Abstract
The three closely related human Ras genes, Hras, Nras, and Kras, are all widely expressed, engage a common set of downstream effectors, and can each exhibit oncogenic activity. However, the vast majority of activating Ras mutations in human tumors involve Kras. Moreover, Kras mutations are most frequently seen in tumors of endodermally derived tissues (lung, pancreas, and colon), suggesting that activated Kras may affect an endodermal progenitor to initiate oncogenesis. Using a culture model of retinoic acid (RA)-induced stem cell differentiation to endoderm, we determined that while activated HrasV12 promotes differentiation and growth arrest in these endodermal progenitors, KrasV12 promotes their proliferation. Furthermore, KrasV12-expressing endodermal progenitors fail to differentiate upon RA treatment and continue to proliferate and maintain stem cell characteristics. NrasV12 neither promotes nor prevents differentiation. A structure-function analysis demonstrated that these distinct effects of the Ras isoforms involve their variable C-terminal domains, implicating compartmentalized signaling, and revealed a requirement for several established Ras effectors. These findings indicate that activated Ras isoforms exert profoundly different effects on endodermal progenitors and that mutant Kras may initiate tumorigenesis by expanding a susceptible stem/progenitor cell population. These results potentially explain the high frequency of Kras mutations in tumors of endodermal origin.
Somatic activating Ras mutations are detected in about 15 to 20% of all human malignancies (1, 10), highlighting the importance of Ras GTPase-mediated signaling pathways in oncogenesis (10, 21, 23, 31). These mutations, which give rise to a protein that is defective for GTP hydrolysis and, therefore, remains constitutively active in a GTP-bound form, have been detected in each of the three closely related human Ras genes—Hras, Nras, and Kras. However, the vast majority of mutations detected in human cancers arise in the Kras gene. This does not appear to reflect clear differences in the biochemical properties of the three Ras proteins, as they each exhibit very similar GTP binding and hydrolysis properties and their interactions with effector targets in vitro are largely indistinguishable (2, 10, 13, 21, 73, 78). In addition, mutationally activated forms of each Ras isoform exhibit transforming activity in cell culture models, such as NIH 3T3 fibroblasts. Notably, Hras is reported to be a considerably more potent oncogene in such models (23, 54, 73, 89) and has been used for the vast majority of cell-based studies of Ras-induced oncogenic transformation thus far. It has also been suggested that tissue-specific expression differences among the Ras genes might explain their association with tumors in distinct tissues, although these three Ras isoforms are all widely expressed (73, 78). Differences in the carboxyl termini of the Ras proteins, which determine their modification by lipids and consequent subcellular membrane distribution, are likely to contribute to their distinct oncogenic potential in vivo (4, 31, 61, 73).
The fact that activating Kras mutations are detected with highest frequency in tumors of endodermally derived tissues, such as those of lung (35%), pancreas (95%), and colon (30%) (1, 11, 21, 31, 88), raises the possibility that mutationally activated Kras exerts its oncogenic activity at the level of an endodermal precursor or stem cell. Indeed, a recent study revealed that activated Kras can expand a bronchoalveolar stem cell population in culture and that this may explain the role of Kras in promoting lung adenocarcinomas in a mouse model (48).
One of the most thoroughly studied in vitro models of stem cell differentiation along the endodermal lineage is the F9 mouse embryonal carcinoma stem cell model (82). Cultured F9 cells express well-established stem cell markers, including Oct3/Oct4, Nanog, SOX2, and SSEA-1, and they undergo self-renewal in vitro (12, 16, 65, 92). When F9 cells are treated with retinoic acid (RA) for 7 to 10 days, the expression of these stem cell markers is lost, the cells stop proliferating, they undergo a morphological transformation, and they begin to express genes associated with early endoderm differentiation, including GATA4 (14, 32). The physiologic relevance of this model is well supported by substantial evidence, indicating a requirement for RA signaling in the differentiation of endodermal tissues during normal vertebrate development (27, 55, 57, 59).
Previous studies have revealed that the expression of mutationally activated Hras (HrasV12) in F9 cells promotes their differentiation to early endoderm in the absence of RA (86, 95). To examine a potentially distinct consequence of activating the other Ras isoforms in endodermal precursors, we have compared the effects of the three different activated Ras genes in this system and identified a dramatic difference in their activities. In striking contrast with the case for HrasV12, expressing KrasV12 promotes an expansion of the F9 population and prevents cell differentiation in response to RA. NrasV12 is essentially inert in this system. This striking difference in biological activity among these closely related Ras proteins reflects differences in their C termini and may account for the high frequency of activating mutant Kras alleles detected in tumors of endodermally derived tissues.
MATERIALS AND METHODS
Primary antibodies.
Primary antibodies used include those raised against Nanog (Bethyl); SSEA1 and SSEA3 (gifts from D. Solter); Oct3/Oct4, pan-ras, and GM130 (Transduction Laboratories); Sox2 and actin (Sigma); pan-ras, Kras, and Hras (Calbiochem); and Raf-1, GATA4, and α-tubulin (Santa Cruz).
Cell culture, transfections, focus formation, and proliferation assays.
F9 cells (a gift from S. Strickland) and PCC4 cells were maintained on gelatin-coated tissue culture dishes in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum (Gibco). To induce endodermal differentiation, F9 or PCC4 cells were seeded sparsely and incubated with 0.2 to 1.0 μM all-trans RA (Sigma). Cells were maintained in the dark, and the medium was replaced every 48 h. F9 or PCC4 cells were transfected using Lipofectamine 2000 or nucleoporated using a Nucleofector II device (Amaxa). For stable transfections, cells were selected for their resistance to G418 (Gibco). For RA-resistant, F9-derived cells, cells were first selected in the appropriate antibiotic for 7 to 10 days after transfection and then incubated with antibiotic plus RA for an additional 7 to 10 days. Dishes were fixed with methanol and stained with Giemsa for documentation. Resistant colonies were either cloned using cloning cylinders or pooled as polyclonal populations and maintained in the presence of the selecting antibiotic, plus or minus RA. The expression vectors used were pEYFP-C1-Kras12V, pEYFP-C1-Hras61L, pEGFP-C3-Nras12D, pEYFP-C1-Hras12V-KrasTail, pEYFP-C1-Kras12V-HrasTail, pcRaf1-HrasTail, and pcRaf1-KrasTail, which were described previously (19, 20); pWP1-HA-KrasG12V, pWP1-HA-KrasG12V-E37G, pWP1-HA-KrasG12V-T35S, pWP1-HA-KrasG12V-Y40C, 2xmyc-NH2-tagged HrasG12V, 3XHA-NH2-tagged KrasG12V, and 3XHA-NH2-tagged NrasG12V in pcDNA3.1 (Invitrogen), which were obtained from the Guthrie Foundation cDNA Resource Center; and pcDNA3.1 with 3XHA as vector control. pMAXGFP (Amaxa) was used to evaluate the efficiency of transfection. The metabolic inhibitors LY294002 and UO126 were used at 10 μM, with solvent dimethyl sulfoxide (DMSO) used as control.
Growth curves were established by plating cells in triplicate or quadruplicate parallel dishes. Cells were counted daily with a hemocytometer to yield cell numbers. Alternatively, relative cell numbers were determined at daily intervals by using SYTO60, a red fluorescent nucleic acid stain (Molecular Probes). Triplicate or quadruplicate dishes were processed for SYTO60 staining, scanned, and quantified with the Li-Cor Odyssey system. Growth curves were generated using Microsoft Excel, with error bars indicating standards of deviation, and P values were generated using one-way analysis of variance. Experiments were performed two to four times.
Immunoblotting.
Cell pellets were collected by removing the medium and washed twice with ice-cold phosphate-buffered saline (PBS), and cells were scraped into a microfuge tube and pelleted at 2,000 × g for 5 min at 4°C. The supernatants were aspirated, and the pellets were then maintained at −20°C. For immunoblotting, pellets were thawed on ice and lysed with Laemmli sample buffer without glycerol or reducing agent, with protease (Sigma) and phosphatase (Calbiochem) inhibitor cocktails, and boiled for 10 min. The supernatants were quantified using detergent-compatible DC protein assay (Bio-Rad) and a VMax kinetic microplate reader (Molecular Devices), with bovine serum albumin as a standard. Concentrations were adjusted with lysis buffer. Laemmli sample buffer (5× concentrated) was then added to the samples. Equal protein concentrations were loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose filters, which were blocked overnight at 4°C. The blots were incubated for 1 h at room temperature with the indicated primary antibodies. Secondary antibodies, conjugated to horseradish peroxidase (Sigma or Cell Signaling Technology), were also incubated for 1 h at room temperature. Signal visualization was achieved using chemiluminescence (Pierce) and BioMax MR film (Kodak). Films were scanned using ScanMaker 8700 (Microtek).
Phase-contrast and immunofluorescence microscopy.
For immunostaining, cells were grown on 12-mm glass coverslips and coated with gelatin or Cell-Tak (Becton Dickinson). The cells were fixed with either 2% paraformaldehyde or −20°C methanol and washed with PBS. Ammonium chloride in PBS was used to diminish autofluorescence. Where necessary, cells were permeabilized with 0.2% Triton X-100 in PBS for 10 min on ice. Blocking was performed with PBS containing 2% serum for 30 min at room temperature. Primary antibodies and secondary antibodies, conjugated to Texas Red-X or Oregon Green (Molecular Probes), were diluted in blocking buffer and incubated with cells for 30 min at room temperature in a humidified chamber. After each incubation, cells were washed three times with PBS. Counterstaining with Hoechst 33258 (Molecular Probes) was performed at the second wash. Coverslips were then mounted on slides using Airvol, visualized using a Zeiss Axioplan2 immunofluorescence microscope, and imaged using an AxioCamMR digital camera and AxioVision 4.5 software. Phase-contrast microscopy was conducted using a Nikon Diaphot inverted microscope and an Olympus SP-350 digital camera.
To assess the subcellular localization of Ras in F9 cells, undifferentiated F9 cells or F9 cells treated with 200 nM RA for 5 days to induce differentiation were plated at 3 × 105 cells per plate into 35-mm dishes containing a 15-mm glass coverslip-covered cutout (MatTek). Cells were transfected 24 h later with either pEYFP-C1-Kras12V, pEYFP-C1-Hras61L, pEYFP-C1-Hras12V-KrasTail, pEYFP-C1-Kras12V-HrasTail, or pEYFP-C1-GalT (galactosyl transferase, a marker for the Golgi) by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours posttransfection, cells were imaged with a Zeiss 510 inverted laser scanning confocal microscope. A minimum of five 0.45-μm z slices were acquired for each cell, and representative images were chosen to display plasma membranes and/or endomembranes. All images shown are representative of more than 90% of cells examined.
