascades in response to growth factor stimulation, one that activates Ral GTPases through the exchange factor's catalytic domain and another that promotes Akt activation through the protein's scaffold function.
Akt binding to RalGDS is indirect, through the scaffold protein JIP1. This conclusion is based on our finding that JIP1 binds to both RalGDS and Akt. In addition depletion of JIP1 from cells suppresses Akt binding to RalGDS as well as PDK1-induced phosphorylation of Akt at T308. JIP1 was first identified as a scaffold for the Jnk kinase cascade (35). Subsequently, JIP1 was found to bind to the PH domain of Akt. In doing so, it replaces the role of PIP3 in altering the conformation of the PH domain, allowing PDK1 to phosphorylate the protein more effectively (17). JIP1 binding to a Ral exchange factor is consistent with the known function of JIP1 as a scaffold for components of the Jnk kinase cascade, since the Ral signaling cascade has been shown to activate Jnk in mammalian cells (8). In addition, Akt has been shown to negatively regulate JIP1/Jnk signaling, suggesting that a negative feedback system is also built into this complex of signaling molecules (16). Whether JIP1 binding to RalGDS participates in these activities remains to be determined.
Although these findings can explain how RalGDS influences Akt phosphorylation at the PDK1 site at threonine 308, they do not explain how RalGDS influences activation of Akt at serine 473. Multiple kinases have been implicated in phosphorylating this site, the most recent of which is the mTor/rictor complex (29). Nevertheless, depletion of RalGDS from MCF-10A or MCF7 cells suppressed both insulin- and EGF-induced phosphorylation of serine 473, and overexpression of RalGDS has the capacity to enhance Akt phosphorylation of this site. The inhibition of serine 473 phosphorylation by RalGDS depletion cannot be due to overall suppression of PI3 kinase activity, which is required for the phosphorylation of both sites on Akt in cells. This is because growth factor stimulation of PKCδ, which is also dependent on PI3-kinase, was not also suppressed by reducing RalGDS levels in cells. How RalGDS influences phosphorylation of S473 remains to be determined.
Why does RalGDS function as a bridge that connects upstream signals to both Ral and Akt activation (Fig. 9)? One explanation may be that both Ral and Akt proteins regulate Forkhead transcription factors, although they appear to do so in opposite directions. In particular, Ral signaling, which is stimulated by the GEF domain of RalGDS, activates FOXO4 through Jnk-mediated phosphorylation at T447 and T451 (7). In contrast, Akt activation, which involves the scaffold function of RalGDS, negatively regulates FOXO proteins through phosphorylation of FOXO1 at Thr24/32 and Ser256/319 and FOXO4 at Thr28 and Ser193/258 (18, 33). FOXO activity is a crucial mediator of the cell cycle and survival. Thus, a growth factor-induced complex between RalGDS, PDK1, and Akt serves as a nexus for signals that control these key transcription factors. An interesting possibility is that the fate of a cell to either arrest in the cell cycle and undergo apoptosis or to proliferate and survive could be driven by specific conditions that tip RalGDS function to favor Ral or Akt activation.
RalGDS is a downstream target of Ras proteins, and a growing body of experiments have highlighted its importance in mediating the effect of Ras on the control of cell proliferation. In Ras-induced cancer cells, RalGDS is thought to contribute significantly to oncogenesis, at least in part, by suppressing apoptosis. For example, knockout of the catalytic domain of RalGDS in mice suppressed tumor formation in a Ras-mediated model of skin cancer due to enhanced cell death (13). RalGDS may have promoted this activity in tumor cells by its ability to activate RalB, which promotes cell survival in many tumor cell types (6). However, this partial knockout may have also suppressed the ability of RalGDS to promote Akt activation.
In a different set of experiments, in which the components necessary to transform primary cells to tumor cells have been studied, Ral-GEFs have been shown to be particularly potent in substituting for Ras (14). In this assay, constitutively activated RalA yielded a phenotype that was qualitatively similar to expression of an activated Ral-GEF; however, its potency was much lower (19). If the Ral-stimulating activity of the Ral-GEF were its main mechanism of action, then expression of a constitutively activated Ral-GTPase should have been more, not less, potent than expression of its exchange factor. This finding, along with many others that show a similar difference between the potencies of Ral-GEFs and Ral GTPases, argues that Ral-GEFs have additional functions besides activation of Ral GTPases. The data presented here confirm this hypothesis in that RalGDS plays a critical role in Akt activation by growth factors (EGF) and hormones (insulin) in cells. It has been shown that insulin activation of Akt does not rely on Ras signaling, which argues that RalGDS may have a broader role in Akt regulation than just through its role as a Ras effector. Moreover, RalGDS is only one member of a family of Ral specific exchange factors. Others include RGL1 and RGL2 (Rlf), both of which also bind JIP1 (data not shown), suggesting that they may also contribute to Akt regulation. Thus, which family member is the predominant regulator of Akt in a particular cell type may depend upon the relative expression levels of Ral-GEF proteins.
