Abstract
Numerous nuclear proteins bind to chromatin by targeting unique DNA sequences or specific histone modifications. In contrast, HMGN proteins recognize the generic structure of the 147-bp nucleosome core particle. HMGNs alter the structure and activity of chromatin by binding to nucleosomes; however, the determinants of the specific interaction of HMGNs with chromatin are not known. Here we use systematic mutagenesis, quantitative fluorescence recovery after photobleaching, fluorescence imaging, and mobility shift assays to identify the determinants important for the specific binding of these proteins to both the chromatin of living cells and to purified nucleosomes. We find that several regions of the protein affect the affinity of HMGNs to chromatin; however, the conserved sequence RRSARLSA, is the sole determinant of the specific interaction of HMGNs with nucleosomes. Within this sequence, each of the 4 amino acids in the R-S-RL motif are the only residues absolutely essential for anchoring HMGN protein to nucleosomes, both in vivo and in vitro. Our studies identify a new chromatin-binding module that specifically recognizes nucleosome cores independently of DNA sequence or histone tail modifications.
The orderly progression of nuclear processes such as transcription, replication, and repair are regulated by precise and specific interactions between nuclear proteins and the chromatin fiber. Numerous chromatin-binding proteins interact with their target through structural motifs that facilitate specific interactions with either DNA or histones. For example, DNA-binding proteins use motifs such as the helix-loop-helix and zinc finger domains to form sequence specific interactions with the DNA (34), while proteins containing chromodomains, bromodomains, and SANT domains bind to chromatin by specially targeting unique modifications in nucleosomal histones (4, 19, 20, 26, 35). A few proteins, such as members of the HMGN nonhistone chromosomal protein family, bind to chromatin by preferentially interacting with the nucleosome itself (5, 10, 18). These proteins bind to nucleosome core particles (CPs) stronger than to either the histones or DNA. Here we identify the amino acids that determine the specific binding of HMGN proteins to nucleosomes and define the limits of the protein motif that confers nucleosome binding specificity to this protein family.
HMGN proteins, one of the three major HMG families, are well-characterized proteins that specifically bind to the 147-bp nucleosome CP, the fundamental building block of the chromatin fiber (5, 10, 11). The interaction of HMGN proteins with nucleosomes stabilizes the structure of the isolated CP, reduces the “compaction” of the higher-order chromatin structure (10), and alters the levels of posttranslational modifications in the tail of the nucleosomal histones (22, 23, 29, 38). Studies with Hmgn1−/− mice and cells indicate that HMGN1 affects the rate of DNA repair (6, 8) and plays a role in developmental processes (7, 17, 18). Significantly, phenotype rescue experiments reveal the phenotypic effects are contingent on the interaction of HMGN1 with chromatin. Thus, elucidation of the major factors that regulate the specific binding of HMGNs to chromatin is relevant to the understanding of the cellular function and mechanism of action of this protein family.
There are four HMGN variants in vertebrate cells, HMGN1, HMGN2, HMGN3, and HMGN4, and their primary structures are evolutionarily conserved (10). HMGNs have a multidomain structure: a bipartite nuclear localization signal, a nucleosomal binding domain (NBD), and a chromatin unfolding domain. The NBD of HMGNs is a 30-amino-acid domain, which has been shown to interact with purified DNA (1) and also to be the minimum peptide that binds specifically to nucleosome CPs (15). Embedded in the NBD is the sequence “RRSARLSA,” an eight-amino-acid motif that is absolutely conserved in all HMGNs (Fig. 1A). This motif is also present in NSBP1, an HMGN-like protein that binds to CPs (33). The two serine residues in this motif have been shown to be important for the interaction of HMGN1 with chromatin (32); however, other regions of the molecule have been shown to contribute to the binding of HMGNs to nucleosomes (15, 30, 37).
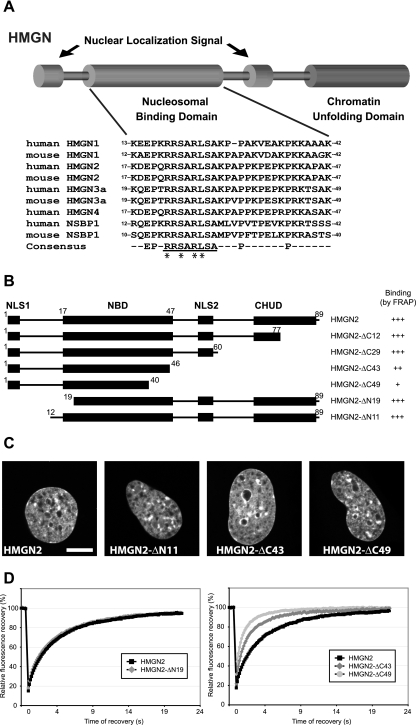
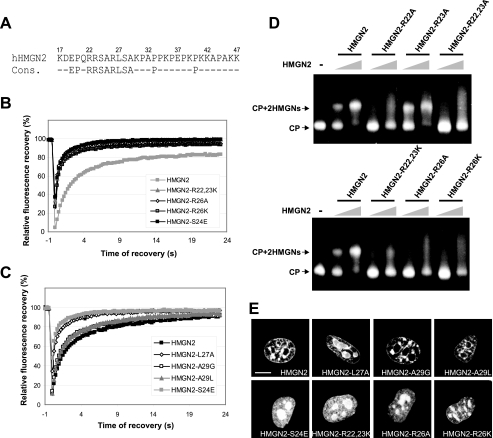
FIG. 1.
Mapping of HMGN2 NBD by FRAP on native chromatin in living cells. (A) Schematic representation of the structure of the HMGN proteins and alignment of the NBD of the mouse and human HMGNs. The numbers at the beginning of each sequence correspond to amino acid position of the first and last residues of the NBD in its respective sequence. The conserved HMGN motif within the consensus sequence is underlined. The amino acid residues that are critical for specific binding to nucleosomes are marked by asterisks (see the text). (B) Outline of the HMGN2 deletion mutants used to map the NBD in living cells. The known functional domains of the protein are indicated by black boxes (NLS, nuclear localization signal; CHUD, chromatin unfolding domain). For FRAP experiments, the proteins were fused to GFP at their C terminus. The name and the binding properties of each mutant are indicated on the right. Binding is rated from “+” (poor binding) to “+++” for wild-type binding (relative values in Table 1). (C) Intranuclear localization of the HMGN2 deletion mutants. Confocal images of live cells expressing the indicated GFP fusion proteins are shown. Scale bar, 10 μm. (D) Quantitative FRAP analysis of N-terminal (left) or C-terminal (right) deletion mutant expressed in MEFs.