RESULTS
F9 cells as a model of stem/progenitor cell differentiation along the endodermal lineage.
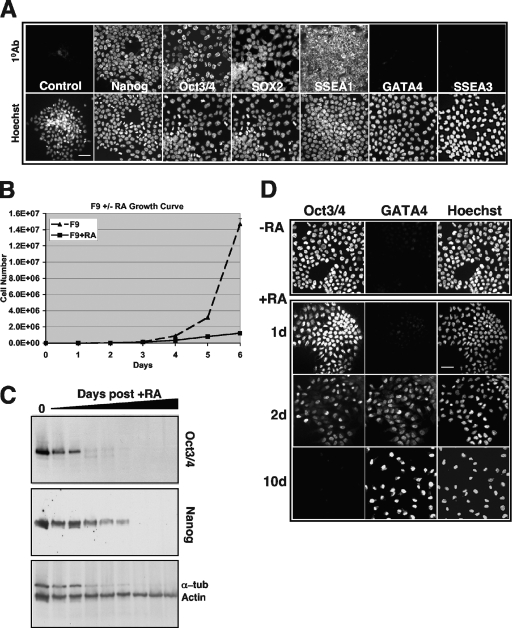
Activating Kras mutations are detected most frequently in tumors of endodermally derived tissues, such as those of lung (35%), pancreas (95%), and colon (30%) (1, 2, 9, 10, 88), suggesting that mutant Kras may initially exert its oncogenic activity in an endodermal progenitor or stem cell. To test the hypothesis that activated Kras can influence the properties of endodermal stem/progenitor cells in a manner distinct from activated Hras or Nras, we exploited the F9 mouse embryonal carcinoma model of stem cell differentiation to endoderm (82). These cells exhibit several stem cell properties, including the ability to undergo self-renewal, the capacity to differentiate along distinct lineages, and the expression of stem cell markers, including Oct3/Oct4, Nanog, SOX2, and SSEA1 (Fig. 1A). In response to continuous treatment with RA, the cells undergo morphological transformation associated with reduced proliferative capacity (Fig. 1B), they lose their stem cell markers, and they begin to express endodermal markers (GATA4 and SSEA3) (Fig. 1C and D and data not shown). Thus, this cell culture model recapitulates aspects of stem cell differentiation to early endoderm during vertebrate development, which also requires RA (27, 55, 57, 59).
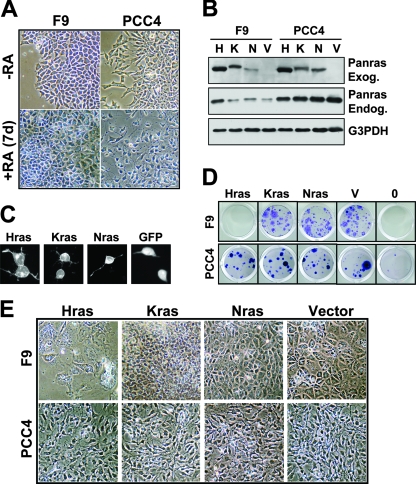
FIG. 1.
F9 cells exhibit stem cell characteristics and can be induced by RA to undergo endodermal differentiation. (A) The upper panels illustrate immunofluorescence micrographs demonstrating that untreated F9 cells express the stem markers Nanog, Oct3/Oct4, SOX2, and SSEA1 and do not express the endodermal markers GATA4 and SSEA3. Control, nonspecific antibody. Lower micrographs illustrate Hoechst counterstaining of the same field. Bar, 20 μM. (B) Growth curve demonstrating the loss of F9 self-renewal (P < 0.001) capacity upon RA treatment (P < 0.001). Error bars represent standard deviations. (C) Immunoblots of F9 cell extracts demonstrating the loss of expression of the stem cell markers Oct3/Oct4 and Nanog as a function of days following RA treatment. Actin and α-tubulin (α-tub) are loading controls. Note that the level of α-tubulin decreases as cells cease self-renewal and start to differentiate, consistent with its decrease in senescent cells (44). (D) Immunofluorescence micrographs of F9 cells to detect stem cell and differentiation markers upon RA treatment. −RA, untreated F9 cells in culture retain expression of stem cell markers such as Oct3/Oct4 and do not differentiate spontaneously or express differentiation markers, such as GATA4. +RA, RA treatment of F9 cells for 1, 2, or 10 days (d) causes the temporal loss of expression of the nuclear stem cell marker Oct3/Oct4 and the gain of expression of the endodermal transcription factor, GATA4, indicative of differentiation to primitive endoderm. Bar, 20 μM.
Distinct activities of Ras isoforms in F9 differentiation.
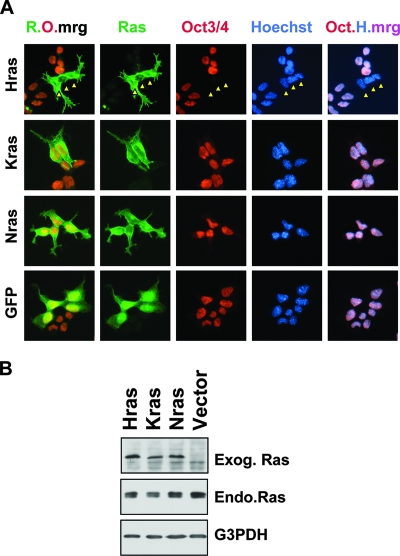
The expression of mutationally activated Hras (HrasV12) in F9 cells is sufficient to promote their differentiation to primitive endoderm in the absence of RA (86, 95). To identify a potentially distinct activity of the other Ras isoforms in F9 differentiation, we established expression vectors for HrasV12, NrasV12, and KrasV12 (Fig. 2). We initially confirmed the functionality of each of these three proteins by demonstrating their focus-forming activities in transfected NIH 3T3 fibroblasts (data not shown), and we then compared their activities in the F9 model system. As was previously reported, we found that HrasV12 promotes the differentiation of F9 cells in the absence of RA. Within 2 days of transfection, HrasV12-transfected cells lose expression of the stem cell markers Oct3/Oct4 (Fig. 2A and 3A) and SSEA1 (data not shown) and exhibit altered morphologies (Fig. 3A). By 7 days posttransfection, they cease proliferating and assume epithelial morphologies (Fig. 3B and data not shown) and they cannot be propagated further. In contrast to HrasV12, F9 cells transfected with NrasV12 or KrasV12 are morphologically indistinguishable from vector-transfected F9 cells (Fig. 2A and 3B) and they continue to express stem cell markers and can be readily passaged (Fig. 2A and data not shown). We observed similar properties among these Ras isoforms following transfection with a range of plasmid concentrations or with different vector backbones and fusion tags, indicating that the observed biological differences do not reflect expression level differences (data not shown).
FIG. 2.
HrasV12, but not KrasV12 or NrasV12, represses the stem cell marker Oct3/Oct4. (A) F9 cells transfected with the indicated Ras-green fluorescent protein (GFP)-fusion plasmids were fixed and processed for immunofluorescence 48 h posttransfection with antibodies against Oct3/Oct4 (red). The Ras proteins are shown in green. Nuclei were counterstained with Hoechst 33258 (blue). Note that the HrasV12-expressing F9 cells (Hras) were already Oct3/Oct4 negative, as indicated by the arrowheads. R.O.mrg, merge of Ras (green) and Oct3/Oct4 (red); Oct.H.mrg, merge (pink) of Oct3/Oct4 (red) and Hoechst (blue). (B) Immunoblots of F9 cell extracts demonstrating the expression of transduced GFP-Ras isoforms (exogenous Ras [Exog. Ras]), which exhibit reduced mobility on SDS-PAGE, compared to that of the endogenous Ras (Endo.Ras). G3PDH, loading control.
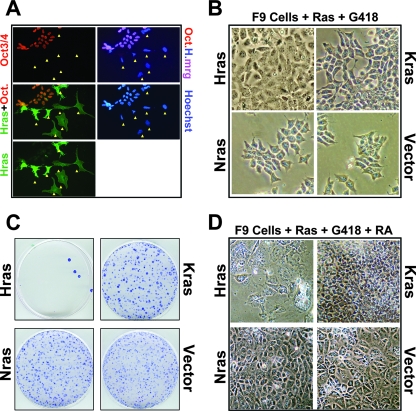
FIG. 3.
HrasV12, KrasV12, and NrasV12 differentially affect endodermal differentiation. (A) Expression of HrasV12 in F9 cells represses Oct3/Oct4 expression and induces morphological alteration. F9 cells transfected with green fluorescent protein (GFP)-HrasV12 were fixed and processed for immunofluorescence 48 h posttransfection with antibodies against Oct3/Oct4. Nuclei were counterstained with Hoechst 33258. Note that the HrasV12-expressing F9 cells (Hras) are already exhibiting altered morphologies as they differentiate into endoderm. Arrows indicate cells that are HrasV12 positive and Oct3/Oct4 negative. Oct.H.mrg, merge of Oct3/Oct4 (red) and Hoechst (blue). (B) Phase-contrast images of F9 cells 10 days after transfection and G418 selection demonstrating the altered morphologies of HrasV12-expressing cells (Hras), while KrasV12- and NrasV12-expressing cells are indistinguishable from vector-transfected cells. (C) F9 cells were transfected with the indicated plasmids, drug selected for 2 weeks, and stained with Giemsa to visualize the drug-resistant colonies selected. Note that HrasV12 expression does not yield stable, G418-resistant colonies, consistent with its induction of differentiation and loss of self-renewal capability. (D) Phase-contrast images of F9 cells transfected with the indicated plasmids and selected with G418 for 1 week and with G418 plus RA for 1 week. Note that HrasV12-transfected cells die and NrasV12-transfected cells, like vector-transfected cells, differentiate, whereas KrasV12-expressing cells completely resist RA-induced differentiation.
Activated Kras promotes stem cell proliferation and blocks differentiation.
We next examined the consequences of sustained expression of the activated Ras isoforms in F9 cells. Two weeks posttransfection, vector-transfected cell colonies can be readily selected, whereas HrasV12-transfected cells fail to yield detectable colonies (Fig. 3C), reflecting HrasV12's ability to promote differentiation and growth arrest (Fig. 3A and B). However, KrasV12- and NrasV12-transfected F9 cells remain undifferentiated and can be readily propagated (Fig. 3B and C and data not shown). Upon RA treatment, vector-transfected and NrasV12-transfected cells differentiate (Fig. 3D) and cannot be propagated beyond two passages (Fig. 4E and data not shown). RA-treated, HrasV12-transduced cells, which are already undergoing HrasV12-induced differentiation, senesce and eventually die (Fig. 3D).