The physiological significance of this newly discovered function of RalGDS is supported by our finding that a mutant RalGDS that lacks its PDK1-associating N terminus loses the ability to activate Akt and promote proliferation NIH 3T3 cells grown in medium with low serum levels. This is despite the fact that this deletion actually enhances the ability of RalGDS to activate Ral proteins in cells (11). Thus, this newly discovered ability of RalGDS to promote Akt activation plays a critical role in the ability of RalGDS to regulate cell proliferation.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Bae, J. Y., S. J. Ahn, W. Han, and D. Y. Noh. 2007. Peroxiredoxin I and II inhibit H2O2-induced cell death in MCF-7 cell lines. J. Cell. Biochem. 1011038-1045. [DOI] [PubMed] [Google Scholar]
- 2.Bellacosa, A., C. C. Kumar, A. Di Cristofano, and J. R. Testa. 2005. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv. Cancer Res. 9429-86. [DOI] [PubMed] [Google Scholar]
- 3.Brymora, A., V. A. Valova, M. R. Larsen, B. D. Roufogalis, and P. J. Robinson. 2001. The brain exocyst complex interacts with RalA in a GTP-dependent manner: identification of a novel mammalian Sec3 gene and a second Sec15 gene. J. Biol. Chem. 27629792-29797. [DOI] [PubMed] [Google Scholar]
- 4.Cantor, S., T. Urano, and L. A. Feig. 1995. Identification and characterization of RalBP1, a potential downstream target of Ral GTPases. Mol. Cell. Biol. 154578-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien, Y., S. Kim, R. Bumeister, Y. M. Loo, S. W. Kwon, C. L. Johnson, M. G. Balakireva, Y. Romeo, L. Kopelovich, M. Gale, Jr., C. Yeaman, J. H. Camonis, Y. Zhao, and M. A. White. 2006. RalB GTPase-mediated activation of the IκB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127157-170. [DOI] [PubMed] [Google Scholar]
- 6.Chien, Y., and M. A. White. 2003. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 4800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Ruiter, N. D., B. M. Burgering, and J. L. Bos. 2001. Regulation of the Forkhead transcription factor AFX by Ral-dependent phosphorylation of threonines 447 and 451. Mol. Cell. Biol. 218225-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Ruiter, N. D., R. M. Wolthuis, H. van Dam, B. M. Burgering, and J. L. Bos. 2000. Ras-dependent regulation of c-Jun phosphorylation is mediated by the Ral guanine nucleotide exchange factor-Ral pathway. Mol. Cell. Biol. 208480-8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essers, M. A., S. Weijzen, A. M. de Vries-Smits, I. Saarloos, N. D. de Ruiter, J. L. Bos, and B. M. Burgering. 2004. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 234802-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fayard, E., L. A. Tintignac, A. Baudry, and B. A. Hemmings. 2005. Protein kinase B/Akt at a glance. J. Cell Sci. 1185675-5678. [DOI] [PubMed] [Google Scholar]
- 11.Feig, L. A. 2003. Ral GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 13419-425. [DOI] [PubMed] [Google Scholar]
- 12.Frankel, P., A. Aronheim, E. Kavanagh, M. S. Balda, K. Matter, T. D. Bunney, and C. J. Marshall. 2005. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 2454-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Garcia, A., C. A. Pritchard, H. F. Paterson, G. Mavria, G. Stamp, and C. J. Marshall. 2005. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell 7219-226. [DOI] [PubMed] [Google Scholar]
- 14.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 162045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jullien-Flores, V., Y. Mahe, G. Mirey, C. Leprince, B. Meunier-Bisceuil, A. Sorkin, and J. H. Camonis. 2000. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 1132837-2844. [DOI] [PubMed] [Google Scholar]
- 16.Kim, A. H., G. Khursigara, X. Sun, T. F. Franke, and M. V. Chao. 2001. Akt phosphorylates and negatively regulates apoptosis signal-regulating kinase 1. Mol. Cell. Biol. 21893-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, A. H., T. Sasaki, and M. V. Chao. 2003. JNK-interacting protein 1 promotes Akt1 activation. J. Biol. Chem. 27829830-29836. [DOI] [PubMed] [Google Scholar]