Most of the information on the interaction of HMGNs with nucleosomes was obtained from in vitro studies with purified nucleosome CPs and isolated proteins. However, in living cells these proteins interact with the chromatin fiber rather than with purified nucleosomes. Furthermore, fluorescence imaging techniques revealed that in the living nucleus the interaction of HMGNs with chromatin is highly dynamic, that these proteins bind transiently to nucleosomes (27) and that HMGN variants compete among themselves, and with histone H1 variants, for nucleosome binding sites (13, 14). These in vivo studies changed the traditional, static view of the organization of structural “architectural” proteins in chromatin (12, 39) and raised the possibility that the in vitro analyses with purified components may not reflect faithfully the in vivo processes occurring in living nuclei.
Here we use fluorescence recovery after photobleaching (FRAP) and whole-cell imaging on a battery of HMGN2 mutants to identify the determinants important for the binding of HMGNs to unperturbed chromatin, in living cells. We compare the in vivo binding of HMGN to native chromatin as assessed by FRAP and imaging, to the in vitro association of HMGN to isolated nucleosomes as determined by mobility shift assays. We find that in most instances the results from the two approaches are very similar, suggesting that the major factors governing the interaction of HMGNs with chromatin in living cells are operative at the level of the single nucleosome. Several regions of the protein cooperate to strengthen the binding affinity of HMGNs to nucleosomes; however, the specific interaction of HMGN with chromatin is contingent on the presence of four amino acid residues, each of which is absolutely necessary for specific binding. The high conservation of the nucleosome binding motif among all HMGNs suggests that it has a biologically important function. We suggest that it serves as an anchoring module that facilitates the specific interaction of HMGNs with chromatin.
MATERIALS AND METHODS
Cell culture and transfection.
Mouse embryonic fibroblasts (MEFs), HeLa and MCF7 cell culture conditions and transfection protocols were described previously (8, 14). Unless otherwise indicated, all experiments were done in MEFs. For all microscopy experiments, cells were plated 72 h before the experiment on coverslips by using glass-bottom petri dishes from MatTek Corp. (Asland, MA) and cultured for the remaining of the experiment in phenol-free Dulbecco modified Eagle medium. Cells were transfected 24 h before the experiment using Fugene 6 reagent (Roche) according to the manufacturer recommendations.
Vectors and mutagenesis.
All HMGN-fluorescent constructs were generated by using the Living Colors fluorescent protein vectors (Clontech). Wild-type HMGN1-green fluorescent protein (GFP) and HMGN2-GFP were described previously (32). Deletion mutants were generated by PCR amplification of the corresponding part of Hmgn2 cDNA and cloning into pEGFP-N2, between the XhoI and BamHI sites. Point mutants were generated by site-directed PCR mutagenesis in the pET vector and subcloned into pEGFP-N2, using XhoI and BamHI sites. All vectors were verified by sequencing.
FRAP.
Quantitative FRAP protocol was as described previously (14, 27), with minor modifications. Briefly, FRAP was performed on a Zeiss LSM 510 confocal microscope, at 37°C, using an 100× objective (NA 1.3). Bleaching was done by using the 458-, 488-, and 514-nm lines of an argon laser and the 543-nm line of an HeNe laser, all set to 100% output. The bleach spot was 3 μm wide and was placed randomly in the nucleus, excluding regions containing nucleoli and large heterochromatin structures. Imaging was performed with the 488-nm line of the argon laser set to 1% attenuation. In a typical experiment, five prebleach images were collected, followed by three 70-ms bleach pulses. After bleaching, images were collected every 150 ms for about 20 s. Each data set consisted of at least 10 cells, and all experiments were performed at least in duplicate. Recovery curves were generated from background-subtracted images and normalized to prebleach images. Preliminary studies indicated that within the range of protein concentrations used in these experiments, the FRAP curves were independent of the cellular levels of HMGN.
Confocal imaging.
Confocal images of mutants were collected at the time of FRAP experiment. The imaging of GFP-expressing cells was as described previously (13).
Preparation of nucleosomes and proteins.
Nucleosome CPs were prepared from chicken red blood cells (3). Wild-type and mutant HMGN proteins were expressed in and purified from E. coli cells as described previously (21).
Electrophoretic mobility shift assay.
CPs were incubated with various concentrations of HMGN2 or mutants in 2× TBE (180 mM Tris, 180 mM boric acid, and 2 mM EDTA [pH 8.3]) containing 1% (wt/vol) Ficoll 400 on ice for 15 min. The complexes were resolved on 5% polyacrylamide gel in 2× TBE, at 4°C. Free and bound CPs were visualized by staining of the nucleosomal DNA with ethidium bromide. Dissociation constant for each HMGN protein was calculated as described previously (30) by using the formula Kd = a[X]/([Y] − 2a[X] − b[X])c[X], where X is the concentration of CP, Y is the concentration of HMGN, a is the fraction of CP-2HMGNs complex, b is the fraction of CP-1HMGNs (at cooperative binding condition, this is 0), and c is fraction of free CP. In gels exhibiting smears, the Kd was estimated by determining the HMGN concentration (i.e., [HMGN]) necessary to shift 50% of either CP or DNA from its original position. At this point 50% of the DNA or CP is complexed with HMGN: Kd = 1/2 [X0][HMGN]/1/2 [X0], where [X0] is the initial concentration of DNA or CP.
Cross-linking and two-dimensional gel electrophoresis.
Purified HMGN2-S28C protein was incubated with the cross-linking reagent S-[N-(4-azidosalicyl)-cysteaminyl]-2-thiopyridyl (AET; Molecular Probes) for 1 h at 4°C. The AET-modified HMGN2-S28C protein was purified from free AET and desalted by chromatography on a Bio-Gel P-6DG spin column (Bio-Rad). For UV-cross-linking experiment, 0.2 μg of the AET modified HMGN2-S28C was incubated with 1 μg of nucleosome CP (the HMGN:CP molar ratio is 2:1, i.e., the optimal binding ratio) in 10 μl of cross-linking buffer (20 mM HEPES-NaOH [pH 7.5], 100 mM NaCl, 10% glycerol) for 15 min at 4°C (36). The mixture, in a volume of 20 μl was irradiated with a 365-nm UV source (Spectroline, model ENF-240C) from a distance of 5 cm for 1 min. After irradiation, 2× sodium dodecyl sulfate (SDS) sample buffer without dithiothreitol (DTT) was added to the mixture. The sample was boiled for 10 min and separated by SDS-15% polyacrylamide gel electrophoresis (PAGE). Gels were subjected to silver staining or Western blot analysis. For two-dimensional PAGE analysis, the first dimension was SDS-15% PAGE without DTT. The lane containing the sample of interest was cut out from the gel, soaked in 2× SDS buffer containing 100 mM DTT for 15 min at 50°C, and then placed on the top of a preparative SDS-15% PAGE gel. Proteins were detected by silver staining.