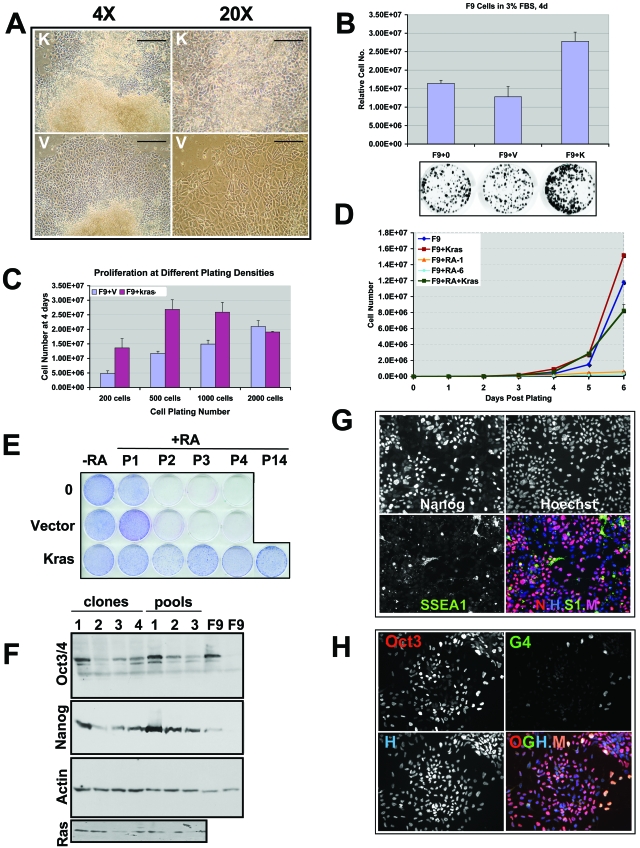
FIG. 4.
KrasV12-expressing F9 cells exhibit enhanced proliferative potential and retain stem cell features in the presence of RA. (A) Phase-contrast micrographs demonstrating the resistance to RA-mediated differentiation of KrasV12-transfected (K), antibiotic-resistant colonies, in contrast to RA-mediated endodermal differentiation of vector-transfected (V), antibiotic-resistant colonies. Note the increased size and polygonal shape of the differentiated cells in response to RA in the vector-transfected colony. Left, magnification, ×4; bar, 200 μm. Right, magnification, ×20; bar, 50 μm. (B) KrasV12-expressing F9 cells exhibit enhanced proliferative potential in reduced serum. F9, F9 plus Vector (F9+V), or KrasV12-expressing F9 (F9+K) cells were plated in medium containing 3% serum (fetal bovine serum [FBS]) for 4 days. Relative cell numbers were determined by using SYTO60 staining, followed by quantification. Parallel plates were fixed and stained with Giemsa to indicate cell growth, and a representative set is shown. Error bars represent standard deviations. P < 0.001. (C) KrasV12-expressing F9 cells exhibit enhanced proliferative potential when plated at low cell density. The indicated number of F9 plus Vector (F9+V) or KrasV12-expressing F9 (F9+Kras) cells was plated and then counted after 4 days. Error bars represent standard deviations. P < 0.001. (D) KrasV12 enhances F9 proliferation in the presence or absence or RA. F9 or RA-resistant KrasV12-expressing F9 cells (maintained in RA) were plated in the presence (F9+RA+Kras) or absence (F9+Kras) of RA and counted on the indicated days. For F9+RA-1, cells were placed in RA at the beginning of the experiment. For F9+RA-6, cells were incubated in RA for 6 days prior to the beginning of the growth curve experiment to demonstrate that the proliferation of naïve F9 cells is very sensitive to continuous RA treatment. Error bars represent standard deviations. P < 0.001. (E) KrasV12 enables indefinite passaging (P) of F9 cells in the presence of RA. F9 (0) or vector-transfected F9 cells can be passaged only two or three times in the presence of RA (+RA), whereas KrasV12 expression confers indefinite passaging potential in the presence (+RA) or absence (−RA) of RA. Shown is passage 14 (P14) in the presence of RA (+RA), and these clones have been maintained beyond passage 25 in RA. Cells were passaged, plated, allowed to grow 1 week, and then fixed and stained with Giemsa at the passage number indicated along the top. (F) Immunoblot demonstrating the expression of the stem cell markers Oct3/Oct4 and Nanog (indicated on the left) in clones or pools of KrasV12-expressing F9 cells in the presence or absence of RA. This demonstration is in contrast to that by untransfected F9 cells, which lose expression of Oct3/Oct4 and Nanog in the presence of RA (indicated in the right two lanes). Actin, loading control. Transduced Ras expression in independent clones and polyclonal, pooled populations of F9 cells transfected with KrasV12 is shown in the bottom panel. (G) KrasV12-expressing F9 cells continue to express the stem cell marker Nanog, but not SSEA1, in the presence of RA. Immunofluorescence micrographs were prepared using antibodies against the indicated antigens. Nuclei were counterstained with Hoechst 33258. N-H-S1-M, merge (M) of Nanog (N), Hoechst (H), and SSEA1 (S1). Letter color indicates fluor of the secondary antibody used. Note that most cells are Nanog positive (pink is merge of Nanog and Hoechst), but SSEA1 negative. (H) KrasV12-expressing F9 cells maintain expression of the stem cell transcription factor Oct3/Oct4 in the presence of RA. Immunofluorescence micrographs were prepared using antibodies against the indicated antigens. Nuclei were counterstained with Hoechst 33258 (H). O-G-H-M, merge (M) of Oct3/Oct4 (O), GATA4 (G), and Hoechst (H). Note that a small number of cells coexpress the stem cell nuclear marker Oct3/Oct4 and the endodermal transcription factor GATA4 (G4) in the presence of RA (O-G-H-M merge; M, peach color). Letter color indicates fluor of the secondary antibody used.
In striking contrast to the case for HrasV12- or NrasV12-transfected F9 cells, KrasV12-transfected cells appear to remain undifferentiated upon RA treatment (Fig. 3D and 4A). Stable KrasV12-expressing cells were expanded for further characterization. They exhibit increased proliferative potential and a reduced serum requirement relative to parental F9 cells (Fig. 4B to D) and can be passaged indefinitely in the presence of RA without obvious consequence (beyond passage 25) (Fig. 4E). Moreover, unlike RA-treated F9 cells, KrasV12-expressing cells cultured long term in RA retain stem cell markers (Fig. 4F to H). Notably, these cells remain RA responsive, as indicated by the fact that RA treatment promotes the loss of SSEA1 (Fig. 4G). We determined that multiple, clonally derived KrasV12-transfected cell lines expressing widely varying levels of KrasV12 protein (Fig. 4F and data not shown) exhibit indistinguishable phenotypes with respect to resistance to differentiation, further confirming that the distinct activities of the Ras isoforms do not reflect expression differences. Thus, KrasV12 is uniquely able to promote the expansion of a population of undifferentiated endodermal stem/progenitor cells.
The ability of activated Kras to promote stem/progenitor cell expansion is lineage restricted.
To determine whether the distinct activities of the activated Ras isoforms are limited to a particular stem/progenitor lineage, we performed analogous studies of PCC4 cells, a mouse embryonic carcinoma stem cell line that responds to RA by differentiating into mesenchyme but not endoderm (Fig. 5A). The expressions of the various activated Ras isoforms fail to induce PCC4 differentiation, and they each yield G418-resistant cells that can be readily propagated (Fig. 5B to D and data not shown). Moreover, RA-treated HrasV12-, KrasV12-, or NrasV12-expressing PCC4 cells remain indistinguishable from vector-transfected cells in that they all differentiate into mesenchymal cells that cannot be serially passaged (Fig. 5E and data not shown). Together, these findings suggest that the lineage-specific context of the progenitor/stem cell determines its response to the various activated Ras proteins and that stem cells partially committed to an endodermal fate (e.g., F9 cells) exhibit very different biological responses to the three mutant Ras isoforms. These findings also suggest that mutationally activated Kras can uniquely expand endodermal stem/progenitor cells.
FIG. 5.
Mesenchymal stem cells are unaffected by expression of activated Ras isoforms. (A) F9 and PCC4 cells differentiate in response to RA, yielding distinct morphologies. Phase-contrast micrographs indicating the morphologies of F9 and PCC4 cells treated (bottom) or untreated (top) with RA for 7 days. (B) F9 and PCC4 cells express transduced Ras genes at comparable levels. Immunoblots of F9 and PCC4 cell extracts demonstrating expression of transduced activated green fluorescent protein (GFP)-Ras isoforms (exogenous Ras [Panras Exog.]), which exhibit reduced mobility under SDS-PAGE, compared to that of the endogenous Ras (Panras Endog.). H, Hras; K, Kras; N, Nras; V, vector-transfected cells. G3PDH: loading control. (C) Ras proteins localize similarly in PCC4 cells. Fluorescence micrographs of PCC4 cells transfected with the indicated activated Ras expression plasmids or GFP. (D) F9 and PCC4 cells were transfected with the indicated activated Ras expression plasmids or vector (V), or were not transfected (0), and were then drug selected for 2 weeks and stained with Giemsa to visualize the drug-resistant colonies. Note that HrasV12 expression does not yield stable, G418-resistant colonies in F9 cells but does so in PCC4 cells and that none of the isoforms produced an obvious phenotype in PCC4 cells. (E) Phase-contrast micrographs indicating the morphologies of F9 and PCC4 cells transfected with the indicated expression plasmids and selected in G418 for 1 week plus G418 and RA for an additional week. Note that none of the Ras isoforms yields a novel phenotype in PCC4 cells, and in all cases, the PCC4 cells differentiate like the vector control in the presence of RA, in contrast to differing phenotypes in F9 cells transfected with the different Ras isoforms. Note that the Hras-transfected F9 cells, which are undergoing differentiation due to Hras expression, are dying in response to RA.
The differing C-terminal domains of HrasV12 and KrasV12 determine their distinct biological activities in F9 differentiation.