- 18.Kops, G. J., N. D. de Ruiter, A. M. De Vries-Smits, D. R. Powell, J. L. Bos, and B. M. Burgering. 1999. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398630-634. [DOI] [PubMed] [Google Scholar]
- 19.Lim, K. H., A. T. Baines, J. J. Fiordalisi, M. Shipitsin, L. A. Feig, A. D. Cox, C. J. Der, and C. M. Counter. 2005. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7533-545. [DOI] [PubMed] [Google Scholar]
- 20.Manning, B. D., and L. C. Cantley. 2007. AKT/PKB signaling: navigating downstream. Cell 1291261-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel, J. J., I. K. Townley, K. L. Dodge-Kafka, F. Zhang, M. S. Kapiloff, and J. D. Scott. 2005. Spatial restriction of PDK1 activation cascades by anchoring to mAKAPα. Mol. Cell 20661-672. [DOI] [PubMed] [Google Scholar]
- 22.Moskalenko, S., D. O. Henry, C. Rosse, G. Mirey, J. H. Camonis, and M. A. White. 2002. The exocyst is a Ral effector complex. Nat. Cell Biol. 466-72. [DOI] [PubMed] [Google Scholar]
- 23.Park, S. H., and R. A. Weinberg. 1995. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene 112349-2355. [PubMed] [Google Scholar]
- 24.Polzin, A., M. Shipitsin, T. Goi, L. A. Feig, and T. J. Turner. 2002. Ral-GTPase influences the regulation of the readily releasable pool of synaptic vesicles. Mol. Cell. Biol. 221714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quilliam, L. A., J. F. Rebhun, and A. F. Castro. 2002. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 71391-444. [DOI] [PubMed] [Google Scholar]
- 26.Rangarajan, A., S. J. Hong, A. Gifford, and R. A. Weinberg. 2004. Species- and cell type-specific requirements for cellular transformation. Cancer Cell 6171-183. [DOI] [PubMed] [Google Scholar]
- 27.Rosse, C., A. Hatzoglou, M. C. Parrini, M. A. White, P. Chavrier, and J. Camonis. 2006. RalB mobilizes the exocyst to drive cell migration. Mol. Cell. Biol. 26727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sablina, A. A., W. Chen, J. D. Arroyo, L. Corral, M. Hector, S. E. Bulmer, J. A. DeCaprio, and W. C. Hahn. 2007. The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell 129969-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarbassov, D. D., D. A. Guertin, S. M. Ali, and D. M. Sabatini. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 3071098-1101. [DOI] [PubMed] [Google Scholar]
- 30.Shipitsin, M., and L. A. Feig. 2004. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol. Cell. Biol. 245746-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song, J. J., and Y. J. Lee. 2005. Dissociation of Akt1 from its negative regulator JIP1 is mediated through the ASK1-MEK-JNK signal transduction pathway during metabolic oxidative stress: a negative feedback loop. J. Cell Biol. 17061-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian, X., G. Rusanescu, W. Hou, B. Schaffhausen, and L. A. Feig. 2002. PDK1 mediates growth factor-induced Ral-GEF activation by a kinase-independent mechanism. EMBO J. 211327-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Horst, A., and B. M. Burgering. 2007. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 8440-450. [DOI] [PubMed] [Google Scholar]
- 34.Wolthuis, R. M., N. D. de Ruiter, R. H. Cool, and J. L. Bos. 1997. Stimulation of gene induction and cell growth by the Ras effector Rlf. EMBO J. 166748-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yasuda, J., A. J. Whitmarsh, J. Cavanagh, M. Sharma, and R. J. Davis. 1999. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol. Cell. Biol. 197245-7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshizaki, H., N. Mochizuki, Y. Gotoh, and M. Matsuda. 2007. Akt-PDK1 complex mediates epidermal growth factor-induced membrane protrusion through Ral activation. Mol. Biol. Cell 18119-128. [DOI] [PMC free article] [PubMed] [Google Scholar]