RESULTS
Mapping the HMGN NBD in living cells.
The structure of the HMGN protein family is highly conserved and consists of distinct functional domains (Fig. 1A). Previous in vitro mobility shift assays suggested that a highly conserved, ∼30-amino-acid region named the NBD is the main region through which HMGNs bind to isolated nucleosomes in vitro (15). To define the HMGN regions that are important for binding to native, unperturbed chromatin in living cells, we constructed a set of expression vectors coding for wild-type and deletion mutants of HMGN2-GFP (Fig. 1B) and determined their relative chromatin-binding activity by FRAP. In the FRAP technique, a small area of a nucleus is irreversibly bleached with a laser beam, and the rate at which the fluorescent signal in the photobleached area recovers is quantified. The FRAP is indicative of the rate at which fluorescent molecules exchange with the photobleached molecules and is directly proportional to the rate at which the molecules migrate throughout the nucleus and inversely proportional to the time that the molecules reside at an immobile binding site such as chromatin (24). For many nuclear proteins, including HMGNs, the contribution of diffusion to the rate of recovery is negligible, and the observed FRAP (“mobility”) is a direct reflection of their chromatin binding affinity. Photobleaching techniques can be used to detect changes in protein mobility and therefore can be used to analyze the in vivo interaction of proteins with chromatin.
We used the time to recover 80% of the initial fluorescence (t80) as a measure of the relative chromatin-binding affinity of the various HMGN2 deletion mutants (Fig. 1B and Table 1). Under the experimental conditions used, the t80 of the wild-type protein was 6.9 s. The FRAP analyses indicated that the N-terminal part of the protein, up to position 19, can be removed without affecting the binding (Fig. 1D) (the t80 value was reduced by 5% [Table 1]). The removal of the C terminus up to position 46, (HMGN2-ΔC43) lowered the FRAP t80 by 60% (Fig. 1D), suggesting that this region plays a role in the binding of HMGN2 to chromatin. The additional deletion of six amino acids at the C terminus of the NBD (HMGN2-ΔC49) further decreased the binding, although without completely abolishing it (t80 = 1.55 s compared to 0.3 s for control GFP).
TABLE 1.
Quantitative FRAP analysis of HMGN deletion and point mutants
| Protein | Mean fluorescence recovery (t80)a ± SD |
|---|---|
| Wild type | 100 ± 18 |
| ΔC12 | 95 ± 23 |
| ΔC29 | 112 ± 29 |
| ΔC43 | 41 ± 15 |
| ΔC49 | 22 ± 5 |
| ΔN19 | 95 ± 24 |
| S24,28E | 10 ± 2 |
| S24E | 9 ± 2 |
| S28E | 34 ± 9 |
| S24,28N | 12 ± 4 |
| S24N | 14 ± 4 |
| P33A | 88 ± 28 |
| P40A | 65 ± 24 |
| L27A | 20 ± 7 |
| A29G | 69 ± 18 |
| A29L | 65 ± 20 |
| R22,23K | 10 ± 4 |
| R26A | 10 ± 3 |
| R26K | 16 ± 6 |
| Control GFP | 4 ± 2 |
Due to small differences in the absolute recovery time of the wild-type proteins in various experiments, the wild-type value for 80% fluorescence recovery (t80) was set as 100% for each experiment, and the mutants were normalized according to this value.
To exclude the possibility that the lower affinity of the ΔC mutants originated from the close proximity of GFP to the NBD, we repeated the experiments with an identical set of deletion mutants in which GFP was fused to the N-terminal of HMGN2. The results obtained with GFP-HMGN proteins (not shown) were the same as those obtained with the HMGN-GFP proteins supporting the conclusion that the NBD of HMGN2, located between residues 19 and 46, is the main domain necessary for the interaction of HMGN2 with nucleosomes in native chromatin.
We previously demonstrated that HMGN proteins that do not bind to chromatin mislocalize to the nucleolus (31). Fluorescence analyses of the cells expressing the various mutants reveal that the intranuclear organization of all of the deletion mutants was identical to that of wild-type HMGN2-GFP (Fig. 1C, compare this to the middle panel of the top row in Fig. 3E). Our findings that both the wild-type protein and the deletion mutants preferentially bound to heterochromatin-rich regions and that they were excluded from nucleoli indicates that they all kept their ability to specifically recognize nucleosomes in chromatin. Thus, both in vitro and in vivo the NBD domain, by itself, can specifically bind to nucleosomes and chromatin.
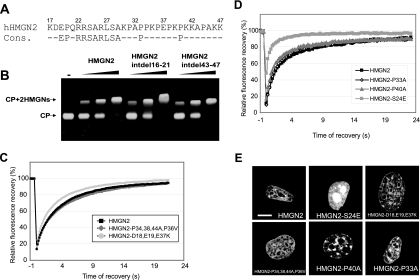
FIG. 3.
Defining the critical region for nucleosome binding specificity within the NBD. (A) HMGN2 NBD and the HMGN NBD consensus sequence. (B) Mobility shift assays of HMGN2 deletion mutants lacking either residues 16 to 21 or residues 43 to 47. Note that these internal deletions did not affect the nucleosome binding. (C) FRAP analysis of the mutants indicated in the legend. (D) FRAP analyses of the point mutants indicated in the legend. (E) Intranuclear localization of the HMGN2 mutants. Confocal images of live cells expressing the indicated GFP fusion protein are shown. Note that the HMGN2-S24E mutant that does not bind chromatin has a very fast FRAP recovery and mislocalizes to the nucleolus (see also Fig. 4). Scale bar, 10 μm.
The NBDs of HMGNs are interchangeable.
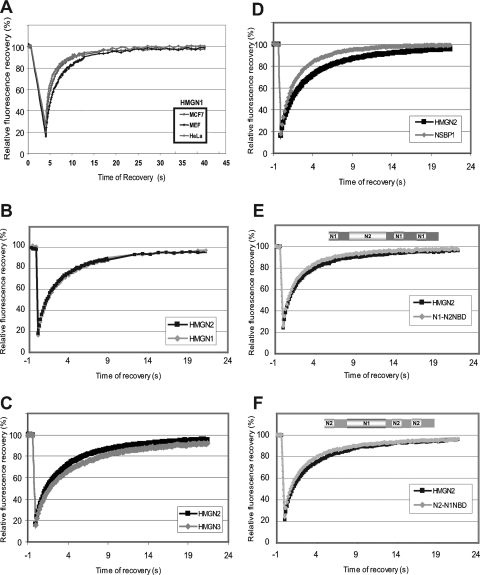
The complement of nonhistone proteins varies among different cell types (40). Since the binding of HMGN to chromatin is highly dynamic and can be influenced by the presence of other chromatin-binding proteins (13, 14, 27), we performed FRAP analyses of HMGN expressed in several cell types. These analyses revealed that the mobilities of HMGN1 (Fig. 2A) and HMGN2 (results not shown) in various cells were indistinguishable, suggesting that the interaction of HMGN with chromatin depends on the intrinsic property of the HMGNs and is not influenced by the minor differences between the chromatin of various cells. The similarity of the FRAP curves and the low standard deviation obtained with repeated experiments in different cells also indicate that in these experiments variations in the levels of ectopically expressed protein did significantly affect the results.