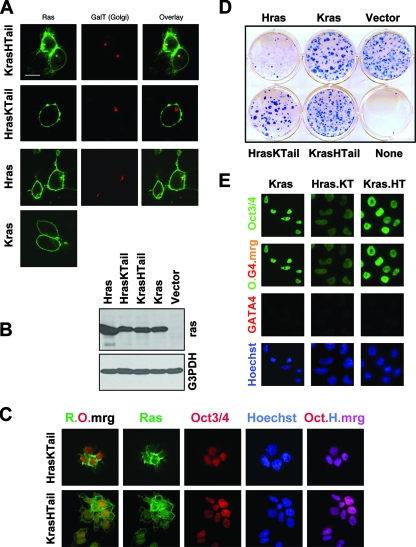
Hras and Kras are highly homologous, except for their carboxy-terminal tails, which determine the association with cellular membranes (4, 19, 31, 73, 81). Kras is largely restricted to the plasma membrane, while Hras localizes to plasma membrane and Golgi. In F9 cells, Kras is similarly restricted to plasma membrane, whereas Hras is detected both at plasma membrane and at Golgi (Fig. 6A). To directly test a role for the Ras carboxyl terminus in the F9 model, we expressed an HrasV12 chimeric protein in which the carboxyl terminus was substituted with that of Kras (Fig. 6B, lane HrasKTail). As expected, HrasKTail no longer localizes to Golgi (Fig. 6A and data not shown). Unlike HrasV12, but similarly to KrasV12, HrasKTail does not repress Oct3/Oct4 expression or induce differentiation (Fig. 6C to E). It is well tolerated in F9 cells, and we could readily establish stably transfected colonies that express stem cell markers (Fig. 6D and E and data not shown). Moreover, RA-treated HrasKTail-expressing cells, like KrasV12-expressing cells, fail to differentiate and they maintain stem cell-like properties, including the expression of Oct3/Oct4 and the ability to be passaged indefinitely (Fig. 6E and data not shown). In a complementary experiment, we examined the properties of a Ras chimera in which the Hras carboxyl terminus was fused to Kras (Fig. 6B, lane KrasHTail). As expected, this chimeric protein demonstrated Golgi localization due to the Hras tail (Fig. 6A). However, unexpectedly, it did not repress Oct3/Oct4 expression or induce F9 differentiation (Fig. 6C to E and data not shown). Thus, while the ability of Hras to induce differentiation seems dependent upon its ability to signal from the Golgi, the ability of Kras to maintain a stem cell phenotype is not strictly dependent on its subcellular localization. However, a critical role for sequences within the Kras C terminus is supported by the additional observation that the expression of an activated version of the less commonly detected Kras isoform, Kras-4A, a splice isoform which differs from the far more abundantly expressed Kras-4B isoform (used in all other studies here) only at the C terminus, fails to promote RA-resistant stem cell maintenance in F9 cells and instead causes apoptosis (data not shown).
FIG. 6.
The differing C-terminal domains of HrasV12 and KrasV12 determine their distinct biological activities in F9 differentiation. (A) Live-cell confocal micrographs of the indicated green fluorescent protein-Ras isoform fusion proteins in transfected F9 cells. Note that HrasV12 and KrasHTail, unlike KrasV12 and HrasKTail, localize to the Golgi, as revealed by GalT staining, in addition to the plasma membrane. The overlay shows the merge (yellow) of Ras and GalT. (B) Immunoblots of F9 cell extracts demonstrating the expression of transduced green fluorescent protein-Ras isoforms, which exhibit reduced mobility on SDS-PAGE gels. G3PDH, loading control. (C) F9 cells transfected with the indicated Ras-green fluorescent protein-fusion plasmids were fixed and processed for immunofluorescence 48 h posttransfection with antibodies against Oct3/Oct4 (red). The Ras proteins are labeled in green. Nuclei were counterstained with Hoechst 33258 (blue). Note that the HrasKTail-expressing F9 cells are Oct3/Oct4 positive, in contrast to Hras (see panel A). R.O.mrg, merge of Ras (green) and Oct3/Oct4 (red). Oct.H.mrg, merge (pink) of Oct3/Oct4 (red) and Hoechst (blue). Letter color indicates fluor of the secondary antibody used or merge color. (D) Expression of HrasV12 with a Kras C terminus (HrasKTail), like KrasV12, but not HrasV12 (also see panel C), is able to generate stable F9 cells. F9 cells were transfected with the indicated expression plasmids, selected with G418, fixed, and stained with Giemsa. (E) F9 cells expressing HrasV12 with the Kras C terminus (Hras.KT) or Kras with the Hras C terminus (Kras.HT), selected in the presence of RA, maintain Oct3/Oct4 and not GATA4 expression, resembling Kras cells, as illustrated with immunofluorescence micrographs. O.G4-mrg, merge (yellow) of Oct3/Oct4 (O) and GATA4 (G4). Nuclei were counterstained with Hoechst 33258 (blue). Letter color indicates fluor of the secondary antibody used or merge color.
PI-3 kinase and MEK are differentially required for endodermal stem/progenitor cell differentiation and stem cell maintenance.
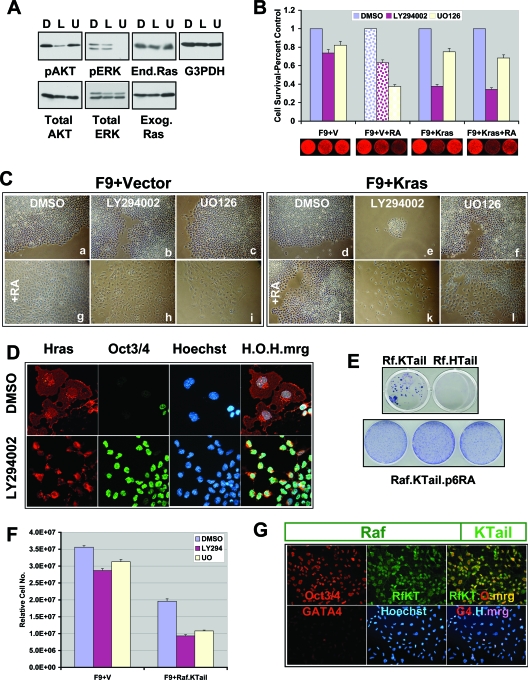
The distinct and seemingly opposing phenotypes associated with HrasV12 and KrasV12 expression in F9 cells prompted us to compare their signaling outputs in the context of F9 differentiation. Ras interacts with multiple downstream effectors via a highly conserved, 9-amino-acid, amino-terminal domain (switch I), encompassing amino acid residues 32 to 40 (91). The three best-established Ras effectors implicated in its oncogenic properties include the Raf kinase, phosphatidylinositol 3-kinase (PI-3 kinase), and the Ral GTPase activator RalGDS (2, 10, 13, 21, 28, 56, 73, 78). Notably, the PI-3 kinase-AKT, Raf-MEK-extracellular signal-regulated kinase, and RalGDS-Ral GTPase pathways have previously been implicated in F9 differentiation by RA or HrasV12 (6, 86, 87) as well as in oncogenic Ras transformation (28, 91). Therefore, we examined the role of these Ras effectors in vector-transfected and KrasV12-expressing F9 cells. Pharmacologic inhibition of PI-3 kinase (with LY294002) (Fig. 7A) does not affect the proliferation of vector-transfected F9 cells or their expression of Oct3/Oct4 (Fig. 7B and C, panel b, and 8B and T). However, PI-3 kinase is required for RA-induced expression of GATA4 and endoderm differentiation (Fig. 7C, panel h, and 8K and W). Moreover, PI-3 kinase is not required for KrasV12-mediated stem cell maintenance (Fig. 8BB and TT), but is required for sustained proliferation (Fig. 7B, Ce, and Ck) and to prevent the loss of stem cell maintenance by RA treatment (Fig. 8KK and WW). In contrast, LY294002-treated F9 cells transiently transfected with HrasV12 maintain Oct3/Oct4 expression and consequently fail to undergo differentiation and growth arrest (Fig. 7D). Together, these results indicate a complex role for PI-3 kinase downstream of both activated Hras and activated Kras that involves distinct requirements in endodermal stem/progenitor cell maintenance, proliferation, and differentiation as well as resistance to the actions of RA.
FIG. 7.
PI-3 kinase and MEK are differentially required for endodermal stem/progenitor cell differentiation and stem cell maintenance. (A) Immunoblots of F9 cell extracts demonstrating the pharmacologic inhibition of PI-3 kinase signaling with LY294002 (L) and MEK signaling with UO126 (U) compared to signaling with solvent DMSO (D). Note that these treatments have no effect on the levels of expression of endogenous (end.) or transduced (exog.) Ras. G3PDH, loading control. (B) Increased sensitivity of KrasV12-expressing F9 cells to LY294002. Vector (V)- or KrasV12-transfected F9 cells were plated in the presence or absence of RA, as indicated along the bottom panels, and representative plates stained with SYTO60 are indicated below the histogram. After 4 days, relative cell numbers, in triplicate or quadruplicate, were determined by quantification of SYTO60 staining. D, endodermally differentiated F9 cells, due to RA treatment. Error bars represent standard deviations. P < 0.001. (C) Phase-contrast micrographs of vector-transfected F9 cells (left) and KrasV12-transfected F9 cells (right) maintained in the presence of the PI-3 kinase inhibitor LY294002 (panels b, e, h, and k), the MEK inhibitor UO126 (panels c, f, i, and l), or solvent DMSO (panels a, d, g, and j), in the absence (panels a to f) or presence (panels g to l) of RA for 8 days. Note the decreased cell number in the presence of LY294002 in cells expressing KrasV12 (panel e compared to d versus panel b compared to a). (D) Immunofluorescence micrographs demonstrating that LY294002 prevents HrasV12 (red)-induced morphological changes and inhibition of Oct3/Oct4 expression (green). H.O.H.mrg, merge of HrasV12 (H), Oct3/Oct4 (O), and Hoechst (H). (E) The top panel shows Giemsa-stained plates illustrating that RA-resistant F9 colonies could be selected from F9 cells transfected with Raf.Kras Tail (Rf.KTail), but not with Raf.Hras Tail (Rf.HTail). The lower panel shows Giemsa-stained plates illustrating that F9 cells (three different isolates of each) expressing Raf.KTail can be passaged indefinitely in the presence of RA (passage 6 is shown). (F) Raf.KTail-expressing cells exhibit increased sensitivity to U0126 and decreased proliferative potential compared to the case for vector-transfected F9 cells. Equal cell numbers were plated at time zero in the presence of solvent DMSO, LY294002 (LY294), or U0126 (UO). Relative cell numbers were determined by using SYTO60 staining and quantified. Error bars represent standard deviations. P < 0.001. (G) Immunofluorescence micrographs demonstrating that F9 cells expressing Raf1 with the Kras C terminus (RfKT), selected in the presence of G418+RA, express Oct3/Oct4 (top), whereas only a few cells express GATA4 (G4) (bottom). Rf-KT-O-mrg, merge (yellow) of Raf1.KTail (RfKT) and Oct3/Oct4 (O). G4-H-mrg, merge (violet) of GATA4 (G) and Hoechst (H). Letter color indicates fluor of the secondary antibody used or merge color. Bar, 20 μm.
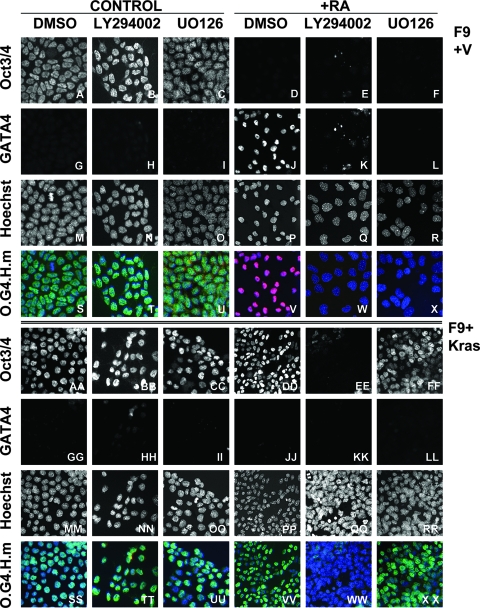
FIG. 8.
MEK is not required for KrasV12 to prevent differentiation and to maintain stem cell features in F9 cells in the presence of RA. Immunofluorescence micrographs of Oct3/Oct4 and GATA4 expression in vector-transfected F9 cells (panels A to X) and KrasV12-transfected F9 cells (panels AA to XX) maintained in the presence of the PI-3 kinase inhibitor LY294002 (panels B, H, N, T, E, K, Q, W, BB, HH, NN, TT, EE, KK, QQ, and WW), the MEK inhibitor UO126 (panels C, I, O, U, F, L, R, X, CC, II, OO, UU, FF, LL, RR, and XX), or solvent DMSO (panels A, G, M, S, D, J, P, V, AA, GG, MM, SS, FF, LL, RR, and XX) in the absence (panels A to U and AA to UU) or presence (panels D to X and DD to XX) of RA. The nuclei were counterstained with Hoechst 33258. O.G4.H.m, merged images of Oct3/Oct4 (green), GATA4 (red), and Hoechst 33258 (blue). Pink depicts the merge of GATA4 (red) and Hoechst (blue), and aqua coloring depicts the merge of Oct3/Oct4 (green) and Hoechst (blue). Note that GATA4 (red) is only expressed in vector-transfected F9 cells treated with RA and DMSO (J and V), yielding the pink color upon merge of GATA4 and Hoechst (V) due to the lack of Oct3/Oct4 staining (green). Note that while MEK is required for RA-mediated repression of Oct3/Oct4 (compare panels F and C) and induction of GATA4 (compare panels L and J) expression and differentiation of vector-transfected F9 cells, MEK is not required for KrasV12 to prevent GATA4 expression (compare panels LL and JJ) and to maintain Oct3/Oct4 expression (compare panels FF and DD) in F9 cells in the presence of RA.
Pharmacologic MEK inhibition (Fig. 7A) does not inhibit stem cell maintenance of vector-transfected F9 cells (Fig. 7B and Cc and 8C and U), but does block RA-induced differentiation to endoderm (Fig. 7B and compare panels Cg and Ci and 8, compare panels J and V with panels L and X, respectively), consistent with a requirement for the endogenous extracellular signal-regulated kinase pathway in RA-mediated F9 differentiation (99). Similarly, MEK inhibition blocks the ability of HrasV12 to induce F9 differentiation (87), suggesting that RA- and HrasV12-induced differentiation of endodermal progenitors involve similar signaling pathways. We determined that MEK inhibition does not affect Oct3/Oct4 expression (stem cell maintenance) of KrasV12-expressing F9 cells, even in the presence of RA (Fig. 8, compare CC and UU with FF and XX), indicating that the actions of KrasV12 in resisting differentiation are MEK independent. However, long-term culture (more than 1 week) of the KrasV12-expressing F9 cells in the presence of the MEK inhibitor leads to growth suppression and a reduction in the number of Oct3-expressing/Oct4-expressing cells (data not shown). These results indicate that like PI-3 kinase, MEK signaling downstream of activated Hras and Kras is utilized for distinct aspects of stem cell maintenance, expansion, and differentiation.
Differential signaling to the Raf-MEK pathway distinguishes the activities of activated Hras and Kras.
Previous studies demonstrated that F9 cells expressing a Raf kinase fusion protein that includes the Hras-derived carboxyl terminus undergo differentiation, indicating that Raf is a key effector of HrasV12-induced differentiation (87). To examine a potential role for compartmentalized Ras-Raf signaling in the differential function of activated Hras and Kras, we tested the effect of forced Raf localization to a subcellular region corresponding to that of Kras by generating a Raf fusion protein containing the carboxyl terminus of Kras (Raf.KTail) and expressing it in F9 cells (Fig. 7G). Raf.KTail-expressing cells, but not Raf.HTail-expressing cells, could be established as stable lines that, like Krasv12- and Hras.KTail-expressing cells, are also refractory to RA-induced differentiation (Fig. 7E), and they maintain Oct3/Oct4 expression (Fig. 7G). Notably, F9 cells expressing the Raf.KTail protein, but not parental F9 cells, exhibit reduced proliferation when maintained in the presence of the MEK inhibitor U0126 (Fig. 7F), implicating a MEK-dependent Kras function in stem/progenitor expansion. Taken together with data presented above indicating a MEK-independent role for KrasV12 in endodermal stem cell maintenance, these findings suggest a bifurcation within this pathway that distinguishes the activities of activated Hras and Kras and reveals both MEK-dependent and MEK-independent functions for KrasV12 in stem cell expansion and stem cell maintenance, respectively.
Analysis of Kras effector mutants reveals a role for RalGDS in stem cell maintenance.
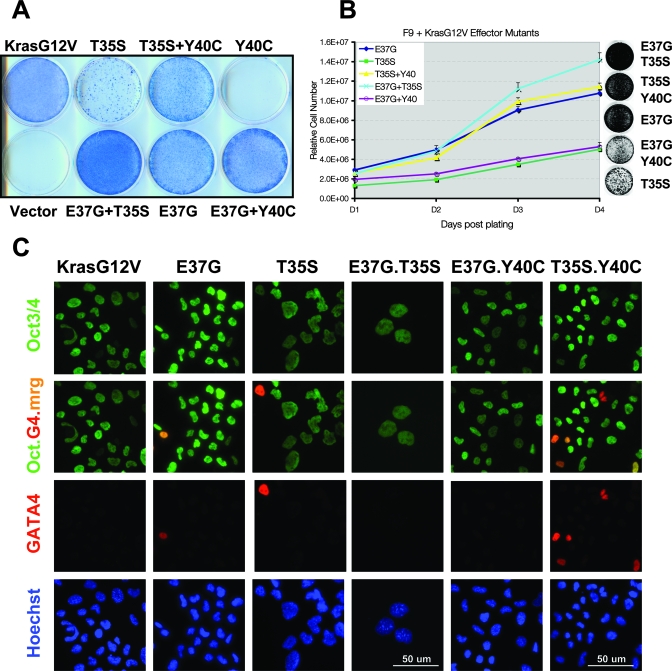
To further analyze the roles of the major Ras effectors in the ability of KrasV12 to promote the expansion and maintenance of endodermal progenitor cells, F9 cells were transfected with various KrasV12 effector domain missense mutants that selectively activate RalGDS (E37G), Raf (T35S), or PI-3 kinase (Y40C) (91). The expression of KrasV12Y40C did not significantly affect F9 differentiation capacity, and transfected cells underwent growth arrest in the presence of RA (Fig. 9A). In contrast, the KrasV12E37G mutant was completely competent for maintenance of Oct3/Oct4 expression (Fig. 9C) and cell proliferation in the presence of RA (Fig. 9A and B). Notably, these same effector mutants behaved analogously in the context of HrasV12-induced myeloid differentiation (64). However, cotransfection of the Y40C with E37G Kras mutants diminished the ability of E37G to promote F9 proliferation, suggesting an antagonism with the RalGDS pathway, but was consistent with the inability of Y40C to resist RA when transfected alone (Fig. 9A and B). Interestingly, the KrasV12T35S mutant was able to maintain Oct3/Oct4 expression (Fig. 9C), but transfected cells exhibited significantly reduced proliferative potential (Fig. 9A and B), consistent with the observations described above using the Raf.KTail chimera. While activation of PI-3 kinase by itself was not sufficient to maintain stem cell properties, the proliferative response to KrasV12T35S expression could be enhanced by cotransfection with the Y40C mutant (Fig. 9A to C), further confirming an important role for PI-3 kinase signaling. Cotransfection of T35C was also able to further increase the proliferative increase associated with E37G expression (Fig. 9A and B), suggesting a cooperative effect of activating these two pathways, which have previously been shown to cooperate in tumor progression (90). Taken together, the studies with pharmacologic inhibitors and Kras effector mutants reveal distinct but important requirements for each of these major Ras effectors in the ability of KrasV12 to promote stem cell expansion and resistance to differentiation.
FIG. 9.
Analysis of Kras effector mutants reveals a role for RalGDS in stem cell maintenance. (A) Giemsa-stained plates illustrating that F9 cells expressing the KrasV12 mutants E37G and T35S, but not Y40C or vector control, can be passaged indefinitely in the presence of RA. Note that the T35S mutant-expressing cells grow more slowly, but can be enhanced by coexpressing the Y40C mutant. (B) The E37G mutant enhances F9 proliferation in the presence of RA. The indicated KrasV12 effector mutant-expressing RA-resistant F9 cells (maintained in RA) were plated in quadruplicate, fixed, and subjected to SYTO60 quantitation on the indicated days postplating (day 1 [D1] to D4). Relative cell numbers are plotted. Error bars represent standard deviations. P < 0.001. (C) Immunofluorescence micrographs demonstrating that F9 cells expressing the indicated effector mutants, in the presence of RA, express Oct3/Oct4, whereas only a few cells express GATA4 (G4). Oct.G4.mrg, merge of Oct3/Oct4 (Oct) and GATA4 (G). Hoechst 33258 was used to counterstain the nuclei. Letter color indicates fluor of the secondary antibody used or merge color. Bar, 50 μm.
DISCUSSION
Mutational activation of Ras is one of the most common oncogenic events detected in human cancers. Although accumulating evidence suggests that Ras activation is an important event in the initiation of tumors of the lung, pancreas, and colon (3, 11, 47, 76, 88) and many of the downstream effectors of Ras have now been identified, its precise role in cancer initiation and progression is still unclear. Moreover, the observation that the vast majority of oncogenic Ras alleles arise in the Kras isoform, as opposed to the very closely related Hras and Nras isoforms, has yet to be explained mechanistically. Here, we provide evidence from cell culture studies that mutationally activated Kras, but not Hras or Nras, can promote the expansion of an endodermal stem/progenitor cell population and prevent differentiation. These observations can potentially explain the relatively high frequency of Kras mutations in human cancer, and they reveal a possible role for Kras activation as an initiating event in tumorigenesis. Moreover, our findings are consistent with recent observations in mouse studies in which it was demonstrated that mutational activation of Kras, but not Nras, promotes expansion of the stem cell compartment of the mouse colon (K. Haigis and T. Jacks, MIT, personal communication), thereby supporting the physiological relevance of these cell culture studies.