FIG. 2.
The NBDs of HMGN1 and HMGN2 are interchangeable. (A) The interaction of HMGN1 with chromatin is dependent on intrinsic properties of HMGN rather than on the type of cell used. FRAP curves of HMGN1 in different cells are shown. (B) Quantitative FRAP analysis of HMGN1 and HMGN2 indicates that their chromatin-binding properties are identical. (C) Quantitative FRAP analysis of HMGN2 and HMGN3 indicates that their chromatin-binding properties are very similar. (D) Quantitative FRAP analysis of HMGN2 and NSBP1 indicates that their chromatin-binding properties are very similar. (E) Quantitative FRAP analysis of a swap mutant in which the NBD of HMGN1 was swapped with that of HMGN2. (F) Quantitative FRAP analysis of a swap mutant in which the NBD of HMGN2 was swapped with that of HMGN1. The FRAP analyses shown in panels B to F were done in MEFs.
The overall structure of all HMGN is very similar and the amino acid sequence of their NBDs is highly conserved; however, the proteins are clearly distinct, especially in amino acid sequence of their C-terminal region. Since in living cells the C-terminal region contributes to the binding of HMGN to chromatin (Fig. 1B and D), we next compared the mobility of various HMGNs in MEFs. Quantitative FRAP analyses reveal that HMGN1-GFP and HMGN2-GFP proteins have identical chromatin residence times in mouse fibroblasts (Fig. 2B). Likewise, although the C termini of HMGN3 and NSBP1 differ significantly from those of HMGN1 and HMGN2, FRAP analyses reveal that their chromatin residence times are very similar (Fig. 2C and D).
The similarity in the FRAP among all members of the HMGN family, taken together with the FRAP analyses of the HMGN2 deletion mutants (Fig. 1) and with sequence conservation of the NBD, suggest that the highly conserved NBD is the major determinant of the interaction of HMGNs with chromatin, in vivo. We therefore tested whether the NBDs are interchangeable and can function independently of their surrounding sequence. To this end, we generated mutants in which the NBDs were swapped between HMGN1 and HMGN2 variants. FRAP data reveal that the mobility of the swap mutant HMGN1-N2NBD-GFP, in which the NBD of N1 was swapped with that of HMGN2, and of HMGN2-N1NBD-GFP, in which the NBD of N1 replaced the NBD of HMGN2, was the same as that of the native proteins (Fig. 2E and F). Our finding that the chromatin residence time of the swap mutants is identical to that of native proteins indicates that both NBDs have similar chromatin binding affinities and that each can act as an independent functional domain.
In summary, although the C-terminal region of HMGN affects the binding affinity of the proteins, the NBD of HMGN is the main region that determines their specific recognition and interaction with nucleosomes. Significantly, the NBD acts as an interchangeable functional domain, independent of the surrounding sequence. These experiments in living cells are in full agreement with previous in vitro mobility shift assays which also indicated that the NBD is the major determinant of the interaction of HMGN with nucleosomes (15).
Identification of the NBD residues critical for the specific binding of HMGN to nucleosomes in chromatin.
Alignment of all of the known human HMGN-like proteins demonstrates that only a few residues are absolutely conserved (Fig. 1A). The alignment contains not only the well-studied HMGN1 and HMGN2 but also HMGN3 and NSBP1, both of which have been shown to bind CPs and produce specific mobility shifts (33, 41).
For further analyses we focused on the NBD of HMGN2 (Fig. 3A) which is the most evolutionarily conserved HMGN protein. Internal deletion of residues 16 to 21 and residues 43 to 47 of HMGN2 did not affect the binding of HMGN2 to nucleosomes (Fig. 3B), suggesting that the highly conserved EP residues (HMGN2 positions 19 and 20) are not crucial for binding. In the C-terminal region of the NBD, spanning amino acids 30 to 47 of HMGN2, there are seven prolines. Since proline residues confer strict steric rigidity to the protein backbone, we next tested whether these prolines affect the binding of HMGN2 to nucleosomes. The proline at position 44 does not play a major role since the deletion mutant HMGN2-Δ43-47 binds to nucleosomes (Fig. 3B). Likewise, the highly conserved prolines at position 33 and 40 are not important for binding specificity since the point mutants HMGN2-P33A and HMGN2-P40A produce FRAP curves that are indistinguishable from those produced by the native proteins (Fig. 3D). In all FRAP experiments the mutant HMGN2-S24E serves as a control for a mutation that abolishes chromatin binding. These mutants also localize to heterochromatin regions and do not mislocalize to the nucleolus (Fig. 3E, compare with HMGN2-S24E mutant). Finally, the FRAP properties and nuclear localization of the quadruple mutant HMGN2-P33,38,44A,P36V in which four prolines were mutated are also indistinguishable from those of the wild-type protein (Fig. 3C and E). The results obtained by FRAP analyses are in full agreement with those obtained by mobility shift and immunofluorescence. We therefore conclude that these proline residues located in the NBD do not play an important role in conferring nucleosome specific binding to HMGN2.
The NBD of HMGN2 contains three negatively charged residues located at positions 18, 19, and 37. FRAP analyses of the triple point mutant HMGN2 D18,E19,E37,K, in which all of the three negative charges were replaced by a positively charged lysine, indicate that these charges have only a marginal effect on the binding affinity of HMGN2 to chromatin (Fig. 3C). In agreement with the FRAP results, confocal microscopy examinations indicate that these mutations do not affect the intranuclear organization of the protein (Fig. 3E).
Taken together with the HMGN sequence alignment (Fig. 1A) and with previous analyses of HMGN1 mutants (30), these results narrow the region required for specific interaction of HMGN2 with chromatin to the sequence RRSARLSA (residues 22 to 29), which is absolutely conserved among all of the members of the HMGN protein family. To study the amino acids necessary for chromatin-binding specificity, we first focused on Ser24 and Ser28 of HMGN2, which are homologous to Ser20 and Ser24 of HMGN1. In HMGN1, these two residues are sites of phosphorylation, a modification that abolishes the interaction of HMGN1with chromatin (32).
Wild-type HMGN2 proteins binds to nucleosome CP and produces a specific band shift containing one molecule of CP and two molecules of HMGN2 (25) (Fig. 4B, left panel). In contrast, the double point mutant protein HMGN2-S24,28E failed to produce specific complexes, and a large excess of this mutant produced a smear that is indicative of nonspecific binding (Fig. 4B, right panel). A nonspecific smear and a large-molecular-weight aggregate is produced when either wild-type HMGN2 or the HMGN2-S24,28E bind to deproteinized DNA(Fig. 4C, left and right panel, respectively). The binding affinity of HMGN2 to nucleosomes (Kd = 0.8 × 10−7 M) is threefold higher than that of the HMGN2-S24,28E mutant. Both the wild-type and the mutant protein have similar affinities for deproteinized DNA. These results suggest that serines 24 and 28 are major determinants for the specific interaction of HMGN2 with CP. Indeed, competitive mobility shift assays in which wild-type HMGN2 is added to a mixture of CP and DNA isolated from CP (CP-DNA) clearly indicate that the wild-type protein binds first to CP and produces CP-2HMGN complexes. In contrast, the HMGN2-S24,28E mutant fails to bind to CPs and binds only to DNA, producing smears (Fig. 4D). In full agreement with these in vitro experiments, FRAP analyses (Fig. 4E) indicates that the chromatin residence time of the HMGN2-S24,28E mutant is significantly shorter (t80 = 0.7 s) than that of the wild-type proteins (t80 = 7 s). Thus, both serine 24 and serine 28, or one of these by itself, are major determinants of the specific binding of HMGN2 to chromatin.
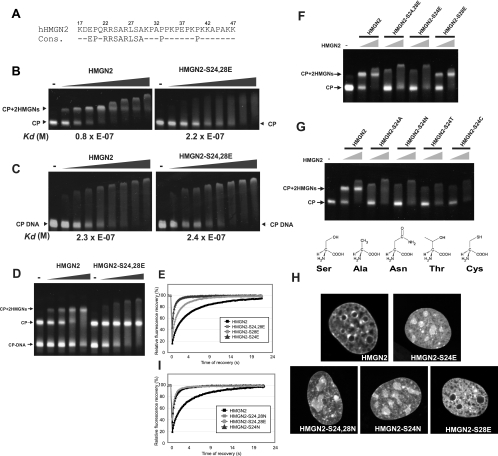
FIG. 4.
Stringent requirement of S24 for specific binding of HMGN2 to chromatin. (A) HMGN2 NBD and HMGN NBD consensus sequence. (B) Mobility shift assay of binding of wild-type and mutant HMGN2 to purified nucleosome CPs indicating that the HMGN2-S20,24E mutant does not bind to nucleosomes. The calculated dissociation constants are shown below the panels. (C) Mobility shift assay indicates equal binding of wild-type and mutant HMGN2 to 147-bp deproteinized DNA, isolated from nucleosome CPs (CP-DNA). Dissociation constants are given below the figure. (D) Competition experiments demonstrating that wild-type HMGN2 binds preferentially to CP, whereas mutant protein binds only CP-DNA. Mobility shift assay results in which either wild-type HMGN2 (left) or mutant HMGN2 (right) were added to an equimolar mixture of CP and CP-DNA are shown. Note that the wild-type protein binds first to CPs, whereas the mutant binds only CP-DNA. (E and I) FRAP analyses demonstrate that mutations of S24 abolishes the binding of HMGN2 to native chromatin. The various point mutants tested are indicated in the legend. (F and G) Mobility shift assays with the HMGN2 mutants indicated on top of the lanes. (H) Intranuclear localization of the HMGN2 mutants. Confocal images of live cells expressing the indicated GFP fusion protein are shown. Note that all of the HMGN2-S24 mutants mislocalize to the nucleolus, whereas the HMGN2-S28 mutant does not.
To test whether both Ser 24 and 28 are critical for the binding of HMGN2 to CPs, we generated single point mutants of HMGN2-GFP proteins in which either S24 or S28 were replaced by glutamic acid. FRAP measurement clearly indicate that only Ser24 is absolutely essential for the binding (Fig. 4E). The t80 of the single point mutant HMGN2-S24E is less than 1 s and similar to that of the double point mutant HMGN2-S24,28E (Fig. 4E and Table 1). The S28E substitution also impacted the mobility of HMGN2 but moderately, with a t80 of about 2.4 s, indicating that it still binds chromatin. The binding capability of these single point mutants was confirmed by electrophoretic mobility shift assays; a specifically shifted band migrating at the CP+2HMGN was observed with HMGN2-S28E but not with the HMGN2-S24E protein, which produced a smear and a large aggregate (Fig. 4F). Glutamic acid is a negatively charged and “larger” amino acid than serine. Therefore, one possible explanation for the loss of CP binding of the HMGN2-S24E mutant is that the negative charge or the steric interference prevent the binding of the HMGN2-S24E mutant to CP. We therefore replaced either only Ser 24 or both Ser 24 and Ser 28 by Asn, resulting in a semiconservative substitution by another neutral, hydrophilic amino acid capable of hydrogen bonding. FRAP analyses (Fig. 4I and Table 1), electrophoretic mobility shift assays (Fig. 4G), and confocal image analyses (Fig. 4H) all indicated that the HMGN2-S24N point mutant does not bind properly to chromatin. The results suggest that Ser 24 plays a major role in the specific interaction of HMGN2 with CPs in chromatin. Indeed, further analyses of point mutants in which Ser 24 was replaced with either Ala, Thr, or Cys, all amino acids that have approximately the same size of side chain as Ser, also abolished the binding of HMGN2 to CP (Fig. 4G).
We next used the same approaches to investigate the importance of each of the three Arg residues located in the NBD. The double point mutant HMGN2-R22,23K did not bind to chromatin (Fig. 5B), did not produce a mobility shift with nucleosomes (Fig. 5D), and mislocalized to nucleoli (Fig. 5E). The single point mutant HMGN2-R22A did not, while the HMGN2-R23A did, bind to nucleosomes (Fig. 5E). Thus, Arg 22 but not Arg 23 is necessary for nucleosome binding. The single point mutants HMGN2-R26K and HMGN2-R26A failed to bind to chromatin as assessed by FRAP (Fig. 5B), mobility shift (Fig. 5D), and image analyses (Fig. 5E). Thus, Arg 26 is necessary for specific nucleosome binding. The Ala at position 25 in HMGN2 corresponds to Ala 21 in HMGN1 (Fig. 1A). The point mutant HMGN1-A21P binds specifically to chromatin (30); therefore, we conclude that this Ala is not necessary for nucleosome binding. In contrast, the leucine at position 27 is necessary since the mutant HMGN2-L27A fails to bind to chromatin (Fig. 5C and Table 1) and mislocalizes to the nucleoli (Fig. 5E). Similar analyses of the HMGN2 point mutants in which Ala 29 was mutated to either Gly or Leu indicate that this position is not necessary for specific nucleosome interaction.