The F9 cell culture model faithfully recapitulates the differentiation of a stem/progenitor cell to early endoderm. The fact that RA treatment can specifically drive this process in culture and that RA signaling is required for the formation of early endoderm in vivo also supports the physiological relevance of the model. Previous studies have demonstrated that mutationally activated Hras is sufficient to promote the differentiation of F9 cells to primitive endoderm in the absence of RA in transfection studies (86, 87, 95). Our findings are consistent with those reports, and this observation can potentially explain why activating Hras mutations are not seen in tumors of endodermal origin, such as pancreatic, lung, and colorectal cancers. Notably, activated Hras has also been reported to promote the differentiation of several other lineages, including adipocytes (8) and neurons (5, 69). However, a role for activated Kras in these settings has been largely unexplored. Our observation that Hras, Nras, and Kras exert very different functions in the context of stem/progenitor cell differentiation suggests that it may be informative to compare the activities of the various Ras isoforms more broadly in the context of differentiation.
The observed differences in differentiation phenotypes associated with distinct Ras isoforms may be related to previous reports implicating tissue-dependent contexts in susceptibility to the transforming activity of particular oncogenes. Thus, lineage-specific factors associated with differentiation programs may contribute to the susceptibility of different tissue types to tumorigenic conversion by various oncogenes and to the resultant transformed phenotype (39, 72). For example, such differences may underlie the observations that activated Nras is associated with myeloid malignancies (66), germ cell tumors (34), congenital melanocytic nevi, and cutaneous melanomas derived from neural crest, but not mucosal melanomas, which are not derived from neural crest (7, 93). In addition, Hras activation is associated with tumorigenesis of the bladder (45, 79, 100) and salivary gland (97, 98), tissues that arise from an ontogenetic transitional zone, a region where endoderm and ectoderm meet. Notably, the expression of activated Hras, Nras, or Kras in PCC4 cells, which can give rise to mesenchymal (but not endodermal) cells in culture, does not result in an observable phenotype. Together, these findings suggest that the distinct tumor types associated with mutational activation of the various Ras isoforms reflect the unique ability of each of the Ras proteins to affect the differentiation program of progenitor or stem cells that differentiate along distinct lineages.
The proposed model for activated Kras function in stem cells or partially committed progenitors, as an initiating step in human oncogenesis, would seemingly imply that “mature” tumors might continue to express stem cell markers. However, while Oct3/Oct4 expression in human tumors has previously been reported (60, 83-85), the expression is typically seen only in a very small fraction of tumor cells (53), possibly cancer stem cells. This can be explained by either of two mechanisms. First, it is possible that a small fraction of cells within a tumor maintain stem cell characteristics and these cells are needed to continuously “replenish” the bulk of the tumor with progeny cells that exhibit a more differentiated phenotype. However, it is also possible that an activated Kras allele is needed to expand the stem cell population early in tumorigenesis, and subsequently, mutant Kras is selected for its ability to contribute to other aspects of tumor progression, while its role in stem cell maintenance is diminished. Indeed, activated Kras has been shown to promote proliferation, survival, and invasive properties in a variety of non-stem cell contexts (29, 31, 38, 70, 71, 74, 78).
It is worth noting that the observed effects of the various mutationally activated Ras proteins on stem cell differentiation do not necessarily reflect a normal requirement for Ras proteins in stem cell maintenance or differentiation. However, the ability of KrasV12 to expand stem cells could be related to the reported studies describing a unique requirement for endogenous Kras, but not Hras or Nras, in mouse embryonic development (46, 49, 68). Despite the ubiquitous expression of Ras isoforms in various tissues, the lack of correlation between expression and malignancy, and the fact that the various isoforms interact with the same constellation of effectors (2, 13, 17, 21, 33, 51, 73, 78), distinct cellular consequences of activating the various Ras family members have been reported (67, 89, 96). This may reflect differential activation of effectors and/or differential signal intensity/duration (24, 25, 62, 63). Indeed, Ras effectors themselves can exhibit seemingly opposing effects, depending on the cellular context (22, 62, 63, 75, 94, 101). Such opposing activities are consistent with our findings that some Ras effectors can mediate distinct (and seemingly opposing) phenotypic consequences in the F9 model.
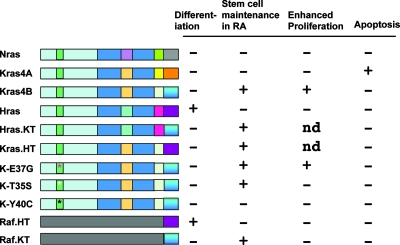
Such differences in isoform-dependent Ras signaling appear to largely involve the differential subcellular localization/processing of Ras and its effectors (15, 18, 30, 35, 41-43, 58, 61, 69, 80). For example, a recent report indicates that oncogenic Hras-induced senescence is mediated by the endoplasmic-reticulum-associated, unfolded protein response (26). Our studies with chimeric Ras isoforms and the Raf effector containing carboxy-terminal motifs that cause distinct subcellular localization of signaling complexes support a critical role for compartmentalized signaling in the differential biological activity of the Hras and Kras isoforms (Fig. 10). However, it is also possible that the carboxyl termini of the various Ras proteins also contribute to their distinct biological effects through unique interactions with cellular proteins that have yet to be identified. Such a possibility is supported by our findings that the Kras chimera containing the extreme C terminus of Hras can maintain stem cell characteristics in RA-treated F9 cells and that the activated Kras-4A splice isoform induces apoptosis (Fig. 10).
FIG. 10.
Structure-function analysis of various Ras mutants and chimeric proteins in F9 cell differentiation, stem cell renewal, proliferation, and apoptosis. Indicated to the left of each wild-type or mutant protein structure is the wild-type or mutated Ras or Raf protein. The light blue rectangle indicates the 100% homologous N-terminal 85 amino acid domain, with the effector (switch I) domain in green. The asterisks mark the sites of effector domain mutations. The dark blue rectangle indicates the 85% homologous region (amino acids 85 to 165), with the internal squares representing the location of amino acid differences and each color representing the Ras isoform source corresponding to the differences. The C-terminal boxes demarcate the hypervariable regions, with the terminal box indicating the 19-amino-acid membrane targeting Tail sequence. The colors indicate the Ras isoform of origin for that domain. The bottom two lines represent Raf (c-Raf) chimeras that include Ras-derived C-terminal tails. Indicated to the right is the presence (+) or absence (−) of the phenotype indicated at the top, encoded by each polypeptide. ND, not determined.
The Raf, PI-3 kinase, and RalGDS proteins are important Ras effectors that contribute to Ras-mediated proliferation and survival or differentiation (50). We observed that inhibiting PI-3 kinase can prevent either differentiation or self-renewal, depending upon the Ras isoform activated. Interestingly, it has also been reported that Ras isoform-dependent transformation potential correlates with PI-3 kinase activation (52). Similarly, we observed that PI-3 kinase activity mediates expression of the POU transcription factor Oct3/Oct4, a homeobox gene, which has direct consequences for self-renewal or differentiation of F9 cells. Moreover, our analysis of Kras effector domain mutants further supports a role for the Raf and PI-3 kinase pathways in endodermal progenitor expansion and reveals an additional important role for RalGDS activation, which has recently emerged as an important Ras effector in the context of human tumorigenesis (37, 40, 77). Interestingly, specific activation of RalGDS has been shown to prevent Hras-mediated differentiation of PC12 and myeloid cells (36, 64).
Our findings with the Raf-KTail chimera and the pharmacologic MEK inhibitor revealed both MEK-independent and MEK-dependent functions of activated Kras in stem/progenitor cells. Interestingly, these findings are consistent with recent studies indicating that KrasV12-induced expansion of mouse colonic epithelial stem cells in vivo appears to be MEK dependent, while maintenance of the undifferentiated state of these cells is MEK independent (K. Haigis and T. Jacks, MIT, personal communication). Taken together, our signaling studies reveal complex and context-dependent roles for three key Ras effectors in the various aspects of stem/progenitor cell maintenance, proliferation, and differentiation (Fig. 10). However, many additional Ras effectors have been identified, and it certainly remains possible that some of those additionally contribute to the distinct functions of activated Hras and Kras in this setting. In summary, the identification of a unique role for Kras in promoting stem/progenitor cell expansion and an inhibition of differentiation to primitive endoderm provides a potential explanation for the high frequency of Kras mutations in tumors of endodermal origin. Finally, the observed unique ability of Kras to expand a stem/progenitor cell population indicates a potentially important role for Kras activation in the initiation of tumorigenesis.
Acknowledgments
We are grateful to S. Strickland for the F9 cells, to D. Solter for providing antibodies to SSEA1 and SSEA3, to M. Rothenberg and A. Singh for the Kras effector constructs, to M. Betson and S. Sharma for critical reading of the manuscript, and to members of the Bernards and Settleman laboratories for helpful discussions.
This work was supported by NIH R01 CA109447-03 to J.S.
Footnotes
Published ahead of print on 2 February 2008.
REFERENCES
- 1.Adjei, A. A. 2001. Blocking oncogenic Ras signaling for cancer therapy. J. Natl. Cancer Inst. 931062-1074. [DOI] [PubMed] [Google Scholar]
- 2.Adjei, A. A. 2001. Ras signaling pathway proteins as therapeutic targets. Curr. Pharm. Des. 71581-1594. [DOI] [PubMed] [Google Scholar]
- 3.Agbunag, C., and D. Bar-Sagi. 2004. Oncogenic K-ras drives cell cycle progression and phenotypic conversion of primary pancreatic duct epithelial cells. Cancer Res. 645659-5663. [DOI] [PubMed] [Google Scholar]
- 4.Apolloni, A., I. A. Prior, M. Lindsay, R. G. Parton, and J. F. Hancock. 2000. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 202475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Sagi, D., and J. R. Feramisco. 1985. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell 42841-848. [DOI] [PubMed] [Google Scholar]
- 6.Bastien, J., J. L. Plassat, B. Payrastre, and C. Rochette-Egly. 2006. The phosphoinositide 3-kinase/Akt pathway is essential for the retinoic acid-induced differentiation of F9 cells. Oncogene 252040-2047. [DOI] [PubMed] [Google Scholar]
- 7.Bauer, J., J. A. Curtin, D. Pinkel, and B. C. Bastian. 2006. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J. Investig. Dermatol. 127179-182. [DOI] [PubMed] [Google Scholar]
- 8.Benito, M., A. Porras, A. R. Nebreda, and E. Santos. 1991. Differentiation of 3T3-L1 fibroblasts to adipocytes induced by transfection of ras oncogenes. Science 253565-568. [DOI] [PubMed] [Google Scholar]
- 9.Bos, J. L. 1988. The ras gene family and human carcinogenesis. Mutat. Res. 195255-271. [DOI] [PubMed] [Google Scholar]
- 10.Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 494682-4689. [PubMed] [Google Scholar]
- 11.Bos, J. L., E. R. Fearon, S. R. Hamilton, M. Verlaan-de Vries, J. H. van Boom, A. J. van der Eb, and B. Vogelstein. 1987. Prevalence of ras gene mutations in human colorectal cancers. Nature 327293-297. [DOI] [PubMed] [Google Scholar]
- 12.Boyer, L. A., T. I. Lee, M. F. Cole, S. E. Johnstone, S. S. Levine, J. P. Zucker, M. G. Guenther, R. M. Kumar, H. L. Murray, R. G. Jenner, D. K. Gifford, D. A. Melton, R. Jaenisch, and R. A. Young. 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122947-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgering, B. M., G. J. Pronk, J. P. Medema, L. van der Voorn, A. M. de Vries Smits, P. C. van Weeren, and J. L. Bos. 1993. Role of p21ras in growth factor signal transduction. Biochem. Soc. Trans. 21888-894. [DOI] [PubMed] [Google Scholar]
- 14.Capo-Chichi, C. D., M. E. Rula, J. L. Smedberg, L. Vanderveer, M. S. Parmacek, E. E. Morrisey, A. K. Godwin, and X. X. Xu. 2005. Perception of differentiation cues by GATA factors in primitive endoderm lineage determination of mouse embryonic stem cells. Dev. Biol. 286574-586. [DOI] [PubMed] [Google Scholar]
- 15.Carey, K. D., R. T. Watson, J. E. Pessin, and P. J. Stork. 2003. The requirement of specific membrane domains for Raf-1 phosphorylation and activation. J. Biol. Chem. 2783185-3196. [DOI] [PubMed] [Google Scholar]
- 16.Chambers, I., and A. Smith. 2004. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene 237150-7160. [DOI] [PubMed] [Google Scholar]
- 17.Chesa, P. G., W. J. Rettig, M. R. Melamed, L. J. Old, and H. L. Niman. 1987. Expression of p21ras in normal and malignant human tissues: lack of association with proliferation and malignancy. Proc. Natl. Acad. Sci. USA 843234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu, V. K., T. Bivona, A. Hach, J. B. Sajous, J. Silletti, H. Wiener, R. L. Johnson II, A. D. Cox, and M. R. Philips. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4343-350. [DOI] [PubMed] [Google Scholar]
- 19.Choy, E., V. K. Chiu, J. Silletti, M. Feoktistov, T. Morimoto, D. Michaelson, I. E. Ivanov, and M. R. Philips. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell 9869-80. [DOI] [PubMed] [Google Scholar]
- 20.Choy, E., and M. Philips. 2001. Green fluorescent protein-tagged Ras proteins for intracellular localization. Methods Enzymol. 33250-64. [DOI] [PubMed] [Google Scholar]
- 21.Colicelli, J. 2004. Human RAS superfamily proteins and related GTPases. Sci. STKE 2004re13. [DOI] [PMC free article] [PubMed]
- 22.Cowley, S., H. Paterson, P. Kemp, and C. J. Marshall. 1994. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 77841-852. [DOI] [PubMed] [Google Scholar]
- 23.Crespo, P., and J. Leon. 2000. Ras proteins in the control of the cell cycle and cell differentiation. Cell. Mol. Life Sci. 571613-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniels, M. A., E. Teixeiro, J. Gill, B. Hausmann, D. Roubaty, K. Holmberg, G. Werlen, G. A. Hollander, N. R. Gascoigne, and E. Palmer. 2006. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444724-729. [DOI] [PubMed] [Google Scholar]
- 25.Deng, Q., R. Liao, B. L. Wu, and P. Sun. 2004. High intensity ras signaling induces premature senescence by activating p38 pathway in primary human fibroblasts. J. Biol. Chem. 2791050-1059. [DOI] [PubMed] [Google Scholar]
- 26.Denoyelle, C., G. Abou-Rjaily, V. Bezrookove, M. Verhaegen, T. M. Johnson, D. R. Fullen, J. N. Pointer, S. B. Gruber, L. D. Su, M. A. Nikiforov, R. J. Kaufman, B. C. Bastian, and M. S. Soengas. 2006. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat. Cell Biol. 81053-1063. [DOI] [PubMed] [Google Scholar]
- 27.Desai, T. J., F. Chen, J. Lu, J. Qian, K. Niederreither, P. Dolle, P. Chambon, and W. V. Cardoso. 2006. Distinct roles for retinoic acid receptors alpha and beta in early lung morphogenesis. Dev. Biol. 29112-24. [DOI] [PubMed] [Google Scholar]
- 28.Downward, J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 311-22. [DOI] [PubMed] [Google Scholar]
- 29.Duursma, A. M., and R. Agami. 2003. Ras interference as cancer therapy. Semin. Cancer Biol. 13267-273. [DOI] [PubMed] [Google Scholar]
- 30.Farrar, M. A., J. Tian, and R. M. Perlmutter. 2000. Membrane localization of Raf assists engagement of downstream effectors. J. Biol. Chem. 27531318-31324. [DOI] [PubMed] [Google Scholar]
- 31.Friday, B. B., and A. A. Adjei. 2005. K-ras as a target for cancer therapy. Biochim. Biophys. Acta 1756127-144. [DOI] [PubMed] [Google Scholar]
- 32.Fujikura, J., E. Yamato, S. Yonemura, K. Hosoda, S. Masui, K. Nakao, J. Miyazaki Ji, and H. Niwa. 2002. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 16784-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furth, M. E., T. H. Aldrich, and C. Cordon-Cardo. 1987. Expression of ras proto-oncogene proteins in normal human tissues. Oncogene 147-58. [PubMed] [Google Scholar]
- 34.Ganguly, S., V. V. Murty, F. Samaniego, V. E. Reuter, G. J. Bosl, and R. S. Chaganti. 1990. Detection of preferential NRAS mutations in human male germ cell tumors by the polymerase chain reaction. Genes Chromosomes Cancer 1228-232. [DOI] [PubMed] [Google Scholar]
- 35.Goetz, C. A., J. J. O'Neil, and M. A. Farrar. 2003. Membrane localization, oligomerization, and phosphorylation are required for optimal raf activation. J. Biol. Chem. 27851184-51189. [DOI] [PubMed] [Google Scholar]
- 36.Goi, T., G. Rusanescu, T. Urano, and L. A. Feig. 1999. Ral-specific guanine nucleotide exchange factor activity opposes other Ras effectors in PC12 cells by inhibiting neurite outgrowth. Mol. Cell. Biol. 191731-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.González-García, A., C. A. Pritchard, H. F. Paterson, G. Mavria, G. Stamp, and C. J. Marshall. 2005. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell 7219-226. [DOI] [PubMed] [Google Scholar]
- 38.Guerra, C., N. Mijimolle, A. Dhawahir, P. Dubus, M. Barradas, M. Serrano, V. Campuzano, and M. Barbacid. 2003. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 4111-120. [DOI] [PubMed] [Google Scholar]
- 39.Gupta, P. B., C. Kuperwasser, J. P. Brunet, S. Ramaswamy, W. L. Kuo, J. W. Gray, S. P. Naber, and R. A. Weinberg. 2005. The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat. Genet. 371047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 162045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hancock, J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 4373-384. [DOI] [PubMed] [Google Scholar]
- 42.Hancock, J. F., and R. G. Parton. 2005. Ras plasma membrane signalling platforms. Biochem. J. 3891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hingorani, S. R., and D. A. Tuveson. 2003. Ras redux: rethinking how and where Ras acts. Curr. Opin. Genet. Dev. 136-13. [DOI] [PubMed] [Google Scholar]
- 44.Jaiswal, A. S., A. S. Multani, S. Pathak, and S. Narayan. 2004. N-methyl-N′-nitro-N-nitrosoguanidine-induced senescence-like growth arrest in colon cancer cells is associated with loss of adenomatous polyposis coli protein, microtubule organization, and telomeric DNA. Mol. Cancer 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johne, A., I. Roots, and J. Brockmoller. 2003. A single nucleotide polymorphism in the human H-ras proto-oncogene determines the risk of urinary bladder cancer. Cancer Epidemiol. Biomarkers Prev. 1268-70. [PubMed] [Google Scholar]
- 46.Johnson, L., D. Greenbaum, K. Cichowski, K. Mercer, E. Murphy, E. Schmitt, R. T. Bronson, H. Umanoff, W. Edelmann, R. Kucherlapati, and T. Jacks. 1997. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 112468-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson, L., K. Mercer, D. Greenbaum, R. T. Bronson, D. Crowley, D. A. Tuveson, and T. Jacks. 2001. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 4101111-1116. [DOI] [PubMed] [Google Scholar]
- 48.Kim, C. F., E. L. Jackson, A. E. Woolfenden, S. Lawrence, I. Babar, S. Vogel, D. Crowley, R. T. Bronson, and T. Jacks. 2005. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121823-835. [DOI] [PubMed] [Google Scholar]
- 49.Koera, K., K. Nakamura, K. Nakao, J. Miyoshi, K. Toyoshima, T. Hatta, H. Otani, A. Aiba, and M. Katsuki. 1997. K-ras is essential for the development of the mouse embryo. Oncogene 151151-1159. [DOI] [PubMed] [Google Scholar]
- 50.Lentzsch, S., M. Chatterjee, M. Gries, K. Bommert, H. Gollasch, B. Dorken, and R. C. Bargou. 2004. PI3-K/AKT/FKHR and MAPK signaling cascades are redundantly stimulated by a variety of cytokines and contribute independently to proliferation and survival of multiple myeloma cells. Leukemia 181883-1890. [DOI] [PubMed] [Google Scholar]