FIG. 5.
Identification of amino acids that determine the specific interaction of HMGN2 with chromatin. (A) HMGN2 NBD and HMGN NBD consensus sequence. (B) FRAP analyses demonstrate the stringent requirement for R22 and R26 for binding. (C) FRAP analyses demonstrate the requirement for L27 but not for A29 for binding. (D) Mobility shift assays demonstrating the requirement for R22 and R26 but not R23 for binding. (E) Confocal imaging of intranuclear localization of HMGN2-GFP or HMGN2-mutant-GFP. Note that mutations in R22, R26, S24, and L27 mislocalize to the nucleolus, whereas mutations at A29 remain associated with chromatin. Scale bar, 10 μm.
Based on these detailed analyses, we conclude that four of the eight amino acids within the octapeptide RRSARLSA, which is a sequence motif conserved among all HMGNs (underlined in Fig. 1A) confer nucleosome-binding specificity to this protein family. Each of these amino acids, which in the HMGN2 sequence are R22;S24;R26;L27 (indicated by asterisks in Fig. 1A), is absolutely necessary for specific binding to nucleosomes. Mutations in any of these will abolish the specific binding to nucleosomes as demonstrated by FRAP, imaging, and mobility assays. Although these four amino acids are necessary for specificity, the entire NBD is the minimum protein domain that binds with any significant affinity to isolated nucleosome cores (37).
Identification of the NBD's nucleosomal target.
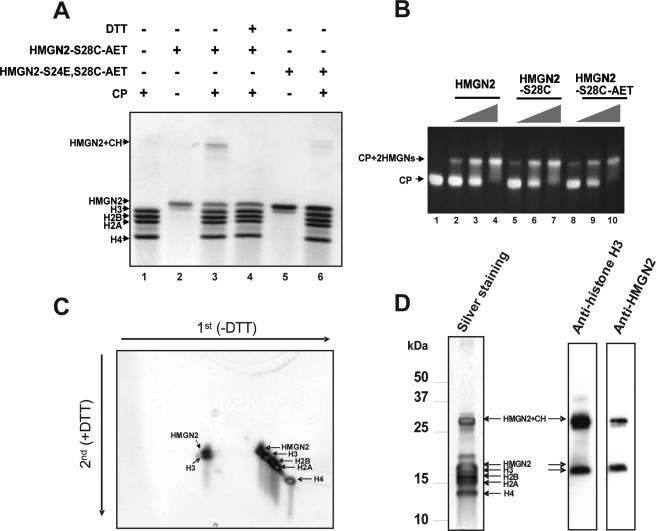
In considering the possible nucleosomal target of the RRSARLSA motif, we note that the affinity of the double point mutant HMGN2-S24,28E for nucleosomes is the same as for purified DNA (Fig. 4B and C). We therefore reasoned that this conserved motif targets the histone octamer component of the nucleosome. To identify the nucleosomal component targeted by the NBD of HMGN2, we constructed the mutant HMGN2-S28C and modified the resulting protein with the bifunctional cross-linker AET. Since HMGN2 does not have any Cys residues, the mutated residue, Cys28, is the only position modified by AET. These modifications did not affect the interaction of the proteins with CP since the binding of both the mutated protein (HMGN2-S28C) and the AET modified HMGN2-S28C (N2-S28C-AET) to CP is very similar to that of the native protein (Fig. 6B). HMGN2-S28C-AET was incubated with CP, the complexes were irradiated by UV to activate the cross-linker, and the samples were fractionated on an SDS-PAGE gel that was run without a reducing agent. A high molecular weight appears only in the CP-HMGN2-S28C-AET complexes (Fig. 6A, lane 3); this band is not detected when only the CP or only the HMGN2-S28C-AET protein alone is treated with the cross-linker (Fig. 6A, lanes 1 and 2). Likewise, this specific band is not produced by the AET modified HMGN2-S24E,S28C (HMGN2-S24E,28C-AET) mutant, which does not bind to nucleosomes (S24 is mutated to E) (Fig. 6A, lanes 5 and 6). The high-molecular-weight band is not present in samples treated with DTT (Fig. 6A, lane 4), providing further proof that the high-molecular-weight band is a result of a cross-link between the modified HMGN2 and the CP.
FIG. 6.
The core region of NBD interacts with histone H3 in the nucleosome. (A) Site-directed cross-linking experiment of CP and AET-modified HMGN2 proteins. CP was incubated with or without AET-modified HMGN2-28C or with HMGN2-S24E,S28C and then irradiated with UV. The mixtures were separated by SDS-15% PAGE, followed by Coomassie blue staining. A high-molecular-weight cross-linked product is visible only in lane 3. CH, core histones. (B) Mobility shift assays of CP with HMGN2, HMGN2-S28C, and AET-modified HMGN2-S28C reveals that the modified protein binds to CP. (C) Two-dimensional PAGE analysis of the cross-linked product. Arrows indicate the direction of the first and second dimensions of electrophoresis. Note that the cross-linked product contained only HMGN2 and H3. (D) Western blot analysis of the cross-linked product. The cross-linked product was separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane, followed by Western blotting with anti-histone H3 or anti-HMGN2.
Two-dimensional electrophoresis of the cross-linked product was used to identify the core histone targeted by the modified HMGN2. The cross-linked product was first fractionated on SDS-PAGE under nonreducing conditions, and the lane containing the fractionated proteins was cut out from the gels, treated with DTT, and refractionated on a preparative SDS gel containing reducing agent. Analysis of the second dimension revealed that the cross-linked product contained only two proteins: HMGN2 and histone H3 (Fig. 6C). Western blot analysis of a duplicate gel run only in the first dimension verified that the cross-linked product contains both H3 and HMGN2 (Fig. 6D). Thus, in the HMGN2-CP complex the NBD of HMGN2 is in close proximity to histone H3.
DISCUSSION
HMGN is the major nuclear protein family known to specifically bind to nucleosome CPs. The physical parameters of interaction of HMGN with isolated nucleosomes in vitro are well characterized; however, the determinants responsible for the specific interaction of HMGNs with nucleosome cores rather than with histones or DNA are still not fully known. Recently developed fluorescent imaging techniques have shown that the interaction of most nuclear proteins, including HMGNs, with chromatin is highly dynamic and that these proteins remain associated with a particular nucleosome for only a few seconds (13, 27, 28). The FRAP analyses provide a new tool to study in detail the properties of proteins in living cells. A major aim of the present study is to define the determinants that are important for the interaction of HMGN proteins with “native” chromatin in the nucleus of living cells.
Previous in vitro analyses identified the NBD, a highly conserved 30-amino-acid region spanning amino acids 17 to 47 of hHMGN2 or amino acids 13 to 42 of hHMGN1 (Fig. 1A), as an important nucleosome-binding module (15, 37); however, this region also binds strongly to purified DNA (1), and therefore it is not clear which determinants specify its unique binding to nucleosome cores. We now show, by FRAP and by confocal imaging on live cells expressing HMGN-GFP and by mobility shift assay, that the NBD contains a core element, RRSARLSA, that determines its specific interaction with nucleosomes both in vivo and in vitro. Thus, deletion of the 19 N-terminal or the 30 C-terminal amino acids of HMGN2 did not affect the chromatin residence time (Fig. 1B and D and Table 1) or the intranuclear localization of the protein (Fig. 1C). Furthermore, both the FRAP curves and the fluorescence images indicate that the HMGN2-ΔC43 deletion mutant, which contains only the 46 N-terminal amino acids, binds specifically to chromatin. Removal of additional six C-terminal amino acids (HMGN2-ΔC49) significantly decreased the binding affinity; however, the protein still recognizes chromatin specifically since the fluorescence images indicate that the protein does not localize to nucleoli; HMGN mutants that do not bind nucleosomes do localize to nucleoli. Furthermore, mobility shift assays with homologous HMGN1 mutants lacking the 55 C-terminal amino acids indicate that these deletion mutants recognize specifically nucleosome cores, albeit with an affinity that is twofold lower than that of the wild-type proteins (38). Taken together, the results indicate that the specificity for nucleosome recognition resides in the NBD, while the C-terminal domain strengthens the affinity of the protein for chromatin.
Interestingly, the wild-type HMGN2 binds purified DNA with an affinity constant that is similar that to the HMGN2-S24,28E mutant, and this mutant interacts with CPs, but with an affinity that is significantly lower than that of the wild-type protein (Fig. 4). These results imply that in living cells HMGNs interact with chromatin not only via highly specific interaction with CPs but also via weaker interactions with the DNA, a possibility consistent with hydroxyradical footprinting, which detected interactions between HMGN and the nucleosomal DNA (2). Thus, in some instances HMGNs could interact with chromatin, perhaps even in a biologically relevant manner (42), without occupying a specific path on the nucleosomal surface.
Most of the available data, however, indicate that the biological function of HMGN is contingent on its specific interaction with chromatin (8, 13, 22, 23, 38). Our detailed in vivo and in vitro studies demonstrate that only a limited region of the NBD determines the specific binding of HMGN2 to nucleosomes. The NBD core element that determines its specific interaction with nucleosomes is the highly conserved sequence RRSARLSA spanning amino acids 22 to 29 in HMGN2 and amino acids 18 to 25 in HMGN1. The alignment of the known NBDs indicates that this sequence is absolutely conserved in all members of the HMGN protein family (Fig. 1A). Significantly, the NBD could be exchanged among HMGN1 and HMGN2 without any effect on the chromatin residence time (Fig. 2), an indication that it can act as an independent functional domain even in the context of the whole protein. We note that there is a very high correlation between the in vitro binding assays performed by mobility shift and the in vivo FRAP analyses on more than 15 different mutants (Fig. 3 to 5). Mutants that by FRAP analyses show a decreased chromatin residence time have lower nucleosome binding affinity as measured by mobility shift assay. The results indicate that the major factors governing the interaction of HMGN with native chromatin in living cells are operative at the level of the single nucleosome, an indication that the “higher”-order chromatin structure does not have a major role in the interaction of HMGNs with chromatin. Whole-cell analyses reveal high concentrations of HMGNs in heterochromatin; most likely this reflects the high local concentration and dense packing of nucleosomes in these regions.
Based on our analyses in which we separately mutated every single amino acid in the conserved RRSARLSA motif, we suggest that only four amino acids, which in the HMGN2 sequence are R22;S24;R26;L27 (asterisks in Fig. 1A) confer nucleosome binding and recognition specificity to the HMGN protein family. A mutation in any single one of these amino acids abolishes the specific binding of the protein to chromatin. The specificity for Ser 24 is absolute; even conservative replacements by structurally similar residues such Cys, Ala, Thr, or Asn abolished the interaction of HMGN with chromatin. Likewise, Lys could not substitute for the two conserved Arg residues, suggesting that it is this specific residue, rather than just the positive charge, that confers nucleosome specificity to the HMGN protein family. The stringent requirements for the four residues in the motif suggest that the consensus sequence of the HMGNs occupies a very exact position in the nucleosomes. Consistent with this possibility, site-directed cross-linking reveals that serine 28 cross-links to a single histone, H3. Similar cross-linking studies with the closely related HMGN1 protein indicates that the amino terminus of the protein is located near H2B, while the C terminus cross-links mainly to the H3 tail (36). The findings that all of the site specific cross-linkers tested target distinct regions of specific histones suggest that the HMGNs are specifically placed on the surface of the nucleosome. Since HMGN do not bind to isolated histones and since the mutant HMGN2-S24E binds to DNA, we suggest that the conserved motif R-S-RL is a protein module that anchors HMGNs to the histone octamer.
The sequence of the four amino acids, R-S-RL, is identical to a canonical serine phosphorylation site (9), which when modified can interact with 14.3.3 proteins (16). Thus, the phosphorylation of this motif (32) and the interaction of the phosphorylated HMGN1 with 14.3.3 (31) may be a mechanism that modulates the interaction of HMGNs with nucleosomes in native chromatin.
Our studies define the limits of a protein motif that regulates the specific binding of the HMGN protein family to nucleosome CPs and identify the amino acid residues that anchor HMGN to nucleosomes in the chromatin of living cells.
Acknowledgments
We thank David Landsman, National Center for Biotechnology Information, National Institutes of Health (NIH), for numerous helpful comments on the manuscript and S. Garfield, Confocal Core Facility, LEC, National Cancer Institute (NCI), for help with imaging experiments.
This research was supported by the intramural program of the NCI, by a JSPS research fellowship for Japanese Biomedical and Behavioral Research at NIH to T.U., and by a Jacob and Lena Joels Foundation visiting professorship from the Hebrew University, Jerusalem, Israel, to M.B.
Footnotes
Published ahead of print on 25 February 2008.