- 51.Leon, J., I. Guerrero, and A. Pellicer. 1987. Differential expression of the ras gene family in mice. Mol. Cell. Biol. 71535-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, W., T. Zhu, and K. L. Guan. 2004. Transformation potential of Ras isoforms correlates with activation of phosphatidylinositol 3-kinase but not ERK. J. Biol. Chem. 27937398-37406. [DOI] [PubMed] [Google Scholar]
- 53.Looijenga, L. H., H. Stoop, H. P. de Leeuw, C. A. de Gouveia Brazao, A. J. Gillis, K. E. van Roozendaal, E. J. van Zoelen, R. F. Weber, K. P. Wolffenbuttel, H. van Dekken, F. Honecker, C. Bokemeyer, E. J. Perlman, D. T. Schneider, J. Kononen, G. Sauter, and J. W. Oosterhuis. 2003. POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res. 632244-2250. [PubMed] [Google Scholar]
- 54.Maher, J., D. A. Baker, M. Manning, N. J. Dibb, and I. A. Roberts. 1995. Evidence for cell-specific differences in transformation by N-, H- and K-ras. Oncogene 111639-1647. [PubMed] [Google Scholar]
- 55.Mark, M., N. B. Ghyselinck, and P. Chambon. 2004. Retinoic acid signalling in the development of branchial arches. Curr. Opin. Genet. Dev. 14591-598. [DOI] [PubMed] [Google Scholar]
- 56.Marshall, C. J. 1996. Ras effectors. Curr. Opin. Cell Biol. 8197-204. [DOI] [PubMed] [Google Scholar]
- 57.Martín, M., J. Gallego-Llamas, V. Ribes, M. Kedinger, K. Niederreither, P. Chambon, P. Dolle, and G. Gradwohl. 2005. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev. Biol. 284399-411. [DOI] [PubMed] [Google Scholar]
- 58.Matallanas, D., I. Arozarena, M. T. Berciano, D. S. Aaronson, A. Pellicer, M. Lafarga, and P. Crespo. 2003. Differences on the inhibitory specificities of H-Ras, K-Ras, and N-Ras (N17) dominant negative mutants are related to their membrane microlocalization. J. Biol. Chem. 2784572-4581. [DOI] [PubMed] [Google Scholar]
- 59.Molotkov, A., N. Molotkova, and G. Duester. 2005. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev. Dyn. 232950-957. [DOI] [PubMed] [Google Scholar]
- 60.Monk, M., and C. Holding. 2001. Human embryonic genes re-expressed in cancer cells. Oncogene 208085-8091. [DOI] [PubMed] [Google Scholar]
- 61.Mor, A., and M. R. Philips. 2006. Compartmentalized Ras/MAPK signaling. Annu. Rev. Immunol. 24771-800. [DOI] [PubMed] [Google Scholar]
- 62.Murphy, L. O., J. P. MacKeigan, and J. Blenis. 2004. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol. Cell. Biol. 24144-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murphy, L. O., S. Smith, R. H. Chen, D. C. Fingar, and J. Blenis. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4556-564. [DOI] [PubMed] [Google Scholar]
- 64.Omidvar, N., L. Pearn, A. K. Burnett, and R. L. Darley. 2006. Ral is both necessary and sufficient for the inhibition of myeloid differentiation mediated by Ras. Mol. Cell. Biol. 263966-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Shea, K. S. 2004. Self-renewal vs. differentiation of mouse embryonic stem cells. Biol. Reprod. 711755-1765. [DOI] [PubMed] [Google Scholar]
- 66.Parikh, C., R. Subrahmanyam, and R. Ren. 2006. Oncogenic NRAS rapidly and efficiently induces CMML- and AML-like diseases in mice. Blood 1082349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Plowman, S. J., and J. F. Hancock. 2005. Ras signaling from plasma membrane and endomembrane microdomains. Biochim. Biophys. Acta 1746274-283. [DOI] [PubMed] [Google Scholar]
- 68.Plowman, S. J., D. J. Williamson, M. J. O'Sullivan, J. Doig, A. M. Ritchie, D. J. Harrison, D. W. Melton, M. J. Arends, M. L. Hooper, and C. E. Patek. 2003. While K-ras is essential for mouse development, expression of the K-ras 4A splice variant is dispensable. Mol. Cell. Biol. 239245-9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qui, M. S., and S. H. Green. 1992. PC12 cell neuronal differentiation is associated with prolonged p21ras activity and consequent prolonged ERK activity. Neuron 9705-717. [DOI] [PubMed] [Google Scholar]
- 70.Rak, J., J. Filmus, G. Finkenzeller, S. Grugel, D. Marme, and R. S. Kerbel. 1995. Oncogenes as inducers of tumor angiogenesis. Cancer Metastasis Rev. 14263-277. [DOI] [PubMed] [Google Scholar]
- 71.Rak, J. W., B. D. St Croix, and R. S. Kerbel. 1995. Consequences of angiogenesis for tumor progression, metastasis and cancer therapy. Anticancer Drugs 63-18. [DOI] [PubMed] [Google Scholar]
- 72.Rassoulzadegan, M., and F. Cuzin. 1987. “Sub-threshold neoplastic states” created in transgenic mice. Oncogene Res. 11-6. [PubMed] [Google Scholar]
- 73.Rebollo, A., and A. C. Martinez. 1999. Ras proteins: recent advances and new functions. Blood 942971-2980. [PubMed] [Google Scholar]
- 74.Rhim, J. S. 1989. Neoplastic transformation of human epithelial cells in vitro. Anticancer Res. 91345-1365. [PubMed] [Google Scholar]
- 75.Ries, S., C. Biederer, D. Woods, O. Shifman, S. Shirasawa, T. Sasazuki, M. McMahon, M. Oren, and F. McCormick. 2000. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell 103321-330. [DOI] [PubMed] [Google Scholar]
- 76.Roberts, M. L., K. G. Drosopoulos, I. Vasileiou, M. Stricker, E. Taoufik, C. Maercker, A. Guialis, M. N. Alexis, and A. Pintzas. 2006. Microarray analysis of the differential transformation mediated by Kirsten and Harvey Ras oncogenes in a human colorectal adenocarcinoma cell line. Int. J. Cancer 118616-627. [DOI] [PubMed] [Google Scholar]
- 77.Rodriguez-Viciana, P., and F. McCormick. 2005. RalGDS comes of age. Cancer Cell 7205-206. [DOI] [PubMed] [Google Scholar]
- 78.Rojas, J. M., and E. Santos. 2002. ras genes and human cancer: different implications and different roles. Curr. Genomics 3295-311. [Google Scholar]
- 79.Schulz, W. A. 2006. Understanding urothelial carcinoma through cancer pathways. Int. J. Cancer 1191513-1518. [DOI] [PubMed] [Google Scholar]
- 80.Sewing, A., B. Wiseman, A. C. Lloyd, and H. Land. 1997. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol. Cell. Biol. 175588-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Silvius, J. R. 2002. Mechanisms of Ras protein targeting in mammalian cells. J. Membr. Biol. 19083-92. [DOI] [PubMed] [Google Scholar]
- 82.Strickland, S., and V. Mahdavi. 1978. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell 15393-403. [DOI] [PubMed] [Google Scholar]
- 83.Tai, M. H., C. C. Chang, M. Kiupel, J. D. Webster, L. K. Olson, and J. E. Trosko. 2005. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis 26495-502. [DOI] [PubMed] [Google Scholar]
- 84.Trosko, J. E., C. C. Chang, B. L. Upham, and M. H. Tai. 2004. Ignored hallmarks of carcinogenesis: stem cells and cell-cell communication. Ann. N. Y. Acad. Sci. 1028192-201. [DOI] [PubMed] [Google Scholar]
- 85.Trosko, J. E., and M. H. Tai. 2006. Adult stem cell theory of the multi-stage, multi-mechanism theory of carcinogenesis: role of inflammation on the promotion of initiated stem cells. Contrib. Microbiol. 1345-65. [DOI] [PubMed] [Google Scholar]
- 86.Verheijen, M. H., R. M. Wolthuis, J. L. Bos, and L. H. Defize. 1999. The Ras/Erk pathway induces primitive endoderm but prevents parietal endoderm differentiation of F9 embryonal carcinoma cells. J. Biol. Chem. 2741487-1494. [DOI] [PubMed] [Google Scholar]
- 87.Verheijen, M. H., R. M. Wolthuis, L. H. Defize, J. den Hertog, and J. L. Bos. 1999. Interdependent action of RalGEF and Erk in Ras-induced primitive endoderm differentiation of F9 embryonal carcinoma cells. Oncogene 184435-4439. [DOI] [PubMed] [Google Scholar]
- 88.Vogelstein, B., E. R. Fearon, S. R. Hamilton, S. E. Kern, A. C. Preisinger, M. Leppert, Y. Nakamura, R. White, A. M. Smits, and J. L. Bos. 1988. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319525-532. [DOI] [PubMed] [Google Scholar]
- 89.Voice, J. K., R. L. Klemke, A. Le, and J. H. Jackson. 1999. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J. Biol. Chem. 27417164-17170. [DOI] [PubMed] [Google Scholar]
- 90.Ward, Y., W. Wang, E. Woodhouse, I. Linnoila, L. Liotta, and K. Kelly. 2001. Signal pathways which promote invasion and metastasis: critical and distinct contributions of extracellular signal-regulated kinase and Ral-specific guanine exchange factor pathways. Mol. Cell. Biol. 215958-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80533-541. [DOI] [PubMed] [Google Scholar]
- 92.Wobus, A. M., and K. R. Boheler. 2005. Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol. Rev. 85635-678. [DOI] [PubMed] [Google Scholar]
- 93.Wong, C. W., Y. S. Fan, T. L. Chan, A. S. Chan, L. C. Ho, T. K. Ma, S. T. Yuen, and S. Y. Leung. 2005. BRAF and NRAS mutations are uncommon in melanomas arising in diverse internal organs. J. Clin. Pathol. 58640-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Woods, D., D. Parry, H. Cherwinski, E. Bosch, E. Lees, and M. McMahon. 1997. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol. Cell. Biol. 175598-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamaguchi-Iwai, Y., M. Satake, Y. Murakami, M. Sakai, M. Muramatsu, and Y. Ito. 1990. Differentiation of F9 embryonal carcinoma cells induced by the c-jun and activated c-Ha-ras oncogenes. Proc. Natl. Acad. Sci. USA 878670-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yan, J., S. Roy, A. Apolloni, A. Lane, and J. F. Hancock. 1998. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J. Biol. Chem. 27324052-24056. [DOI] [PubMed] [Google Scholar]
- 97.Yoo, J., and R. A. Robinson. 2000. H-ras gene mutations in salivary gland mucoepidermoid carcinomas. Cancer 88518-523. [DOI] [PubMed] [Google Scholar]
- 98.Yoo, J., and R. A. Robinson. 2000. ras gene mutations in salivary gland tumors. Arch. Pathol. Lab. Med. 124836-839. [DOI] [PubMed] [Google Scholar]
- 99.Yoshida-Koide, U., T. Matsuda, K. Saikawa, Y. Nakanuma, T. Yokota, M. Asashima, and H. Koide. 2004. Involvement of Ras in extraembryonic endoderm differentiation of embryonic stem cells. Biochem. Biophys. Res. Commun. 313475-481. [DOI] [PubMed] [Google Scholar]
- 100.Zhang, Z. T., J. Pak, H. Y. Huang, E. Shapiro, T. T. Sun, A. Pellicer, and X. R. Wu. 2001. Role of Ha-ras activation in superficial papillary pathway of urothelial tumor formation. Oncogene 201973-1980. [DOI] [PubMed] [Google Scholar]
- 101.Zhu, J., D. Woods, M. McMahon, and J. M. Bishop. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 122997-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]