REFERENCES
- 1.Abercrombie, B. D., G. G. Kneale, C. Crane-Robinson, E. M. Bradbury, G. H. Goodwin, J. M. Walker, and E. W. Johns. 1978. Studies on the conformational properties of the high-mobility-group chromosomal protein HMG 17 and its interaction with DNA. Eur. J. Biochem. 84173-177. [DOI] [PubMed] [Google Scholar]
- 2.Alfonso, P. J., M. P. Crippa, J. J. Hayes, and M. Bustin. 1994. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J. Mol. Biol. 236189-198. [DOI] [PubMed] [Google Scholar]
- 3.Ausio, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206451-463. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 5.Bianchi, M. E., and A. Agresti. 2005. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 15496-506. [DOI] [PubMed] [Google Scholar]
- 6.Birger, Y., F. Catez, T. Furusawa, J. H. Lim, M. Prymakowska-Bosak, K. L. West, Y. V. Postnikov, D. C. Haines, and M. Bustin. 2005. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 656711-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birger, Y., J. Davis, T. Furusawa, E. Rand, J. Piatigorsky, and M. Bustin. 2006. A role for chromosomal protein HMGN1 in corneal maturation. Differentiation 7419-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birger, Y., K. L. West, Y. V. Postnikov, J. H. Lim, T. Furusawa, J. P. Wagner, C. S. Laufer, K. H. Kraemer, and M. Bustin. 2003. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 221665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 2941351-1362. [DOI] [PubMed] [Google Scholar]
- 10.Bustin, M. 2001. Chromatin unfolding and activation by HMGN(*) chromosomal proteins. Trends Biochem. Sci. 26431-437. [DOI] [PubMed] [Google Scholar]
- 11.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 195237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bustin, M., F. Catez, and J. H. Lim. 2005. The dynamics of histone H1 function in chromatin. Mol. Cell 17617-620. [DOI] [PubMed] [Google Scholar]
- 13.Catez, F., D. T. Brown, T. Misteli, and M. Bustin. 2002. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3760-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catez, F., H. Yang, K. J. Tracey, R. Reeves, T. Misteli, and M. Bustin. 2004. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 244321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crippa, M. P., P. J. Alfonso, and M. Bustin. 1992. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J. Mol. Biol. 228442-449. [DOI] [PubMed] [Google Scholar]
- 16.Fu, H., R. R. Subramanian, and S. C. Masters. 2000. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40617-647. [DOI] [PubMed] [Google Scholar]
- 17.Furusawa, T., J. H. Lim, F. Catez, Y. Birger, S. Mackem, and M. Bustin. 2006. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol. Cell. Biol. 26592-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hock, R., T. Furusawa, T. Ueda, and M. Bustin. 2007. HMG chromosomal proteins in development and disease. Trends Cell Biol. 1772-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 20.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 21.Lim, J. H., F. Catez, Y. Birger, Y. V. Postnikov, and M. Bustin. 2004. Preparation and functional analysis of HMGN proteins. Methods Enzymol. 375323-342. [DOI] [PubMed] [Google Scholar]
- 22.Lim, J. H., F. Catez, Y. Birger, K. L. West, M. Prymakowska-Bosak, Y. V. Postnikov, and M. Bustin. 2004. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell 15573-584. [DOI] [PubMed] [Google Scholar]
- 23.Lim, J. H., K. L. West, Y. Rubinstein, M. Bergel, Y. V. Postnikov, and M. Bustin. 2005. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 243038-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell. Biol. 2444-456. [DOI] [PubMed] [Google Scholar]
- 25.Mardian, J. K., A. E. Paton, G. J. Bunick, and D. E. Olins. 1980. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science 2091534-1536. [DOI] [PubMed] [Google Scholar]
- 26.Marmorstein, R. 2001. Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell. Biol. 2422-432. [DOI] [PubMed] [Google Scholar]
- 27.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404604-609. [DOI] [PubMed] [Google Scholar]
- 28.Phair, R. D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D. T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 246393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Postnikov, Y. V., G. I. Belova, J. H. Lim, and M. Bustin. 2006. Chromosomal protein HMGN1 modulates the phosphorylation of serine 1 in histone H2A. Biochemistry 4515092-15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Postnikov, Y. V., D. A. Lehn, R. C. Robinson, F. K. Friedman, J. Shiloach, and M. Bustin. 1994. The cooperative binding of chromosomal protein HMG-14 to nucleosome cores is reduced by single point mutations in the nucleosomal binding domain. Nucleic Acids Res. 224520-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prymakowska-Bosak, M., R. Hock, F. Catez, J. H. Lim, Y. Birger, H. Shirakawa, K. Lee, and M. Bustin. 2002. Mitotic phosphorylation of chromosomal protein HMGN1 inhibits nuclear import and promotes interaction with 14.3.3 proteins. Mol. Cell. Biol. 226809-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prymakowska-Bosak, M., T. Misteli, J. E. Herrera, H. Shirakawa, Y. Birger, S. Garfield, and M. Bustin. 2001. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 215169-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shirakawa, H., D. Landsman, Y. V. Postnikov, and M. Bustin. 2000. NBP-45, a novel nucleosomal binding protein with a tissue-specific and developmentally regulated expression. J. Biol. Chem. 2756368-6374. [DOI] [PubMed] [Google Scholar]
- 34.Struhl, K. 1989. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem. Sci. 14137-140. [DOI] [PubMed] [Google Scholar]
- 35.Taverna, S. D., H. Li, A. J. Ruthenburg, C. D. Allis, and D. J. Patel. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 141025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trieschmann, L., B. Martin, and M. Bustin. 1998. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc. Natl. Acad. Sci. USA 955468-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trieschmann, L., Y. V. Postnikov, A. Rickers, and M. Bustin. 1995. Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional enhancement domain distinct from the nucleosomal binding domain. Mol. Cell. Biol. 156663-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueda, T., Y. V. Postnikov, and M. Bustin. 2006. Distinct domains in high mobility group N variants modulate specific chromatin modifications. J. Biol. Chem. 28110182-10187. [DOI] [PubMed] [Google Scholar]
- 39.Van Holde, K. E. 1988. Chromatin. Springer-Verlag, New York, NY.
- 40.Wakabayashi, K., S. Wang, G. Hord, and L. S. Hnilica. 1973. Tissue-specific nonhistone chromatin proteins with affinity for DNA. FEBS Lett. 3246-51. [DOI] [PubMed] [Google Scholar]
- 41.West, K. L., Y. Ito, Y. Birger, Y. Postnikov, H. Shirakawa, and M. Bustin. 2001. HMGN3a and HMGN3b, two protein isoforms with a tissue-specific expression pattern, expand the cellular repertoire of nucleosome-binding proteins. J. Biol. Chem. 27625959-25969. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, N., and U. Hansen. 2007. HMGN1 modulates estrogen-mediated transcriptional activation through interactions with specific DNA-binding transcription factors. Mol. Cell. Biol. 278859-8873. [DOI] [PMC free article] [PubMed] [Google Scholar]